Giacomello and Scorrano discuss work from Mattie et al. redefining mitofusin topology and questioning the current models of mitochondrial fusion.
Abstract
Mitofusins are outer membrane proteins essential for mitochondrial fusion. Their accepted topology posits that both N and C termini face the cytoplasm. In this issue, Mattie et al. (2018. J. Cell Biol. https://doi.org/10.1083/jcb.201611194) demonstrate instead that their C termini reside in the intermembrane space. These findings call for a revision of the current models of mitochondrial fusion.
Mitochondria are dynamic organelles whose morphological changes are strictly coupled to the regulation of bioenergetics as well as of cell signaling, with key implications for cellular function and dysfunction. Steady-state mitochondrial morphology results from the net balance between organelle fission and fusion. Fission is controlled by the dynamin-related protein Drp1, whereas fusion depends on three GTPases: mitofusins 1 and 2 (Mfn1 and Mfn2), located in the outer mitochondrial membrane (OMM), and optic atrophy 1 (Opa1), anchored to the inner mitochondria membrane (IMM; Pernas and Scorrano, 2016). Despite 20 yr of intense research, the molecular details of the processes of fission and especially fusion are still unclear. In a commonly accepted model, mitochondrial fusion depends on the stepwise fusion of the OMM followed by that of the IMM (Meeusen et al., 2004). Mechanistically, in the absence of SNARE proteins that mediate fusion of other organelles, Mfns are believed to self-suffice to the process: they mediate tethering of mitochondria in trans, and then the GTPase-induced conformational change leads to mixing of the two OMMs. Structurally, this process has been inferred from the supposed structural similarity between Mfns and their cyanobacteria homologue bacterial dynamin-like protein (BDLP). Despite considerable genetic divergence, Mfns are believed to be organized like BDLP: both the GTPase and GTPase effector domains are cytosolic because of the existence of two transmembrane domains. In one model, tethering of organelles in trans and their fusion would be driven by oligomerization of the effector domain that includes two heptad-repeat domains, HR1 and HR2, in Mfns (Qi et al., 2016). In a second model, Mfns can shuffle between a “closed” inactive conformation, where HR2 interacts in cis with HR1 from the same Mfn molecule, and an open profusion one, where HR2 is extended to interact with an HR2 domain from a different Mfn2 molecule on an in-trans mitochondria (Franco et al., 2016). Irrespective of how these two models explain interaction, tethering, and promotion of fusion, they both rely on the presence of the HR1 and HR2 on the same cytosolic face of the OMM. This concept was established based on the (weak) homology between Mfns and the yeast counterpart Fzo1p, organized in such a manner and on experiments of Mfn2 topology performed using antibodies directed toward the N and C terminus of the protein (Rojo et al., 2002).
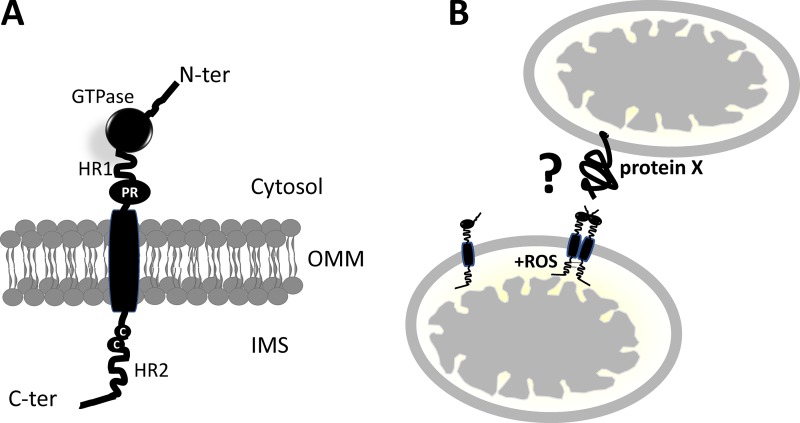
In this issue, Mattie et al. provide conclusive evidence that the C terminus of Mfn, containing the HR2 domain, localizes in the mitochondrial intermembrane space (IMS; Fig. 1 A). This new topological analysis suggests that models of mitochondrial fusion based on the coexistence of HR1 and 2 on the cytosolic face of the OMM shall be revised and offers a mechanistic basis for OMM–IMM fusion coordination as well as for its regulation by mitochondria-derived reactive oxygen species (ROS). Mattie et al. (2018) used bioinformatic analysis to show the fundamental divergence between yeast and metazoan Mfns. Next, they performed classical experiments of protease protection to show that the C terminus of the protein is accessible to the protease only after hypotonic shock of the organelle, a condition known to expose IMS residues to the action of the protease. Indeed, they showed that a high–molecular weight polyethylene glycol derivative that reacts only with free sulfhydryl of Cys residues can bind to Mfn on inside-out outer membrane vesicles prepared by sonication. To analyze the functional consequence of this new topology, they explored the possibility that the Cys residues in Mfn are sensitive to changes in ROS, known inducers of mitochondrial fusion (Shutt et al., 2012). In a set of elegant experiments, they prove that this region is required for Mfn-dependent ROS-induced mitochondrial fusion (Fig. 1 B). Indeed, increased levels of oxidized glutathione associated with cell stress promote the formation of disulphide bridges between adjacent Mfn molecules, leading to high–molecular weight Mfn complexes. Notably, truncation of C-terminal residues 602–757 disrupts Mfn2 fusogenic activity, suggesting that the protein–protein interaction occurring at the IMS are fundamental for fusion. Interestingly, Mattie et al. (2018) note that the cysteine residues involved in these disulphide bonds are located at the C terminus and consequently propose that Mfn oligomerization occurs in the IMS—a major variant from all the models of mitochondrial fusion proposed so far (Fig. 1 B).
Figure 1.
Schematic of revised Mfn topology. (A) Mfns are characterized by a GTPase, a coiled-coil (HR1), and a proline-rich (PR) domain (heptad repeats; HR) facing the cytosol, a transmembrane domain spanning the OMM, and a second coiled-coil (HR) C-terminal domain protruding into the IMS. The cysteine residues sensitive to changes in ROS also face the IMS. (B) Oligomerization of Mfns occurs through the formation of disulphide bridges. Whether Mfn partners with itself or with a yet-undiscovered protein (protein X) to mediate mitochondria docking in trans, necessary for fusion to occur, remains to be defined.
The work by Mattie et al. (2018) revolutionizes the way we think of Mfn function and of mitochondrial fusion in general. Although the notion that the C terminus is responsible for Mfn docking at the OMM is already established, the topology deriving from a single transmembrane domain is completely novel and opens a handful of questions. First, which are the OMM mediators of mitochondria docking in trans necessary for fusion? Does this rely on interactions between HR1 domains, or is it mediated by GTPase head-to-head interaction? As a corollary, we are left with the question of what drives membrane proximity to promote fusion. One possibility is that tethering depends on GTPase domain architecture. Indeed, the R94Q mutant of Mfn2 is as efficient as the WT molecule in driving mitochondrial fusion but does not restore the Mfn2-mediated ER–mitochondria tethering (de Brito and Scorrano, 2008).
Second, which functions rely on the IMS Mfn domain? Mattie et al. (2018) elegantly show that this domain transduces changes in ROS levels into changes in mitochondrial fusion via conserved Cys residues that drive Mfn oligomerization. Interestingly, Mfn1 is required to drive Opa1-dependent mitochondrial fusion, and Mfns interact with Opa1 (Cipolat et al., 2004). One possibility is that these interactions rely on Cys residues, which are also retrieved in the C terminus of Opa1, and that they are influenced by redox conditions, highlighting how mitochondrial function can be coordinately regulated by mitochondrial respiration (and hence ROS generation). In addition, the long IMS domain of Mfns might also interact with other proteins involved not only in shape but also, for example, in biogenesis, like CHCHD4, the mammalian orthologue of Mia40, which interacts with AIF1 (Hangen et al., 2015).
Third, why is the topology of Mfns so different in yeast and vertebrates? This topological difference is not limited to Mfns; for example, the core mitochondrial cristae organizing system (MICOS) component MIC60 is highly divergent between yeast and mammals, justifying its interaction with the bifunctional mitochondrial fusion-cristae organizing protein Opa1 in the latter organisms (Glytsou et al., 2016). One possibility as evolutionary trigger that pushed for MIC60 and Mfn divergence between yeast and higher metazoans might have been the recruitment of mitochondria in vertebrate apoptosis, where cristae remodeling and mitochondrial fission accompany cytochrome c release (Pernas and Scorrano, 2016). Interestingly, this divergence in topology can also explain how Opa1-dependent IMM and Mfn-dependent OMM fusion are coordinated; it is tempting to speculate that they physically interact to coordinate not only fusion but also cristae remodeling and maintenance of OMM morphology.
In conclusion, the study by Mattie et al. (2018) highlights how careful inspection of crystallized concepts even in a young field like mitochondrial dynamics can lead to unexpected and paradigm-shifting discoveries. Exciting future research will elucidate how the new model of Mfn organization can explain the processes of mitochondrial tethering, fusion, docking, and even the pathophysiology of Charcot-Marie-Tooth IIa, caused by Mfn2 mutations occurring in both the cytoplasmic and the IMS domain of the molecule.
Acknowledgments
Work in the authors' laboratories is supported by Telethon (GGP15192), Associazione Italiana per la Ricerca sul Cancro (IG19991), and Fondation Leducq (TNE004015) to L. Scorrano and DiBio PRID Seed 2017 and a Fondazione Cariparo Starting Grant 2016 AIFbiol to M. Giacomello.
The authors declare no competing financial interests.
References
- Cipolat S., Martins de Brito O., Dal Zilio B., and Scorrano L.. 2004. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 101:15927–15932. 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito O.M., and Scorrano L.. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456:605–610. 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- Franco A., Kitsis R.N., Fleischer J.A., Gavathiotis E., Kornfeld O.S., Gong G., Biris N., Benz A., Qvit N., Donnelly S.K., et al. 2016. Correcting mitochondrial fusion by manipulating mitofusin conformations. Nature. 540:74–79. 10.1038/nature20156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glytsou C., Calvo E., Cogliati S., Mehrotra A., Anastasia I., Rigoni G., Raimondi A., Shintani N., Loureiro M., Vazquez J., et al. 2016. Optic Atrophy 1 Is Epistatic to the Core MICOS Component MIC60 in Mitochondrial Cristae Shape Control. Cell Reports. 17:3024–3034. 10.1016/j.celrep.2016.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangen E., Féraud O., Lachkar S., Mou H., Doti N., Fimia G.M., Lam N.V., Zhu C., Godin I., Muller K., et al. 2015. Interaction between AIF and CHCHD4 Regulates Respiratory Chain Biogenesis. Mol. Cell. 58:1001–1014. 10.1016/j.molcel.2015.04.020 [DOI] [PubMed] [Google Scholar]
- Mattie S., Riemer J., Wideman J.G., and McBride H.M.. 2018. A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 10.1083/jcb.201611194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J.M., and Nunnari J.. 2004. Mitochondrial fusion intermediates revealed in vitro. Science. 305:1747–1752. [DOI] [PubMed] [Google Scholar]
- Pernas L., and Scorrano L.. 2016. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu. Rev. Physiol. 78:505–531. 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- Qi Y., Yan L., Yu C., Guo X., Zhou X., Hu X., Huang X., Rao Z., Lou Z., and Hu J.. 2016. Structures of human mitofusin 1 provide insight into mitochondrial tethering. J. Cell Biol. 215:621–629. 10.1083/jcb.201609019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo M., Legros F., Chateau D., and Lombès A.. 2002. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115:1663–1674. [DOI] [PubMed] [Google Scholar]
- Shutt T., Geoffrion M., Milne R., and McBride H.M.. 2012. The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 13:909–915. 10.1038/embor.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]