Abstract
Measurement, identification, and quantitation of endogenous peptides in tissue samples by mass spectrometry (MS) contribute to our understanding of the complex molecular mechanisms of numerous biological phenomena. For accurate results, it is essential to arrest the postmortem degradation of ubiquitous proteins in samples prior to performing peptidomic measurements. Doing so ensures that the detection of endogenous peptides, typically present at relatively low levels of abundance, is not overwhelmed by protein degradation products. Heat stabilization has been shown to inactivate the enzymes in tissue samples and minimize the presence of protein degradation products in the subsequent peptide extracts. However, the efficacy of different heat treatments to preserve the integrity of full-length endogenous peptides has not been well documented; prior peptidomic studies of heat stabilization methods have not distinguished between the full-length (mature) and numerous truncated (possible artifacts of sampling) forms of endogenous peptides. We show that thermal sample treatment via rapid conductive heat transfer is effective for detection of mature endogenous peptides in fresh and frozen rodent brain tissues. Freshly isolated tissue processing with the commercial Stabilizor T1 heat stabilization system resulted in the confident identification of 65% more full-length mature neuropeptides compared to widely used sample treatment in a hot water bath. This finding was validated by a follow-up quantitative multiple reaction monitoring MS analysis of select neuropeptides. The rapid conductive heating in partial vacuum provided by the Stabilizor T1 effectively reduces protein degradation and decreases the chemical complexity of the sample, as assessed by determining total protein content. This system enabled the detection, identification, and quantitation of neuropeptides related to 22 prohormones expressed in individual rat hypothalami and suprachiasmatic nuclei.
Introduction
Neuropeptides are a diverse class of cell-to-cell signaling molecules in the central and peripheral nervous systems. They are derived from larger precursor proteins, prohormones, after enzymatic cleavage by prohormone convertases at enzyme-specific sequences, with these expected cleavage sites referred to as conventional cleavage sites.1–3 Neuropeptides participate in a broad range of physiological and pathological processes—pain sensation, food intake, circadian rhythm, depression, tissue regeneration, and drug addiction—by acting as neuromodulators, neurotransmitters, and hormones.4–11 Significant advances in the study of neuropeptides have been achieved in the past three decades, in part due to rapid progress in mass spectrometry (MS) instrumentation and software tools for data analysis.4,12–15
Even with these advancements, peptide detection, including discovery of new molecules, remains challenging in complex biological samples. Some of the challenges are technical, including unknown recovery rates for peptide extraction and rapid postmortem peptide degradation16 during sampling, as well as matrix-related ion signal suppression during the measurement process. Fast postmortem degradation of ubiquitous proteins into polypeptides is especially problematic when analyzing the neuropeptidome.17 The resultant degradation products increase the chemical complexity and dynamic range of concentration of sample constituents, with this concentration range at times exceeding the analytical performance of many characterization systems, and hence, obscuring the detection of low-abundance endogenous peptides. Prior work has shown that the peptides originating from degraded proteins constitute as much as 95% of the total peptide identifications in brain tissue samples.18–20 This extensive degradation is due in part to postmortem cerebral hypoxia, which induces many intracellular and extracellular events, including neuronal cell death via apoptosis – an enzymatic self-destruction process. The enzymes involved digest proteins into peptides ex-vivo, thus changing the peptidome of the sampled tissue.21
To prevent chemical protein degradation during tissue sampling, several tissue stabilization approaches have been developed,12,18,20,22–24 with heat stabilization being one of the most effective methods of sample preparation for MS experiments.25 Numerous studies since the 1990s26 have shown that heat stabilization of tissues arrests postmortem protein degradation,27,28 improves the detection of endogenous peptides by MS,29 and preserves post-translational modifications (PTMs).27,30–33 For example, experiments conducted by Stingl et al.21 and Segerström et al.34 demonstrated that heat stabilization effectively arrests the ex-vivo peptidase activity, thereby preserving the chemical sample composition as compared to samples from unstabilized tissues. Various means of heat stabilization, such as incubation in a hot water bath,8,35,36 animal sacrifice by focused microwave irradiation (FMR),26,29,37 microwaving of dissected tissue,18 and laser-guided rapid conductive heating in a commercial device, the Denator Stabilizor T1 (ST1),38,39 were previously investigated in a variety of neural and neuroendocrine tissues.
Rapid conductive sample heating in a partially deoxidized environment25,40 using a scanning laser to determine accurate sample size is designed to overcome the technical limitations of prior thermal tissue stabilization methods. This includes tissue size/volume limits and compromised brain morphology for FMR, and uneven and irreproducible temperatures inside dissected tissue caused by conventional microwave ovens.20,21,25 The ST1 benchtop instrument heats fresh and frozen samples quickly and uniformly by rapid conductive heating through metallic blocks pressed against the sample. This technique has been reported to successfully retain the endogenous proteome and peptidome close to the in vivo state, as assessed in previous studies33,40–42 including analysis of the mature neuropeptide complement in crustaceans39 and snap-frozen mammalian tissues.38
Several studies have compared the performance of the ST1 with other methods of thermal stabilization.25,34,39 However, there have been limited efforts to comprehensively evaluate the efficiency of the device in preserving the full-length neuropeptide complement in mammals. Segerström et al.34 studied the relative levels of only a few select neuropeptides, and Skold et al.16 researched specific peptide markers of the protein degradation process in fresh and frozen tissues associated with different heat stabilization methods. Stingl and co-workers21 tested various tissue stabilization methods in combination with urea-based peptide extraction for neuropeptide detection in the rat brain. However, they did not distinguish between full-length neuropeptides, intermediate processing and/or linker peptides derived from the prohormones and, most importantly, numerous truncated versions of neuropeptides often resulting from sequential one amino acid loss that is commonly observed in peptidomics measurements.25 The distinction is important as full-length peptides are produced, accumulated, and released from cells in-vivo,1–3 and are more likely to be biologically active, whereas randomly truncated forms often observed in the MS measurements could be sampling artifacts caused by the loss of the sample cellular integrity and release of lysosomal proteases, or by chemical degradation under the extreme pH conditions typical for peptide extract preparation. Ladder sequence truncation, as evidenced in the supplemental data set by Secher et al.42 for example, is a well-known form of peptide chemical degradation under extreme pH that had been used for peptide sequencing before the advent of tandem MS (MS/MS)-capable instrumentation.43
As mentioned above, previous studies evaluated the performance of the ST1 relative to other heat stabilization methods; however, our work is distinct in a several aspects. First, most of the previous studies either evaluated the stabilization efficacy based on protease activity, overall number of peptide identifications, or identification rates of a select set of full-length neuropeptides.21,28,44,45 In contrast, we compared the efficiencies of the heat stabilization methods on the entire repertoire of mature full-length neuropeptides. We also determined quantitative differences in the detection of known peptides in individual suprachiasmatic nuclei (SCN) treated with both approaches and analyzed using multiple reaction monitoring (MRM).
For the few studies that indeed considered the entire mature neuropeptide complement, they either used the invertebrate animal model crustaceans,39 or used a mammalian model but only to evaluate the performance of the ST1 on snap-frozen tissues. Moreover, hot water treatment, perhaps the most simple and cost-effective way of stabilizing the sample, which has been used previously and proved effective for tissue stabilization,38 was evaluated here in comparison to the ST1 in a mammalian model for the first time. Questions as to whether the ST1 and hot water work equally well on fresh and frozen tissues, and how ST1 and hot water treatment compare with each other for both fresh and frozen tissue samples from a mammalian brain model appear not to have been examined. Hence, to address this specific lack of information, we performed the current study using the rat hypothalamus.
Why use the hypothalamus for peptide characterization? The hypothalamus plays an important role in controlling various body functions and activities, such as temperature regulation, the sleep-wake cycle, hunger, mood, different aspects of behavior, endocrine system activity, growth, and metabolism. Not surprisingly, both the hypothalamus and its sub-structural region, the SCN, have been well investigated via peptidomics approaches by us and others,35,38,46–51 creating an effective model for peptidomic methodology development and comparison.
Here, using both qualitative and quantitative liquid chromatography (LC)–MS measurements, we demonstrate the important advantages of laser-guided rapid conductive heating by the ST1 device compared to a hot water bath as a sample treatment for both fresh and frozen tissues, specifically for investigation of the full-length neuropeptide content of brain tissue.
Materials and methods
Chemicals
Reagents were obtained from either Sigma-Aldrich Chemical Co. (St. Louis, MO, USA) or Thermo Fisher Scientific (Waltham, MA, USA).
Animals
Male Sprague-Dawley rats (N=24), 27–47 days old, were obtained from Envigo (Indianapolis, IN, USA). For comparative experiments, animals were matched by age into three sets, one set of N=12 and two sets of N=6. The animal procedures were carried out according to protocols approved by the Institutional Animal Care and Use Committee, University of Illinois at Urbana-Champaign, and in full accordance with the ARRIVE guidelines.52
Peptide extraction
The brain was dissected 30 s after decapitation; the hypothalami or SCN were isolated according to stereotaxic coordinates in the rat brain atlas53 and stabilized either using the Denator Stabilizor ST1 system (Denator, Uppsala, Sweden) (N=3), or a hot water bath (95°C) (N=3), within 3 min of sacrifice.
For the rapid heat sample stabilization in the ST1 device, each isolated hypothalamus was transferred into a Maintainor Tissue card (Denator) and processed at 95°C using the quick fresh compression mode; processing time was 35 s. Since the sample geometry is measured by a laser scan, this measurement automatically calibrates the heating process, including the heating time, without intervention from an operator. Due to similar sizes of the individual samples used in our measurements as well as their small sizes (just several millimeters in one dimension), the time of treatment was identical (35 s) and sufficient to achieve 95°C. Stabilized hypothalami were transferred into clean microcentrifuge tubes and placed on ice until peptide extraction.
For sample treatment with the hot water bath, each isolated hypothalamus was transferred into a clean microcentrifuge tube containing 300 μL of LC–MS grade water preheated in a 95°C water bath. The hypothalami were incubated in the water bath at 95°C for 10 min, in accordance with prior peptidomics work on rat brain tissues,22 and then placed on ice until peptide extraction. LC–MS grade solvents and reagents were used to prepare all of the peptide extraction and elution buffers unless specified otherwise.
After thermal stabilization, the hypothalamic samples were homogenized on ice and subjected to a three-stage peptide extraction procedure consisting of ice-cold water, acidified acetone (40:5:5/acetone: water: acetic acid), and water containing 0.25% (v/v) acetic acid.31 The extracts from the three stages were combined, creating a single pooled sample, desalted and pre-concentrated using a Pierce C18 spin column (Thermo Scientific, Waltham, MA, USA).
Total protein assay
Measurements of desalted hypothalamus peptide extracts from one set of animals, N=6 (N=3 with ST1+N=3 with hot water bath), were performed. Total protein content was measured in 20% of the volume of the peptide extract eluent collected from the desalting spin column using a commercial Pierce BCA protein assay kit (Thermo Scientific) and an Epoch microplate spectrophotometer (BioTek, Winooski, VT, USA). The remaining eluent was dried in a SpeedVac (Genevac, Ipswich, Suffolk, UK) and stored at −20°C until analysis.
Hypothalamus peptide measurement by nano-electrospray ionization-nanoLC-MS/MS
Each of the dried hypothalamus samples was reconstituted in 10 μL of water supplemented with 0.1% (v/v) formic acid (FA) prior to LC–MS analysis. Next, two distinct LC–MS analytical platforms were used for peptide characterization.
Platform 1 consisted of a Dionex Ultimate 3000 nanoLC system (Thermo Scientific) coupled to an Impact HD QqTOF mass spectrometer (Bruker Daltonics, Billerica, MA, USA). This system was used for qualitative characterization of the chemical complexity of samples exposed to either of the thermal sample treatments. The remaining peptide eluent from each hypothalamus in the total protein assay mentioned above was used on Platform 1. The sample was loaded onto a PepMap 100 pre-column trap (C18, 5 μm, 100 Å) for 3 min. The trap was then switched in-line with an analytical PepMap RSLC column (75 μm×15 cm, C18, 2 μm, 100Å) (Thermo Scientific), and peptides were separated at a uniform flowrate of 300 nL/min using a multi-step gradient. Water with 0.1% FA, and acetonitrile (ACN) with 0.1% FA, were used as solvents A and B, respectively. The gradient conditions were: 0–3 min, 4–4% B; 3–50 min, 4–50% B; 50–55 min, 50–95% B; 55–58 min, 95–95% B; 58–65 min, 95–4% B; 65–75 min, 4–4% B. Data were acquired over a full scan range of m/z 300–3000. Collision-induced dissociation (CID)-assisted MS/MS acquisition was performed in a data-dependent manner. The top five most-intense peaks were selected for fragmentation from the full scan. Dynamic exclusion of precursor ions was 60 s.
Platform 2 consisted of an Eksigent 1D NanoLC plus system (Dublin, CA, USA) equipped with a PicoFrit ProteoPep II column (C18, 5 μm, 100Å) (New Objective, Woburn, MA, USA) hyphenated to an 11 Tesla Fourier transform (FT) ion cyclotron resonance mass spectrometer (LTQ-FT Ultra, Thermo Fisher). This system was used with a different animal set for assessment of neuropeptide identification rates. Peptide extraction was performed via the three-stage peptide extraction procedure as described above for all four sample groups described below. Platform 2 solvents A (water with 0.1% FA) and B (ACN with 0.1% FA) were used at a 300 nL/min flow rate with the following gradients: 0–80 min, 30% B; 80–105 min, 30–45% B; 105–120 min, 45–60% B; 120–125 min, 60–85% B; 125–130 min, 85–85% B; 130–145 min, 85–0% B. The MS acquisition was set to scan at m/z 300–2000 with an m/z 10 precursor isolation window for CID. Data-dependent precursor selection was restricted to the top three most-intense ions. Dynamic exclusion was enabled with three repeat counts and a 120 s exclusion duration.
Bioinformatic peptide identification from the MS/MS data
Data from both LC–MS platforms described above have been used for qualitative peptide analysis in the hypothalamus. PEAKS software (version 7.5, Bioinformatics Solutions Inc., Waterloo, ON, Canada) was used for peptide identification. The data from the Bruker Impact instrument were converted to Mascot generic format using Bruker Data Analysis (version 4.2) before loading into the PEAKS software. Data from the Thermo LTQ-FT instrument were directly loaded into PEAKS for identification. The PEAKS workflow included creation of de novo sequence tags that were then queried against a rat proteome database (33,475 protein entries) downloaded from UniProt using the PEAKS DB search protocol.54 The search parameters included: 20 ppm precursor mass tolerance, 0.1 Da fragment mass tolerance, no enzymatic cleavage, and variable PTMs, including acetylation, amidation, phosphorylation, half-disulfide bond, pyroglutamination, and Met oxidation. The maximum number of variable PTMs per peptide was 3. Filtering criteria of a 1% false discovery rate was set for the final peptide spectral matches.
Statistical analysis of peptide profiles in the hypothalamus extracts
The data from Platform 2 were used for comparisons of the detection of neuropeptides in relation to the method of thermal stabilization. Comparisons were made between the methods of treatment (the ST1 device or hot water bath) and tissue storage (fresh or frozen tissue) using a set of 12 animals (N=12) assayed via Platform 2 as described above. For these comparisons, the individual hypothalami (N=12) were divided into four experimental groups, each containing three biological replicates. Group 1 - snap-frozen immediately after isolation and stabilized by the ST1; Group 2 - freshly isolated and stabilized with the ST1; Group 3 - snap-frozen immediately after isolation and stabilized in a hot water bath; Group 4 - freshly isolated and stabilized in a hot water bath. Settings for the ST1 system and hot water bath, and the peptide extraction procedures, were identical to those described above. The peptide identification results from PEAKS were exported in .csv format into Microsoft Excel. The average number of peptide identifications for each group was determined by calculating the mean of the number of unique peptides among the technical replicates (+/− SD). To test whether the values of identified peptides were significantly different between the groups, a paired Student’s t-test was performed. For the statistical analysis of just the neuropeptides, the peptides formed during enzymatic processing of mono/di or tribasic prohormone cleavage sites were first sorted out, followed by the same statistical analysis procedure as described above. For calculating the overlap in neuropeptide detection between the groups, neuropeptide identifications from all the three technical replicates of a group were merged together, with duplicates removed. Effectively, peptides identified in at least one of the three technical replicates were considered representative of the whole group.
MRM analysis of select neuropeptides in the SCN extracts
A second set of six rats were used in this experiment. The SCN areas from left and right brain hemispheres (N=6×2=12) were isolated and heat-treated individually using either the ST1 system (N=6) or hot water (N=6) within 3 min after animal sacrifice. Peptides extracts were prepared using the methods described above and analyzed individually.
After sample desalting and analyte pre-concentration, peptides extracts were reconstituted in 30 μL of 0.1% FA and analyzed with an Advance UHPLC system (Bruker Daltonics) coupled to an EVOQ triple quadrupole mass spectrometer (Bruker Daltonics). A Kinetex column packed with 1.7 μm C18 resin (Phenomenex, Torrance, CA, USA) was used with a flow rate of 300 μL/min for peptide separation. The peptide signals were identified by selecting four transition ions (the most-intense one as quantifier, remaining three as qualifiers). MRM channels for proSAAS[42–59] (monoisotopic mass=1783.97, dominant charge state: +2, fragment m/z 341.6 used as quantifier, and m/z 498.8, 569.8 and 892.0 used as qualifier ions), proMCH[131–143] (monoisotopic mass=1447.69, dominant charge state: +2, fragment m/z 228.2 used as quantifier, and m/z 375.2, 503.3 and 659.3 used as qualifier ions), neurosecretory protein VGF [488–507] (monoisotopic mass=1914.02, dominant charge state: +3, fragment m/z 576.9 used as quantifier, and m/z 605.8, 703.9 and 816.9 used as qualifier ions), and proSAAS[62–75] (monoisotopic mass=1366.72, dominant charge state: +2, fragment m/z 599.3 used as quantifier, and m/z 618.8, 389.2 and 880.5 used as qualifiers ions). MS Data Review (Version 8.2, Bruker Daltonics) was used for data analysis. The peak area of the most-intense fragment ion signal was calculated. An unpaired Student’s t-test (p<0.05) was performed to verify that the peak areas corresponding to the targeted peptides were significantly different between treatments.
Results and Discussion
Our goal here is to develop protocols for characterizing the peptides within a tissue using LC–MS, as opposed to characterizing the contents of individual cells,55,56 spatially localizing peptides within tissue with MS imaging (MSI),55,56 or in preparing living tissues for dynamic measurements such as measuring peptide release.57,58 Approaches for preparing tissues for spatial and temporal measurements are distinct from the chemical characterization of tissue presented here.
For tissue characterization by LC–MS, compelling evidence accumulated over several decades has established heat stabilization as an effective and necessary procedure for halting postmortem protein degradation during tissue sampling procedures.20,23,25–27,30,32,34,37,39,41,45,59–61 To avoid redundancy with previous studies, we did not test and compare the detection of peptides in untreated tissue samples relative to heat-treated ones. Our investigation focuses on comparing the effectiveness of different heat treatment approaches specifically for improved discovery and characterization of the physiologically relevant neuropeptide complement. Towards this goal, we measured and compared total peptide/protein recovery during extraction (N=6), the number of detected chemically unique peptides (N=6), the number of mature, full-length neuropeptides among the identified peptides in both fresh and frozen tissues (N=12), and quantified select neuropeptide levels resulting from the two different heat stabilization methods by MRM (N=6).
Advantages of rapid conductive heating for full-length neuropeptide recovery from fresh tissue
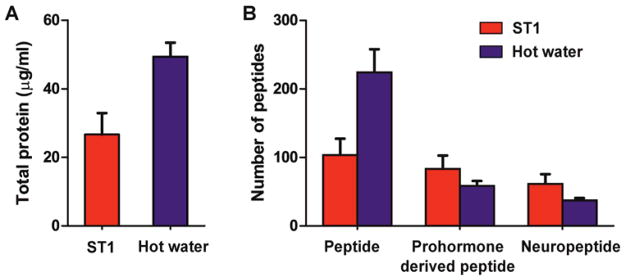
We used the BCA total protein assay to investigate the amount of detectable peptides and proteins extracted from fresh rat hypothalami treated with the ST1 and hot water-based heat stabilization methods; a significant difference in extracted polypeptide amounts was observed between the examined datasets (Student’s t-test, p=0.006). As shown in Figure 1A, the concentration of total peptide and protein in extracts from hot water-stabilized hypothalami (49 μg/mL) was two-fold higher than ST1-stabilized hypothalami (27 μg/mL). These results indicate that sample exposure to hot water extracts an overall higher amount of soluble analytes. This observation correlates well with the results of our LC–MS/MS analysis that detected an average of 204 and 104 peptides per sample, in hot water- and ST1-treated samples, respectively. However, despite a higher total number of chemically unique peptides, there were on average 30% fewer prohormone-derived peptides in the hot water-treated samples compared to the ST1-treated samples (Figure 1B). Over 70% of the total peptides observed in the hot water-treated samples are from cytosolic, mitochondrial, structural, and other ubiquitous proteins. These results suggest that hot water stabilization reduces postmortem degradation of cytosolic proteins less efficiently than ST1 stabilization, and results in a broader chemical dynamic range. This in turn interferes with the detection of prohormone-derived peptides, which are usually present in lower amounts.
Figure 1.
Influence of the two different tissue heat treatments on results of total protein and peptidomic LC–MS/MS measurements of corresponding analyte extracts. (A) Total protein content of the samples as determined by the BCA test (p=0.0061, N=3). (B) Average total number of peptides, prohormone-derived peptides, and neuropeptides identified in the hot water-treated hypothalamic samples. Extracts were split as follows: 20% were used in total protein measurements and 80% for LC–MS/MS qualitative analysis (N=3). Error bars represent standard deviation.
Analysis of prohormone-derived peptides in the two groups of freshly isolated hypothalami revealed that only 58 prohormone-derived peptides per sample were detected in the hot water-treated samples, but 84 peptides, corresponding to a 45% increase, were identified in the ST1-treated samples. Full-length prohormone-derived peptides formed by cleavage at the canonical basic amino acid cleavage sites were evaluated by examining the prohormone sequence adjacent to the N- and C-termini of the identified neuropeptides. Of the 58 prohormone-derived peptides detected in the samples treated with a hot water bath, on average, 37 of the peptides were flanked by dibasic, monobasic, or tribasic cleavage sites. In contrast, in the ST1 treated samples, 61 full-length neuropeptides were detected, which again is an increase of 65% compared to the hot water approach. Moreover, vasopressin-neurophysin 2-copeptin, proenkephalin-B, and tachykinin-3 prohormones were exclusively identified in the rapid conductive-heat stabilized samples, leading to more comprehensive characterization of the neuropeptide content in hypothalamic tissue (Table 1). Our observed improved detection of endogenous neuropeptides with the ST1 system may be due to two reasons: first, the rapid conductive-heating procedure is highly effective in arresting the enzymatic activity that degrades the peptides21 and second, the method results in fewer protein degradation products and a reduction in the chemical complexity of the sample.
Table 1.
Prohormones detected in extracts of ST1- and hot water-treated samples.
| Prohormone | ST1 | Hot Water |
|---|---|---|
| Cerebellin-1 | X | X |
| Chromogranin-A | X | |
| Cocaine- and amphetamine-regulated transcript protein | X | X |
| Corticoliberin | X | X |
| Neuroendocrine protein 7B2 | X | |
| Neuropeptide S | X | |
| Neurosecretory protein VGF | X | X |
| Neurotensin/neuromedin N | X | |
| Pituitary adenylate cyclase-activating polypeptide | X | X |
| Proenkephalin-A | X | X |
| Proenkephalin-B | X | |
| Progonadoliberin-1 | X | X |
| Secretogranin-1 | X | X |
| Pro-MCH | X | X |
| ProSAAS | X | X |
| Protachykinin-1 | X | X |
| Prothyroliberin | X | X |
| Secretogranin-2 | X | X |
| Secretogranin-3 | X | X |
| Somatostatin | X | X |
| Tachykinin-3 | X | |
| Vasopressin-neurophysin 2-copeptin | X |
Prohormones detected in analyte extracts of different biological replicates (N=3) for each of the sample treatment procedures were combined, with peptide duplicates removed. A prohormone was considered only if it had at least one unique full-length peptide sequence; full length is defined as a peptide that is flanked by known basic cleavage sites in the prohormone.
Advantages of rapid conductive heating for full-length neuropeptide recovery from frozen tissue
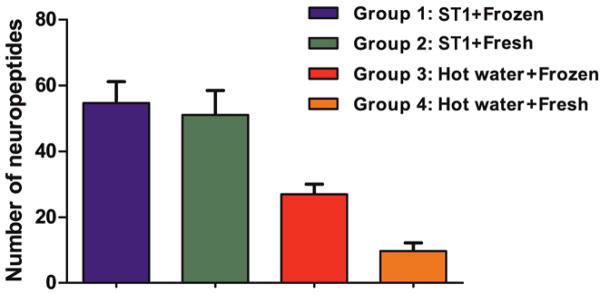
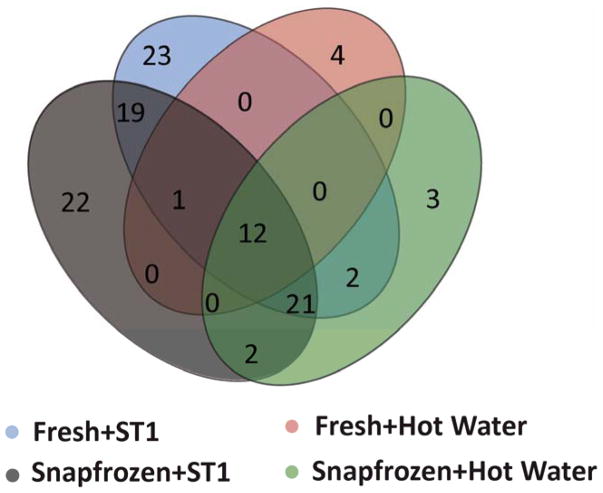
In addition to the improved detection of mature neuropeptides using the ST1 for fresh tissue in the current study, previous investigations have demonstrated the advantages of heat stabilization in terms of reducing enzyme activity, decreasing protein solubility preservation of PTMs, and enabling endogenous peptide quantitation.25,30,45,62 The advantages were observed for both fresh and frozen tissues collected from different animal models. We were also interested in knowing whether or not full-length endogenous neuropeptides can be as effectively recovered from tissues frozen for transportation or storage, followed by ST1 treatment. Thus, using a different set of animals, we evaluated the performance of the two heat treatments for LC–MS detection and identification of neuropeptides in both frozen and fresh rodent brain samples by comparing the effects of freezing, followed by the two heat stabilization methods. Qualitative LC–MS/MS analyses of the four groups (G1–4) of samples described in the methods section were performed and the corresponding metrics compared. The average number of full-length neuropeptides detected per sample was higher in the ST1-treated samples than the hot water-treated samples, regardless of tissue status, fresh or frozen (group 1 vs. 3, p=1.9e-3; group 2 vs. 4, p=6.1e-4) (Figure 2). Additionally, no differences in the number of prohormone-derived peptides were observed between fresh and frozen hypothalami samples treated with the ST1 system. A summation of unique identifications from triplicate runs (redundant identifications removed) is represented in a Venn diagram (Figure 3). Briefly, for the ST1-treated samples, the presence of 77 and 78 neuropeptides in fresh and frozen samples, respectively, were observed; 53 neuropeptides were found in both types of ST1-treated samples. Twenty-five full-length neuropeptides with unique sequences and endogenous cleavage sites were specific for frozen, and 24 were specific for fresh, ST1-stabilized samples. Our results show that rapid conductive heat-based stabilization, but not incubation in hot water, leads to LC–MS detection of representative hypothalamus neuropeptide profiles previously characterized by other approaches, and it is true for both frozen and fresh tissues. The finding is in agreement with a different study that investigated enzyme inactivation and PTM quantitation in the frozen mouse striatum,25 which showed that the activity of enzymes such as phosphatase and cytochrome C oxidase is significantly lowered by heat treatment with the ST1 compared to no heat treatment. Moreover, we observed that the hot water treatment of frozen tissues performs significantly better (p=8.7e-4) than the same treatment of fresh tissues. This information could be useful to researchers who wish to use the inexpensive hot water treatment, helping ensure that they achieve the best possible results.
Figure 2.
The number of neuropeptides identified per sample by LC–MS/MS in four hypothalamus sample groups (N=3). Error bars represent standard deviation.
Figure 3.
Overlap between the data sets containing full-length endogenous neuropeptides detected in extracts of hypothalami processed with four different sample treatment protocols. Data collected from triplicate biological samples, merged, and duplicate peptide entries removed.
Interestingly, the effects of heat stabilization on the detection of the neuropeptide complement has been tested using an invertebrate model and reached a different conclusion. Li’s group39 demonstrated that there was no difference between the rapid conductive heat or hot water sample treatments for MS-based neuropeptide identification in blue crab pericardial organ samples. However, they did report an increased complexity of the mass spectra acquired from the hot water-treated sample. The discrepancy in Li’s finding with numerous reports from mammalian studies may be due to the anatomical differences between the mammalian brain and an invertebrate neurohemal gland. Such differences may relate to extensive myelination of vertebrate axons; for example, degradation products of myelin basic protein are overwhelmingly detected in mammalian brain tissue extracts.63,64 Accordingly, the LC–MS analysis of the crab neurohemal gland, comprised of unmyelinated neural processes, is less likely to benefit from sample heat treatment in the same way as mammalian tissue. Another factor increasing the observed chemical complexity of mammalian brain tissue extracts is contamination from blood proteins, hemoglobin in particular.18,64 The mammalian brain is a highly vascularized organ with a significant volume of blood present in the tissue, whereas the crab neurohemal gland has lower vascularization; in addition, its open vascular system anatomy allows effective hemolymph drainage during dissection. Therefore, the advantage of heat treatment may be lessened in an invertebrate model like the crab due to its less complex anatomy and histology.
Targeted quantitative assessment of neuropeptide levels in analyte extracts from thermally stabilized tissues
To quantify neuropeptide level differences according to the method of thermal stabilization used, we monitored three neuropeptides previously observed in the SCN.8 The SCN was selected because of our experience in characterizing it’s peptidome35,51 and our demonstrated ability to dissect a well-defined brain nucleus. Small samples such as these pose a distinct set of isolation/sampling requirements compared to larger anatomical regions.
Since the in-vivo prohormone-related peptide amounts are relatively low in the SCN to begin with because of its small size, significant quantitative differences in measured peptide levels due to analyte degradation or interference with targeted peptide detection may be expected. Relative measured levels of little SAAS (proSAAS[42–59]) and MCH precursor-related neuropeptide-glutamic acid-isoleucine (proMCH[131–143]) were higher in the ST1-treated SCN samples compared to those exposed to hot water (Figure 4); the measured differences were significant (p<0.05). Interestingly, no differences between the levels of neuroVGF[488–507] were observed based on the heat treatment method used. We speculate that this peptide is less labile; it is known that peptides of different structure are enzymatically degraded and hydrolyzed at different rates.65 As one example, in the presence of aminopeptidase M, peptides with N-terminal proline, such as neuroVGF[488–507], have an extended lifetime compared to other peptides.66,67 Overall data suggest that processing SCN tissue with the ST1 system conserves full-length neuropeptides more effectively than tissue incubation in a hot water bath. This assumes equal peptide recovery from the heat-treated tissues during follow-up extraction. In addition to differences in peptide signal intensity, the MRM mass chromatograms have a higher background in hot water-exposed sample extracts, perhaps due to the increased sample complexity, which affects peptide quantitation accuracy (Figure S1).
Figure 4.
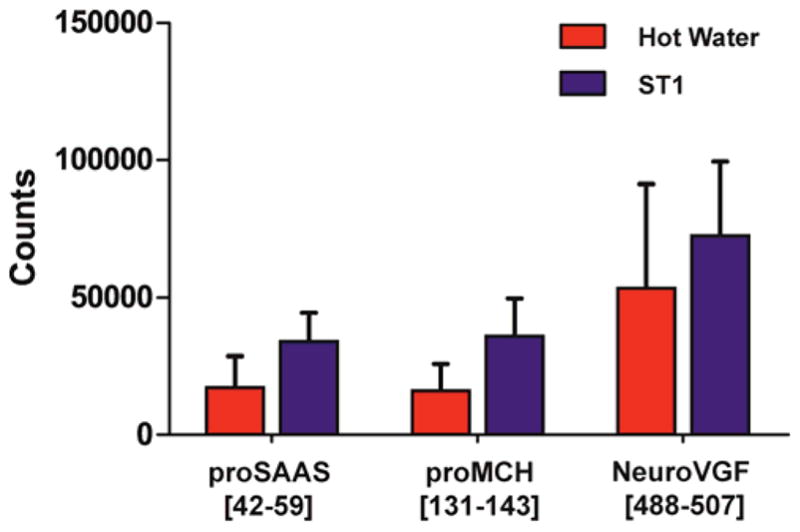
Comparison of relative levels of selected neuropeptides detected in extracts of ST1- and hot water-stabilized SCN samples. Integrated peak areas on the MRM channels of proSAAS[42–59], proMCH[131–143], and neuroVGF [488–507] are calculated and compared (proSAAS paired comparison, p=0.018; neuroVGF paired comparison, p=0.012). Error bars represent standard deviation.
A sequential ladder type of amino acid chain truncation of endogenous neuropeptides at unconventional sites is often documented in peptidomic studies. This phenomenon could be a sign of native catabolic processing or ex-vivo neuropeptide degradation. As an example from our results, a truncated form of the mature peptide proSAAS[62–76], proSAAS[62–75], was detected exclusively in hot water-exposed SCN samples (Figure S2), which suggests that peptide degradation continues during hot water treatment. Although only select peptides were investigated by MRM for level differences related to type of thermal stabilization, the results hint to an explanation of why a lower number of full-length neuropeptides were detected in samples subjected to stabilization in hot water. Fewer degraded proteins and peptides were detected in the extracts of rapid ST1 heat-stabilized samples, including both whole hypothalamus and SCN, which resulted in improved endogenous peptide detection, identification, and quantitation.
Differences between rapid ST1 conductive heat and hot water bath sample stabilization
We demonstrate that the rapid ST1 conductive heat stabilization of samples performed at controlled pressure is more effective than the hot water treatment for successful characterization of mature neuropeptides in the hypothalamus and its anatomically defined nucleus, the SCN. Several features of the ST1 device allow uniform and controlled heating of the tissue above 90ºC, while not exceeding 95ºC anywhere in the sample. This precision is attained by accurate sample size determination via a scanning laser and automatic adjustment of heating temperature and time, important parameters for irreversible inactivation of proteolytic enzymes, without significant increase in temperature-dependent degradation of peptides and proteins.4 Uniform and rapid heat transfer is achieved by establishing a vacuum-assisted seal between a sample-containing membranous chamber and a heating block, which rapidly deforms the sample into a thin geometry that facilitates rapid heat transfer. Especially with larger samples, a hot water bath heats tissue gradually, at a rate inversely proportional to sample size. The treatment time could be another disadvantage of the water bath approach. In established protocols, hot water incubations are relatively long (10 min) to ensure deep heat transfer within the treated tissue, while the ST1 requires less than a minute. Hence, the kinetics of thermal enzyme deactivation in the tissue sample is likely to be slower in a hot water bath, leading to greater protein and peptide degradation. Collectively, the above-mentioned technical features of the ST1 device results in improved LC–MS detection of full-length mature peptides.
We reached this conclusion by comparing the percentage of full-length neuropeptides amongst the total peptides reported for the hypothalamus in the current literature. Recently, Secher et al.42 reported on an analytical framework for peptide characterization using the longest peptide variant (LPV) method, where peptides were assembled into LPVs to account for the sequential degradation ladder sequences due to non-specific enzymatic activity. Using this approach, 14,416 unique peptide sequences were grouped into 2,835 LPVs, of which 356 were derived from prohormone precursors. About 25% of the 356 prohormone-derived LPVs have basic cleavage sites whereas with the ST1 approach, we detected 65% of prohormone-derived peptides with C/N termini basic cleavage sites. Though the mass spectrometric platforms used in both studies are different, we posit that the principal factor contributing to this dissimilarity could be the difference in the peptide extraction method used. Secher et al.42 used urea, which is a preferred choice for protein extraction as it helps dissolve the protein, but may not be optimal for peptides, especially as peptides are smaller and more easily dissolvable compared to proteins. Moreover, the otherwise insoluble proteins would now be soluble, with a potential to degrade during sample processing, and thus can mask some of the trace-level endogenous peptides.
Does rapid heat sample stabilization have an advantage over other sample stabilization approaches?
Not tested here, but known in the peptidomics community, focused microwave radiation (FMR) has been proven effective in arresting enzymatic activities.32 Unfortunately, FMR is not available for most laboratories as it requires highly specialized and expensive equipment, and FMR is difficult to use for samples larger than the rat brain.20 This is because as the organ of interest gets bigger in size, the heating becomes more uneven, leading to non-reproducible results. Moreover, FMR treatment changes the texture of the brain tissue, compromises brain morphology, and makes immediate precise isolation of small defined internal brain regions such as the SCN difficult.
Other more practical methods of sample conservation, such as freezing and conventional microwave oven irradiation, have been reported for peptide and protein analysis. Freezing alone does not stop peptide and protein degradation and is less effective than heat, likely because some enzymatic activity reactivates during sample thawing, while cellular integrity is compromised.21,25 Conventional microwave ovens have been used for treatment of different tissues, including smaller brain regions, demonstrating improved outcomes for LC–MS peptidomics analysis.26,27,37 The approach has several of the same limitations as FMR, but does not provide as fast and uniform tissue heating. Additionally, microwave oven-assisted irradiation has been reported to have lower uniformity in heating in frozen samples as it heats a few pockets of thawed tissue to a high temperature, leaving the remaining frozen tissue almost unaffected.25
In order to allow time for dissection, especially of defined small brain structures, without using heat treatments, options such as “precooling” the brain by perfusing cold saline into the organ before dissection,36 injecting cold modified Gey’s buffer salt solution into the body cavity immediately after euthanasia,36,64 or fast sample freezing are effective. Although these methods reduce postmortem analyte degradation and are useful when precise anatomical areas need to be isolated, they do not prevent degradation without the need for additional stabilization methods.
Conclusions
Thermal tissue stabilization during sampling has been shown to moderate postmortem protein degradation. With multiple methodological approaches available for raising sample temperature, our goal was to determine the best approach for improving qualitative and quantitative neuropeptide measurements in the brain by capturing the endogenous state of the neuropeptidome for biochemical analysis. Of course, when the goals of the experiment are to preserve the spatial localization of the peptides for mass spectrometry imaging, heat stabilization using the ST1 is not appropriate, and so other approaches are required. For neuropeptide characterization, we find that rapid, uniform heat transfer into tissues with a short exposure time in a vacuum environment provided by a commercially available device about doubles the full-length neuropeptide identification via LC–MS-MS, and significantly reduces the detection of truncated peptide forms, in comparison to simple hot water treatment. Even though hot water stabilization has certain advantages, such as being inexpensive, easier to use, and compatibility with frozen tissues, it is not as efficient as the laser-guided, rapid conductive heat stabilization approach; more full-length neuropeptides in their endogenous form were detected with the ST1 device for the samples used here. This contrasts with the results obtained in similar experiments performed by Li’s group at UW-Madison on crustacean pericardial organs;39 such differences may indicate that the efficiency of a heat stabilization method may vary for different animal models. In addition, by reducing the number of degraded proteins and peptides, important identity and abundance details previously obscured are uncovered. As a result, a more accurate picture of the in vivo chemical composition of the tissue of interest can be measured, and a correlation between neuropeptides to certain behaviors or phenotypes established more easily, when LC–MS is integrated into a multidisciplinary study. Use of the fast, well-controlled, and effective heating procedure described here enables the detection of physiologically active peptides present in trace amounts.
Supplementary Material
Acknowledgments
The project described was supported by Award No. P30 DA018310 from the National Institute on Drug Abuse, Award No. R01 NS031609 from the National Institute of Neurological Disorders and Stroke, and Award No. CHE-16-06791 from the National Science Foundation. The authors thank Dr. Eric Jansson for his help in the initial Stabilizor T1 use and setup. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
References
- 1.Garden RW, Shippy SA, Li L, Moroz TP, Sweedler JV. Proc Natl Acad Sci U S A. 1998;95:3972–3977. doi: 10.1073/pnas.95.7.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hummon AB, Huang HQ, Kelley WP, Sweedler JV. J Neurochem. 2002;82:1398–1405. doi: 10.1046/j.1471-4159.2002.01070.x. [DOI] [PubMed] [Google Scholar]
- 3.Li L, FPD, Rubakhin SS, Romanova EV, Jing J, Alexeeva VY, Dembrow NC, Weiss KR, Vilim FS, Sweedler JV. J Neurochem. 2001;77:1569–1580. doi: 10.1046/j.1471-4159.2001.00360.x. [DOI] [PubMed] [Google Scholar]
- 4.Fricker LD, Lim J, Pan H, Che FY. Mass Spectrom Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- 5.Hillebrand JJG, de Wied D, Adan RAH. Peptides. 2002;23:2283–2306. doi: 10.1016/s0196-9781(02)00269-3. [DOI] [PubMed] [Google Scholar]
- 6.Hook V, Bandeira N. J Am Soc Mass Spectrom. 2015;26:1970–1980. doi: 10.1007/s13361-015-1251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kormos V, Gaszner B. Neuropeptides. 2013;47:401–419. doi: 10.1016/j.npep.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Lee JE, Atkins N, Jr, Hatcher NG, Zamdborg L, Gillette MU, Sweedler JV, Kelleher NL. Mol Cell Proteomics. 2010;9:285–297. doi: 10.1074/mcp.M900362-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schank JR, Ryabinin AE, Giardino WJ, Ciccocioppo R, Heilig M. Neuron. 2012;76:192–208. doi: 10.1016/j.neuron.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong TH, Romanova EV, Roberts-Galbraith RH, Yang N, Zimmerman TA, Collins JJ, 3rd, Lee JE, Kelleher NL, Newmark PA, Sweedler JV. J Biol Chem. 2016;291:8109–8120. doi: 10.1074/jbc.M115.709196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Haes W, Van Sinay E, Detienne G, Temmerman L, Schoofs L, Boonen K. Biochim Biophys Acta. 2015;1854:812–826. doi: 10.1016/j.bbapap.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Boonen K, Landuyt B, Baggerman G, Husson SJ, Huybrechts J, Schoofs L. J Sep Sci. 2008;31:427–445. doi: 10.1002/jssc.200700450. [DOI] [PubMed] [Google Scholar]
- 13.Buchberger A, Yu Q, Li L. Annu Rev Anal Chem (Palo Alto Calif) 2015;8:485–509. doi: 10.1146/annurev-anchem-071114-040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvatovich P, Mischoff R. Eur J Mass Spectrom. 2010;16:101–121. doi: 10.1255/ejms.1050. [DOI] [PubMed] [Google Scholar]
- 15.Romanova EV, Dowd SE, Sweedler JV. Curr Opin Chem Biol. 2013;17:801–808. doi: 10.1016/j.cbpa.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE. Proteomics. 2007;7:4445–4456. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 17.Finoulst I, Pinkse M, Van Dongen W, Verhaert P. J Biomed Biotechnol. 2011;2011:245291. doi: 10.1155/2011/245291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Che FYY, Lim J, Pan H, Biswas R, Fricker LD. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Richter R, Schulz-Knappe P, Schrader M, Ständker L, Jürgens M, Tammen H, Forssmann WG. J Chromatogr B. 1999;726:25–35. doi: 10.1016/s0378-4347(99)00012-2. [DOI] [PubMed] [Google Scholar]
- 20.Svensson M, Sköld K, Svenningsson P, Andren PE. J Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 21.Stingl C, Söderquist M, Karlsson O, Borén M, Luider TM. J Proteome Res. 2014;13:2807–2817. doi: 10.1021/pr401232e. [DOI] [PubMed] [Google Scholar]
- 22.Dowell JA, Vander Heyden W, Li L. J Proteome Res. 2006;5:3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell V, Feyereisen K, Bouret S, Leroy D, Beauvillain JC. J Histochem Cytochem. 2001;49:901–910. doi: 10.1177/002215540104900710. [DOI] [PubMed] [Google Scholar]
- 24.Scopes RK. Protein Purification. Springer; New York, NY: 1994. pp. 310–345. [DOI] [Google Scholar]
- 25.Svensson M, Boren M, Skold K, Falth M, Sjogren B, Andersson M, Svenningsson P, Andren PE. J Proteome Res. 2009;8:974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 26.Theodorsson E, Stenfors C, Mathé AA. Peptides. 1990;11:1191–1197. doi: 10.1016/0196-9781(90)90151-t. [DOI] [PubMed] [Google Scholar]
- 27.Galli C, Racagni G. Methods Enzymol. 1982;86:635–642. doi: 10.1016/0076-6879(82)86234-4. [DOI] [PubMed] [Google Scholar]
- 28.Goodwin RJA, Lang AM, Allingham H, Borén M, Pitt AR. Proteomics. 2010;10:1751–1761. doi: 10.1002/pmic.200900641. [DOI] [PubMed] [Google Scholar]
- 29.Nylander I, Stenfors C, Tan-No K, Mathé AA, Terenius L. Neuropeptides. 1997;31:357–365. doi: 10.1016/s0143-4179(97)90072-x. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed MM, Gardiner KJ. J Neurosci Methods. 2011;196:99–106. doi: 10.1016/j.jneumeth.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kultima K, Sköld K, Borén M. J Proteomics. 2011;75:145–159. doi: 10.1016/j.jprot.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 32.O’Callaghan JP, Sriram K. J Neurosci Methods. 2004;135:159–168. doi: 10.1016/j.jneumeth.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Rountree CB, Van Kirk CA, You H, Ding W, Dang H, VanGuilder HD, Freeman WM. Proteome Sci. 2010;8:61–61. doi: 10.1186/1477-5956-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segerström L, Gustavsson J, Nylander I. Biopreserv Biobank. 2016;14:172–179. doi: 10.1089/bio.2015.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JE, Zamdborg L, Southey BR, Atkins N, Jr, Mitchell JW, Li M, Gillette MU, Kelleher NL, Sweedler JV. J Proteome Res. 2013;12:585–593. doi: 10.1021/pr300605p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanova EV, Rubakhin SS, Ossyra JR, Zombeck JA, Nosek MR, Sweedler JV, Rhodes JS. J Neurochem. 2015;135:1038–1048. doi: 10.1111/jnc.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathè AA, Stenfors C, Brodin E, Theodorsson E. Life Sci. 1990;46:287–293. doi: 10.1016/0024-3205(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 38.Colgrave ML, Xi L, Lehnert SA, Flatscher-Bader T, Wadensten H, Nilsson A, Andren PE, Wijffels G. Proteomics. 2011;11:1264–1276. doi: 10.1002/pmic.201000423. [DOI] [PubMed] [Google Scholar]
- 39.Sturm RM, Greer T, Woodards N, Gemperline E, Li L. J Proteome Res. 2013;12:743–752. doi: 10.1021/pr300805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boren M. Methods Mol Biol. 2011;717:91–100. doi: 10.1007/978-1-61779-024-9_5. [DOI] [PubMed] [Google Scholar]
- 41.Ahnoff M, Cazares LH, Skold K. Bioanalysis. 2015;7:1885–1899. doi: 10.4155/bio.15.122. [DOI] [PubMed] [Google Scholar]
- 42.Secher A, Kelstrup CD, Conde-Frieboes KW, Pyke C, Raun K, Wulff BS, Olsen JV. Nat Commun. 2016;7:11436. doi: 10.1038/ncomms11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergman T. EXS. 2000;88:133–144. doi: 10.1007/978-3-0348-8458-7_9. [DOI] [PubMed] [Google Scholar]
- 44.Rossbach U, Nilsson A, Falth M, Kultima K, Zhou Q, Hallberg M, Gordh T, Andren PE, Nyberg F. J Proteome Res. 2009;8:1091–1098. doi: 10.1021/pr800669g. [DOI] [PubMed] [Google Scholar]
- 45.Scholz B, Skold K, Kultima K, Fernandez C, Waldemarson S, Savitski MM, Soderquist M, Boren M, Stella R, Andren P, Zubarev R, James P. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M900229-MCP200. M900229mcp900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atkins N, Jr, Mitchell JW, Romanova EV, Morgan DJ, Cominski TP, Ecker JL, Pintar JE, Sweedler JV, Gillette MU. PLoS One. 2010;5:e12612. doi: 10.1371/journal.pone.0012612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dube MG, Kalra PS, Crowley WR, Kalra SP. Brain Res. 1995;690:275–278. doi: 10.1016/0006-8993(95)00644-6. [DOI] [PubMed] [Google Scholar]
- 48.Qu D, Ludwig DS, Gammeltoft S, Piper M, Pelleymounter MA, Cullen MJ, Mathes WF, Przypek J, Kanarek R, Maratos-Flier E. Nature. 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- 49.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, Arch JRS, Buckingham RE, Haynes AC, Carr SA, Annan RS, McNulty DE, Liu WS, Terrett JA, Elshourbagy NA, Bergsma DJ, Yanagisawa M. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 50.Samson WK, Lumpkin MD, McCann SM. Endocrinology. 1986;119:554–560. doi: 10.1210/endo-119-2-554. [DOI] [PubMed] [Google Scholar]
- 51.Southey BR, Lee JE, Zamdborg L, Atkins N, Mitchell JW, Li M, Gillette MU, Kelleher NL, Sweedler JV. Anal Chem. 2014;86:443–452. doi: 10.1021/ac4023378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 7. Academic Press; 2013. p. 472. [Google Scholar]
- 54.Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, Ma B. Mol Cell Proteomics. 2012;11:M111.010587. doi: 10.1074/mcp.M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Comi TJ, Do TD, Rubakhin SS, Sweedler JV. J Am Chem Soc. 2017;139:3920–3929. doi: 10.1021/jacs.6b12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Do TD, Comi TJ, Dunham SJB, Rubakhin SS, Sweedler JV. Anal Chem. 2017;89:3078–3086. doi: 10.1021/acs.analchem.6b04819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hatcher NG, Atkins N, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Proc Natl Acad Sci U S A. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee CY, Fan Y, Rubakhin SS, Yoon S, Sweedler JV. Sci Rep. 2016;6:26940. doi: 10.1038/srep26940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boren M. Methods Mol Biol. 2014;1295:21–32. doi: 10.1007/978-1-4939-2550-6_2. [DOI] [PubMed] [Google Scholar]
- 60.Kanaoka Y, Hirai K, Ishiko O. J Obstet Gynaecol Res. 2005;31:359–367. doi: 10.1111/j.1447-0756.2005.00303.x. [DOI] [PubMed] [Google Scholar]
- 61.Smejkal G, Rivas-Morello C, Chang J, Freeman E, Trachtenberg A, Lazarev A, Ivanov A, Kuo W. Electrophoresis. 2011;32:2206–2215. doi: 10.1002/elps.201100170. [DOI] [PubMed] [Google Scholar]
- 62.Robinson AA, Westbrook JA, English JA, Boren M, Dunn MJ. Proteomics. 2009;9:4433–4444. doi: 10.1002/pmic.200900287. [DOI] [PubMed] [Google Scholar]
- 63.Heijs B, Tolner EA, Bovée JVMG, van den Maagdenberg AMJM, McDonnell LA. J Proteome Res. 2015;14:5348–5354. doi: 10.1021/acs.jproteome.5b00849. [DOI] [PubMed] [Google Scholar]
- 64.Tillmaand EG, Yang N, Kindt CA, Romanova EV, Rubakhin SS, Sweedler JV. J Am Soc Mass Spectrom. 2015;26:2051–2061. doi: 10.1007/s13361-015-1256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mentlein R, Lucius R. Brain Res Protoc. 1997;1:237–246. doi: 10.1016/s1385-299x(96)00035-9. [DOI] [PubMed] [Google Scholar]
- 66.Johnsen AH. Anal Biochem. 1991;197:182–186. doi: 10.1016/0003-2697(91)90376-5. [DOI] [PubMed] [Google Scholar]
- 67.Yoshimoto T, Tsuru D. J Biochem. 1983;94:619–622. doi: 10.1093/oxfordjournals.jbchem.a134396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.