Abstract
An imbalance between oxidants and antioxidants is considered a major factor in the development of pulmonary vascular diseases. Oxidative stress seen in pulmonary vascular cells is regulated by increased expression of prooxidant enzymes (e.g., nicotinamide adenine dinucleotide phosphate reduced oxidases) and/or decreased production of antioxidants and antioxidant enzymes (e.g., superoxide dismutases). We and others have shown that expression of antioxidant genes in pulmonary artery cells is regulated by epigenetic mechanisms. In this study, we investigate the regulation of oxidative stress in pulmonary artery cells using inhibitors of histone deacetylases (HDACs). Human pulmonary artery endothelial cells (HPAECs) and human pulmonary artery smooth muscle cells were exposed to an array of HDAC inhibitors followed by analysis of anti- and prooxidant gene expression using quantitative RT-PCR and quantitative RT-PCR array. We found that exposure of HPAECs to scriptaid, N-[4-[(hydroxyamino)carbonyl]phenyl]-α-(1-methylethyl)-benzeneacetamide, and trichostatin A for 24 hours induced expression of extracellular superoxide dismutase (EC-SOD) up to 10-fold, whereas expression of the prooxidant gene NADPH oxidase 4 was decreased by more than 95%. We also found that this differential regulation of anti- and prooxidant gene expression resulted in significant attenuation in the cellular levels of reactive oxygen species. Induction of EC-SOD expression was attenuated by the Janus kinase 2 protein kinase inhibitor AG490 and by silencing Janus kinase 2 expression. Augmentation of EC-SOD expression using scriptaid was associated with increased histone H3 (Lys27) acetylation and H3 (Lys4) trimethylation at the gene promoter. We have determined that oxidative stress in pulmonary endothelial cells is regulated by epigenetic mechanisms and can be modulated using HDAC inhibitors.
Keywords: oxidative stress, superoxide dismutase, histone acetylation, endothelial cells, NADPH oxidases
Clinical Relevance
Pulmonary hypertension and pulmonary vascular remodeling are known to be regulated by an imbalance of oxidants and antioxidants and by epigenetic changes in the vascular wall, although the precise signaling pathways involved have not been studied in detail. This study evaluates the role of specific histone modifications in regulation of major pro- and antioxidant enzymes in human pulmonary artery endothelial cells.
Increased oxidative stress due to imbalances between prooxidants and antioxidants are associated with cardiovascular pathologies, including pulmonary arterial hypertension and pulmonary vascular remodeling. Oxidative stress is characterized by increased production of superoxide radicals, hydrogen peroxide, and nitric oxide (NO) and/or decreased levels of antioxidants and antioxidant enzymes. Reactive oxygen species (ROS) are produced in the lungs and vascular tissues due to up-regulation of NADPH oxidase (NOX) expression (1), tissue hypoxia, and ischemia (2, 3) or as a result of inflammatory cascade activation (4). NOX4 is the predominant NOX isoform in pulmonary arteries responsible for production of ROS (5). Increases in oxidative stress cause direct damage to cells, modulate numerous signaling cascades, and activate or inactivate transcription factors, leading to alterations in normal pattern of gene expression in lung tissue (6). The most important line of cellular defense against ROS is the superoxide dismutase (SOD) family of enzymes. Three unique and highly compartmentalized mammalian SODs have been biochemically and molecularly characterized to date: SOD1 or Cu,Zn-SOD, SOD2 or Mn-SOD, and SOD3 or extracellular superoxide dismutase (EC-SOD).
EC-SOD expression has been localized to specific cells and tissues, with the highest expression levels occurring in the lung, heart, kidney, and vasculature. In the vessel wall, EC-SOD is expressed at the highest level, comprising up to half of the total vascular SOD, with activity levels approximately 10-fold higher than in other tissues (7, 8). This extremely high level of EC-SOD in the vascular wall suggests that it plays a critical role in the regulation of vascular tone by controlling the extracellular redox state at or near the juncture of endothelial and smooth muscle cells. In healthy vessels, EC-SOD is produced almost exclusively by smooth muscle cells (7), whereas little EC-SOD synthesis has been detected in endothelial cells (9). Vascular EC-SOD is mostly localized in the space between endothelium and smooth muscle cells, where its concentration is up to 3,000 times higher than in the surrounding spaces (10). At this important location, EC-SOD regulates the concentration of superoxide radicals and therefore helps regulate the flow of endothelium-derived NO that diffuses to the smooth muscle layer stimulating vessel relaxation (8). It is known that many cardiovascular pathophysiological conditions are associated with an increased production of superoxide radicals that can undergo extremely rapid reaction with NO to produce peroxynitrite anion (ONOO−) (11). Thus, in vessel walls, EC-SOD not only regulates the pool of bioavailable NO but also prevents the formation of highly toxic peroxynitrite.
Despite the obviously important role EC-SOD plays in protection against oxidative stress, remarkably little is known about factors that regulate EC-SOD gene expression. It has been shown that basal and inducible transcription of the murine EC-SOD gene is regulated, at least in part, by proximal and distal promoter elements, where Sp1/Sp3 transcription factors interact with the former (12) and where Ets, Kruppel, and MZF-1 transcription factors interact with the latter (13). Sp1/Sp3 transcription factors also play an important role in the regulation of the newly identified promoter for human EC-SOD (14). In addition, EC-SOD expression in pulmonary and cancer cells appears regulated by epigenetic factors, such as DNA methylation and histone acetylation (15–17).
DNA methylation and histone modifications play an important role in the regulation of vascular remodeling. Several histone deacetylase (HDAC) inhibitors were tested for attenuation of pulmonary vascular remodeling. One of the most studied HDAC inhibitors is trichostatin A (TSA). Recent data indicate that selective inhibitors of class I HDACs (HDAC1–3 and 8) reduce pulmonary arterial pressure and vascular wall thickening (18). The different families of mammalian HDACs are grouped in four distinct classes: class I (HDAC1–3 and 8), class II (HDAC4–7, 9, and 10), class III sirtuins (SIRT 1–7), and class IV (HDAC11) (19, 20). Acetylation of lysine residues on histones may activate transcription through neutralization of the basic charge of these residues and through the recruitment of bromodomain-containing protein complexes, which may include other histone acetyl transferases and chromatin-remodeling enzymes.
In the present study, we have identified histone acetylation and methylation as novel epigenetic factors regulating EC-SOD expression in endothelial cells. Moreover, we show that HDAC inhibitors regulate overall oxidative stress burden in pulmonary endothelium most likely by differential control of EC-SOD and NOX4 expression.
Materials and Methods
ROS Measurement
HPAECs were seeded onto a 24-well plate at a density of 1 × 105 cells/well in 500 μl of medium. The next day, cells were incubated with HDAC-42 (1 μM), scriptaid (8 μM), TSA (1.5 μM), or DMSO (as control) for 24 hours. HPAECs were loaded with 10 μM of CM-H2DCFDA (Invitrogen, Carlsbad, CA) dissolved in Endothelial Growth medium (Cell Applications, San Diego, CA) for 30 minutes. At the end of incubation, the medium containing dihydrodichlorofluorescein was aspirated, cells were washed once with complete medium, and then complete medium with HDAC inhibitors was added. Cells were incubated for an additional 4 hours, washed twice with PBS, and observed using an Olympus IX-70 fluorescent microscope (Olympus, Center Valley, PA) with excitation and emission set at 490 and 530 nm (FITC filter), respectively. Fluorescence of oxidized 2,7-dichlorofluorescein (DCF) in cells was captured with a Retiga2000 digital camera (Qimaging, Surrey, BC, Canada). Fluorescence intensity was calculated using ImageJ software (National Institutes of Health, Bethesda, MD). To increase levels of ROS in HPAECs, cells were exposed to 250 nM of phorbol 12-myristate 13-acetate (PMA) for 24 hours. Quantitative detection and analysis of DCF fluorescence was performed using a Synergy HT microplate fluorometer (Synergy, Reading, PA).
MTT Assay
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) reagent was used to analyze cell viability and proliferation. Yellow MTT is reduced to purple formazan in the mitochondria of living cells. The reduction occurs only when mitochondrial reductase enzymes are active and therefore reflects the number of viable cells in the assay. HPAECs were seeded onto a 96-well plate at a density of 1.5 × 104 cells per well. The next day, cells were exposed to HDAC-42 (1 μM), scriptaid (8 μM), TSA (1.5 μM), or DMSO (as control) for 24 hours. After exposure, 10 μl of 5 mg/ml of MTT solution in DMSO was added to each well and incubated for 4 hours in a CO2 incubator at 37°C. The medium was removed, and 100 μl of DMSO was added to each well. Plates were incubated on a rotating platform for 5 minutes at room temperature. The optical density at 540 nm was measured using a SpectraMAX 250 plate reader (Molecular Devices, Sunnyvale, CA).
EC-SOD Immunoprecipitation
HPAECs were grown in 100-mm dishes to near confluency and then exposed to either DMSO or 10 μM scriptaid for 24 hours. Cells were washed with ice-cold PBS and lysed in RIPA buffer (50 mM Tris-HCl [pH 8.0], 0.15 M NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40) with protease and phosphatase inhibitors. Immunoprecipitation was performed using ImmunoCruz IP/WB Optima C System (SantaCruz Biotechnology, Santa Cruz, CA) and EC-SOD–specific antibodies (sc-32219). After several washes with ice-cold PBS, bound proteins were eluted from the beads by heating samples in loading buffer with denaturant SDS. Immunoprecipitated EC-SOD was detected using rabbit polyclonal anti–EC-SOD antibodies produced in our laboratory.
Results
Only HDAC Inhibitors Class 1 and 2 Induce Expression of EC-SOD
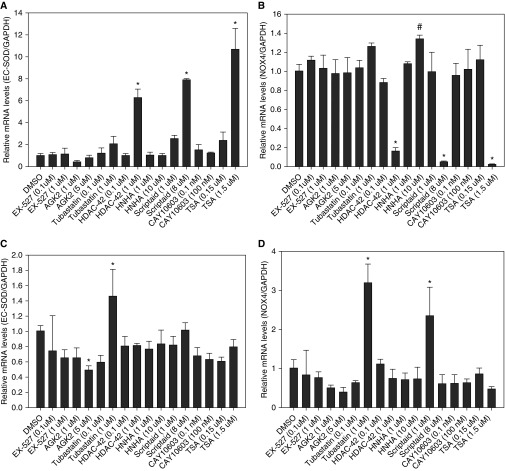
To analyze the effects of HDAC inhibitors on expression of the main antioxidant genes present in pulmonary artery endothelial cells, we exposed cells to an array of HDAC inhibitors. We found that scriptaid, HDAC-42, and TSA, which are all related to HDAC class 1 and 2 inhibitors, were very potent activators of EC-SOD gene expression. EC-SOD mRNA levels were induced up to 11-fold after 24 hours of incubation using these inhibitors (Figure 1A). At the same time, exposure of cells to these three HDAC inhibitors almost completely abrogated expression of the major prooxidant gene NOX4 (Figure 1B). EX527 and AGK2, specific inhibitors of SIRT 1 and 2, did not show any significant effect on expression of EC-SOD and NOX4 genes in HPAECs. Similarly, no gene expression effects were seen for tubastatin and CAY10603, specific inhibitors of HDAC6. Interestingly, these same HDAC inhibitors were unable to induce EC-SOD expression or reduce NOX4 expression in human pulmonary artery smooth muscle cells (Figures 1C and 1D).
Figure 1.
Effect of histone deacetylase (HDAC) inhibitors on expression of extracellular superoxide dismutase (EC-SOD) and NADPH oxidase (NOX) 4 genes in pulmonary artery cells. Cells were incubated with the indicated concentrations of HDAC inhibitors for 24 hours. Total RNA was isolated from human pulmonary artery endothelial cells (HPAECs) (A and B) or human pulmonary artery smooth muscle cells (C and D), and gene-specific messenger RNA (mRNA) levels were analyzed using quantitative RT-PCR. EC-SOD and NOX4 mRNA levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. The results are shown as mean ± SD (for primer sequences, see Table 1). *P < 0.001 and #P < 0.05 when compared with cells treated with DMSO (one-way ANOVA and Bonferroni test).
Table 1.
Primer Sequences for Quantitative RT-PCR
| Gene Name | Gene Bank Number | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Amplicon Size (bp) |
|---|---|---|---|---|
| EC-SOD | NM_003102 | TGCCCCGCGTCTTCAG | CCAAACATTCCCCCAAAGG | 71 |
| NOX4 | NM_016931 | TGGCTGCCCCTCTGGTGAATG | CAGCAGCCCTCCTGAAACATGC | 280 |
| LSD1 | NM_015013 | CAAGTGTCAATTTGTTCGGG | TTCTTTGGGCTGAGGTACTG | 217 |
| SMCX | NM_001282622 | GGCCAAAGACAAGACTCTGC | CCGTAGCCTCATGGTCATCT | 186 |
| RBP2 | NM_001042603 | CCTCCATTTGCCTGTGAAGT | CCTTTGCTGGCAACAATCTT | 208 |
| Sp1 | NM_138473 | TTGAAAAAGGAGTTGGTGGC | TGCTGGTTCTGTAAGTTGGG | 427 |
| Sp3 | NM_003111 | CCAGGATGTGGTAAAGTCTA | CTCCATTGTCTCATTTCCAG | 568 |
| GAPDH | NM_002046 | CCATGTTCGTCATGGGTGTGA | CATGGACTGTGGTCATGAGT | 152 |
| CyPB | NM_000942 | CCAACGCAGGCAAAGACACCAA | GCTCTCCACCTTCCGCACCA | 131 |
| NOX1 | NM_013955 | CACAAGAAAAATCCTTGGGTCAA | GACAGCAGATTGCGACACACA | 110 |
| NOX2 | NM_000397 | GTCACACCCTTCGCATCCATTCTCAAGTCAGT | CTGAGACTCATCCCAGCCAGTGAGGTAG | 225 |
| NOX3 | NM_015718 | TCTTCAACCTGGAACGCTAC | GACGCCTGCTATTGTCCTTA | 164 |
| NOX5 | NM_024505 | AACTTCTGGAAGTGGCTGCT | GAGGAGATGAGTGACCTTGGA | 196 |
| HDAC1 | NM_004964 | ATCGGTTAGGTTGCTTCA | TCATTCGTGTTCTGGTTAGTC | 265 |
| HDAC2 | NM_001527 | ACACAATCCGTAATGTTGCTCG | CACAGGTAGTCGTCCTGGTCCAAGGAT | 261 |
| HDAC3 | NM_003883 | ACGTGGGCAACTTCCACTAC | GACTCTTGGTGAAGCCTTGC | 219 |
| HDAC8 | NM_018486 | GGATCCCATGTGCTCCTTTA | ATAGCCTCCTCCTCCCAAAA | 106 |
| JAK2 | NM_004972 | TCACCAACATTACAGAGGCCTACTC | GCCAAGGCTTTCATTAAATATCAAA | 89 |
Definition of abbreviations: CyPB, cyclophilin B; EC-SOD, extracellular superoxide dismutase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HDAC, histone deacetylase; JAK, Janus kinase; LSD1, lysine-specific histone demethylase 1; NOX, NADPH oxidase; RBP2, retinoblastoma binding protein-2; SMCX, lysine (K)-specific demethylase 5C; Sp, specificity protein.
Next, we analyzed the time course of HDAC inhibitors regulation of EC-SOD and NOX4 genes. For EC-SOD, the maximal induction was observed after 72 hours incubation (Figure 2A). The highest inhibition of NOX4 gene expression was detected after 24 hours for scriptaid and HDAC-42 and at 48 hours for TSA (Figure 2B). To determine the optimal concentration of HDAC inhibitors that was required for maximal induction of EC-SOD, cells were exposed to different concentration of scriptaid and HDAC-42. We found that scriptaid at a concentration of 16 μM induced EC-SOD mRNA levels up to 32-fold after 24 hours of incubation (Figure 2C). Very similar but reciprocal effects were observed on NOX4 mRNA levels (Figure 2D).
Figure 2.
Analysis of time-course and concentration effects of HDAC inhibitors on gene expression in HPAECs. (A and B) Cells were exposed to scriptaid (8 μM), N-[4-[(hydroxyamino)carbonyl]phenyl]-α-(1-methylethyl)-benzeneacetamide [(S)-HDAC-42] (1 μM), and trichostatin A (TSA) (1.5 μM) for 24, 48, and 72 hours. Control cells were exposed to vehicle (DMSO) for the indicated time. (C and D) Cells were exposed to different concentrations of scriptaid (SA) or (S)-HDAC-42 for 24 hours. Control cells were exposed to DMSO. Total RNA was isolated, and gene-specific mRNA levels were analyzed using quantitative RT-PCR. EC-SOD (A and C) and NOX4 (B and D) mRNA levels were normalized to GAPDH expression. (E) Analysis of EC-SOD protein levels in cell lysates using immunoprecipitation (IP) and Western blot of HPAECs exposed to 10 μM scriptaid for 24 hours. (F) Analysis of NOX4 protein levels using Western blot of HPAECs exposed to 5 μM scriptaid for 24 and 48 hours. Digital adjustment of brightness and contrast was applied to the whole blot images using Adobe Photoshop (Adobe). (G) Quantitative analysis of Western blot depicted in F. The results are shown as mean ± SD. *P < 0.001, **P < 0.01, and #P < 0.05 and when compared with cells treated with DMSO at the corresponding time (one-way ANOVA with Bonferroni post test).
Differential effects of scriptaid on EC-SOD and NOX4 protein levels in HPAECs were analyzed using Western blot. As expected, exposure of HPAECs to 10 μM scriptaid increases protein levels of EC-SOD (Figure 2E) but attenuated NOX4 protein levels (Figures 2F and 2G).
Treatment of HPAECs with HDAC Inhibitors Attenuates Oxidative Stress
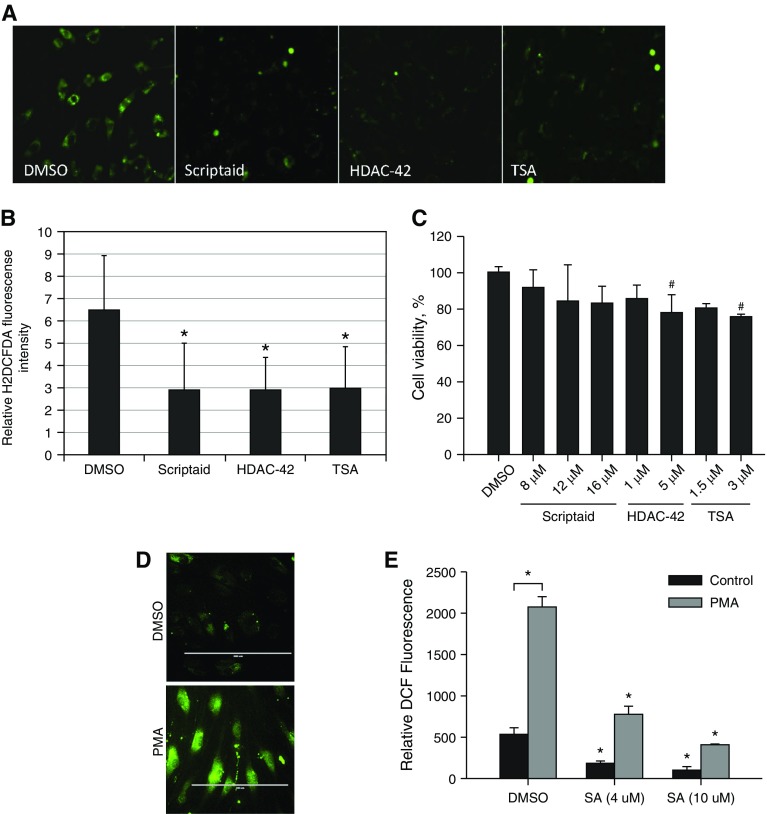
The analysis of gene expression in response to HDAC inhibitors in HPAECs indicated that expression of the major antioxidant gene EC-SOD was significantly up-regulated, whereas expression of the major prooxidant enzyme in pulmonary vasculature, NOX4, was almost completely abrogated. These results imply that HDAC inhibitors have the potential to reduce oxidative stress in HPAECs. To analyze this possibility, HPAECs were exposed to HDAC inhibitors (scriptaid, HDAC-42, and TSA), and then levels of ROS were measured using a highly fluorescent marker of oxidative stress. Treatment with all three HDAC inhibitors significantly attenuated ROS levels in HPAECs, as detected using fluorescent microscopy and fluorescent image analysis (Figures 3A and 3B). The level of emitted fluorescence was reduced by half after treatment. At the same time, HPAECs did not show any significant reduction in cell viability after exposure to the same treatment (Figure 3C). To investigate the antioxidant effects of scriptaid in a pathologically relevant in vitro model, HPAECs were exposed to inflammatory agonist PMA alone or in combination with increasing concentrations of scriptaid for 24 hours. Exposure to PMA induced DCF fluorescence from 532.3 ± 70.5 RFU to 2,074.0 ± 123.5 RFU, whereas scriptaid attenuated ROS-induced DCF fluorescence signal in dose-dependent manner (Figures 3D and 3E).
Figure 3.
Attenuation of reactive oxygen species levels in HPAECs after treatment with HDAC inhibitors. (A) HPAECs were exposed to indicated HDAC inhibitors or DMSO for 24 hours. Cells were loaded with dihydrodichlorofluorescein (H2DCF) for 30 minutes and then incubated with inhibitors for an additional 4 hours. H2DCF is converted to highly fluorescent 2,7-dichlorofluorescein (DCF) by reactive oxygen species (DMSO, scriptaid [8 μM], HDAC-42 [1 μM], and TSA [1.5 μM]). Images were subjected to a uniform adjustment of brightness and contrast before analysis. (B) Quantitative analysis of cell fluorescence. *P < 0.001 (one-way ANOVA with Bonferroni post test; n = 14). (C) Effect of HDAC inhibitors on the viability of HPAECs. Cells were exposed to the indicated concentrations of HDAC inhibitors for 24 hours. Cell viability was determined using MTT assay as described in Materials and Methods. #P < 0.05 determined using one-way ANOVA with Bonferroni post test. (D) HPAECs were exposed to 250 nM of phorbol 12-myristate 13-acetate (PMA) for 24 hours. Cells were loaded with H2DCF for 30 minutes and then incubated for additional 30 minutes. DCF fluorescence was visualized by fluorescent microscopy. Scale bars = 200 μm. (E) HPAECs were exposed to DMSO or to 250 nM of PMA for 24 hours. SA was added to the incubation medium at the indicated concentration at the time of PMA addition. Cells were loaded with H2DCF for 30 minutes, and then DCF fluorescent signal was detected using microplate fluorometry. *P < 0.01 (one-way ANOVA with Bonferroni post test; n = 3). H2DCFDA, dihydrodichlorofluorescein diacetate.
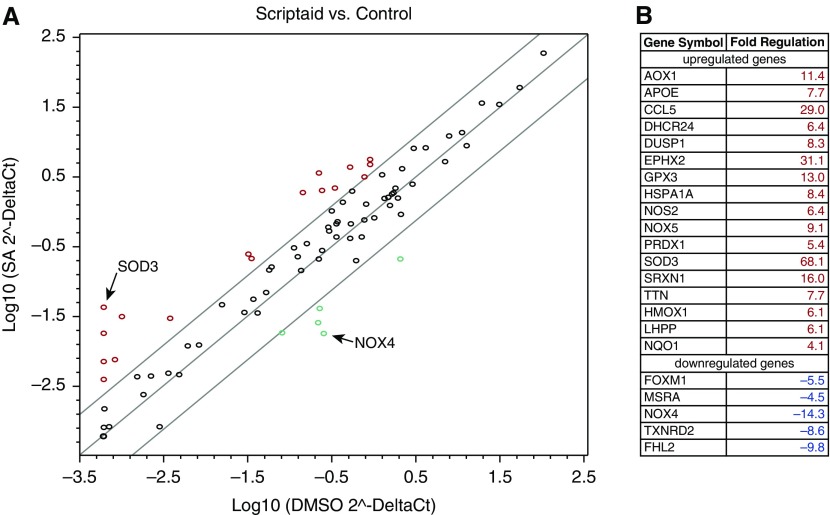
Furthermore, we investigated the effect of scriptaid on the expression of genes involved in regulation of oxidative stress. As expected, NOX4 and EC-SOD genes were the most up- and down-regulated genes among more than 80 genes analyzed (Figure 4). Although scriptaid induced the expression of prooxidant gene NOX5 up to 9.1-fold, the overall expression levels of NOX5 gene were more than 2,000-fold lower compared with NOX4 mRNA levels (see Table E1 in the online supplement). Thus, the increase in NOX5 expression levels can be considered to have negligible effects on the redox balance in HPAECs. These data indicate that HDAC inhibitors reduce the oxidative stress seen in endothelial cells, likely by modifying expression of anti- and prooxidant enzymes.
Figure 4.
Quantitative RT-PCR array analysis of oxidative stress–related genes. (A) HPAECs were exposed to either DMSO or 10 μM scriptaid for 24 hours. Synthesis of cDNA was performed using the RT2 First Strand kit, and PCR was performed using the RT2 Profiler PCR Array. Samples from SA-treated HPAECs were compared with DMSO-treated cells. (B) List of up- and down-regulated genes was determined using RT-PCR array.
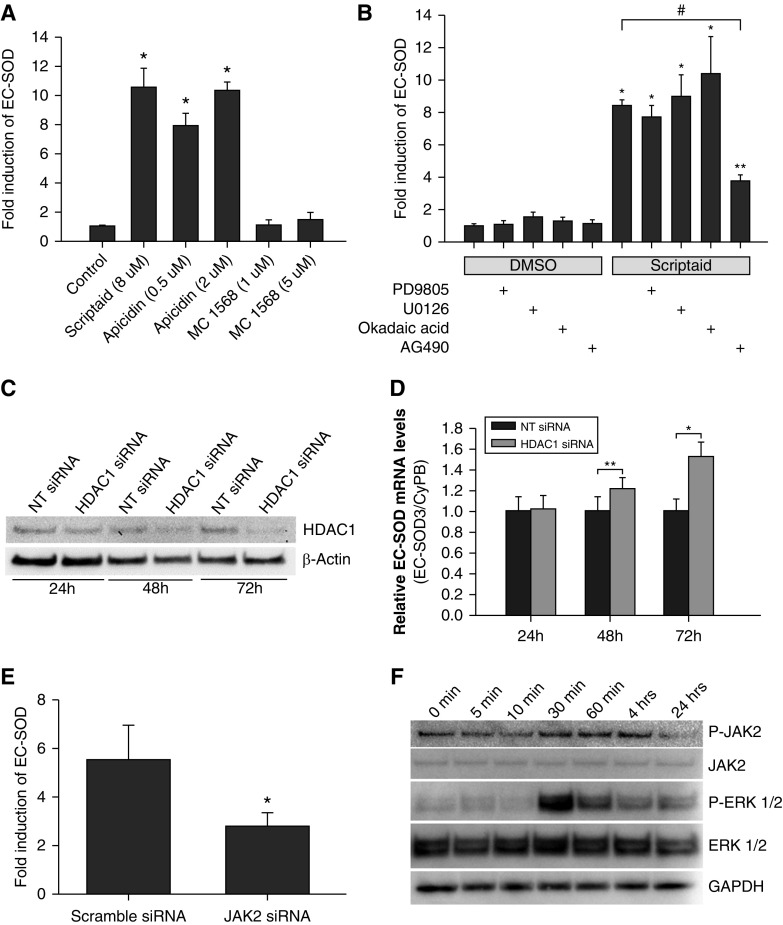
Effect of Selective HDAC Inhibitors and HDAC1 Silencing on EC-SOD Expression
To determine which isoforms of HDACs are responsible for induction of EC-SOD gene expression in HPAECs, we exposed cells to two highly selective inhibitors: apicidin, an inhibitor of class 1 HDAC (HDAC1, HDAC2, HDAC3, and HDAC8), and MC 1568, an inhibitor of class 2 HDAC (HDAC4, HDAC5, HDAC7, and HDAC9). EC-SOD gene expression was induced only with apicidine to the same extent as with scriptaid, whereas MC 1568 compound had no effect on EC-SOD mRNA levels (Figure 5A). In addition, we found that the potent and highly selective inhibitor of bromodomain and extra-terminal (BET) bromodomain (JQ1) did not significantly change EC-SOD expression (data not shown). We found that a specific inhibitor of the Janus kinase 2 (JAK2) protein, AG490, attenuated induction of EC-SOD by scriptaid by more than 50% (from 8.4- to 3.7-fold) (Figure 5B). On the other hand, the phosphatase inhibitors okadaic acid, PD98059, and U0126 did not produced any significant effects on EC-SOD expression or its induction by scriptaid.
Figure 5.
Regulation of EC-SOD expression in HPAECs. (A) HPAECs were exposed to the indicated HDAC inhibitors for 24 hours. (B) HPAECs were exposed to PD98059 (10 μM), U0126 (5 μM), okadaic acid (5 nM), or AG490 (25 μM) for 30 minutes before exposure either to DMSO or scriptaid (8 μM) for 24 hours. (C) Analysis of HDAC1 protein levels in HPAECs transfected with nontargeted (NT) small interfering RNA (siRNA) or siRNA specific for HDAC1. (D) Time-dependent effects of HDAC1 silencing on EC-SOD expression in HPAECs. (E) HPAECs were transfected with either scrambled siRNA or Janus kinase 2 (JAK2)-specific siRNA for 48 hours. Then, cells were incubated in complete medium with or without scriptaid for 24 hours. Total RNA was isolated, and EC-SOD–specific mRNA levels were analyzed using quantitative RT-PCR and normalized to GAPDH expression. The results are shown as mean ± SD. (F) Western blot was performed to detect changes in the phosphorylation of JAK2 and extracellular signal-regulated protein kinases 1 and 2 (ERK1/2) after exposure of HPAECs to scriptaid. *P < 0.001 and **P < 0.05 when compared with cells treated with DMSO; #P < 0.001 when compared with cells treated with only scriptaid (one-way ANOVA with Bonferroni post test).
Class I HDAC consists of four members: HDAC1, HDAC2, HDAC3, and HDAC8. To determine which isoform is expressed at the highest levels in HPAECs and most likely involved in regulation of EC-SOD expression, we performed quantitative RT-PCR. HDAC1 showed the highest expression levels among all four members of this class (Figure E1) and was chosen as a candidate for silencing using small interfering RNA (siRNA).
Next, we analyzed the effects of HDAC1 silencing on EC-SOD gene expression in HPAECs. The level of HDAC1 protein was significantly attenuated by siRNA technology in a time-dependent manner (Figure 5C). The reciprocal increase in EC-SOD gene expression was observed at the same time points (Figure 5D). These data confirm that expression of EC-SOD gene in HPAECs is regulated, at least in part, by HDAC1 protein.
To elucidate the molecular mechanisms responsible for AG490-dependent attenuation of EC-SOD gene induction by scriptaid, we reduced expression of JAK2 using siRNA technology. Transfection of JAK2-specific siRNA significantly decreased levels of JAK2 expression in HPAECs at mRNA and protein levels (Figure E2A and E2B). This specific attenuation of JAK2 expression reduced EC-SOD induction by scriptaid from 5.49 ± 1.42-fold to 2.77 ± 0.57-fold (P = 0.037) (Figure 5E). In addition, we analyzed the effects scriptaid on the phosphorylation status of JAK2, extracellular signal-regulated protein kinases 1 and 2 (ERK1/2) and STAT3. We found that scriptaid induces phosphorylation of JAK2 at tyrosine 1,007 and 1,008 and phosphorylation of ERK1/2 at threonine 202 and tyrosine 204 starting 30 minutes after exposure (Figure 5F). Interestingly, the phosphorylation status of these proteins returned to normal levels at 24 hours after exposure. These data indicate that scriptaid exposure increases phosphorylation of JAK2 and ERK1/2 at least during early stages of activation. Thus, AG490 inhibitor can attenuate scriptaid-induced EC-SOD expression through inhibition of JAK2 and ERK1/2 phosphorylation and activation.
HDAC Inhibitors Increase Histone Acetylation but Do Not Induce Sp1/Sp3 Expression
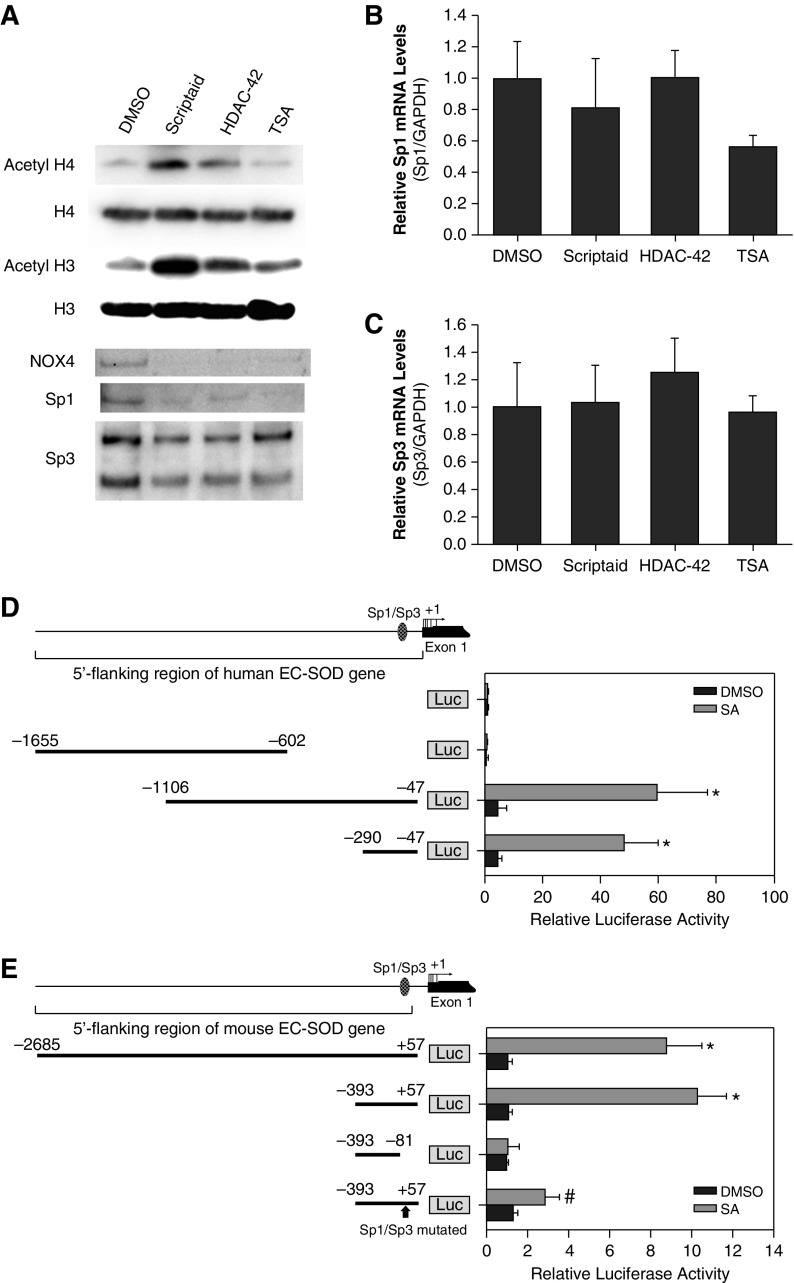
The effects of HDAC inhibitors on acetylation status of histones H3 and H4 were analyzed using Western blot. Scriptaid was the most potent inhibitor to increase acetylation status of histones, whereas HDAC-42 and TSA showed only mild effects (Figure 6A). It has been shown previously that Sp1/Sp3 transcription factors play important roles in EC-SOD basal and inducible expression. Thus, we analyzed the effects of HDAC inhibitors on Sp1/Sp3 mRNA and protein levels. We found that exposure to scriptaid, HDAC-42, or TSA only slightly attenuated levels of these transcription factors and had no significant effects on their mRNA levels (Figures 6B and 6C).
Figure 6.
Effect of HDAC inhibitors on the expression of specificity protein (Sp) 1/Sp3 and histone acetylation. (A) Western blot of histone modification, NOX4, and Sp1/Sp3 protein levels in HPAECs exposed to the indicated HDAC inhibitors. The same amount of protein was loaded in each well. Digital adjustment of brightness and contrast was applied to the whole blot images using Adobe. (B and C) Analysis of Sp1 and Sp3 expression in response to HDAC inhibitors. HPAECs were incubated with HDAC-42 (1 μM), scriptaid (8 μM), or TSA (1.5 μM) for 24 hours. Levels of mRNA were analyzed using quantitative RT-PCR and normalized to GAPDH expression. Data are presented as mean ± SD. (D and E) Identification of scriptaid responsive promoter elements within the EC-SOD gene. 5′-Regions of the human (A) and mouse (B) EC-SOD gene were cloned in front of luciferase (Luc) reporter gene in pGL3-Basic vector. The promoter–reporter constructs were transiently transfected into HPAECs, and firefly luciferase activity was measured 18 hours later. Reporter activity was normalized to Renilla luciferase activity produced by the cotransfection of control plasmid pRL-CMV. Results shown as mean ± SD from at least two independent transfection experiments, each performed in quadruplicate. The hatched oval represents the Sp1/Sp3 consensus binding site. *P < 0.001; #P < 0.01 (t test).
Effect of Scriptaid on Activation of EC-SOD Proximal Promoter
We investigated the role of cis-elements located in the 5′-flanking region of the EC-SOD gene that might direct induction of EC-SOD gene expression by scriptaid in HPAECs. Transient transfection of HPAECs with the wild-type pGL3-hSOD3(−1,106/−47) reporter plasmid after exposure to scriptaid for 20 hours showed marked induction of the reporter activity (Figure 6D). The 5′-flanking region truncated to only 240 bp was still responsive to scriptaid treatment, suggesting that scriptaid responsive cis-elements are located in this region. In addition, we performed similar experiments using promoter-reporter constructs derived from mouse EC-SOD gene. As we expected, treatment with scriptaid induced reporter expression up to 10-fold (Figure 6E). Next, we determined whether the scriptaid-responsive element colocalized with the Sp1/Sp3 binding site in the mouse EC-SOD promoter region. Mutation of a functional Sp1/Sp3 binding site that we have previously shown to regulate basal promoter activity, pGL3-mSOD3(−208/+242)mut(+93/+96), significantly attenuated promoter activity induced by scriptaid in HPAECs (Figure 6E). The same effect was observed with a plasmid bearing a deleted Sp1/Sp3 binding site. In addition, we analyzed binding of Sp1 to the EC-SOD promoter before and after scriptaid exposure. Treatment with 8 μM of scriptaid for 8 hours did not change occupancy of the EC-SOD promoter by Sp1 transcription factor (Figure 7A). Prolonged exposure of HPAECs to scriptaid for 24 hours did not change Sp1 abundance at the proximal promoter as well (data not shown). These data indicate that the Sp1/Sp3 binding site located between nucleotides +93 and +96 is the main scriptaid-responsive element in the EC-SOD promoter region. These results indicate that HDAC inhibitors activate the EC-SOD promoter mostly through the putative Sp1/Sp3 binding site but do not change the expression levels of these trans-factors or their binding to the promoter.
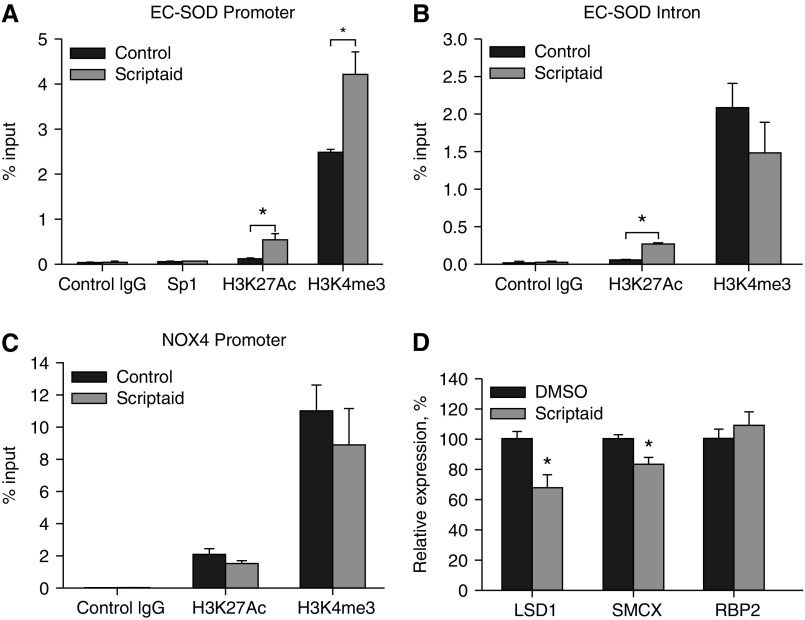
Figure 7.
Analysis of histone acetylation and methylation at the EC-SOD and NOX4 promoters. HPAECs were exposed to DMSO (control) or scriptaid (8 μM) for 8 hours. Binding of Sp1 transcription factor and histone H3 acetylated at lysine 27 (H3K27Ac) and trimethylated at lysine 4 (H3K4 me3) were analyzed using chromatin immunoprecipitation assay with corresponding antibodies. Corresponding nonimmune IgG were used as control. Abundance of purified DNA fragments was analyzed using quantitative PCR with primers specific for the EC-SOD promoter (A), the EC-SOD intron region (B), and the NOX4 promoter (C). (D) Analysis of histone demethylases expression in HPAECs after exposure to scriptaid (8 μM) for 24 hours (see Table 2). *P < 0.01. LSD1, lysine-specific histone demethylase 1; RBP2, retinoblastoma binding protein-2 also known as lysine (K)-specific demethylase 5A (KDM5a); SMCX, lysine (K)-specific demethylase 5C.
Table 2.
Primer Sequences for Chromatin Immunoprecipitation Quantitative PCR
| Amplification TargetName | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Amplicon size (bp) |
|---|---|---|---|
| EC-SOD promoter | GGCCTGCTTTTCCTCCCTGA | CAGCCAGCCCAGGAACGCAG | 128 |
| NOX4 promoter | AGGACCGAGGGTCAAAGACT | GTCTGGGCAGCTGAGTGG | 144 |
| EC-SOD intron | AAAACCAGACATCTGATGTG | AGGATTAGTTCAGCTGGAGT | 160 |
Definition of abbreviations: EC-SOD, extracellular superoxide dismutase; NOX, NADPH oxidase.
Methylation and Acetylation of Histones at the EC-SOD and NOX4 Promoters
To investigate the acetylation and methylation status of histones at the EC-SOD and NOX4 promoters, we performed chromatin immunoprecipitation assays with antibodies specific for histone H3 acetylated at Lys 27 (H3K27Ac) and histone H3 trimethylated at Lys 4 (H3K4 me3). First, we found that, in untreated cells, acetylated histones mostly associated with the promoter of highly expressed NOX4 gene (2% of input), whereas their presence at the EC-SOD promoter and intron regions was significantly lower (0.12%) (Figures 7A–7C). Treatment of HPAECs with scriptaid for 8 hours significantly increased acetylation of histones at the EC-SOD promoter from 0.12 to 0.55%, whereas there was no effect on acetylation of histones associated with the NOX4 promoter. Next, we analyzed methylation status of histone H3 at both gene promoters. We were specifically interested in H3K4 me3 modification because it had been associated with actively transcribed chromatin regions. We identified a significantly higher level of H3K4 me3 modification at the Nox4 promoter (12% input) compared with the EC-SOD promoter (2.8% input). Moreover, treatment of HPAECs with scriptaid significantly increased methylation of histone H3 at the EC-SOD promoter, whereas it had no effect on histone H3 located at the NOX4 promoter or at the EC-SOD intron region. These data indicate that, in addition to altering the acetylation status of histones at the EC-SOD promoter, scriptaid increases the methylation of histone H3 at Lys4. Because the steady state of histone methylation depends on the balance between activities of histone methylases and histone demethylases, we analyzed expression levels of three demethylases that catalyze the removal of the methyl groups from H3 Lys 4. We found that scriptaid significantly attenuated expression of two isoforms of histone demethylases (LSD1 and SMCX), whereas it had no effect on RBP2 demethylase expression (Figure 7D). These data indicated that the increased methylation status of histone H3 after scriptaid treatment is possibly due to attenuation of histone demethylase expression. The detailed exploration of molecular mechanisms involved in this regulation needs further experimental investigation.
Discussion
In this study, we identified differential regulation of the prooxidant gene NOX4 and the major antioxidant enzyme EC-SOD by class 1 HDAC inhibitors in HPAECs. Moreover, we found that exposure of HPAECs to these inhibitors attenuated oxidative stress. Up-regulation of EC-SOD expression was attributed to the promoter-specific acetylation and methylation of histones. Analysis of the wide array of HDAC inhibitors indicated that only three inhibitors (scriptaid, HDAC-42, and TSA) were able to induce EC-SOD expression and attenuate NOX4 mRNA levels. These three inhibitors have relatively broad specificity targeted toward HDAC class 1 and 2. On the other hand, specific inhibitors of HDAC6, tubastatin, and CAY10603, as well as inhibitors of HDAC class 3 (sirtuins), likely have no effect on the expression of these two genes. Using more specific HDAC inhibitors, we identified that only HDAC class 1 inhibitors play a role in differential regulation of EC-SOD and NOX4 genes, whereas HDAC class 2 inhibitors do not appear to be involved in this process. It has been shown that HDACs are not redundant in their biological activity. Class 1 HDACs are involved in regulation of cell proliferation and apoptosis, whereas class 2 HDACs appear to be important in regulation of tissue-specific functions. Moreover, exposure of HPAECs to scriptaid affected the expression of other genes involved in regulation of oxidative stress. The effects of scriptaid might have broad, pleiotropic effects. For example, several other antioxidant genes were strongly up-regulated, such as glutathione peroxidase 3, sulfiredoxin 1, and epoxide hydrolase 2. Interestingly, the ATOX 1 gene involved in the delivery of copper to the active site of EC-SOD was also up-regulated after exposure to scriptaid (21). Thus, attenuation of oxidative stress by scriptaid might not be attributed solely to EC-SOD and NOX4 modulation.
Our data indicate that scriptaid attenuates ROS production in endothelial cells. These observations are in line with recently published studies indicating suppression of oxidative stress in vitro and in vivo by the histone deacetylase inhibitor β-hydroxybutyrate (22). In this study, oxidative stress was attenuated through up-regulation of FOXO3a and MT2 genes: mice treated with β-hydroxybutyrate became more resistant to oxidative stress. Ryu and colleagues showed that the HDAC inhibitor suberoylanilide hydroxamic acid abrogated neuronal cell death induced by oxidative stress in vitro and in vivo via augmentation of Sp1 acetylation (23). Swingler and colleagues demonstrated that induction of MMP28 gene by HDAC inhibitors are mediated through acetylation of Sp1/Sp3 via HDAC 1 (24). We found that Sp1 and Sp3 transcription factors regulate basal and inducible expression of EC-SOD in pulmonary cells (12, 14). Thus, acetylation of Sp1/Sp3 transcription factors, in response to HDAC inhibitors, is a plausible molecular mechanism for regulating the induction of EC-SOD in endothelial cells. The molecular mechanisms responsible for attenuation of NOX4 expression by scriptaid in transformed cells and HUVECs were described recently (25). We also found some differences between our results and data from Siuda and colleagues (25). We found that exposure of HPAECs to scriptaid did not significantly change the acetylation and methylation status of histone H3 at the NOX4 promoter, whereas their data indicated increased acetylation of histones at this location. These discrepancies could be attributed to the different cell types used in our studies (primary HPAECs versus HUVECs and HUVEC-derived EA.hy 926 cells). Our data are in line with these studies and with our previous reports of EC-SOD induction by TSA in various transformed lung cell lines (12, 16).
Increases in the concentration of ROS have been detected in transformed cells exposed to variety of HDAC inhibitors, including TSA, vorinostat, and sodium butyrate (26, 27). It has been proposed that acute toxicity of HDAC inhibitors can be attributed to the generation of ROS in transformed cells, whereas primary cells are more resistant due to significantly lower ROS levels. In our experimental setting, we measured ROS after exposure to scriptaid for 24 hours, when expression of EC-SOD was significantly induced and NOX4 expression was attenuated.
Our data indicate that the JAK2/STAT3 and p44/42 ERK1/2 signaling pathways might be involved in the regulation of EC-SOD gene expression because scriptaid induces phosphorylation of ERK1/2 and JAK2 protein kinases and because exposure to AG490 attenuated scriptaid-induced EC-SOD induction by more than 50%. AG490 is a tyrosine kinase inhibitor of JAK2, EGFR, and ERK1/2 that belongs to the family of tyrphostins. JAK2-mediated phosphorylation activates STAT3 transcription factor. Activated STAT3 translocates to the nucleus, where it binds to DNA and regulates gene transcription. This observation is in line with the previously published observation that the STAT3 pathway may be important in transducing HDAC-initiated signaling in activated renal interstitial fibroblasts (28). In addition, it has been shown that TSA inhibits STAT3-dependent transcriptional activity induced by platelet-derived growth factor (29). On the other hand, exposure to okadaic acid does not affect induction of EC-SOD by scriptaid in HPAECs, whereas a similar exposure to okadaic acid attenuated induction of CYP46A1 gene by TSA in neuroblastoma cells (30).
We found that exposure of endothelial cells to scriptaid not only increases histone acetylation but also significantly affects trimethylation of histones H3 at lysine 4. These modifications were specific for the EC-SOD promoter region and did not occur at the NOX4 promoter or the EC-SOD intron region. The single lysine residue can be variably methylated to mono-, di, and trimethylated states. Activated promoters are enriched in trimethylated H3 lysine 4 (H3Kme3) residues (31), whereas di- and trimethylated histone H3 lysine 9 residues are strongly correlated with transcriptional repression (32). The methylation of histones in response to exposure to histone deacetylase inhibitors has been described previously. It has been shown that class I–specific HDAC inhibitor increases H3K4 me2 and H3K4 me3 levels in rat cortical neurons and astrocytes and regulate heat-shock protein expression (33). Different cross-talk mechanisms were suggested between histone acetylation and histone methylation systems. The activity of H3K4 methyl transferase MLL4 was stimulated by acetylated peptides (34) or HDAC inhibitors (35). It has been shown that histone deacetylase inhibitors up-regulate histone H3 lysine 4 methylation (36). This effect was attributable to suppression of the RBP2 and JARID1 family of histone demethylases, including PLU-1, SMCX, and LSD1. The LSD1 gene encodes a flavin-dependent monoamine oxidase, which can demethylate mono- and dimethylated lysines, specifically histone 3, and lysines 4 and 9 (H3K4 and H3K9). The SMCX gene encodes histone demethylase, which specifically demethylates lysine 4 on histone 3. LSD-1, also known as KDM1A, produces hydrogen peroxide, which oxidizes bases in targeted promoters and enhancers (37).
Data presented in our work indicate that HDAC inhibitors can potentially be used to alleviate pulmonary vascular diseases associated with endothelial oxidative stress. Indeed, several histone deacetylase inhibitors were tested for prevention or attenuation of hypertension development or heart failure. The beneficial effects of TSA in preventing heart failure have been shown in mice subjected to aortic constriction (38). Treatment with TSA and scriptaid, another HDAC inhibitor, for 3 weeks significantly reduced cardiac hypertrophy and cardiomyocyte size in a pressure-overload mouse model (39). Recent data indicate that selective inhibitor of class I HDACs (HDAC1, -2, and -3) reduced pulmonary arterial pressure and vascular wall thickening in a rat model of hypoxia-induced pulmonary arterial hypertension (18). Similar results were observed when animals were treated with valproic acid, a class I HDAC inhibitor, and suberoylanilide hydroxamic acid (vorinostat), an inhibitor of class I, II, and IV HDACs (40).
In summary, our study demonstrates a new approach to modify the levels of ROS in pulmonary endothelial cells. Our results show that HDAC inhibitors are potent modulators of ROS levels in pulmonary endothelium, most likely through differential regulation of EC-SOD and NOX4 enzymes. We also demonstrate that up-regulation of EC-SOD gene expression by scriptaid is associated with acetylation and methylation of specific lysines at H3 histones located in close proximity to the proximal promoter. Although the exact mechanism of EC-SOD up-regulation by HDAC inhibitors is yet to be defined, our studies indicate a potential role for the JAK2/STAT signaling pathway in the regulation of EC-SOD expression in pulmonary vasculature. Further studies are in progress to unravel the role of epigenetic signaling pathways in regulating oxidative stress and ROS production in the vascular endothelium.
Footnotes
This work was supported by a grant from the American Lung Association Biomedical Research and by a University of Louisville Intramural Research Incentive grant.
Author Contributions: Conception and design: I.N.Z. Analysis and interpretation: I.N.Z. and R.J.F. Drafting the manuscript for important intellectual content: I.N.Z. and R.J.F.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0260OC on March 6, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Li S, Tabar SS, Malec V, Eul BG, Klepetko W, Weissmann N, Grimminger F, Seeger W, Rose F, Hanze J. NOX4 regulates ROS levels under normoxic and hypoxic conditions, triggers proliferation, and inhibits apoptosis in pulmonary artery adventitial fibroblasts. Antioxid Redox Signal. 2008;10:1687–1698. doi: 10.1089/ars.2008.2035. [DOI] [PubMed] [Google Scholar]
- 2.Hoshikawa Y, Ono S, Suzuki S, Tanita T, Chida M, Song C, Noda M, Tabata T, Voelkel NF, Fujimura S. Generation of oxidative stress contributes to the development of pulmonary hypertension induced by hypoxia. J Appl Physiol. 2001;90:1299–1306. doi: 10.1152/jappl.2001.90.4.1299. [DOI] [PubMed] [Google Scholar]
- 3.Al-Mehdi AB, Zhao G, Dodia C, Tozawa K, Costa K, Muzykantov V, Ross C, Blecha F, Dinauer M, Fisher AB. Endothelial NADPH oxidase as the source of oxidants in lungs exposed to ischemia or high K+ Circ Res. 1998;83:730–737. doi: 10.1161/01.res.83.7.730. [DOI] [PubMed] [Google Scholar]
- 4.Voelkel NF, Cool C, Lee SD, Wright L, Geraci MW, Tuder RM. Primary pulmonary hypertension between inflammation and cancer. Chest. 1998;114:225S–230S. doi: 10.1378/chest.114.3_supplement.225s. [DOI] [PubMed] [Google Scholar]
- 5.Sturrock A, Cahill B, Norman K, Huecksteadt TP, Hill K, Sanders K, Karwande SV, Stringham JC, Bull DA, Gleich M, et al. Transforming growth factor-beta1 induces Nox4 NAD(P)H oxidase and reactive oxygen species-dependent proliferation in human pulmonary artery smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2006;290:L661–L673. doi: 10.1152/ajplung.00269.2005. [DOI] [PubMed] [Google Scholar]
- 6.Kunsch C, Medford RM. Oxidative stress as a regulator of gene expression in the vasculature. Circ Res. 1999;85:753–766. doi: 10.1161/01.res.85.8.753. [DOI] [PubMed] [Google Scholar]
- 7.Stralin P, Karlsson K, Johansson BO, Marklund SL. The interstitium of the human arterial wall contains very large amounts of extracellular superoxide dismutase. Arterioscler Thromb Vasc Biol. 1995;15:2032–2036. doi: 10.1161/01.atv.15.11.2032. [DOI] [PubMed] [Google Scholar]
- 8.Oury TD, Day BJ, Crapo JD. Extracellular superoxide dismutase: a regulator of nitric oxide bioavailability. Lab Invest. 1996;75:617–636. [PubMed] [Google Scholar]
- 9.Marklund SL. Expression of extracellular superoxide dismutase by human cell lines. Biochem J. 1990;266:213–219. doi: 10.1042/bj2660213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson K, Marklund SL. Binding of human extracellular-superoxide dismutase C to cultured cell lines and to blood cells. Lab Invest. 1989;60:659–666. [PubMed] [Google Scholar]
- 11.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelko IN, Folz RJ. Sp1 and Sp3 transcription factors mediate trichostatin A-induced and basal expression of extracellular superoxide dismutase. Free Radic Biol Med. 2004;37:1256–1271. doi: 10.1016/j.freeradbiomed.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Zelko IN, Folz RJ. Myeloid zinc finger (MZF)-like, Kruppel-like and Ets families of transcription factors determine the cell-specific expression of mouse extracellular superoxide dismutase. Biochem J. 2003;369:375–386. doi: 10.1042/BJ20021431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelko IN, Mueller MR, Folz RJ. Transcription factors sp1 and sp3 regulate expression of human extracellular superoxide dismutase in lung fibroblasts. Am J Respir Cell Mol Biol. 2008;39:243–251. doi: 10.1165/rcmb.2007-0378OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelko IN, Stepp MW, Vorst AL, Folz RJ. Histone acetylation regulates the cell-specific and interferon-gamma-inducible expression of extracellular superoxide dismutase in human pulmonary arteries. Am J Respir Cell Mol Biol. 2011;45:953–961. doi: 10.1165/rcmb.2011-0012OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zelko IN, Mueller MR, Folz RJ. CpG methylation attenuates Sp1 and Sp3 binding to the human extracellular superoxide dismutase promoter and regulates its cell-specific expression. Free Radic Biol Med. 2010;48:895–904. doi: 10.1016/j.freeradbiomed.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teoh-Fitzgerald ML, Fitzgerald MP, Jensen TJ, Futscher BW, Domann FE. Genetic and epigenetic inactivation of extracellular superoxide dismutase promotes an invasive phenotype in human lung cancer by disrupting ECM homeostasis. Mol Cancer Res. 2012;10:40–51. doi: 10.1158/1541-7786.MCR-11-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavasin MA, Demos-Davies K, Horn TR, Walker LA, Lemon DD, Birdsey N, Weiser-Evans MC, Harral J, Irwin DC, Anwar A, et al. Selective class I histone deacetylase inhibition suppresses hypoxia-induced cardiopulmonary remodeling through an antiproliferative mechanism. Circ Res. 2012;110:739–748. doi: 10.1161/CIRCRESAHA.111.258426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zschoernig B, Mahlknecht U. SIRTUIN 1: regulating the regulator. Biochem Biophys Res Commun. 2008;376:251–255. doi: 10.1016/j.bbrc.2008.08.137. [DOI] [PubMed] [Google Scholar]
- 20.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh S, Ozumi K, Kim HW, Nakagawa O, McKinney RD, Folz RJ, Zelko IN, Ushio-Fukai M, Fukai T. Novel mechanism for regulation of extracellular SOD transcription and activity by copper: role of antioxidant-1. Free Radic Biol Med. 2009;46:95–104. doi: 10.1016/j.freeradbiomed.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu H, Lee J, Olofsson BA, Mwidau A, Dedeoglu A, Escudero M, Flemington E, Azizkhan-Clifford J, Ferrante RJ, Ratan RR. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci USA. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swingler TE, Kevorkian L, Culley KL, Illman SA, Young DA, Parker AE, Lohi J, Clark IM. MMP28 gene expression is regulated by Sp1 transcription factor acetylation. Biochem J. 2010;427:391–400. doi: 10.1042/BJ20091798. [DOI] [PubMed] [Google Scholar]
- 25.Siuda D, Zechner U, El Hajj N, Prawitt D, Langer D, Xia N, Horke S, Pautz A, Kleinert H, Forstermann U, et al. Transcriptional regulation of Nox4 by histone deacetylases in human endothelial cells. Basic Res Cardiol. 2012;107:283. doi: 10.1007/s00395-012-0283-3. [DOI] [PubMed] [Google Scholar]
- 26.Ruefli AA, Ausserlechner MJ, Bernhard D, Sutton VR, Tainton KM, Kofler R, Smyth MJ, Johnstone RW. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci USA. 2001;98:10833–10838. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis M, Rosato RR, Brault L, Osbild S, Battaglia E, Yang XH, Grant S, Bagrel D. The histone deacetylase inhibitor sodium butyrate induces breast cancer cell apoptosis through diverse cytotoxic actions including glutathione depletion and oxidative stress. Int J Oncol. 2004;25:1701–1711. [PubMed] [Google Scholar]
- 28.Pang MY, Kothapally J, Mao HP, Tolbert E, Ponnusamy M, Chin YE, Zhuang SG. Inhibition of histone deacetylase activity attenuates renal fibroblast activation and interstitial fibrosis in obstructive nephropathy. Am J Physiol-Renal. 2009;297:F996–F1005. doi: 10.1152/ajprenal.00282.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catania A, Iavarone C, Carlomagno SM, Chiariello M. Selective transcription and cellular proliferation induced by PDGF require histone deacetylase activity. Biochem Biophys Res Commun. 2006;343:544–554. doi: 10.1016/j.bbrc.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Nunes MJ, Moutinho M, Milagre I, Gama MJ, Rodrigues E. Okadaic acid inhibits the trichostatin A-mediated increase of human CYP46A1 neuronal expression in a ERK1/2-Sp3-dependent pathway. J Lipid Res. 2012;53:1910–1919. doi: 10.1194/jlr.M027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 32.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Marinova Z, Leng Y, Leeds P, Chuang DM. Histone deacetylase inhibition alters histone methylation associated with heat shock protein 70 promoter modifications in astrocytes and neurons. Neuropharmacology. 2011;60:1109–1115. doi: 10.1016/j.neuropharm.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 35.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 36.Huang PH, Chen CH, Chou CC, Sargeant AM, Kulp SK, Teng CM, Byrd JC, Chen CS. Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Mol Pharmacol. 2011;79:197–206. doi: 10.1124/mol.110.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 38.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 39.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao L, Chen CN, Hajji N, Oliver E, Cotroneo E, Wharton J, Wang D, Li M, McKinsey TA, Stenmark KR, et al. Histone deacetylation inhibition in pulmonary hypertension: therapeutic potential of valproic acid and suberoylanilide hydroxamic acid. Circulation. 2012;126:455–467. doi: 10.1161/CIRCULATIONAHA.112.103176. [DOI] [PMC free article] [PubMed] [Google Scholar]