Abstract
Background
Increasing evidence suggests that dysregulation of phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) plays an important role in carcinogenesis. However, the relationship between PIK3CA expression and gastric cancer (GC) prognosis remains controversial.
Methods
We searchedPubMed, Embase, Web of Science, and the Cochrane Library databases for relevant studies up to June 30, 2017. Primary outcomes were hazard ratio (HR), odds ratio (OR), and 95% confidence intervals (CI) for association with overall survival and clinicopathological features.
Results
Eleven studies comprising 2481 GC patients were analyzed. Pooled analysis showed that PIK3CA upregulation was significantly associated with worse overall survival (HR = 1.79, 95% CI 1.42–2.27, p< 0.001) at the protein (HR = 1.94, 95% CI 1.52–2.47, p< 0.001) but not the gene (HR = 1.57, 95% CI 0.92–2.69, p= 0.097) level. PIK3CA gene mutation did not correlate with overall survival (HR = 1.05, 95% CI 0.83–1.34, p= 0.666) but was significantly associated with poor tumor differentiation (OR = 0.37, 95% CI 0.17–0.76, p= 0.011).
Conclusion
High PIK3CA protein expression predicted poor prognosis in GC, whereas PIK3CA gene amplification or mutation did not. Moreover, PIK3CA mutation was an indicator of poorly differentiated tumors.
Keywords: PIK3CA, gastric cancer, prognosis, alteration, meta-analysis
INTRODUCTION
Gastric cancer (GC) is the fourth most common malignant neoplasm worldwide, and it was estimated to be the second leading cause of cancer-related death in 2014 [1]. Despite recent breakthroughs in the diagnosis and treatment of GC, its prognosis remains unfavorable [2]. Therefore, understanding the biological alterations associated with GC, especially those leading to the dysregulation of signaling pathways, might help to predict patient prognosis and identify novel therapeutic targets.
The phosphatidylinositol 3-kinase (PI3K) pathway is one of the most commonly activated and altered signaling pathways in cancer, including GC [3–5]. Since the PI3K pathway plays an essential role in several cellular processes, including cell growth, metabolism, and survival, it is not surprising that its dysregulation is also of critical importance to pathological processes such as the development, progression, and metastasis of cancer [6, 7]. A key step in the PI3K/AKT pathway is the generation of phosphatidylinositol-3, 4, 5-trisphosphate (PIP3) by PI3K. The PI3K family exists as three subfamilies, one of which is composed of a p110 catalytic subunit coupled to a regulatory subunit. p100α, the gene product of phosphatidylinositol-4, 5-bisphosphate3-kinase, catalytic subunit alpha (PIK3CA), is of particular importance in signaling through the canonical PI3K pathway [8, 9]. Recently, large-scale next-generation sequencing studies have identified mutations in PIK3CA in Epstein–Barr virus (EBV)-associated and microsatellite instability (MSI)-associated GCs [10]. However, the prognostic value of PIK3CA alterations at both the protein and transcriptional levels in GC patients remains unclear and controversial.
Therefore, we conducted a meta-analysis of published studies to elucidate the precise relationship between PIK3CA dysregulation and the prognosis and other clinicopathological features of GC patients.
RESULTS
Eligible studies and general characteristics
A total of 582 studies were retrieved from a search of PubMed, Embase, Web of Science, and Cochrane Library online databases using established strategies (see Materials and Methods). After a review of the titles and abstracts, 167 duplicates and 54 non-English language studies were excluded. Of the remaining 361 studies, 235 were excluded because they did not meet the eligibility criteria of our analysis. Another 44 studies did not report patient survival, 51 focused on components of the PI3K/AKT pathway other than PIK3CA, and 19 did not examine PIK3CA expression in cancer tissues. Finally, a total of 11 studies were included in our meta-analysis (Figure 1).
Figure 1. Flow chart of study selection process.
The general characteristics of the included studies are presented in Table 1. The 11 studies comprised 2481 patients for pooling analysis (median 208, range 79–568). Two studies were from Europe (UK and Italy) and the remaining nine were from East Asia (four from China, two from Japan, two from Korea, and one from Singapore). The various techniques used for the qualitative and quantitative detection of PIK3CA included polymerase chain reaction (PCR) followed by direct sequencing or pyrosequencing of amplicons, quantitative real-time PCR (qPCR), and immunohistochemistry (IHC) to detect protein. All studies were scored above 5 stars except Y Sukawa’s and M Liang’s reports according the Newcastle–Ottawa Scale (NOS) [22], which indicated a relatively high risk of bias [23, 24] (Supplementary Table 1).
Table 1. Major characteristics of the selected studies.
| No. | First author | Year | Number | Sex(M/F) | Age | Region | TNM Stagea | Alteration type | Detection method | Treatment | Follow up | HR estimate | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | S Barbi [11] | 2010 | 264 | 175/89 | 67.4(median) | Italy | I-IV | Gene mutation(exon 9,20) | Direct sequencing | R0 Only | NR | Provided(M) | 7 |
| 2 | J Shi [12] | 2012 | 131 | 102/29 | NR | China | I-IV | Gene mutation(exon 9,20) and amplification | Direct sequencing and qPCR | R0/R1 | NR | Provided(M) | 7 |
| 3 | Y Sukawa [13] | 2012 | 231 | 157/74 | 71(25-91) | Japan | IB-IV | Gene mutation(exon 9,20) | Pyrosequence | NR | NR | FC | 5 |
| 4 | AFC Okines [14] | 2013 | 337(303)b | 263/74 | 63(median) | UK | I-IIIC | Gene mutation(exon 9,20) | Direct sequencing | R0 Only/R0+Chemotherapy | NR | FC | 9 |
| 5 | ML Chong [15] | 2013 | 79 | 64/15 | 53% (≥69) | Singapore | I-IV | Gene amplification | qPCR | R0 Only | 15(1-131) | Provided(U) | 8 |
| 6 | H Lee [16] | 2015 | 110 | 83/27 | 50% (≥60) | Korea | I-III | Gene mutation (exon9,20) and Amplification | Direct sequence and qPCR | R0 Only | 82.2(3.7-158.8) | FC | 7 |
| 7 | M Liang [17] | 2015 | 107 | NR | NR | China | I-IV | Protein overexpression | IHC | R0/R1 | NR | FC | 5 |
| 8 | K Harada [18] | 2016 | 208 | 148/60 | NR | Japan | I-IV | Gene mutation(exon 9,20) | Pyrosequencing | R0 Only | 60 | FC | 7 |
| 9 | M Dong [19] | 2016 | 568 | 396/172 | 52.3% (≥60) | China | I-IV | Protein overexpression | IHC | R0/R1 | 37.2(0-93.8) | FC | 7 |
| 10 | SH Jang [20] | 2016 | 178 | 127/51 | 68%(≥55) | Korea | I-IV | Protein overexpression | IHC | R0+Chemotherapy | 58(0-95) | Provided(M) | 7 |
| 11 | JW Kim [21] | 2017 | 302 | 202/100 | 20.2%(≥70) | Korea | I-IV | Gene mutation | qPCR(exon 1,4,7,9,20) | R0/R1 | NR | FC | 7 |
aAmerican Joint Committee on Cancer 7th edition TNM stage system. b The actual number of patients analyzing PIK3CA status and survival was 303. FC: figure calculation; HR: hazard ratio; IHC: immunohistochemistry; M/F: male to female ratio; M: multivariate analysis; NOS: Newcastle–Ottawa Scale; NR: not reported; qPCR: quantitative polymerase chain reaction; U: univariate analysis; R0: radical resection; R1: palliative surgery.
Association between PIK3CA overexpression in GC tissue and patient prognosis
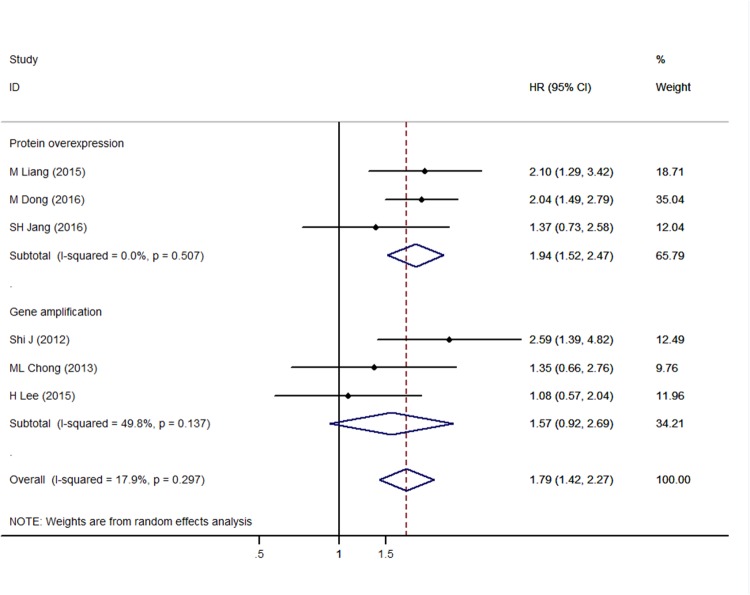
Six studies provided sufficient data to analyze the relationship between overall survival (OS) and PIK3CA expression level. The pooled analysis showed that high PIK3CA expression was associated with worse OS (hazard ratio [HR] =1.79, 95% confidence intervals [CI] 1.42–2.27, p < 0.001). Although no significant heterogeneity was observed (inconsistency index [I2] = 17.9%, p= 0.297), judging the presence of potential bias sources, a random-effects model was applied to make the conclusion more conservative and reliable [25] (Figure 2).
Figure 2. Forest plot of hazard ratios for overall survival of gastric cancer patients and PIK3CA overexpression stratified by PIK3CA alteration measurement.
We then performed a subgroup analysis of studies that examined overexpression at the gene and protein level. The results showed that OS was significantly associated with PIK3CA protein overexpression (HR = 1.94, 95% CI 1.52–2.47, p< 0.001) but not PIK3CA gene amplification (HR = 1.57, 95% CI 0.92–2.69, p= 0.097) (Figure 2).
When it refers to the different treatment settings, subgroup analysis showed that in the radical resection and palliative surgery mixed group [12, 17, 19], the association between PIK3CA overexpression and overall survival remains significant (HR = 2.13, 95% CI 1.67–2.72, p < 0.001), while in the radical resection-only group [15, 16, 20], it was not significant (HR = 1.25, 95% CI 0.86-1.83, p=0.246), which might indicate a more predominant role of PIK3CA overexpression predication on survival in those patients with late-stage diseases.
Furthermore, the associations remained significant in both the univariate (HR=1.74, 95%CI 1.31-2.33, p < 0.001) and multivariate (HR=1.89, 95%CI 1.01-3.53, p=0.046) analysis models (Table 2, Left section).
Table 2. Subgroup analysis of PIK3CA status and patients’ survival.
| Subgroup | PIK3CA overexpression | PIK3CA mutation | ||||||
|---|---|---|---|---|---|---|---|---|
| Studies (Patients) | HR (95%CI) | Heterogeneity | Studies (Patients) | HR (95%CI) | Heterogeneity | |||
| I2 (%) | P | I2 (%) | P | |||||
| Method | ||||||||
| IHC | 3(853) | 1.94(1.52-2.47) | 0 | 0.507 | / | / | / | / |
| qPCR and direct sequence | 3(320) | 1.57(0.92-2.69) | 49.8 | 0.137 | 5(1110) | 1.04(0.79-1.38) | 0.0 | 0.840 |
| Pyrosequence | / | / | / | / | 2(439) | 1.10(0.68-1.78) | 0.0 | 0.476 |
| Treatment | ||||||||
| R0 | 3(367) | 1.25(0.86-1.83) | 0 | 0.850 | 4(885) | 0.99(0.71-1.38) | 0.0 | 0.889 |
| R0/R1 | 3(806) | 2.13(1.67-2.72) | 0 | 0.796 | 2(433) | 1.07(0.69-1.65) | 0.0 | 0.351 |
| NR | / | / | / | / | 1(231) | 1.27(0.68-2.37) | / | / |
| Race | ||||||||
| Mongolian | 6(1173) | 1.79(1.42-2.27) | 17.9 | 0.297 | 5(982) | 1.08(0.79-1.47) | 0 | 0.845 |
| Caucasian | / | / | / | / | 2(567) | 1.02(0.69-1.51) | 0 | 0.466 |
| Analysis method | ||||||||
| Multivariate | 2(309) | 1.89(1.01-3.53) | 49.4 | 0.160 | 2(395) | 1.17(0.78-1.77) | 0 | 0.453 |
| Univariate | 4(864) | 1.74(1.31-2.33) | 26.5 | 0.253 | 5(1154) | 1.00(0.74-1.35) | 0 | 0.906 |
| HR evaluation | ||||||||
| Provided | 3(388) | 1.72(1.11-2.64) | 23.1 | 0.272 | 2(395) | 1.17(0.78-1.77) | 0 | 0.453 |
| Calculated | 3(785) | 1.80(1.28-2.53) | 40.5 | 0.186 | 5(1154) | 1.00(0.74-1.35) | 0 | 0.906 |
HR: hazard ratio; R0: radical resection; R1: palliative surgery; NR: not reported; CI: confidence interval; I2: inconsistency index.
Association between PIK3CA mutation in GC tissue and patient prognosis
A total of seven studies were pooled for the analysis of the effect of PIK3CA mutation on GC patient prognosis. The results showed that the presence of mutations (mainly in exons 9 and 20) did not correlate significantly with OS (HR = 1.05, 95% CI 0.83–1.34, p = 0.666) (Figure 3).
Figure 3. Forest plot of hazard ratios for overall survival of gastric cancer patients with PIK3CA gene mutations.
A subgroup analysis was also conducted due to the potential existence of study bias (Table 2, Right section). However, no obvious heterogeneity was observed and the association remains insignificant either in the subgroup of different statistical analysis or HR evaluation methods, which indicated a stable meta-analysis result within these subgroups.
Association between PIK3CA dysregulation and clinicopathological features of GC patients
The associations between PIK3CA dysregulation (overexpression and mutation) and major clinicopathological features are shown in Table 3. Of the characteristics analyzed, the only significant association was between the presence of mutated PIK3CA and tumor differentiation grade (well/moderate vs. poor/undifferentiated, odds ratio [OR] 0.37, 95% CI 0.17–0.79, p= 0.011).
Table 3. Meta-analysis of the association between PIK3CA alterations and clinicopathological features of gastric cancer patients.
| PIK3CA overexpression Stratification | No. of studies | No. of patients | Pooled OR (95%CI) | Heterogeneity | |
|---|---|---|---|---|---|
| I2(%) | P-value | ||||
| Gender (M/F) | 3 | 877 | 1.03 (0.77-1.39) | 0 | 0.380 |
| Lymph node metastasis (N0/N1-3) | 3 | 877 | 2.23 (0.53-9.36) | 91.1 | <0.001 |
| Distant metastasis (M1/M0) | 2 | 1326 | 3.11 (0.35-27.42) | 79 | 0.029 |
| TNM stage (I+II/III+IV)a | 3 | 877 | 0.52 (0.09-2.96) | 95 | <0.001 |
| Differentiation (Well+Moderate/Poor) | 2 | 308 | 1.31 (0.20-8.74) | 88 | 0.004 |
| Tumor size (<5cm vs ≥5cm) | 2 | 699 | 4.43 (0.55-35.43) | 85.8 | 0.008 |
| Lauren classification (Intestinal/diffuse) | 2 | 661 | 0.97 (0.16-6.04) | 95.8 | <0.001 |
| PIK3CA mutation Stratification | No. of studies | No. of patients | Pooled OR (95%CI) | Heterogeneity | |
| I2(%) | P-value | ||||
| Gender (M/F) | 6 | 1200 | 0.93 (0.64-1.36) | 0 | 0.822 |
| Lymph node metastasis (N0/N1-3) | 6 | 1183 | 0.95 (0.63-1.42) | 0 | 0.762 |
| TNM stage (I+II/III+IV)a | 3 | 614 | 0.66 (0.39-1.12) | 0 | 0.511 |
| Differentiation (Well+Moderate/Poor) | 2 | 386 | 0.37 (0.17-0.79) | 0 | 0.522 |
| Lauren classification (Intestinal/diffuse) | 5 | 1009 | 0.76 (0.41-1.43) | 52.7 | 0.076 |
| Microsatellite instability (MSI/MSS) | 2 | 566 | 0.68 (0.15-3.06) | 82.9 | 0.016 |
aAmerican Joint Committee on Cancer 7th edition TNM stage system. CI: confidence interval; I2: inconsistency index; MSI: microsatellite instability.
MSS: microsatellite stability; OR: odds ratio.
Publication bias
Potential publication bias was assessed in the association of OS with PIK3CA alteration type. As shown in Figure 4, no significant bias was detected in the included studies. For the overexpression studies, p= 0.260 for Begg’s test and p= 0.271 for Egger’s test, and for the mutation studies, p= 0.881 for Begg’s test and p = 0.22 for Egger’s test.
Figure 4.
Funnel plot evaluation of potential publication bias on the effect of PIK3CA overexpression (A) and mutation (B) on overall survival of gastric cancer patients.
DISCUSSION
The PI3K/AKT pathway is an important therapeutic target because its components, including PIK3CA, are frequently activated in cancer cells; indeed, several PI3K pathway inhibitors are currently in clinical trials [26, 27]. Therefore, identifying patients who might benefit from targeted therapy is of great importance [28, 29]. An increasing number of studies are focused on PIK3CA dysregulation and its potential impact in GC, one example being the MAGIC trial [30], highlighting the urgent need to elucidate more precisely the contribution of PIK3CA alterations to GC. To the best of our knowledge, this meta-analysis of 11 studies comprising 2481 GC patients represents the first comprehensive systematic evaluation of the prognostic value of PIK3CA alterations detected at the protein and gene levels. The results suggest that PIK3CA could represent a useful biomarker for predicting patient survival.
In theory, alterations in PIK3CA activity or expression could lead to downstream activation of PI3K/AKT pathway and promote the growth and invasion of GC cells [7, 31]. Our pooled analysis showed that PIK3CA dysregulation is associated with poorer tumor differentiation and worse OS among GC patients, indicating that PIK3CA is a prognostic biomarker for GC patients. However, the significant association between PIK3CA mutation and tumor differentiation should be interpreted with caution because of the small sample size (two studies comprising 386 patients) and need more studies to confirm. Furthermore, our results showed no significant association between PIK3CA mutational status and patient prognosis. This might be attributed to the fact that gene mutation of a single component does not necessarily reflect the status of an entire signaling pathway in terms of dysregulation and functional impact. Therefore, our results suggest that the detection of gene amplification and/or protein overexpression is a more direct and promising strategy to predict GC patient prognosis than gene mutation analysis.
The most recent NCCN guidelines for gastric cancer have described MSI and EBV status as potential molecular biomarkers for targeting treatment strategies in GC patients [32]. Although mutant PIK3CA is more common in MSI than in other GC types [10], our analysis failed to confirm this, possibly because of the small number of studies that analyzed MSI association (two studies comprising 566 patients). There were also insufficient data to analyze the association between PIK3CA mutation and EBV positivity, so further studies are necessary to evaluate this relationship.
Several limitations to our study should be noted. First, although we observed no obvious heterogeneity (Figures 2 and 3), the studies used different designs, PCR primers, and IHC antibodies, which might have contributed to heterogeneity. Therefore, a random-effects model was applied to make our results more conservative. Second, we obtained insufficient data to analyze the relationship between PIK3CA and disease-free survival, which reduces the predictive value of PIK3CA status on the outcomes of GC patients. Third, the included studies did not investigate the relationship between PIK3CA gene amplification and protein overexpression, which might be critical for selecting the appropriate method of analysis for different GC patients. Furthermore, association analysis of some clinicopathological features (Table 3) included only two to three studies, and therefore should be interpreted with caution; additional studies are required to confirm the conclusion. It is also noticeable that the patients from the MAGIC trial of AFC Okines’ study included a small portion of esophageal cancer patients (38 out of 337), which might add to the bias of our results. Finally, we limited the analysis to English language publications, which might have led to selection bias in our meta-analysis.
In conclusion, we found that overexpression of PIK3CA protein, but not the presence of PIK3CA gene amplification or mutations, predicted worse prognosis for GC patients. However, our findings, as well as other important variables such as the association of PIK3CA alterations with GC molecular subtypes, require further verification in more rigorous studies with consistent and standardized methodology.
MATERIALS AND METHODS
Literature search
The search was conducted by consulting PubMed, Embase, Web of Science, and Cochrane Library online databases from inception to June 30, 2017. The search terms were “gastric/stomach cancer” or “gastric/stomach neoplasm” and “PIK3CA” or “phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit alpha” and “prognosis” or “survival.” Article language was limited to English. The reference section of all relevant articles was examined to identify additional related studies. Two researchers (HL and YG) independently assessed the eligibility of the identified studies, and disagreements were resolved by discussion or consultation with a third researcher.
Inclusion criteria
Studies were selected according to the following inclusion criteria: (1) inclusion of pathologically confirmed GC patients; (2) focus on PIK3CA alterations, at either the protein or gene level; and (3) inclusion of survival data according to the intra-tumor PIK3CA alteration status.
Exclusion criteria
The exclusion criteria were (1) studies published in languages other than English, (2) replicate or overlapping publications, (3) analysis of only cancer cell lines, and (4) studies with a small sample size (≤20 patients).
Quality assessment
The quality of the included studies was assessed independently by two researchers using the Newcastle–Ottawa Scale [22]. This scale is an eight-item instrument that assesses patient population and selection, study comparability, and outcomes. We considered a study high-quality if it was awarded six or more stars [23].
Data extraction
The following items were extracted from the included studies: study title, name of the first author, publication year, region, PIK3CA alteration type, detection method, and clinicopathological characteristics (i.e., gender, TNM stage, and tumor differentiation). The HRs, ORs, and 95% CIs for the association of PIK3CA status with OS and clinicopathological features were collected from the studies. If HRs or ORs were not directly reported, survival data were calculated using the methods described by Parmar et al. [33] and Tierney et al. [34].
Statistical analysis
Meta-analysis was performed using Stata version 12.0 software (StataCorp, College Station, TX, USA). A significant heterogeneity was observed when P < 0.05 or I2 > 50%, and a random-effects model was then applied. Otherwise, a fixed-effects model was used.
SUPPLEMENTARY MATERIALS TABLE
Acknowledgments
We thank Anne M. O’Rourke, PhD and Clare Cox, PhD for editing the English text of a draft of this manuscript.
Author contributions
Z.H and D.L. design this study. J.C and Y.G. searched, evaluated and collected the studies. D.W. and S.L extracted the data. H.L and S.C performed the meta-analysis. Hui L. and Y.Q. helped the whole process. Y.Q and T.Z. contributed to the study enrollment and revision. H.L and Y.G wrote the manuscript. All authors reviewed this manuscript.
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
ETHICAL APPROVAL AND INFORMED CONSENT
No studies with human participants were performed by any of the authors of this study. All the analyses were based on previously published studies.
FUNDING
This work was supported by Municipal Science funds of Xingtai (No. 2015ZC202).
REFERENCES
- 1.Stewart BW. World Cancer Report 2014. International Agency for Research on Cancer. 2015 [PubMed] [Google Scholar]
- 2.Nilsson M. Postgastrectomy follow-up in the West: evidence base, guidelines, and daily practice. Gastric Cancer. 2017;20:135–40. doi: 10.1007/s10120-016-0654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 4.Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S, Jr, Duval A, Carneiro F, Machado JC, Hamelin R, Seruca R. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–54. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willems L, Tamburini J, Chapuis N, Lacombe C, Mayeux P, Bouscary D. PI3K and mTOR signaling pathways in cancer: new data on targeted therapies. Curr Oncol Rep. 2012;14:129–38. doi: 10.1007/s11912-012-0227-y. [DOI] [PubMed] [Google Scholar]
- 7.Matsuoka T, Yashiro M. The Role of PI3K/Akt/mTOR Signaling in Gastric Carcinoma. Cancers (Basel) 2014;6:1441–63. doi: 10.3390/cancers6031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 9.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 10.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbi S, Cataldo I, De Manzoni G, Bersani S, Lamba S, Mattuzzi S, Bardelli A, Scarpa A. The analysis of PIK3CA mutations in gastric carcinoma and metanalysis of literature suggest that exon-selectivity is a signature of cancer type. J Exp Clin Cancer Res. 2010;29:32. doi: 10.1186/1756-9966-29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G, Ji M, Xu L, He N, Shi B, Hou P. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. doi: 10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukawa Y, Yamamoto H, Nosho K, Kunimoto H, Suzuki H, Adachi Y, Nakazawa M, Nobuoka T, Kawayama M, Mikami M, Matsuno T, Hasegawa T, Hirata K. Alterations in the human epidermal growth factor receptor 2-phosphatidylinositol 3-kinase-v-Akt pathway in gastric cancer. World J Gastroenterol. 2012;18:6577–86. doi: 10.3748/wjg.v18.i45.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okines AF, de Castro DG, Cunningham D, Chau I, Langley RE, Thompson LC, Stenning SP, Saffery C, Barbachano Y, Coxon F, Middleton G, Ferry D, Crosby T, et al. Biomarker analysis in oesophagogastric cancer: results from the REAL3 and TransMAGIC trials. Eur J Cancer. 2013;49:2116–25. doi: 10.1016/j.ejca.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Chong ML, Loh M, Thakkar B, Pang B, Iacopetta B, Soong R. Phosphatidylinositol-3-kinase pathway aberrations in gastric and colorectal cancer: meta-analysis, co-occurrence and ethnic variation. Int J Cancer. 2014;134:1232–8. doi: 10.1002/ijc.28444. [DOI] [PubMed] [Google Scholar]
- 16.Lee H, Hwang IS, Choi IJ, Kang YN, Park KU, Lee JH. Are PIK3CA mutation and amplification associated with clinicopathological characteristics of gastric cancer. Asian Pac J Cancer Prev. 2015;16:4493–6. doi: 10.7314/apjcp.2015.16.11.4493. [DOI] [PubMed] [Google Scholar]
- 17.Liang M, Shi B, Liu J, He L, Yi G, Zhou L, Yu G, Zhou X. Downregulation of miR203 induces overexpression of PIK3CA and predicts poor prognosis of gastric cancer patients. Drug Des Devel Ther. 2015;9:3607–16. doi: 10.2147/DDDT.S85525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada K, Baba Y, Shigaki H, Ishimoto T, Miyake K, Kosumi K, Tokunaga R, Izumi D, Ohuchi M, Nakamura K, Kiyozumi Y, Kurashige J, Iwatsuki M, et al. Prognostic and clinical impact of PIK3CA mutation in gastric cancer: pyrosequencing technology and literature review. BMC Cancer. 2016;16:400. doi: 10.1186/s12885-016-2422-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong M, Wang HY, Zhao XX, Chen JN, Zhang YW, Huang Y, Xue L, Li HG, Du H, Wu XY, Shao CK. Expression and prognostic roles of PIK3CA, JAK2, PD-L1, and PD-L2 in Epstein-Barr virus-associated gastric carcinoma. Hum Pathol. 2016;53:25–34. doi: 10.1016/j.humpath.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Jang SH, Kim KJ, Oh MH, Lee JH, Lee HJ, Cho HD, Han SW, Son MW, Lee MS. Clinicopathological significance of elevated PIK3CA expression in gastric cancer. J Gastric Cancer. 2016;16:85–92. doi: 10.5230/jgc.2016.16.2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JW, Lee HS, Nam KH, Ahn S, Kim JW, Ahn SH, Park DJ, Kim HH, Lee KW. PIK3CA mutations are associated with increased tumor aggressiveness and Akt activation in gastric cancer. Oncotarget. 2017;8:90948–58. doi: 10.18632/oncotarget.18770. https://doi.org/10.18632/oncotarget.18770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Zhang D, Feng N, Chen G, Liu J, Chen G, Zhu Y. Increased intake of vegetables, but not fruit, reduces risk for hepatocellular carcinoma: a meta-analysis. Gastroenterology. 2014;147:1031–42. doi: 10.1053/j.gastro.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Gu WJ, Wang F, Tang L, Liu JC. Single-dose etomidate does not increase mortality in patients with sepsis: a systematic review and meta-analysis of randomized controlled trials and observational studies. Chest. 2015;147:335. doi: 10.1378/chest.14-1012. [DOI] [PubMed] [Google Scholar]
- 25.Borenstein M, Hedges LV, Higgins JP, Rothstein HR. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res Synth Methods. 2010;1:97–111. doi: 10.1002/jrsm.12. [DOI] [PubMed] [Google Scholar]
- 26.Zhang CH, Awasthi N, Schwarz MA, Schwarz RE. The dual PI3K/mTOR inhibitor NVP-BEZ235 enhances nab-paclitaxel antitumor response in experimental gastric cancer. Int J Oncol. 2013;43:1627–35. doi: 10.3892/ijo.2013.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong CH, Loong HH, Hui CW, Lau CP, Hui EP, Ma BB, Chan AT. Preclinical evaluation of the PI3K-mTOR dual inhibitor PF-04691502 as a novel therapeutic drug in nasopharyngeal carcinoma. Invest New Drugs. 2013;31:1399–408. doi: 10.1007/s10637-013-0007-z. [DOI] [PubMed] [Google Scholar]
- 28.Farran B, Müller S, Montenegro RC. Gastric cancer management: kinases as a target therapy. Clin Exp Pharmacol Physiol. 2017;44:613–22. doi: 10.1111/1440-1681.12743. [DOI] [PubMed] [Google Scholar]
- 29.Joo MK, Park JJ, Chun HJ. Recent updates of precision therapy for gastric cancer: Towards optimal tailored management. World J Gastroenterol. 2016;22:4638–50. doi: 10.3748/wjg.v22.i19.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ, Falk SJ, Iveson TJ, Smith DB, Langley RE, Verma M, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355:11–20. doi: 10.1056/NEJMoa055531. [DOI] [PubMed] [Google Scholar]
- 31.Sonohara F, Inokawa Y, Hayashi M, Kodera Y, Nomoto S. Epigenetic modulation associated with carcinogenesis and prognosis of human gastric cancer. Oncol Lett. 2017;13:3363–8. doi: 10.3892/ol.2017.5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology. Gastric Cancer, Version 2. 2017 doi: 10.6004/jnccn.2017.0146. https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–34. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.