Abstract
Non-alcoholic steatohepatitis (NASH) is commonly associated with obesity, type 2 diabetes, and/or hypertriglyceridemia, while alcoholic steatohepatitis (ASH) is associated with alcohol abuse. Both NASH and ASH patients can develop cirrhosis and hepatocellular carcinoma (HCC) if left untreated. However, the rate of tumorigenesis in NASH and ASH appears to be different. Individuals with NAS H progress to HCC at a rate of 0.5% annually (Lindenmeyer et al., 2017), when individuals with ASH progress to HCC at a rate of 3–10% annually (Schwartz et al., 2012). Thus, the objective of our study is to determine if there are differences in NASH versus ASH in the levels of different proteins expressed involved in cancer development. The method used was measuring the proteins expressed in liver biopsied sections from NASH and ASH patients using immunohistochemical staining with fluorescent ant ibodies and then quantitating the fluorescence intensity morphometrically. The 20 proteins tested are parts of the Ingenuity Canonical Pathway of Molecular Mechanisms of Cancer and include: RAP2B, NAIP, FYN, PAK6, SUV39H1, GNAI1, BAX, E2F3, CKDN2B, BAK1, BCL2, DIABLO, RASGRF2, GNA15, PIK3CB, BRCA1, MAP2K1, BIRC3, CDK2, and ATM. In ASH, the proteins that showed upregulated levels of expression were SUV39H1, E2F3, BCL2, BAK1, BIRC3, and GNAI1. In NASH, the proteins that showed upregulated levels of expression were BAK1 and GNAI1 and the protein that showed downregulated level of expression was BCL2. Additionally, levels of expression for SUV39H1, E2F3, BCL2, BAK1, BIRC3, and GNAI1 were significant upregulated in ASH compared to NASH. These results showed significant differences in ASH compared to normal liver, and significant differences in ASH compared to NASH. Thus, we conclude that there are more proteins involved in tumorigenesis in ASH compared to NASH and in ASH compared to normal liver, which is consistent with the known tumor development rate in ASH and NASH.
Keywords: Tumorigenesis, Alcoholic Hepatitis, Pre-cancer Gene Expression, Hepatocellular Carcinoma (HCC)
Introduction
Molecular Mechanisms of Cancer Pathway is one of the Ingenuity Canonical Pathways developed by Qiagen Corporation (Table 1) (Liu et al., 2015). These pathway categories are adopted by science research communities world-wide in the quest to uncover cancer pathogenesis. In this particular pathway, the 20 proteins involved are RAP2B, NAIP, FYN, PAK6, SUV39H1, GNAI1, BAX, E2F3, CDKN2B, BAK1, BCL2, DIABLO, RASGRF2, GNA15, PIK3CB, BRCA1, MAP2K1, BIRC3, CDK2, and ATM. In our study, six of these proteins showed significant differences in protein levels’ expression between ASH and NASH and control liver. These proteins are discussed below.
Table 1.
Molecular Mechanisms of Cancer Pathway (bolded)
| © 2000–2014 QIAGEN. All rights reserved. | |
|---|---|
| Canonical Pathways | |
| Ingenuity Canonical Pathways | Molecules |
| BRCA1-mediated tumor suppression | CDNK1A,FANCG,BRCA2,SLC19A1,BRCC3,RFC5,E2F3,BRCA1,FANCA,ATM |
| Cell Cycle: G1/S Checkpoint Regulation | NRG1,HDAC11,SUV39H1,E2F3,CDKN1A,CDKN2B,CDK2,ATM |
| p70S6K Signaling | PLCD3,CD19,IL2RG,SYK,GNAI1,PIK3CB,SFN,PLCL1,MAP2K1,ATM |
| Tec Kinase Signaling | TEC,FYN,STAT5A,GNG11,PAK6,GNA15,GNG2,GNAI1,STAT2,PIK3CB,FRK,ATM |
| DNA Double-Strand Break Repair by Homologous Recombination | BRCA2,BRCA1,ATM |
| Apoptosis Signaling | DIABLO,NAIP,BAX,TNFRSF1B,BIRC3,MAP2K1,BAK1,BCL2 |
| Fatty Acid α-oxidation | BCO2,PTGS2,ALDH3A1 |
| CCR3 Signaling in Eosinophils | GNG11,PAK6,GNG2,PLA2G5,GNAI1,PIK3CB,LIMK2,MAP2K1,ATM |
| Prostate Cancer Signaling | SUV39H1,PIK3CB,CREB5,MAP2K1,CDK2,ATM,BCL2 |
| p53 Signaling | SIRT1,PIK3CB,BAX,SFN,BRCA1,CDK2,ATM,BCL2 |
| GADD45 Signaling | BRCA1,CDK2,ATM |
| IL-9 Signaling | STAT5A,IL2RG,PIK3CB,ATM |
| IL-8 Signaling | NOX4,GNG11,FLT4,GNG2,GNAI1,PIK3CB,LIMK2,BAX,PTGS2,MAP2K1,ATM,BCL2 |
| UVA-Induced MAPK Signaling | PLCD3,PARP16,PIK3CB,RPS6KA5,PLCL1,SMPD3,ATM |
| Interferon Signaling | STAT2,BAX,BAK1,BCL2 |
| Role of Tissue Factor in Cancer | FYN,STAT5A,GNA15,PIK3CB,RPS6KA5,LIMK2,FRK,ATM |
| Molecular Mechanisms of Cancer | RAP2B,NAIP,FYN,PAK6,SUV39H1,GNAI1,BAX,E2F3,CDKN2B,BAK1,BCL2,DIABLO,RASGRF2,GNA15,PIK3CB,BRCA1,MAP2K1,BIRC3,CDK2,ATM |
| Natural Killer Cell Signaling | FYN,PAK6,SYK,PIK3CB,HCST,MAP2K1,INPP5D,ATM |
| G-Protein Coupled Receptor Signaling | FYN,PTGIR,PDE3A,GNAI1,PDE1A,CREB5,HRH1,GNA15,ADRA2A,PDE4D,DUSP4,PIK3CB,MAP2K1,ATM,HTR2A |
SUV39H1 is a histone methyltransferase involved in regulating transcription and promoting cell growth. SUV39H1 overexpression plays important roles in HCC development and progression (Chiba et al., 2014). SUV39H1 and ESET function to methylate Histone H3 to allow it to progress with transcription through formation of histone H3 lysine 9 trimethylation (H3K9me3). Interestingly, only SUV39H1 knockdown, but not ESET knockdown, reduces H3K9me3 levels and impairs HCC cell growth and sphere formation. Thus, SUV39H1 could be a potential pharmacological inhibition target in preventing hepatocellular carcinoma development.
E2F3 is a member of the E2F family of transcription factors and plays a crucial role in the control of cell cycle. E2F3’s two cousins E2F1 and E2F2 are negatively regulated by pRb, a well-known tumor suppressor. Myc, a popular onco-protein, activates E2F3. E2F3’s copy number gains are frequently observed in HCC and almost all HCC samples have increased expression of E2F3 (Kent et al., 2017)
BCL2 inhibits apoptosis along the intrinsic mitochondrial apoptosis pathway. BCL2 is part of a large family of proteins involved both in preventing apoptosis and promoting it. BCL2 protects mitochondrial cell membranes in the events of cytotoxic injuries. Bcl-2 may play a role in hepatocarcinogenesis as an inhibitor of apoptosis of tumor cells (El-Emshaty et al., 2016).
BAK1 is a member of the BCL2 protein family that functions as a pro-apoptotic regulator. BAK and BAX form homo-oligomers within the mitochondrial membrane, resulting in the release of cytochrome c, which activates Apaf1 and results in caspase 9 activation. BAK1 works with other pro-apoptotic molecules in the mitochondria, including DIABLO (Cory et al., 2002). BAK1’s upregulation induces apoptosis of HCC cells. (To et al., 2011).
BIRC3 is a member of the inhibitor of apoptosis (IAP) family of proteins. BIRC3 inhibits apoptosis by binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2. BIRC3’s DNA amplification has been observed in mouse HCC (Zender et al., 2006).
GNAI1 functions as a transducer downstream of G protein-coupled receptors in numerous signaling cascades. GNAI1 can regulate cell proliferation and differentiation, assist platelet aggregation, and act as receptors in multiple cancers. GNAI1 is significantly down-regulated in HCC compared with normal liver (Yao et al. 2012). GNAI1 is hypothesized to function as an inhibitor of HCC migration and invasion.
The other 14 proteins’ functions and significance are summarized in Table 2.
Table 2.
Function and significance of 14 other proteins in the Molecular Mechanisms of Cancer Pathway
| Protein | Function/Significance | Reference |
|---|---|---|
| RAP2B | RAP2B is a member of RAS oncogene family and is highly expressed in HCC tissue. | Zhang et al., 2017 |
| NAIP | NAIP is a member of the Inhibitors of apoptosis proteins (IAPs) family. NAIP mRNA has a slightly lower expression level in cancer tissue (FC =0.67) | Augello et al., 2009 |
| FYN | FYN is a non-receptor tyrosine kinase that belongs to the Src family kinases (SFKs). FYN’s expression is observed in various cancers. | Elias et al., 2015 |
| PAK6 | PAK6 is a member of the p21-activated kinases. PAK6 might be a tumor suppressor gene in HCC. | Liu et al., 2015 |
| BAX | BAX is a pro-apoptotic member of the BCL2 family. Increased expression is correlated with good prognosis in HCC. | Garcia et al., 2002 |
| CDKN2B | CDKN2B are potent inhibitors of the cyclin D complex and function as tumor suppressors. Gene deletion is found in 13% HCC. | Lin et al., 1998 |
| DIABLO | DIABLO is a pro-apoptogenic mitochondrial protein. Expression is reduced in HCC. | Bao et al., 2006 |
| RASGRF2 | RASGRF2 is a nucleotide exchange factor that activates the RAS pathway. The protein expression is reduced in lung cancer. | Chen et al., 2006 |
| GNA15 | GNA15 is a guanine nucleotide binding protein involved in prostate cancer development. | Haibi et al., 2013 |
| PIK3CB | PIK3CB is a member of the phosphoinositide 3-kinases. PIK3CB is most likely upregulated in HCC. | Lang et al., 2012 |
| BRCA1 | BRCA1 is a protein involved in repairing damaged DNA and has low expression in HCC. | Ferroudj et al., 2016 |
| MAP2K1 | MAP2K1 is a mitogen-activated protein kinase and is upregulated in HCC. | Janku et al., 2014 |
| CDK2 | CDK2 is a cyclin-dependent kinase that its expression is elevated in HCC. | Li et al., 2002 |
| ATM | ATM is a serine/threonine protein kinase that phosphorylates proteins involved in repairing damaged DNA. ATM is impaired in HCC. | Liu et al., 2017 |
Methods
Formalin-fixed paraffin-embedded biopsies of 8 to 12 alcoholic hepatitis livers, 1 to 5 NASH livers, and 3 normal livers were obtained from Harbor-UCLA Medical Center and from the Long Beach Veterans Affairs’ clinical trial in treatment of alcoholic hepatitis. The study was carried out according to the principals of the Declaration of Helsinki and was designated as exempt by our institutional ethics review board. The data was analyzed anonymously. The slides were double stained for ubiquitin plus one of the twenty proteins tested using a fluorescent labeled antibody. Texas Red (Millipore, Temecula, CA) was used to detect ubiquitin. Either donkey-anti mouse or anti rabbit Alex Fluor (Jackson Labs, West Grove, PA) were used as the second antibody to detect the protein. The staining of all the samples was done at the same time to provide accurate comparison between groups.
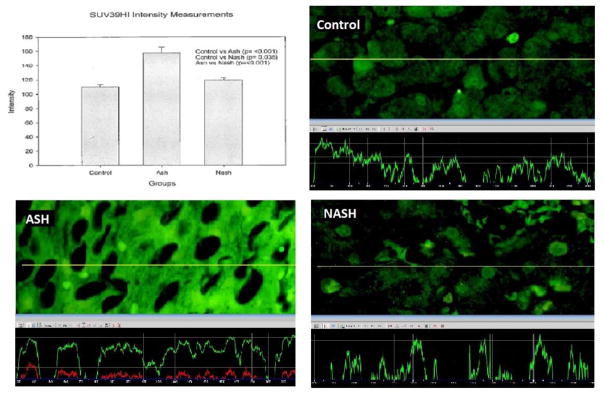
We measured the intensity of the fluorescent staining in three different areas on each slide with 40x magnifications and 800ms standard exposure time by using a Nikon 400 fluorescent microscope. The Nikon morphometric system was used to quantitate the florescent intensity (See figure 1). The mean, standard error, and statistical differences of data achieved from the Nokia were analyzed by Graph pad statistical software. Control versus ASH, control versus NASH, and ASH versus NASH were compared by unpaired t-test. Only p <0.05 was considered statistically significant.
Figure 1.
SUV39H1’s levels of expression was measured using Nikon 400 fluorescent microscope. The yellow tracer line converted the fluorescence intensity into the green line in the graph.
Results
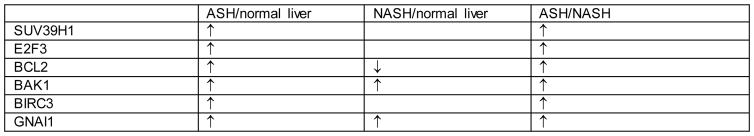
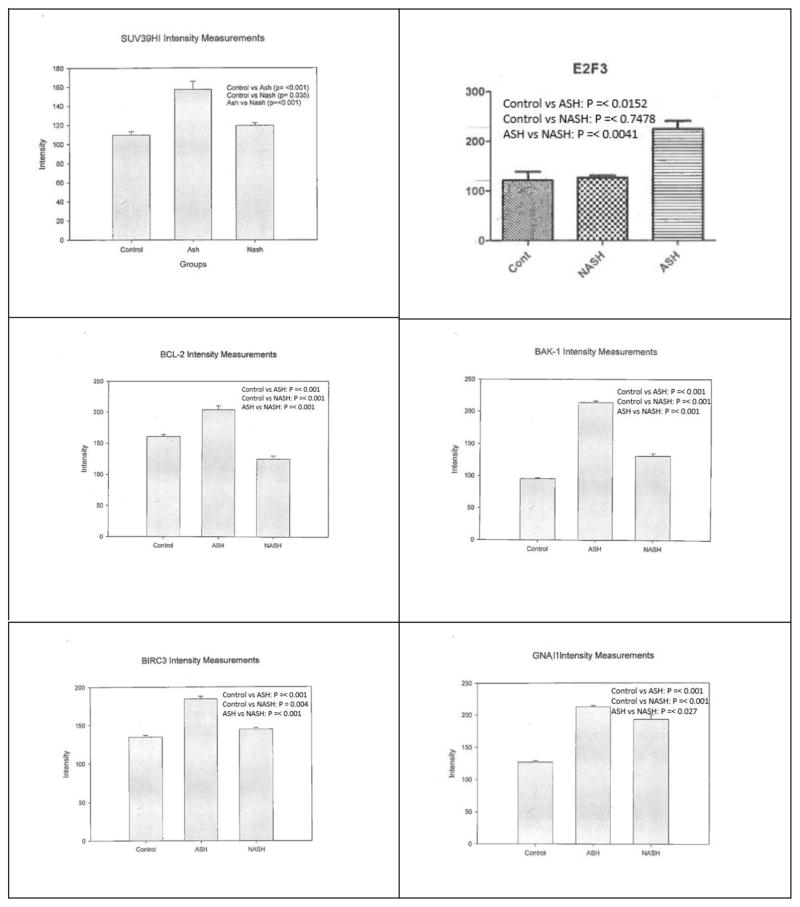
Statistical significant differences between the groups were observed for SUV39H1, E2F3, BCL2, BAK1, BIRC3, and GNAI1 as summarized in figure 2. The measurements and statistical significance for each protein was summarized in figure 3. Changes observed for the other 14 proteins (Table 2) were not statistically significant.
Figure 2.
Proteins’ levels of expression’s comparison table
Figure 3.
Means with SD of intensity measurements for each protein’s levels of expression. Significant differences were observed between control versus ASH, control versus NASH, and/or ASH versus NASH
Discussion and Conclusion
Tumorigenesis invariably involves mutations of oncogenes or tumor suppressor genes. When an oncogene is mutated, the amounts of the respectively transcribed onco-protein product usually increase, which promotes cell growth and division, creating a monoclonal population. When a tumor suppressor gene is mutated, the cells are liberated from its growth suppressing effects and are able to proliferate. When a cell’s DNA is mutated or damaged, the amount of tumor suppressor proteins are elevated, promoting apoptosis and destroying the cells before tumor develops. Thus, the mutation of tumor suppressor genes may even be more important than mutation of pro-oncogenes in tumorigenesis.
In ASH compared to control liver, 4 out of the 6 proteins elevated (SUV39H1, E2F3, BCL2, BIRC3) function in assisting tumorigenesis in HCC (see Introduction above). The other two elevated proteins function to prevent tumorigenesis with BAK1 promoting cell apoptosis and GNAI1 inhibiting tumor migration. BAK1 and GNAI1, thus, could be part of the cell mechanisms to hinder tumor growth. BAK1 could also increase cell death for exhausted hepatocytes due to hepatocyte over-activity in alcoholics (Masouminia et al., 2016). However, if BAK1 and GNAI1 become mutated, their protective activity will be lost.
In NASH compared to control liver, BAK1 and GNAI1 are elevated while BCL2 is downregulated. Interestingly, BAK1 and GNAI1 are commonly downregulated in HCC while BCL2 is upregulated. And as mentioned above, BAK1 and GNAI1 are important in preventing cancer growth in hepatocellular carcinoma. This may suggest that not only NASH has a low rate of tumorigenesis, but NASH also has a high rate of antagonistic effect on tumorigenesis.
Levels of 6 proteins (SUV39H1, E2F3, BCL2, BAK1, BIRC3, GNAI1) were significantly upregulated in ASH compared to NASH. This is consistent with the fact that the rate of tumorigenesis is higher in ASH compared to NASH. At the very least, this could support the idea that ASH and NASH have two different pathogenic mechanisms, although these mechanisms could converge when cancer develops.
In conclusion, our data is consistent with the rate of tumorigenesis observed in ASH. Our data does not show strong evidence for tumorigenesis in NASH. This could suggest that NASH develops tumor at a much slower rate than previously suspected. Our data also shows that ASH and NASH are most likely two distinct entities and further research is needed to define the differences.
Acknowledgments
This study was funded by NIH/AAA grant # UO-21898-05.
Part of the study will be presented as a poster at Experimental Biology in San Diego in 2018.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Augello C, Caruso L, Maggioni M, et al. Inhibitors of apoptosis proteins (IAPs) expression and their prognostic significance in hepatocellular carcinoma. BMC Cancer. 2009;9:125. doi: 10.1186/1471-2407-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Gui S, Lin M. Relationship between expression of Smac and Survivin and apoptosis of primary hepatocellular carcinoma. Hepatobiliary Pancreat. Dis Int. 2006;5(4):580–3. [PubMed] [Google Scholar]
- Chen H, Suzuki M, Nakamura Y, et al. Aberrant methylation of RASGRF2 and RASSF1A in human non-small cell lung cancer. Oncol Rep. 2006;15(5):1281–5. [PubMed] [Google Scholar]
- Chiba T, Saito T, Yuki K, et al. Histone lysine methyltransferase SUV39H1 is a potent target for epigenetic therapy of hepatocellular carcinoma. Int J Cancer. 2015;136(2):289–298. doi: 10.1002/ijc.28985. [DOI] [PubMed] [Google Scholar]
- Cory S, Adams J. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2(9):647–56. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- El-Emshaty H, Saad E, Toson E, et al. Apoptosis and cell proliferation: correlation with BCL-2 and P53 oncoprotein expression in human hepatocellular carcinoma. Hepatogastroenterology. 2014;61(133):1393–401. [PubMed] [Google Scholar]
- Elias D, Ditzel H. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol Res. 2015;100:250–4. doi: 10.1016/j.phrs.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Ferroudj S, Yildiz G, Bouras M, et al. Role of Fanconi anemia/BRCA pathway genes in hepatocellular carcinoma chemoresistance. Hepatol Res. 2016;46(12):1264–1274. doi: 10.1111/hepr.12675. [DOI] [PubMed] [Google Scholar]
- Garcia E, Lawson D, Cotsonis G, et al. Hepatocellular carcinoma and markers of apoptosis (bcl -2, bax, bcl-x): prognostic significance. Appl Immunohistochem Mol Morphol. 2002;10(3):210–7. doi: 10.1097/00129039-200209000-00004. [DOI] [PubMed] [Google Scholar]
- Haibi C, Sharma P, Singh R, et al. Differential G protein subunit expression by prostate cancer cells and their interaction with CXCR5. Mol Cancer. 2013;12:64. doi: 10.1186/1476-4598-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, Kaseb A, Tsimberidou A, et al. Identification of novel therapeutic targets in the PI3K/AKT/mTOR pathway in hepatocellular carcinoma using targeted next generation sequencing. Oncotarget. 2014;5(10):3012–22. doi: 10.18632/oncotarget.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent L, Bae S, Tsai S, et al. Dosage-dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. J Clin Invest. 2017;127(3):830–842. doi: 10.1172/JCI87583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun. 2012;426(2):247–52. doi: 10.1016/j.bbrc.2012.08.075. [DOI] [PubMed] [Google Scholar]
- Li K, Ng I, Fan S, et al. Activation of cyclin-dependent kinases CDC2 and CDK2 in hepatocellular carcinoma. Liver. 2002;22(3):259–68. doi: 10.1046/j.0106-9543.2002.01629.x. [DOI] [PubMed] [Google Scholar]
- Lin W, Chen H, Huang G, et al. Infrequent mutations and no methylation of CDKN2A (p16/MTS1) and CDKN2B(p15/MTS2) in hepatocellular carcinoma in Taiwan. Eur J Cancer. 1998;34(11):1789–95. doi: 10.1016/s0959-8049(98)00189-0. [DOI] [PubMed] [Google Scholar]
- Lindenmeyer C, McCullough A. The natural history of nonalcoholic fatty liver disease – an evolving view. Clin Liver Dis. 2018;22(1):11–21. doi: 10.1016/j.cld.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Gong M, French B, et al. Aberrant modulation of the BRCA1 and G1/S cell cycle pathways in alcoholic hepatitis patients with Mallory Denk bodies revealed by RNA sequencing. Oncotarget. 2015;6(40):42491–503. doi: 10.18632/oncotarget.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu Y, Meng L, et al. Synergistic antitumor effect of sorafenib in combination with ATM inhibitor in hepatocellular carcinoma cells. Int J Med Sci. 2017;14(6):523–529. doi: 10.7150/ijms.19033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu Y, Liu H, et al. Tumor suppressive function of p21-activated kinase 6 in hepatocellular carcinoma. J Biol Chem. 2015;290(47):28489–501. doi: 10.1074/jbc.M115.658237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masouminia M, Samadzadeh S, Mendoza A, et al. Upregulation of autophagy components in alcoholic hepatitis and nonalcoholic steatohepatitis. Exp Mol Pathol. 2016;101(1):81–88. doi: 10.1016/j.yexmp.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J, Reinus J. Prevalence and natural history of alcoholic liver disease. Clin Liver Dis. 2012;16(4):659–66. doi: 10.1016/j.cld.2012.08.001. [DOI] [PubMed] [Google Scholar]
- To A, Chen G, Chan U, et al. ZBP-89 enhances Bak expression and causes apoptosis in hepatocellular carcinoma cells. Biochim Biophys Acta. 2011;1813(1):222–30. doi: 10.1016/j.bbamcr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Liang L, Zhang Y, et al. GNAI1 suppresses tumor cell migration and invasion and is post-transcriptionally regulated by Mir-320a/c/d in hepatocellular carcinoma. Cancer Biol Med. 2012;9(4):234–41. doi: 10.7497/j.issn.2095-3941.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector M, Xue W, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125(7):1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Duan H, Yang Y. Knockdown of Rap2B inhibits the proliferation and invasion in hepatocellular carcinoma cells. Oncol Res. 2017;25(1):19–27. doi: 10.3727/096504016X14685034103914. [DOI] [PMC free article] [PubMed] [Google Scholar]