Abstract
SERINC5(S5) is a multi-span transmembrane protein that potently blocks the infectivity of HIV-1 produced by human T-cells. The ability of S5 to restrict infectivity correlates with its presence in the virion, but the exact mechanism by which S5 restricts HIV-1 is unknown. Here we tested whether the core from HIV-1 virions containing S5 is delivered to the cytoplasm. Using the “fate of the capsid” assay, we demonstrated that the viral core of S5-restricted HIV-1 does not reach the cytoplasm of target cells, suggesting a block in the delivery of the core to the cytoplasm. In agreement with evidence suggesting that the viral determinants for S5 restriction map to the envelope of HIV-1, we observed that S5 induces conformational changes to the HIV-1 envelope. Further, we demonstrated that S5 localizes to detergent-resistant membranes (DRMs), as has been shown previously for the HIV-1 envelope in producer cells. In order to identify the determinants of S5 restriction, we explored the ability of all human SERINC proteins to restrict HIV-1. In contrast to human S5, we observed that human SERINC2(S2) did not restrict HIV-1, and was inefficiently incorporated into HIV-1 virions when compared to S5. Experiments using S5-S2 chimeric proteins revealed two functional domains for restriction: one necessary for S5 incorporation into virions, which does not seem to be necessary for restriction, and a second one necessary to change the HIV-1 envelope conformation, localize to DRMs, and block infection.
Keywords: SERINC5, SERINC2, HIV-1, restriction, envelope, core, DRMs
INTRODUCTION
Nef is a lentiviral accessory protein essential for viral replication and induction of AIDS-like symptoms in Rhesus macaques by a pathogenic SIV strain (Kestler et al., 1991). In 1994, Chowers and colleagues reported that primate lentiviral accessory protein Nef can enhance the infectivity of HIV-1 virions produced in human T cells (Chowers et al., 1994). Their experiments demonstrated that Nef increases HIV-1 replication in human T-cells. Similar results were reported subsequently by other investigators (Aiken and Trono, 1995; Schwartz et al., 1995). In 2015, two groups reported independently that the human proteins SERINC3 (S3) and SERINC5 (S5) are the HIV-1 restriction factors switched off by Nef (Rosa et al., 2015; Usami et al., 2015). S3 and S5 are members of the serine incorporator (SERINC) family of proteins, which is highly conserved among eukaryotes (Inuzuka et al., 2005).
Mechanistic studies have led to the following findings regarding the ability of S3 and S5 to restrict HIV-1 (Rosa et al., 2015; Usami et al., 2015): a) S3 and S5 are incorporated into the HIV-1 viral particle, which correlates with blockage of infectivity, and b) the ability of Nef to overcome restriction correlates with decreased incorporation of SERINC proteins into the virus. Although both reports suggested that there is a virus-cell fusion defect, Rosa and colleagues noted that the fusion pores between viruses and cells are formed (Rosa et al., 2015).
Early studies suggested that the envelope protein of HIV-1 is the determinant for the Nef-sensitive HIV-1 restriction imposed by T cells (Lai et al., 2011; Usami and Gottlinger, 2013). In agreement with these findings, recent evidence also linked the HIV-1 envelope to the restriction observed by S5 (Beitari et al., 2017; Sood et al., 2017). Altogether, these experiments suggested that S5 might be changing the conformation of the envelope through direct interaction or by changing the lipid composition of the membrane. So far no evidence has shown an interaction between S5 and the envelope of HIV-1. Although it has been suggested that expression of SERINC proteins increases phosphatidylserine synthesis in bacteria (Inuzuka et al., 2005), recent evidence showed that S5 does not alter the lipid composition of the HIV-1 virion (Trautz et al., 2017). This showed that much is left to be investigated regarding the mechanism used by S5 to block HIV-1. Although primate evolutionary studies showed that there are no notable sequence signatures for positive selection in S5 and S3 (Murrell et al 2016), studies have shown that the Nef-mediated SERINC5 antagonism may determine the ability of primate lentiviruses to spread within natural hosts (Heigele et al., 2016).
Previous investigations showed that S5-containing HIV-1 particles do not undergo reverse transcription, suggesting that S5-containing HIV-1 viruses might not be able to deliver the viral core into the cytoplasm of target cells; therefore, we investigated whether the core of S5-containing HIV-1 is actually released into the cytoplasm of target cells. Using the “fate of the capsid” assay, we found that the HIV-1 core from S5-containing particles is not released into the cytoplasm of target cells, implying a defect in cytoplasmic delivery of the HIV-1 core. The viral determinants for S5 restriction have been mapped to the HIV-1 envelope, suggesting that S5 is critically affecting the function of the HIV-1 envelope. The presence of S5 in the viral particle was found to induce conformational changes in the HIV-1 envelope that correlated with restriction. Consistently with this, it was found that S5 localizes to detergent-resistant membranes (DRMs), as has been shown for the HIV-1 envelope in producer cells, suggesting that S5 and HIV-1 envelope co-localization in DRMs is important for restriction. To define in greater detail the determinants for this restriction, we sought to identify SERINC proteins that do not restrict HIV-1, and for this purpose, evaluated the ability of all human SERINC proteins to restrict HIV-1. We observed that human S2 did not restrict HIV-1, and was incorporated only inefficiently into HIV-1 when compared to S5. These results identified S2 as a suitable protein with which to construct chimeras that might help identify determinants for restriction. Gratifyingly, S5-S2 chimeric proteins revealed two important domains for restriction: one necessary for incorporation into viral particles, and a second domain necessary to change the HIV-1 envelope conformation, localize to DRMs, and block HIV-1 infection.
RESULTS
Ability of human SERINC proteins to restrict HIV-1 infection
In order to begin our investigations on the mechanism by which S5 blocks HIV-1, we sought to find human SERINCs that differentially restrict HIV-1. The simultaneous study of the five human SERINC proteins will help defining the requirements for restriction. For this purpose, we tested the ability of all human SERINC proteins to restrict HIV-1 (Fig. 1). We challenged TZM-bl GFP-reporter cells with HIV-1SF162 particles produced in the presence of increasing concentrations of the indicated SERINC proteins (Fig.1). At 48 h post-challenge, infection was determined by measuring the percentage of GFP-positive cells, and the results were used to calculate fold-restriction. At the same time, producer cells were analyzed for expression of SERINC proteins and GAPDH using anti-FLAG and anti-GAPDH antibodies, respectively. Similarly, SERINCs and p24 expression was analyzed in partially purified virions (using a 20% sucrose cushion). Detection of SERINCs required the use of a modified Western blot protocol described in Methods.
Figure 1. Ability of human SERINC proteins to restrict HIV-1.
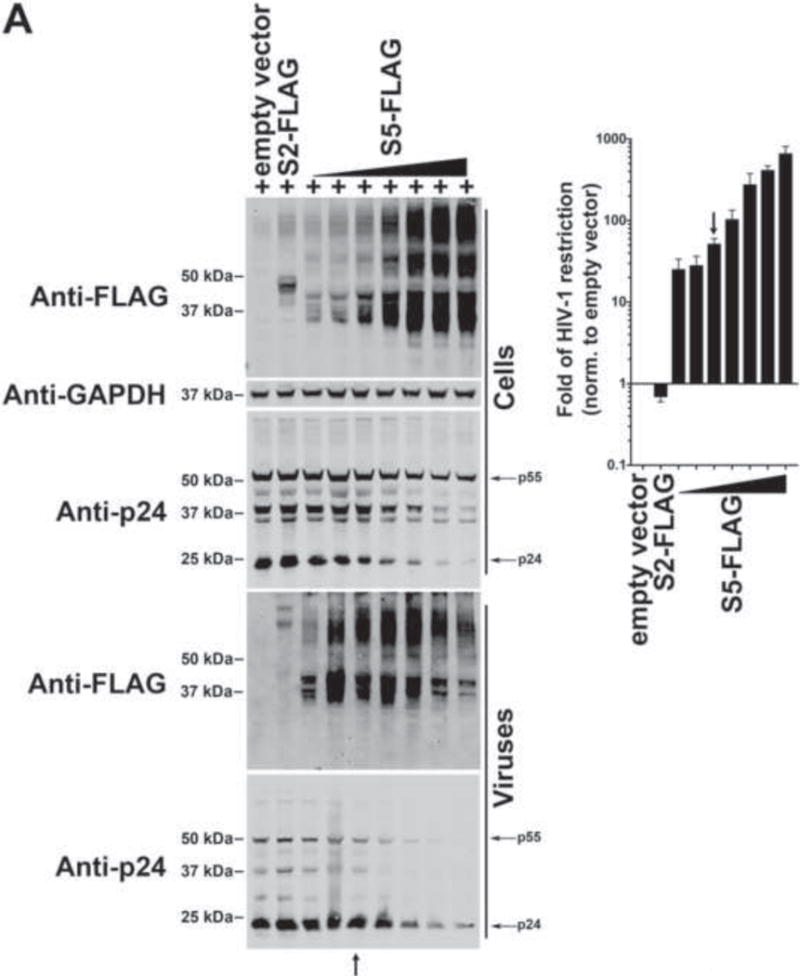


To test the ability of S5 (A), S2 (B), S4 (C), S3 (C), and S1 (C) to restrict HIV-1, HIV-1ΔNef particles expressing the SF162 envelope (HIV-1SF162) in the presence of increasing amounts of the indicated SERINC protein were produced. Viruses and producer cells were harvested 48 hours post-transfection. Producer cells (Cells) were lysed and analyzed for expression of the indicated SERINC protein, p24 and GAPDH by Western blotting using anti-FLAG, anti-p24 and anti-GAPDH antibodies (left panels), respectively. Produced HIV-1SF162 viruses (Viruses) were partially purified using a 20% sucrose cushion and analyzed for expression of the indicated SERINC protein and p24 using anti-FLAG and anti-p24 antibodies (left panels), respectively. At the same time, TZM-bl GFP-reporter cells were challenged with the different HIV-1SF162 viruses. At 24 h post-challenge, infection was determined by measuring the percentage of GFP-positive cells (right panel). Fold-restriction is defined as the ratio of %infection by viruses produced in the presence of empty vector to %infection by viruses produced in the presence of the indicated SERINC protein (right panel). The fold of HIV-1 restriction shown is the average of 3 independent experiments. Black arrows point to the experiments where the levels of SERINC expression did not affect virus production as measured by p24. Experiments were repeated at least three times and the Western blot from a representative example is shown. (C) The fold of HIV-1 restriction at SERINC levels that do not affect particle release (black arrow) for three independent experiments with standard deviation is shown. In addition, accession number, molecular weight and number of amino acids for each human SERINC protein is illustrated.
The use of increasing concentrations of S5 tagged with a FLAG epitope (S5-FLAG) revealed two blocks for HIV-1 infection (Fig. 1A): 1) an HIV-1 infectivity block of ~10–40 fold was observed in released virions when low levels of S5-FLAG were detectable in producer cells (note: block did not affect viral production), and 2) an artifactual block of HIV-1 infectivity of >40 fold observed when high levels of S5-FLAG were expressed in virus- producing cells, which affected virus production and/or release as measured by blotting for p24. These results indicated that high S5 expression inhibits virion production, causing the latter to be an overexpression artifact; as shown in producer cells there was a decrease in Gag expression and maturation in producer cells when high levels of S5 were used (Fig. 1A). Therefore, all experiments in this manuscript will assess HIV-1 restriction using S5 levels that do not affect viral particle release, as restriction in endogenous T-cells was originally described (Chowers et al., 1994). Of note with regard to these experiments, S5-FLAG was shown to incorporate into HIV-1 virions (Fig. 1A). As shown in Figure 1A, a black arrow points to the experiments where the levels of SERINC expression did not affect virus production as measured by p24, which also indicates the restriction fold we are considering for S5. This type of titration assays will be used through out this work to understand restriction of the different SERINC variants.
Similarly, the ability of different concentrations of S2 tagged with a FLAG epitope (S2-FLAG) to block the infectivity of released HIV-1 particles was tested. As shown in Fig. 1B, expression of S2 at different levels in producer cells did not negatively affect virion production or infectivity of released virions, which is in agreement with recent findings (Sood et al., 2017). Instead, we observed that S2 expression in producer cells was observed to enhance the infectivity of released HIV-1 by as much as two-fold. Our results suggested that S2 incorporates less efficiently into HIV-1 when compared to S5 (Fig. 1B). Because of its inability to block the infectivity of released viruses, S2 serves as a useful control for experiments designed to elucidate how S5 affects HIV-1 infection.
Next we tested restriction of HIV-1 infection by S4, the closest homologue to S5, using increasing amounts of S4 (Fig. 1C and Fig. S1). Interestingly, expression of S4 tagged with a FLAG epitope (S4-FLAG) potently blocked infectivity of released viruses (Fig. 1C). Similarly to S5, the use of high S4 expression resulted in a decrease of Gag expression and maturation (Fig. S1). As a control we included S2-FLAG, which does not restrict the infectivity of released virions. These results showed that at similar levels of total expression S4 blocks infectivity of HIV-1 as potently as S5 (Fig. S1).
Expression of S3 tagged with a FLAG epitope (S3-FLAG) in producer cells diminished the infectivity of released HIV-1 virions two- to three-fold (Fig. 1C), demonstrating that S3 is a less potent restriction factor than S5, as shown earlier (Rosa et al., 2015; Usami et al., 2015).
Expression of S1 tagged with a FLAG epitope (S1-FLAG) in producer cells blocked the infectivity of released HIV-1 virions only minimally (Fig. 1C), indicating that S1 has limited or no activity against HIV-1.
S5 topology and contribution of the different loops to HIV-1 restriction
To understand the contribution of the S5 loops to restriction, we first sought to understand the membrane topology of S5. Members of the SERINC family were originally described as membrane proteins containing 10–11 transmembrane domains (Inuzuka et al., 2005). The model simulated by TOPCONS (http://topcons.cbr.su.se) predicts that S5 contains ten transmembrane domains (TM), five extracellular loops, and four intracellular loops (L) (Fig. 2A). Based on this model, we explored the membrane topology of S5 using flow cytometry by testing surface (fixed) and total (fixed & permeabilized) expression of S5 variants. S5 proteins containing a FLAG epitope peptide inserted into each of the nine loops (L1–9) were used. As shown in Figure 2A, S5 tagged with a C-terminal FLAG epitope showed poor surface detection (fixed) when compared to total expression (fixed & permeabilized), suggesting that the C-terminus of S5 resides within the intracellular compartment. By contrast S5 containing a FLAG epitope in L7 (S5-L7-FLAG) or L9 (S5-L9-FLAG) showed similar surface detection versus total expression, suggesting that L7 and L9 reside in the extracellular compartment. In a similar manner, we showed that L2, L4, L8 and the C-terminal tail of S5 face toward the intracellular compartment (Fig. 2A). The remaining constructs were poorly expressed, preventing us from drawing firm conclusions. Overall, however, our observations were in agreement with the notion that L7 and L9 are facing out toward the extracellular compartment, whereas the L2, L4, L8 and the C-terminal region of S5 face toward the intracellular compartment. To corroborate these results, immunofluorescence staining experiments were performed using anti- FLAG antibodies. S5-L7-FLAG and S5-L9-FLAG showed a similar staining pattern when compared to wild-type S5 (Fig. 2B). Taken together, the results were consistent with the topology predicted by the TOPCONS model.
Figure 2. S5 topology and contribution of the different loops to HIV-1 restriction.



(A) The theoretical membrane topology for human S5 (top left panel) predicted by the software TOPCONS is shown. N and C indicates the N- and C-terminus, respectively. The protein loops (L) are numbered L1-L9. Similarly, the transmembrane (TM) regions are numbered TM1-TM10. The residues spanning the TM regions are shown. Surface (fixed) and total expression (fixed & permeabilized) of the indicated S5 variants containing a FLAG epitope peptide (red circle) inserted into each of the nine loops (L1-L9) of S5 were analyzed by flow cytometry using anti-FLAG antibodies (red line). As a control, flow cytometric analysis was performed using an Isotype-matched control (blue line). The percentage of FLAG-positive cells for the illustrated experiment is presented. The standard deviation of three independent experiments is shown (right panels). (B) HeLa cells expressing the indicated S5 variants containing a FLAG epitope tag were imaged by fluorescence microscopy using anti-FLAG antibodies (red). Nuclei were stained with DAPI (blue). (C) The ability of the indicated S5 variants to restrict HIV-1 was measured. The fold of HIV-1 restriction at SERINC levels that do not affect particle release for three independent experiments with standard deviation is shown.
To explore the role of S5 extracellular and intracellular loops as determinants for restriction, we tested the HIV-1 restriction ability of S5 variants containing a FLAG epitope peptide inserted on L2, L4, L7, L8 and L9. These experiments are based on the notion that insertion of a FLAG tag will disrupt the normal function of the Loops. Interestingly, expression of S5-L2-FLAG, S5-L7-FLAG and S5-L8-FLAG, in producer cells potently blocked the infection of released HIV-1 particles (Fig. 2C). By contrast S5-L4 and S5-L9 exhibited an attenuated ability to block HIV-1 infection (~4 fold) when compared to wild type S5 (Fig. 2C). Although the expression of some of these proteins was poorly detectable by Western blotting, we have ensured detection of expression by flow cytometry using anti-FLAG antibodies in fixed/permeabilized cells. Since we are using increasing levels of S5 variants in producer cells, we are considering restriction only when viruses did not exhibit a viral particle release/budding defect. As controls, we have used wild type S5 and S2. These experiments suggested that insertion of a tag in loops 2, 7, or 8 does not affect the ability of S5 to restrict HIV-1. However, disruption of L4 or L9 attenuated the ability of S5 to block HIV-1.
S5 blocked delivery of HIV-1 cores into the cytosol of target cells
Rosa and colleagues showed that S5-containing HIV-1 particles do not undergo reverse transcription, suggesting that S5-containing HIV-1 viruses might not be able to deliver the viral core into the cytoplasm of target cells (Rosa et al., 2015). To test this hypothesis, we explored the fate of the core in virions produced in the presence of S5 during infection. To this end, we challenged TZM-bl GFP-reporter cells with HIV-1SF162 viruses produced in the presence of S5, and performed the “fate of the capsid” assay (Fig. 3A). HIV-1SF162 viruses used in the infections were normalized with respect to p24 (Fig. 3B). As previously described (Yang et al., 2014), the “fate of the capsid” assay reveals 1) the amount of capsid delivered into the cytoplasm of target cells (INPUT), 2) the fraction of total capsid that is forming cores (PELLET), and 3) the amount of soluble capsid in the cytoplasm (SOLUBLE). As shown in Figure 3A, HIV-1SF162 delivered HIV-1 capsid into the cytoplasm of target cells, as measured at 8 h post-infection (INPUT). However, infection by HIV-1SF162 viruses performed in the presence of the small-molecule drug TAK-779 completely prevented capsid delivery into the cytoplasm, as shown by a very faint band that represents the background of the assay (INPUT). TAK-779 is a CCR5 antagonist that prevents fusion of viral membrane with the cellular membrane (Baba et al., 1999). Remarkably, HIV-1SF162 viruses produced in the presence of S5 poorly delivered HIV-1 capsid into the cytoplasm of cells (INPUT), in a manner resembling infection by HIV-1SF162 in the presence of TAK-779 (INPUT). As a control, we measured the infectivity of the viruses used in the “fate of the capsid” assay (Fig. 3C). Taken together the experiments showed that HIV-1SF162 virions containing S5 are defective in the ability to deliver the core into the cytoplasm of target cells. Furthermore, the finding that TAK-779 could phenotypically recapitulate restriction caused by S5 was a notable finding of this work.
Figure 3. S5 prevents the delivery of HIV-1 cores to the cytosol of target cells.
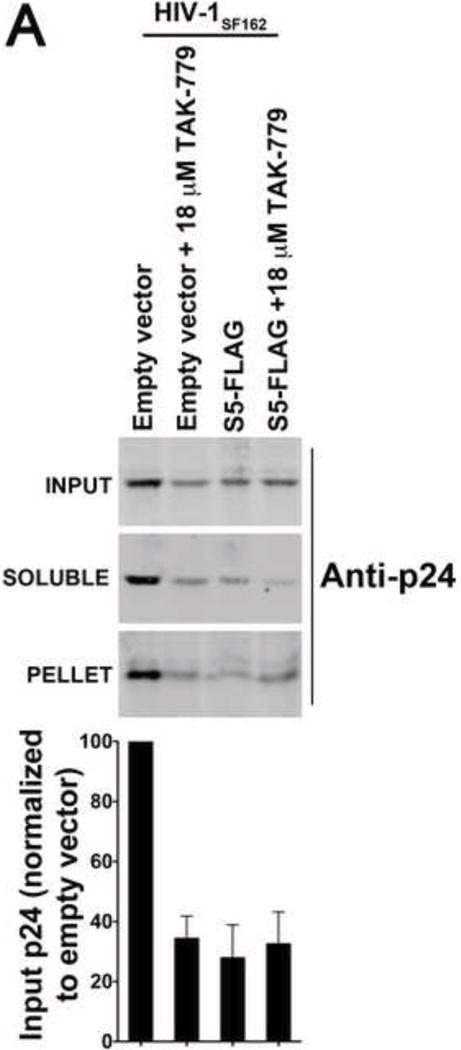

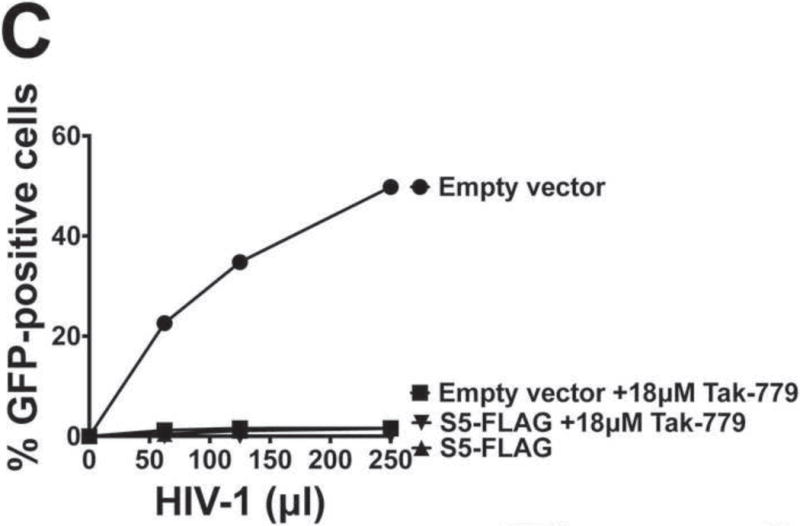
Human TZM-bl GFP-reporter cells were challenged with normalized amounts of HIV-1SF162 that do or do not contain S5. Eight hours post-infection cells were harvested and processed as described in Methods. Briefly, cells were incubated in a hypotonic buffer and homogenized using a Dounce homogenizer. A post-nuclear supernatant (INPUT) was obtained by pelleting the nuclear fraction. The INPUT represents the total capsid delivered into the cytoplasm of the target cell. The post-nuclear supernatant was then spun on 50% sucrose cushion to separate the pelletable capsid (PELLET) from the soluble capsid (SOLUBLE), as described in Methods. The PELLET represents the capsid that is forming HIV-1 cores. The SOLUBLE fraction represents the soluble capsid. INPUT, SOLUBLE and PELLET fractions were analyzed by Western blot using anti-p24 antibodies (A upper panel). As a control, similar infections were also performed in the presence of the inhibitor TAK-779, which is a CCR5 antagonist that prevents the fusion of viral membrane with the cellular membrane. The standard deviation of the INPUT for three independent experiments is shown (A lower panel). For these experiments viruses that do or do not contain S5 were normalized using p24 levels (B). Infectivity of viruses used in the fractionation assay is shown (C).
S5 changes the conformation of the HIV-1 envelope glycoprotein
Previous work suggested that the envelope glycoprotein of HIV-1 is the viral determinant for the Nef-sensitive HIV-1 restriction observed in Jurkat T cells: 1) HIV-1 virions produced in the presence of Nef protein showed increased sensitivity to neutralizing antibodies when compared to virions without Nef (Lai et al., 2011), and 2) experiments demonstrated that the variable regions of gp120 are the viral determinants for Nef-sensitive HIV-1 restriction in Jurkat T cells (Usami and Gottlinger, 2013). We initially investigated whether envelope incorporation and/or maturation were affected in HIV-1 particles containing S5. As others have shown, envelope incorporation and maturation are not affected by the presence of S5 (Sood et al., 2017; Usami et al., 2015). Thus we hypothesized that S5 alters the conformation of the envelope. To test this hypothesis we measured whether the presence of S5 in the virion alters their sensitivity to neutralizing antibodies that target the viral envelope. In the initial experiments we used HIV-1 virions produced in human HEK293T cells. As shown in Figure 4A, experiments using neutralizing antibodies against the membrane proximal external region (MPER) of gp41, such as 4E10, 2F5 and 10E8, showed that S5-containing virions were more sensitive to neutralization when compared to HIV-1SF162 variants containing S2. Milder effects were observed when using the anti-gp41 antibodies 240-D and 246-D (Fig. 4A). Interestingly, the anti-gp120 antibody 2G12 also showed different neutralization patterns when comparing HIV-1 particles containing S5 versus S2. Other antibodies such as PG9 and NC-1 did not affect the infectivity of HIV-1SF162 variants containing S5 (Fig. 4B). By contrast, highly neutralizing antibodies such as 3BNC117 and VRCO1 (against the CD4-binding site of the HIV-1 envelope), potently neutralized HIV-1SF162 viruses containing either S5 or S2 (Fig. 4B), as has been shown for other HIV-1 viruses (Scheid et al., 2011). These experiments supported the notion that S5 induces conformational reorganization of the HIV-1 envelope. To determine whether the effect of S5 on the HIV-1 envelope glycoprotein also occurs when using endogenous levels of S5, we performed similar neutralization experiments using HIV-1SF162 virions produced in Jurkat TAg cells, which express endogenous levels of S5 and S3. As controls, we used HIV-1SF162 virions produced in Jurkat TAg cells knocked out for expression of S5 and S3 (S3/S5 KO). As expected, HIV-1SF162 containing endogenous levels of S5 and S3 likewise affected the conformation of the HIV-1 envelope glycoprotein (Fig. 4C).
Figure 4. S5 affects the conformation of the HIV-1 envelope.




(A) The ability of different HIV-1 neutralizing antibodies to block HIV-1SF162 viruses produced in the presence S5 was measured. HIV-1SF162 viruses produced in human HEK293T cells in the presence of S5, S2, or empty vector were incubated with increasing amounts of the indicated neutralizing antibody for 1 h at 37 °C. The virus- antibody mixture was then used to infect TZM-bl GFP-reporter cells. Infectivity was determined by measuring the percentage of GFP-positive cells twenty-four hours post-infection. Infection values were normalized to the infection of viruses that were not incubated with neutralizing antibodies. (B) Similar neutralization experiments were performed using HIV-1SF162 viruses with anti-gp41 and anti-gp120 antibodies, which either don´t neutralize SF162 viruses or with anti-gp120 antibodies, which strongly neutralize SF162 viruses. (C) Neutralization experiments were also performed using HIV-1SF162 viruses produced in human Jurkat TAg cells that do or do not express S3 and S5 (S3/S5 KO). (D) HIV-1SF162 viruses produced in HEK293T (top) or Jurkat TAg S3/S5 KO cells (bottom) in the presence of S5, S2, or empty vector were used to challenge TZM-bl GFP-reporter cells in the presence of increasing concentrations of the small molecule inhibitor 484. Infectivity was determined by measuring the % of GFP-positive cells 24 h post-infection. Infection values were normalized to the infection of viruses that were not incubated with the drug. Infections were performed in triplicates and standard deviations are shown.
We next tested the novel HIV-1 entry inhibitor 18A, which prevents CD4-induced disruption of quaternary structures at the trimer apex of the HIV-1 envelope, as well as exposure of the gp41 HR1 coiled coil (Herschhorn et al., 2014). For this purpose, we tested the sensitivity of HIV-1 virions containing S5 with increasing concentrations of the drug 484, which is a next-generation version of 18A (Herschhorn et al., 2014). As shown in Figure 4D, HIV-1 virions containing S5 were more sensitive to 484 than those containing S2. Similar differences were observed between virions produced in Jurkat TAg cells and those produced in Jurkat TAg S3/S5 knockouts (Fig. 4D). Taken together, the results suggested that the presence of S5 alters in some way the three-dimensional shape of the HIV-1 envelope consequentially affecting fusion, which is likely to inhibit the delivery of the viral core to the cytoplasm of target cells.
S5 localizes to detergent-resistant membranes (DRMs)
The foregoing results suggested that S5 can alter the shape and/or conformation of the envelope glycoproteins, thereby preventing delivery of the HIV-1 core to the cytoplasm. This raised the possibility that S5 is changing the conformation of the HIV-1 envelope by direct physical interaction. Several lines of evidence have suggested that HIV-1 budding can occur in DRMs (Bhattacharya et al., 2004; Bhattacharya et al., 2006; Nguyen and Hildreth, 2000; Ono and Freed, 2001), which has been used as a working description for lipid rafts when sphingolipids and GPI-anchored proteins were insoluble in Triton X-100 at 4 °C, and floated to a characteristic density following equilibrium density gradient centrifugation (Lingwood and Simons, 2007). Therefore, we reasoned that if interaction occurs between S5 and the HIV-1 envelope, it might be taking place in DRMs. Accordingly, we examined whether S5 localizes to DRMs. Flotation assays were performed in S5- and S2-expressing human cells using a discontinuous sucrose density gradient (5–40%) as previously described (Diaz-Griffero et al., 2005; Scherer et al., 1994). Interestingly, S5 was detected in fractions near the top of the sucrose gradient, consistently with localization to DRMs (Fig. 5A). In contrast, S2 was detected in fractions nearer to the bottom of the gradient when compared to S5, indicating that S2 localizes to membrane microdomains of different density when compared to S5. As a control for protein that localizes to DRMs, we used caveolin-1-GFP (Scherer et al., 1994). To control for proteins that do not localize to DRMs, we used the tumor virus protein S3 fused to GFP(TVBS3-GFP), which is known not to localize to DRMs (Diaz-Griffero et al., 2005). Similar results were obtained when the experiments were performed in cells producing HIV-1 virions (Fig. 5B). Here we also explored the presence of the HIV-1 envelope in the different fractions. As shown in Figure 5B, a small amount of HIV-1 envelope cofractionates with S5, which is in agreement with findings showing partial localization of envelope in DRMs (Yang et al., 2010). Taken together, the experiments indicated that S5 localizes to DRMs.
Figure 5. S5 localizes to detergent-resistant membranes (DRMs).


(A) HEK293T cells expressing S5-FLAG or S2-FLAG were lysed and fractionated using a flotation gradient as described in Methods. Cells were homogenized in a Triton X-100 detergent solution and fractionated by discontinuous sucrose gradient centrifugation (5–40%). Gradient fractions 1–12 were analyzed by Western blotting using anti-FLAG antibodies. As positive controls, similar analysis was performed using HEK293T cells separately transfected with Caveolin-1 fused to GFP (Caveolin-1-GFP), which localizes to DRMs. As a negative control, we studied the membrane localization of the receptor TVBS3 fused to GFP (TVBS3-GFP), which does not localize to DRMs. Caveolin-1-GFP and TVBS3-GFP expression was detected by Western blotting using anti-GFP antibodies. Fractions for S2 and S5 from three independent experiments were quantified using a Li-Cor instrument, and they are shown as % of total FLAG protein with standard deviations (lower panel). (B) Similar fractionation experiments were performed in human HEK293T cells producing HIV-1SF162 viruses in the presence of S5-FLAG or S2-FLAG. The DNA mix for each transfection included 8 μg NL4–3ΔNef, 0.5 μg SF162, plus either empty vector, S2-FLAG, S5-FLAG, caveolin-1-GFP, or TVB-1. Forty-eight hours post-transfection cells were lyzed and fractionated as described in Methods. Fractions were analyzed by Western blotting using anti-FLAG, anti-GFP, or anti-Env antibodies. Fractions for S2 and S5 from three independent experiments were quantified using a Li-Cor instrument, and they are shown as % of total FLAG protein with standard deviations (lower panel).
S5 residues 203–411 containing regions TM5-TM9 are important for HIV-1 restriction
In this section, we further explored the S5 determinants for HIV-1 restriction. For this purpose, we constructed chimeric proteins using S5 and S2. Fortuitously, the lengths of these proteins were suitable for chimera construction; S5 and S2 are 461 and 464 amino acids in length, respectively. Furthermore, TOPCONS software predicted that S5 and S2 should possess similar membrane topology (Fig. 6A). In contrast to S5, we have shown that S2 does not restrict HIV-1; therefore, S5-S2 chimeric proteins could perhaps provide useful information regarding restriction determinants.
Figure 6. S5 residues 203–411 containing regions TM5-L5-TM6-L6-TM7-L7-TM8-L8-TM9 are important for HIV-1 restriction.


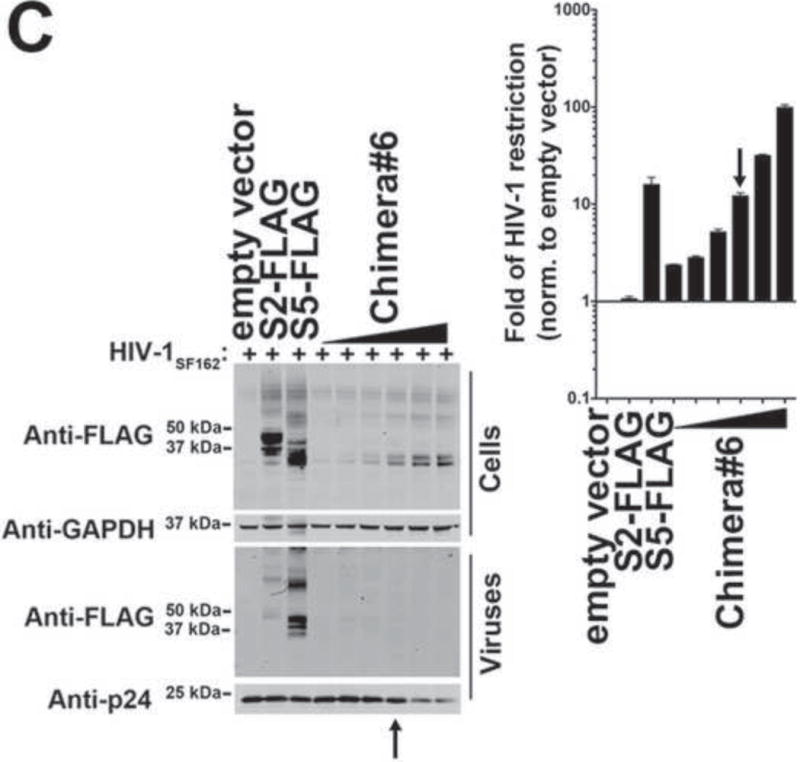
(A) The ability of the indicated S5-S2 chimeric proteins to restrict HIV-1 is shown as fold-restriction. The amino acid region of each chimera is shown (right panel). The fold-restriction is defined as the ratio of % infection by viruses produced in the presence of empty vector to % infection by viruses produced in the presence of the indicated S5-S2 chimera. The HIV-1 fold-restriction where expression of the chimera did not affect viral production is shown with a standard deviation from six independent experiments. Schematic representations of the chimeras are shown, with protein regions of S5 and S2 colored in orange and black, respectively. The ability of S5-S2 chimera#5 (B) and chimera#6 (C) to restrict HIV-1 is shown. Black arrows point to the experiments where the levels of chimera expression did not affect virus production as measured by p24. Experiments were repeated at least three times and the Western blot from a representative example is shown.
As shown in Figure 6A, a series of S5–S2 chimeric proteins were constructed and their ability to restrict the infectivity of HIV-1 was measured. As before, chimeras were studied at expression levels that do not affect particle production in producer cells. These experiments showed that chimera#5 (Fig. 7B) and chimera#6 (Fig. 7C) potently restricted HIV-1 infection, suggesting that they are likely to contain important determinants for restriction. By contrast, chimeras#1–4 poorly restricted HIV-1 infection when expressed in producer cells (Fig. 6A). Overall, these experiments indicated that S5 residues 203–411, which cover the sequences between TM5 and TM9 (Fig. 6A), do indeed contain important determinants for restriction.
Figure 7. The S5 region L5-TM6-L6 is required for incorporation into HIV-1 virions.
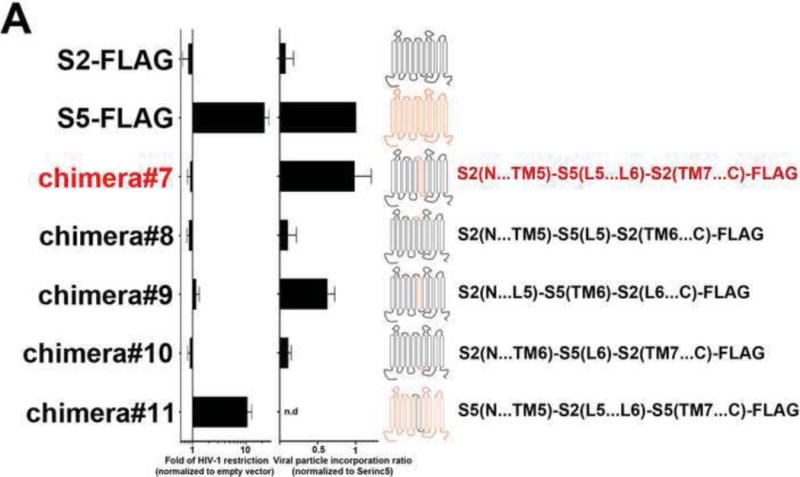

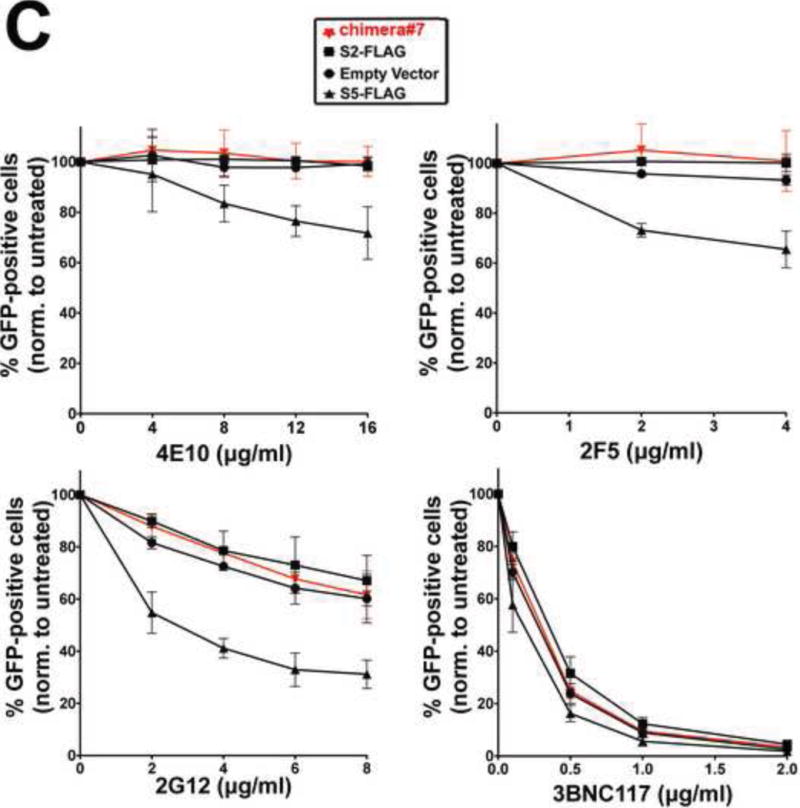


(A) Ability of S2 proteins containing S5 regions to restrict HIV-1 infection and be incorporated into virions. S2 proteins containing individual elements (L5, TM6 or L6), or the entire L5-TM6-L6 region of S5, were analyzed for the ability to restrict HIV-1 (fold-restriction) and be incorporated into virions (viral particle incorporation ratio). Fold-restriction is defined as the ratio of %infection by viruses produced in the presence of empty vector to %infection by viruses produced in the presence of the indicated S5-S2 chimera. The HIV-1 fold-restriction where expression of the chimera did not affect viral production is shown with a standard deviation from six independent experiments. Incorporation of the chimera into viral particles is defined as the ratio of protein incorporated into virions to the protein expressed in producer cells. Viral particle incorporation ratio where expression of the chimera did not affect viral production is shown with a standard deviation from three independent experiments. Schematic representations of S2 proteins containing S5 regions are shown with protein regions of S2 and S5 colored in black and orange, respectively. (B) The ability of chimera#7 and chimera#9 to restrict HIV-1 is shown. Black arrows point to the experiments where the levels of chimera expression did not affect virus production as measured by p24. Experiments were repeated at least three times and the Western blot from a representative example is shown. (C) The ability of the indicated HIV-1 neutralizing antibodies to block HIV-1SF162 viruses produced in the presence of chimera#7 was measured. Viruses produced in the presence of the indicated SERINC proteins were incubated with increasing amounts of the indicated neutralizing antibody for 1 hr at 37 °C. The virus-antibody mixture was then used to infect TZM-bl GFP-reporter cells. Infectivity was determined by measuring the percentage of GFP-positive cells at 24 h post-infection. Infection values were normalized to the infection of virions that were not incubated with neutralizing antibodies. Infections were performed in triplicates and standard deviations are shown. (D) Human 293T cells expressing the indicated proteins were lysed in Triton X-100 and fractionated on a discontinuous sucrose gradient (5–40%) as described in Methods. As controls, human HEK293T cells expressing caveoilin-1 fused to GFP (Caveolin-1-GFP) and tumor virus B receptor (TVBS3-GFP) were analyzed as probes for proteins that did or did not localize to DRMs, respectively. Twelve fractions (1–12) were analyzed per sample using anti-FLAG or anti-GFP antibodies. Fractions for the indicated proteins from three independent experiments were quantified using a Li-Cor instrument, and they are shown as % of total FLAG protein with standard deviations (lower panel). (E) The ability of chimera#11 to block HIV-1 infection was measured, as described above.
Region L5-TM6-L6 is required for the ability of S5 to be incorporated into HIV-1 virions
Previous work has correlated the presence of S5 in the virion with restriction (Rosa et al., 2015; Usami et al., 2015), and we therefore sought to identify the protein determinants for the ability of S5 to be incorporated into HIV-1. We have already shown that S2 incorporates less efficiently into HIV-1 than S5 does. Our earlier experiments had demonstrated that the protein segments containing determinants for HIV-1 restriction by S5 are located between TM5 and TM9 (Fig. 6A upper panel). Therefore, we created several S2 variants containing different components of the TM5…TM9 region of S5, and tested them for incorporation into the viral particle and restriction. As shown in Figures 7A and 7B, the S2 protein containing the S5 region L5-TM6-L6 (chimera#7) did not restrict HIV-1 infection but did incorporate into virions as efficiently as the wild-type S5. Remarkably, the experiments mapped the ability of S5 to be incorporated into virions to the S5 region L5-TM6-L6. To narrow down the minimally requisite domain for S5 incorporation, we constructed S2 chimeras containing individually L5, TM6 or L6 (chimera#8, 9 and 10) (Fig. 7A). Interestingly, the S2 chimera containing the TM6 region of S5 increased S2 incorporation into virions by a factor of six (chimera#9)(Fig. 7A). The experiments suggested that, in the context of S2, the TM6 region of S5 suffices by itself to effect incorporation. By contrast, S2 chimeric proteins containing the S5 regions L5 or L6 neither incorporated into virions nor restricted infection (chimera#8 and 10) (Fig. 7A and Fig. S2).
Next we asked whether incorporation of a SERINC protein such as S2 into the virion changes the conformation of the envelope. As shown in Figure 7C, the neutralization pattern of HIV-1SF162 viruses produced in the presence S2 protein containing the S5 region L5-TM6-L6 (chimera#7) did not change when compared to those produced in the presence of S2 or empty vector when using neutralizing antibodies 4E10, 2F5, and 2G12. By contrast, the neutralization pattern of HIV-1SF162 produced in the presence of S5 changed when compared to S2 (Fig. 7C). As a control, we showed that all viruses were neutralized when using the antibody 3BNC117. This experiment showed that incorporation of a SERINC protein into HIV-1 is not sufficient by itself to change the conformation of the viral envelope, suggesting that an additional domain is probably required in order to bring about conformational change in envelope glycoproteins.
We next tested whether S2 containing the protein segment L5-TM6-L6 of S5 (chimera#7) localized to DRMs. As shown in Fig. 7D, this chimera, which had the ability to be efficiently incorporated into HIV-1, did not localize to a membrane microdomain similar to S5. Our results showed that the S2 protein containing the S5 region L5-TM6-L6 behaved like wild-type S2.
Having identified a region of S5 that enabled S2 to be efficiently incorporated into HIV-1 virions, we proceeded to test whether this region is important for the ability of S5 to restrict HIV-1SF162 particles. To this end, the restriction ability of an S5 variant containing the S2 region L5-TM6-L6 (chimera#11) was assessed (Figs. 7A and E). The variant containing the S2 region L5-TM6-L6 fully restricted HIV-1 when compared to wild-type S5, suggesting either that this domain is not required for restriction or that S5 utilizes redundant mechanisms for incorporation into HIV-1 virions.
The S5 region L5-TM6-L6-TM7-L7 is required in order to alter virion neutralizability, localization to DRMs, delivery of the HIV-1 core to the cytoplasm and infectivity
The foregoing observations using chimera#7 suggested that the S5 region L5-TM6-L6 is important for its ability to be incorporated into HIV-1 virions. In order to discern the elements that are important for restriction, we began by adding N- or C-terminal elements of S5 to chimera#7 (Fig. 8A). Chimeric proteins whose S5 region was extended toward the C-terminus (chimera#14–18) were more restrictive than those with N-terminal extensions (chimera#12–13). Remarkably, the S2 chimeric proteins containing the region L5-TM6-L6-TM7-L7 (chimera#15) potently restricted HIV-1 when compared to wild-type S5 (Figs. 8A and B). In agreement, longer C-terminal extensions likewise restricted HIV-1 (Fig. 8A). By contrast, however, none of the N-terminal extensions blocked infection (Fig. 8A). Taken together, the results demonstrated that the S5 region L5-TM6-L6-TM7-L7 is sufficient by itself to bring about restriction.
Figure 8. The S5 region L5-TM6-L6-TM7-L7 is required for it’s the ability to restrict the infectivity of HIV-1.





(A) HIV-1 restriction ability of S2 proteins containing S5 regions. S2 proteins containing the L5-TM6-L6 region of S5 were extended using S5 elements toward the N- and C-terminus of the protein. The new variants were then tested for the ability to block HIV-1 infection. The HIV-1 fold-restriction where expression of the chimera did not affect viral production is shown with a standard deviation from six independent experiments. Schematic representations of S2 proteins containing S5 regions are shown, and regions of S2 and S5 colored in black and orange, respectively. (B) The ability of S2 containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) to block HIV-1 infection is shown. The fold of HIV-1 restriction shown is the average of 3 independent experiments. Black arrows point to the experiments where the levels of chimera expression did not affect virus production as measured by p24. Experiments were repeated at least three times and the Western blot from a representative example is shown. (C) The ability of the indicated HIV-1 neutralizing antibodies to block HIV-1SF162 viruses produced in the presence of the S2 protein containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) was measured. HIV-1SF162 viruses produced in the presence of the indicated SERINC proteins were incubated with increasing amounts of the indicated neutralizing antibody for 1 hr at 37 °C. The virus-antibody mixture was then used to infect TZM-bl GFP-reporter cells. Infectivity was determined by measuring the percentage of GFP-positive cells at 24 h post-infection. Infection values were normalized to the infection of viruses that were not incubated with neutralizing antibodies. (D) S2 protein containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) localized to DRMs in a manner similar to wild-type S5. (E) S2 protein containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) prevented the delivery of HIV-1 cores to the cytosol as measured by the fate of the capsid. The standard deviation of the INPUT for three independent fate of the capsid experiments is shown (lower panel).
We next tested whether S2 containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) changes the conformation of the envelope as measured by antibody neutralization assays. Incorporation of S2 containing the S5 region L5-TM6-L6 (chimera#7) did not affect the neutralization sensitivity of the viral particle when compared to wild-type S5 (Fig. 8C). However, expression of the S2 protein containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) did change the neutralization pattern of HIV-1 in a manner similar to that of virions containing S5 (Fig. 8C). Taken together, the results showed that gain of restriction correlates with a change in neutralization pattern.
We next tested whether the S2 chimeric protein containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) would localize to DRMs. As shown in Fig. 8D, chimera#15 was detected in fractions near the top of the gradient, as in the case of wild-type S5. The results correlated the ability of S5 to restrict HIV-1 with localization to DRMs. Finally, we tested whether chimera#15 prevents the delivery of the HIV-1 core to the cytoplasm. As shown in Figure 8E, chimera#15 prevents delivery of the HIV-1 core to the cytoplasm as illustrated by the fate of the capsid assay (INPUT). Remarkably, these experiments showed that chimera#15 has gained the properties of a fully functional S5. In sum, what we did in these experiments was to map the restriction domain of S5 to the region L5-TM6-L6-TM7-L7, and to show that HIV-1 restriction by S5 correlates with DRM localization, conformational change of the HIV-1 envelope glycoprotein, and inhibition of HIV-1 core delivery into the cytoplasm.
DISCUSSION
While conducting these experiments we encountered two challenging issues regarding the study of S5 as an inhibitor of HIV-1. First, two different blocks were observed depending upon the level of S5 expression in producer cells. High S5 expression in producer cells artefactually blocked release of HIV-1 virions, whereas low expression inhibited the infectivity of the released particles, a scenario more similar to restriction of HIV-1ΔNef by T-cells (Chowers et al., 1994; Rosa et al., 2015; Usami et al., 2015). We believe this is an important distinction that will allow us to study the HIV-1 restriction by S5 that is closer to the restriction observed in T-cells. It is also important to point out that using high levels of S5 expression generates an artefactual block where the levels of HIV-1 Gag expression and processing are affected. This suggested that titration of S5 variants is important in order to differentiate the real restriction from the artefactual block caused by high levels of S5 expression. Secondly, detection of SERINC proteins expression by Western blot required the use of a special protocol as detailed in Methods. Because of Wstern blot detection issues, we used, in some cases, flow cytometry in fixed/permeabilized cells to detect expression of SERINC proteins.
S5 has a complex membrane topology. The software TOPCONS (http://topcons.cbr.su.se) predicts that it is a transmembrane protein that spans the membrane ten times. We tested this model by separately inserting a FLAG epitope in each of the nine loops (L1–9) of S5 and measuring surface expression of the protein using anti-FLAG antibodies. These experiments revealed that L7 and L9 are extracellular. This is in agreement with a previous observation suggesting that L7 is an extracellular loop (Usami et al., 2015). Furthermore, our results suggested that L2, L8, and the C-terminal tail of S5 face the intracellular compartment. Overall, by validating the TOPCONS model, this work affords a better, though still incomplete, understanding of the topology of S5. Interestingly S5-L4-FLAG and S5-L9-FLAG were diminished in their ability to restrict HIV-1 suggesting a role for these loops in HIV-1 restriction.
This work shows that Incorporation of S5 into HIV-1 prevents delivery of the core to the cytoplasm. Rosa and colleagues showed that the fusion pore is formed in the presence of S5, suggesting a block after formation of the fusion pore, possibly involving pore size expansion (Rosa et al., 2015). One possibility is that S5 blocks either enlargement of the pore blocking the delivery of the viral content to the cytoplasm. While this manuscript was under review investigators have also suggested that S5 interferes with the ability of the virus to form small fusion pores (Sood et al., 2017). Any of the aforementioned blocks would prevent delivery of the viral core to the cytoplasm. To test whether the core is, in fact, released to the cytoplasm, we measured the amount of capsid delivered to the cytoplasm by virions containing S5. Our experiments demonstrated that S5 indeed prevents the capsid delivery to the cytoplasm. Furthermore, use of the inhibitor TAK-779 during infection resulted in phenotypic recapitulation of the restriction caused by S5, which suggested that the S5 block is before delivery of the HIV-1 core into the cytoplasm.
Previous studies suggested that the envelope of HIV-1 is the viral determinant for the Nef-sensitive HIV-1 restriction observed in Jurkat T cells (Lai et al., 2011; Usami and Gottlinger, 2013). By performing neutralization experiments with conformation-sensitive antibodies, we demonstrated that HIV-1 virions containing S5 are more sensitive to HIV-1 neutralizing antibodies when compared to S2-containing virions. In agreement with this, S5 incorporation into virions changed sensitivity to the small-molecule inhibitor 18A (Herschhorn et al., 2014). Although the neutralization sensitivity to antibodies 4E10 and 2F5 were similar when using viruses produced in HEK293T and Jurkat cells, we observed that the difference in sensitivity to antibody 2G12 and drug 484 was less pronounced in the case of viruses produced in Jurkat cells. One possibility for this difference is that exogenous expression of S5 in HEK293T cells is able to reach higher levels when compared to endogenous expression in Jurkat cells. Altogether, these experiments suggested that S5 directly or indirectly affects the conformation of the viral envelope glycoproteins. Usami and colleagues established that the variable regions 1 and 2 of the HIV-1 envelope are the viral determinants for restriction (Usami and Gottlinger, 2013); this work suggested the possibility that S5 might be physically interacting directly with the HIV-1 envelope, thereby modifying its conformation. While this manuscript was under revision, two independent reports showed that the presence of S5 sensitizes HIV-1 to neutralizing antibodies using different HIV-1 envelopes (Beitari et al., 2017; Sood et al., 2017). Interestingly, one of these groups has argued that S5 changes the envelope conformation based upon accelerated envelope inactivation (Sood et al., 2017).
S5 amino acids 203–411 are important for its ability to block HIV-1 infection. To find determinants for the ability of S5 to block HIV-1, we performed restriction studies using chimeric proteins combining S5 and S2. In contrast to S5, S2 did not block HIV-1 infection. Interestingly, several of these chimeric proteins potently blocked HIV-1 infection. These experiments suggested that the amino acid region 203–411 contains key elements for restriction. Next we reasoned that these key determinants are likely to participate in two requisite steps for restriction: i) incorporation into virions, and ii) an as yet undefined effector function necessary to block infectivity. Others and we have observed that S5 does not inhibit the infectivity of HIV-1 pseudotyped with the VSV-G envelope (Rosa et al., 2015; Usami et al., 2015), suggesting that S5 incorporation into virions may not suffice to prevent infection. In addition, S5 incorporation is independent of the presence of envelope in virus-like particles (data not shown). On the other hand, elegant experiments using chimeric HIV-1 envelopes have demonstrated that the variable regions of the envelope are determinants of the ability of S5 to block HIV-1 infection (Usami and Gottlinger, 2013). Overall this suggests that S5 restriction requires a target, the viral envelope, to exert an effector function that can block infection.
The ability of S5 to be incorporated into HIV-1 particles maps to the L5-TM6-L6 region (residues 203–411). Unlike S5, S2 poorly incorporates into HIV-1 virions, suggesting that S5 might contain the sequence determinants that promote virion incorporation. The use of S5-S2 chimeric proteins revealed that the region L5-TM6-L6 of S5 can strongly promote S2 incorporation into HIV-1, but not restriction. The S2 chimera containing the L5-TM6-L6 region of S5 (chimera#7) became an important tool in the search for restriction determinants. Thus, we extended the S5 components of this chimeric protein toward the N- and C-terminus. Remarkably, we found a chimeric protein that potently restricted HIV-1 infection: S2 protein containing the S5 region L5-TM6-L6-TM7-L7 (chimera#15) blocked HIV-1 infection as potently as wild-type S5. Furthermore, S2 protein containing this S5 region also gained the ability to alter the conformation of the HIV-1 envelope when compared to the protein containing the S5 region L5-TM6-L6. These findings suggested that the region TM7-L7 contributes to the ability of S5 to change the viral envelope conformation and block infection.
S5 localization to DRMs correlated with its ability to restrict HIV-1. Our findings suggested that S5, like the positive control caveolin-1, localizes to DRMs. In addition, S2 protein containing S5 regions sufficient for virion particle incorporation and restriction likewise localized to DRMs. These results linked S5 localization to DRMs to restriction, raising the possibility that S5 affects the HIV-1 envelope in the producer cell, and might not be necessary in order for SERINC5 to be incorporated into the viral particle. The creation of an S5 variant that cannot be incorporated into virions might allow future testing of this hypothesis.
Our investigations support a model in which S5 can trigger a change of envelope conformation in DRMs in either the producer cell, or the viral particle, or in both. Our results link restriction to a change of envelope conformation. Changes of envelope conformation could perhaps lead to a restricted virus by interfering with the normal functions of envelope. Although this is an attractive idea, we have observed that S5 affects membrane fusion two- to threefold in a syncytium assay when compared to S2 (data not shown). This two- to threefold defect in membrane fusion may only partly explain the infectivity defect, which is up to 40-fold. The literature suggests that mutations in the HIV-1 envelope causing a syncytium formation defect only poorly decrease viral infectivity (Kowalski et al., 1991). The fact that pore formation by S5-containing virions has been observed suggested the possibility of a block subsequent to pore formation. Here we showed that HIV-1 virions containing S5 are unable to deliver the core to the cytoplasm, which is consistent with a block on either pore expansion or core delivery to the cytoplasm. S5 might be preventing pore expansion by changing the rigidity of the plasma membrane in the membrane regions where hemifusion occurs. Alternatively, S5 might be adversely affecting release of the HIV-1 core into the cytoplasm by creating a stronger physical association between the viral membrane and the core.
METHODS
Constructs, Cells, Antibodies, and Drugs
SERINC variants analyzed in this work were codon-optimized and cloned in pcDNA3.1 or LPCX. TZM-bl GFP-reporter cells were a kind gift from Dr. M. Pizzato. Human embryonic kidney HEK293T cells were obtained from ATCC (CRL-3216). Neutralizing and anti-p24 antibodies were obtained from the NIH AIDS repository. Mouse monoclonal M2 anti-FLAG (Sigma Aldrich, F1804), mouse monoclonal anti-GAPDH IgG (clone 6C5) (Ambion, AM4300), mouse IgG1 isotype control (Thermo Scientific, MA1–10405), Alexa Fluor 488 goat anti-mouse IgG (H+L) (Life Technologies, A11001), and Dye680LT goat anti-mouse IgG (H+L) antibody (Li-Cor Biosciences, 926–68020) were used in the immunoblotting experiments.
Restriction analysis of SERINC proteins
To test the ability of the different wild-type or mutant SERINC proteins, we produced HIV-1 viral particles in the presence or absence of the indicated SERINC variants by co-transfecting HEK293T cells with 8 μg HIV-1 NL4–3ΔenvΔNef, 2 μg of the HIV-1 envelope SF162, and increasing amounts of the SERINC variants. Producer cells were analyzed for expression of the indicated variant and GAPDH by Western blotting with anti-FLAG and anti-GAPDH antibodies, respectively. Produced virions were partially purified using a 20% sucrose cushion and analyzed for the presence of the indicated SERINC variant and p24 by Western blotting with anti-FLAG and anti-p24 antibodies, respectively. Viral infectivity was determined by challenging TZM-bl GFP-reporter cells and measuring the percentage of infected cells by flow cytometry.
Detection of SERINC variants by Western blots
1×106 HEK293T cells expressing the indicated SERINC variant were harvested, washed in PBS1X, and disrupted by freezing at −20 °C. Cell pellets were thawed at room temperature and lysed for 1 h on ice in 500 μl of DM lysis buffer (0.5 % (w/v) n-Decyl-ß-D-maltopyranoside in 20 mM Tris-Cl, pH 7.5, 10 % (v/v) glycerol + protease inhibitors and Benzonase). Insoluble material was removed by centrifugation for 1 hr at 14,000 rpm at 4 °C, and the supernatant mixed with SDS-PAGE sample buffer and incubated at 37 °C for 15 min before SDS-PAGE analysis. Nu-PAGE 10 % Bis-Tris gels (Life Technologies NP0315) were pre-run at 60V for 15 min before loading. Nu-PAGE gels were run at 60V for 2 h. Proteins were transferred to a nitrocellulose membrane using a semi-dry transfer apparatus (BioRad TransBlot3D Semi Dry Transfer cell) and SDS-free transfer buffer (20% (v/v) methanol in 25 mM TrisCl pH 7.5 + 250 mM glycine), for 1 h at 25 V. To make the protein accessible for antibody detection the membrane was washed once in PBS +0.1 % (v/v) Tween-20, stripped for 15 mins at 55 °C in 62.5 mM TrisCl pH 6.8, 2 % SDS (w/v) and 100 mM ß-mercaptoethanol. Stripping buffer was removed by 2 × 10 min washes in PBS +0.1 % (v/v) Tween-20, followed by 20 min blocking with 5 % (w/v) skim milk powder in PBS +0.1 % (v/v) Tween-20. Membranes were probed using anti-FLAG and anti-p24 antibodies to detect the SERINC variant and p24, respectively. Fluorescence images were acquired using the LI-COR Odyssey imager in the 680 nm channel.
Detection of surface and total expression of S5 variants by flow cytometry
To explore the topology of S5, flow cytometry was used to measure the surface (fixed) and total expression (fixed & permeabilized) of S5 variants containing a FLAG epitope peptide inserted into each of the nine loops (L1–9) of S5. Cells transiently expressing the S5 variants were washed in PBS1X and fixed in 1.5 % (w/v) paraformaldehyde. Permeabilized (0.1 % (w/v) saponin for 30 min) or non-permeabilized cells were stained with mouse anti-FLAG and anti-mouse-Alexa-Fluor-488 antibodies. Similar staining was performed using an isotype-matched control. Expression was detected with a BDFACSCalibur™ flow cytometer.
Fractionation of detergent-resistant membranes (DRMs)
1×106 HEK293T cells transiently expressing the indicated protein were washed in PBS1X and re-suspended in ice-cold Triton X-100 buffer (25 mM MES, pH 6.5, 150 mM NaCl, 0.1 % (v/v) Triton X-100, protease inhibitors). Cells were lysed using a Dounce homogenizer (5-strokes with pestle size A). Homogenates were diluted 1:2 with 80 % (w/v) sucrose in 25 mM MES, pH 6.5 +150 mM NaCl, and 1.5 ml of this mixture was overlaid with 1.5 ml 30 %(w/v) sucrose and 1.5 ml of 5 % (w/v) sucrose in 25 mM MES, pH 6.5 +150 mM NaCl. These gradients were centrifuged using a SW55ti rotor (Beckman) for 18 h at 44,500 rpm and 4 °C without applying the brake to end spinning. After centrifugation, the gradient was separated into 12 fractions of 375 μl each. Fractions were spotted onto a nitrocellulose membrane using a BioDot Microfiltration Apparatus (BioRad), and the membranes were separately blotted with anti-FLAG and anti-GFP antibodies.
Antibody neutralization assays
Virions containing or lacking S5 were incubated with increasing amounts of the indicated anti-HIV-1 neutralizing antibodies for 1 hr at 37 °C, and the virus-antibody mixtures were used to challenge TZM-bl GFP-reporter cells. Forty-eight hours later, infection was determined by measuring the percentage of GFP-positive cells by flow cytometry.
Fate of the capsid assay
TZM-bl GFP-reporter cells were infected with HIV-1SF162 viruses that do or do not contain S5. As a control, similar infctions were performed in the presence of the drug TAK-779. After an incubation time of 8 hours, cells were detached with Pronase for 5 min on ice and washed 3 times with ice-cold PBS. Cell pellets were resuspended in hypotonic buffer [10 mM tris-HCl pH8; 10 mM KCl; 1 mM EDTA] and incubated for 15 min on ice. Cells were lysed in a 7.0 ml Dounce homogenizer with pestle B. Cellular debris were cleared by centrifugation for 7 minutes at 3000 rpm. The cleared lysate was layered onto a 50% sucrose (weight: volume) cushion in 1× PBS and centrifuged at 125,000 × g for 2 hours at 4°C in a Beckman SW41 rotor. Input, soluble and pellet fractions were analyzed by Western blotting using anti-HIV-1 p24 antibody.
Supplementary Material
Highlights.
SERINC5 prevents the delivery of HIV-1 cores to the cytoplasm of target cells.
SERINC5 contains specific domains important for viral incorporation and restriction.
HIV-1 restriction by SERINC5 correlates with localization to DRMs and a change in envelope conformation.
Acknowledgments
We thank the NIH AIDS repository and other indicated donors for generously providing the following reagents: TAK-779, anti-HIV-1 gp41 monoclonal antibodies (4E10 and 2F5), anti-HIV-1 gp120, monoclonal antibody (2G12) from Polymun Scientific, monoclonal antibody (10E8) from Dr. Mark Connors, anti-HIV-1 gp120 monoclonal antibody (3BNC117) from Dr. Michel C. Nussenzweig, HIV-1 p24 monoclonal antibody (183-H12-5C) from Dr. Bruce Chesebro and Kathy Wehrly, anti-HIV-1 gp41 monoclonal antibody (NC-1) from Dr. Shibo Jiang, Anti-HIV-1 gp120 Monoclonal (PG9), Anti-HIV-1 gp120 Monoclonal (VRC01), from Dr. John Mascola, and anti-HIV-1 gp41 monoclonal antibody (240-D) from Dr. Susan Zolla-Pazner. The work was supported by an R01 grant from the NIH (AI087390, to F.D.-G).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci U S A. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitari S, Ding S, Pan Q, Finzi A, Liang C. Effect of HIV-1 Env on SERINC5 Antagonism. J Virol. 2017;91 doi: 10.1128/JVI.02214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya J, Peters PJ, Clapham PR. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J Virol. 2004;78:5500–5506. doi: 10.1128/JVI.78.10.5500-5506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya J, Repik A, Clapham PR. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J Virol. 2006;80:5292–5300. doi: 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Jackson AP, Brojatsch J. Cellular uptake of avian leukosis virus subgroup B is mediated by clathrin. Virology. 2005;337:45–54. doi: 10.1016/j.virol.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Sturzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, Hahn BH, Kirchhoff F. The Potency of Nef-Mediated SERINC5 Antagonism Correlates with the Prevalence of Primate Lentiviruses in the Wild. Cell Host Microbe. 2016;20:381–391. doi: 10.1016/j.chom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschhorn A, Gu C, Espy N, Richard J, Finzi A, Sodroski JG. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat Chem Biol. 2014;10:845–852. doi: 10.1038/nchembio.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka M, Hayakawa M, Ingi T. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem. 2005;280:35776–35783. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- Kestler HW, 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- Kowalski M, Bergeron L, Dorfman T, Haseltine W, Sodroski J. Attenuation of human immunodeficiency virus type 1 cytopathic effect by a mutation affecting the transmembrane envelope glycoprotein. J Virol. 1991;65:281–291. doi: 10.1128/jvi.65.1.281-291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai RP, Yan J, Heeney J, McClure MO, Gottlinger H, Luban J, Pizzato M. Nef decreases HIV-1 sensitivity to neutralizing antibodies that target the membrane-proximal external region of TMgp41. PLoS Pathog. 2011;7:e1002442. doi: 10.1371/journal.ppat.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Detergent resistance as a tool in membrane research. Nat Protoc. 2007;2:2159–2165. doi: 10.1038/nprot.2007.294. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PE, Lisanti MP, Baldini G, Sargiacomo M, Mastick CC, Lodish HF. Induction of caveolin during adipogenesis and association of GLUT4 with caveolin-rich vesicles. J Cell Biol. 1994;127:1233–1243. doi: 10.1083/jcb.127.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Danos O, Heard JM. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem. 2017;292:6014–6026. doi: 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautz B, Wiedemann H, Luchtenborg C, Pierini V, Kranich J, Glass B, Krausslich HG, Brocker T, Pizzato M, Ruggieri A, Brugger B, Fackler OT. The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J Biol Chem. 2017;292:13702–13713. doi: 10.1074/jbc.M117.797332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Gottlinger H. HIV-1 Nef responsiveness is determined by Env variable regions involved in trimer association and correlates with neutralization sensitivity. Cell Rep. 2013;5:802–812. doi: 10.1016/j.celrep.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Wu Y, Gottlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Ai LS, Huang SC, Li HF, Chan WE, Chang CW, Ko CY, Chen SS. The cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein gp41 harbors lipid raft association determinants. J Virol. 2010;84:59–75. doi: 10.1128/JVI.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Luban J, Diaz-Griffero F. The fate of HIV-1 capsid: a biochemical assay for HIV-1 uncoating. Methods Mol Biol. 2014;1087:29–36. doi: 10.1007/978-1-62703-670-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


