Abstract
This study was designed to investigate the brain proteome of the Ts65Dn mouse model of Down syndrome. We profiled the cerebellum and hippocampus proteomes of six and twelve-month-old trisomic and disomic mice by Difference Gel Electrophoresis. We quantified levels of 2082 protein spots and identified 272 (170 unique UniProt accessions) by mass spectrometry. Four identified proteins are encoded by genes trisomic in the Ts65Dn mouse. Three of these (CRYZL11, EZR, and SOD1) were elevated with p value < 0.05, and two proteins encoded by disomic genes (MAPRE3 and PHB) were reduced. Inter-gel comparisons based on age (six versus twelve-month) and brain region (cerebellum versus hippocampus) revealed numerous differences. Specifically, 132 identified proteins were different between age groups, and 141 identified proteins were different between the two brain regions. Our results suggest that compensatory mechanisms exist which ameliorate the effect of trisomy in the Ts65Dn mice. Differences observed during aging may play a role in the accelerated deterioration of learning and memory seen in Ts65Dn mice.
Keywords: Aging, Down syndrome, Alzheimer’s disease, Ts65Dn, Brain, Proteome
1. Introduction
Down syndrome (DS) is the most common chromosomal condition diagnosed in the United States, affecting about 1 in 700 live births (Parker et al., 2010; Presson et al., 2013). Incidence is similar in England and Wales, although Morris et al. note a drop of ~1.1% in live births from 1989/90 to 2007/8 due to prenatal screening, and subsequent termination of pregnancies (Morris and Alberman, 2009). However, prenatal diagnosis and termination of pregnancy is offset by increasing maternal age and improved survival of children with DS (Irving et al., 2008). Incidence in the Netherlands is about 1 in 625 births (Weijerman et al., 2008). Data for the developing world is largely unavailable. The DS phenotype is due to partial or complete trisomy of chromosome 21 (Hsa21), implicating overexpression of some or all Hsa21 genes, resulting in altered protein levels that are at least in part, responsible for the DS phenotype. DS neuropathology is generally characterized by structural, cellular, and anatomic defects in specific areas of the brain. Notably, neocortical structures, the hippocampus, and cerebellum are all disproportionately small. These brain changes become more pronounced with age (Lott, 2012; Raz et al., 1995).
DS is the most common genetic cause of significant intellectual disability, individuals with DS have the highest incidence of dementia in people under 60 years of age worldwide, and individuals with DS constitute the largest well-defined group of people afflicted with early onset Alzheimer’s disease (AD) (Hartley et al., 2015). AD is currently the sixth most common cause of death in the USA (Miniño et al., 2010). Almost all individuals with DS develop AD-like pathology by age 40, and 70% or more develop dementia by age 60 (Hartley et al., 2015; Mann, 1988). Krinsky-McHale et al. report an unusual case of a 70-year-old man with Down syndrome who did not exhibit decline in cognitive or functional capacities even though he had full trisomy of chromosome 21 (Krinsky-McHale et al., 2008). The life expectancy of an individual with DS now approaches 60 years, and as life expectancy improves, the number of people with DS that also have AD is likely to increase (Glasson et al., 2002). Worldwide, the number of AD cases is predicted to rise to about 115 million by 2050 (Wimo and Prince, 2010). The association between DS and AD is important because studies of DS provide insights relevant to AD and its causes. Understanding the mechanisms underlying the neurodegenerative changes in DS, and implementing approaches to prevent, delay, or treat them, are increasingly important health issues (Bittles et al., 2007; Hartley et al., 2015).
Several mouse models of DS are currently available (Vacano et al., 2012). The best characterized and most widely used of these is the Ts65Dn mouse [Ts(1716)65Dn (Davisson et al., 1990)]. The Ts65Dn mouse is trisomic for roughly 100 protein-coding genes syntenic to well-curated Hsa21 genes [based on the Mmu16 breakpoint in Ts65Dn (Duchon et al., 2011) and MGI (Mouse Genome Informatics) annotation (Bult et al., 2016)], and is disomic for about 16 Hsa21/Mmu16 protein-coding genes (Yu et al., 2010). Additionally, the Ts65Dn mouse is trisomic for up to 60 Mmu17 genes (Duchon et al., 2011; Reinholdt et al., 2011), although only ~28 encode proteins. Some researchers have suggested that the Ts65Dn mouse should be replaced by models that better recapitulate human trisomy 21, and this has fueled the development of such models (Vacano et al., 2012). Analyses employing these mouse models are likely to contribute greatly to our understanding of DS. However, we believe that at the present time, given the substantial body of published work employing this model, the high degree of similarity in phenotypic features to DS features, and the relatively incomplete characterization of the newer DS mouse models, the Ts65Dn mouse is the best mouse model of DS. The Ts65Dn mouse exhibits features in common with people with DS including loss of functional basal forebrain cholinergic neurons, structural and cellular abnormalities in the hippocampus and cerebellum, degeneration of the hippocampus, learning and memory disability, increased impairment of learning and memory with age, and numerous other developmental and age related abnormalities (Aldridge et al., 2007; Holtzman et al., 1996; Insausti et al., 1998; Kirsammer et al., 2008; Kurt et al., 2004; Levine et al., 2009; Patterson and Costa, 2005; Reeves et al., 1995; Vacano et al., 2012). Ts65Dn mice also exhibit reduced cerebellum and hippocampus volumes (Baxter et al., 2000; Insausti et al., 1998; Lorenzi and Reeves, 2006; Roper et al., 2006). The Ts65Dn learning and memory deficits correlate with a hippocampal dysfunction that worsens as the mice age (Hyde and Crnic, 2001). Ts65Dn mice exhibit motor difficulties that resemble those observed in individuals with DS and these are consistent with cerebellar abnormalities (Hampton et al., 2004; Latash et al., 2002). A recent study by Olmos-Serrano et al. demonstrates longitudinal loss of aspects of cognition (learning and memory) in Ts65Dn mice as they age from 2 months to 11 months of age, and includes an extensive discussion of the use of this mouse as a model for cognitive decline in DS and as a model for features of AD (Olmos-Serrano et al., 2016b). While Ts65Dn mice do not develop the plaques and tangles characteristic of AD, they are trisomic for the APP gene, and at 10- and 12-months of age they exhibit elevated levels of APP and soluble APP metabolites (Choi et al., 2009). In addition, MRI studies reveal in vivo cholinergic changes potentially relevant to early onset AD in the brains of Ts65Dn mice (Chen et al., 2009). Basal forebrain cholinergic neurons are important for memory function, and progressively degenerate in DS, idiopathic AD, and in Ts65Dn mice (Lockrow et al., 2012). Ts65Dn mice have increased levels of phosphorylated tau (at Ser202) in the hippocampus and cerebellum, which are normalized after a reduction in Dyrk1A copy number (García-Cerro et al., 2017). It is now clear from studies using a wide variety of agents, that learning and memory abnormalities in Ts65Dn mice can be ameliorated via therapeutic intervention (Gardiner, 2010; Möhler, 2012; Ruparelia et al., 2012). However, clinical trials on people with DS have had only limited success (Gupta et al., 2016). It has been suggested that combinations of pharmacological and/or other treatments may be more effective (Gardiner, 2015). Clearly, identifying more targets for therapeutic intervention is a priority, and a primary motivation for this study.
Gene dosage studies in both mice and humans show that RNA expression levels of some Hsa21 genes in diploid individuals vary substantially by more or less than the 150% expression expected in trisomy (Aït Yahya-Graison et al., 2007; Prandini et al., 2007; Sultan et al., 2007). In many cases, protein expression correlates poorly with gene expression, and changes in protein abundance may be more directly relevant to brain development and function than changes in gene expression (Schwanhäusser et al., 2011). In a recent study of a set of proteins thought to be involved in learning and memory in DS and mouse models of DS, Spellman et al. report that protein levels are inconsistent with previously reported mRNA levels (Spellman et al., 2013).
Targeted studies employing reverse phase protein arrays (RPPA) in brain regions of Ts65Dn and euploid mice at 6 and 12 months of age compared levels of proteins in pathways hypothesized or known to be involved in brain function and cognition, including proteins in four pathways/processes; MAP kinase signaling, NMDA receptors, NUMB protein interactions, and the apoptosis pathway (Ahmed et al., 2017; 2014). Numerous significant changes in these proteins were observed due to trisomy, and changes seemed to be more significant at 12 months than at 6 months. Fernandez et al. carried out a targeted analysis of synapse proteins in the Ts65Dn cerebrum and found only modest differences in protein levels (Fernandez et al., 2009). Numerous studies of individual proteins encoded by genes on chromosome 21 clearly demonstrate altered levels of expression and link these changes to the Ts65Dn phenotype. These studies focused on specific proteins or selected subsets of proteins based on hypotheses regarding their role in DS.
There have been several studies aimed at characterizing “global” gene and/or protein expression. For example, studies by microarray or serial analysis of gene expression (SAGE), including one study focusing on the cerebellum (Saran, 2003), demonstrate changes in several hundred genes in trisomic mice (Chrast et al., 2000; Saran, 2003; Sommer and Henrique-Silva, 2008). Olmos-Serrano et al. performed transcriptome analysis of three brain regions (cerebral neocortex, hippocampus, and cerebellar cortex) from DS and euploid individuals over the course of development, and found genome wide alteration in the expression of many genes. Principal Component Analysis (PCA) indicated segregation between DS and euploid samples, but even greater segregation by brain region and developmental period. Additionally, they demonstrated co-dysregulation of genes associated with oligodendrocyte differentiation and myelination, and validated these by cross-species comparison with Ts65Dn mice (Olmos-Serrano et al., 2016a).
Two dimensional gel electrophoresis (2DGE) has been used to detect alterations in protein levels in both fetal and adult DS brain (G. Lubec, 2013; Sun et al., 2011). Many of the differentially abundant proteins identified in these studies are Hsa21 gene products, but these are not consistently elevated, and the expression of some proteins encoded by genes on other chromosomes is also altered significantly. Harris and colleagues employed a variant of 2DGE, difference gel electrophoresis (DiGE), to quantify protein differences in adult brains from individuals with DS (Harris et al., 2007). All proteins with altered levels were more abundant in brains from individuals with DS, but none were Hsa21 gene products.
These “global” methods identify and measure relatively abundant proteins. Rabilloud and Lelong point out that 2DGE based methods are highly unlikely to detect transcription factors because of their low level of abundance (Rabilloud and Lelong, 2011), and this is supported by a study of mouse brain proteins that demonstrated transcription factor protein levels are generally very low (Wang et al., 2006). However, abundant proteins play significant roles in cell structure and function, and it is important to determine whether levels of these proteins change with trisomy and age in various brain regions, especially in regions of the brain known to be affected in DS. Challenges in the proteomic analysis of DS brain include variability in samples due to differences in post-mortem interval, age of individuals, pre-mortem events, and sample acquisition and preparation. Many studies have shown that expression of specific genes and proteins is altered in DS and in various mouse models of DS (for example, App, Kcnj6, Dyrk1a), and in several cases, these genes/proteins have been shown to affect the Ts65Dn phenotype. The abundance of these proteins is generally low, and would be difficult to detect using 2DGE based methods. For example, Wang et al. semi-quantitatively analyzed levels of 7792 mouse brain proteins, and only detected 7 proteins encoded by genes trisomic in Ts65Dn mice, and these were all present at relatively low levels (Wang et al., 2006).
Our goal was to test the hypothesis that the phenotype differences observed in Ts65Dn mice by ploidy (trisomy versus disomy) and age should be accompanied by measurable differences in protein levels. We focused on cerebellum and hippocampus because, as discussed above, both are disproportionately small in DS and Ts65Dn mice, and because learning and memory deficits due to hippocampal dysfunction and motor difficulties due to cerebellar dysfunction are present in DS and Ts65Dn mice. We believe these two neocortical regions are the most significant to the DS and Ts65Dn phenotypes. The study reported here is novel in its approach of assessing differences in protein levels by ploidy in the context of age and brain region, and by method: we believe this is the only study employing euthanasia by high energy focused microwave radiation, and one of very few employing DiGE.
2. Materials and Methods
2.1 Tissue samples
Male Ts65Dn and disomic controls were obtained from the Ts65Dn breeding colony at the Eleanor Roosevelt Institute by crossing Ts65Dn females (The Jackson Laboratory, stock No: 001924) with B6EiC3SnF1/J males (The Jackson Laboratory, stock No: 001875), and from Dr. Tarik Haydar. Males were used exclusively for experiments since: a) Ts65Dn mice are difficult to breed, producing small litters with only ~25% trisomic pups, b) Ts65Dn males are sterile, and cannot be employed in colony maintenance, c) the majority of past work on the Ts65Dn mice has been done on males, and d) including females would have required (roughly) doubling the required time and resources for experiments. Mice were cytogenetically tested to evaluate trisomy (Davisson et al., 1990). All procedures regarding the care and husbandry of these animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Denver, in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Mice were shipped to the University of North Dakota, Grand Forks, via World Courier, housed at the University of North Dakota’s Center for Biomedical Research, and allowed at least 5 days’ acclimation prior to sacrifice. Mice were sacrificed via head-focused high-energy microwave irradiation. We had previously demonstrated that this method fixes brain chemistry at time of death and preserves the in situ state of proteins (Hunsucker et al., 2008). Additionally, it is a humane and rapid method for euthanizing small laboratory rodents (Euthanasia, 2013). Cerebellum and hippocampus were dissected immediately after irradiation, frozen on a metal plate atop dry-ice within 2 minutes, and stored at −80°. All procedures regarding the care and euthanasia of these animals were approved by the IACUC of the University of North Dakota, in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The number and type of samples are listed below.
2.2 Preparation of hippocampal and cerebellar protein extracts
Tissues were homogenized as described previously (Hunsucker et al., 2008), except the sonication time was three 15-second bursts (Branson Sonifier Cell Disruptor 185, setting 3) separated by 30-second incubations on ice. Samples were then centrifuged (16,000 × g for 25 minutes), and the supernatants were collected, frozen on dry ice, and stored at −80 °C. The extracts were diluted 4-fold with TBS (50 mM Tris, pH 7.5, 150 mM NaCl) and the proteins were precipitated by the addition of methanol/chloroform (Wessel and Flügge, 1984). The protein pellets were dried in a centrifuge under reduced pressure (Speedvac, Savant), then allowed to rehydrate overnight in reaction buffer (7 M urea, 2 M thiourea, 4% w/v CHAPS). The protein sample was supplemented with 10 mM DTT, incubated (2 hours, room temperature), homogenized and centrifuged (16,000×g, 15 minutes, room temperature). The re-solubilized protein was collected and protein concentrations were determined (Bradford, 1976). Each sample was diluted to 5 mg/ml of total protein with reaction buffer containing 10 mM DTT, flash frozen with liquid N2, and stored at −80 °C until analysis.
2.3 Labeling with Cy dyes
Labeling reactions used 50 µg of protein, either from individual samples, or from the pool of all samples (a combination of equal volumes of protein from all the individual samples which serves as an internal standard for inter-gel comparisons). A 10 µl aliquot containing 50 µg of protein in reaction buffer was supplemented with 1 µl of 300 mM Tris, pH 8.5, and the final pH was checked with pH paper to ensure optimal labeling. One µl of 200 µM Cy dye (200 pmol) in anhydrous dimethylformamide (Aldrich) was added to the protein and the reaction allowed to proceed [30 minutes, 4 °C, in the dark (Unlü et al., 1997)]. The reaction was quenched by addition of 1 µl of 10 mM lysine and the samples combined (e.g., one sample labeled with Cy5, one sample labeled with Cy3, and the internal standard sample labeled with Cy2). This mixture was supplemented with reaction buffer and with 0.1 M hydroxyethyl disulfide (5.4 µl, Destreak, GE Healthcare), 1% broad range Pharmalytes 3–10 NL (GE Healthcare), and 0.003% bromophenol blue to give a final volume of 450 µl for 2D gel electrophoresis.
2.4 2D gel electrophoresis
Disomic and trisomic samples were paired and run together on analytical gels (for example, the 8 disomic six-month-old cerebellum samples were paired with the 8 trisomic six-month-old cerebellum samples and run on 8 separate gels), and each gel was also loaded with the pooled standard. DiGE was performed as described previously (Hunsucker et al., 2008).
2.5 Scanning of labeled images
Analytical gels were scanned at 100 µm resolution on a Typhoon 9400 Variable Mode Laser Imager (GE Healthcare) with filter settings for Cy3 at excitation 532 nm and emission 580 nm with bandpass 30 nm; scanning for Cy5 was at excitation 633 nm and emission 670 nm with bandpass 30 nm and scanning for Cy2 was at excitation 468 nm and emission 520 nm with bandpass 40 nm.
2.6 Image analysis
Progenesis SameSpots software (Nonlinear Dynamics) was used for spot detection and relative quantification of proteins based on fluorescence images. Spot boundaries were detected automatically, peak volumes were measured for each protein spot in the three fluorescent channels (Cy2, Cy3 and Cy5) and the normalized volume ratios were calculated to directly compare volumes of each protein spot between channels. Spot volumes for each sample are provided in Supplementary Data (“Spot Volumes” tab). A two-tailed T-test was used to calculate probabilities associated with relative fold-changes.
2.7 Spot excision, enzymatic digestion, and protein identification
A list of candidate spots for identification was prepared: the criteria for spot selection was based on whether the spot had good boundaries and was well resolved from neighboring spots. Protein identification was based on preparative gels loaded with 50 µg of the pooled internal standard labeled with Cy2 (for visualization) and 750 µg of unlabeled pooled internal standard (an adequate amount of protein for identification by mass spectrometry). Preparative gels were prepared as described above except that isoelectric focusing was for 133,000 volt-hours. Gels were stained with Lava Purple Total Protein Stain (Fluortechnics) and scanned at excitation 532 nm and emission 610 nm with bandpass 30 nm. Protein spots were matched to the preparative gels, excised, subjected to in-gel tryptic digestion and proteins were identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Protein spot excision and in-gel enzymatic digestion were automated (Ettan Spot Picker fitted with a 2.0 mm picking head and Ettan Spot Digester; GE Healthcare) as previously described (Hunsucker et al., 2008). All digests were analyzed by MALDI-TOF MS (Voyager DE-PRO, Applied Biosystems) as previously described (Hunsucker et al., 2008). Spectra were collected over the range m/z 500–5000. Peptide mass fingerprints were internally calibrated to monoisotopic trypsin autolysis peaks. Spectra were processed using ProTS Data (Efeckta Technologies) to generate a peak list that was then submitted to Mascot Daemon (Matrix Science Ltd.) for database searching. Spectral pre-processing included defining the baseline, noise, and signal-to-noise ratio as well as monoisotopic peak selection. A signal-to-noise ratio in ProTS Data > 4 was required for inclusion in the peak list. Search parameters were as follows: taxonomy selection, Mammalia (49,887 sequences); fixed modification, carbamidomethylation of cysteines; trypsin as the enzyme allowing for one missed cleavage and ± 50 ppm peptide mass tolerance. Searches were not constrained by pI or MW. The minimal requirements for protein identification have been described (Hunsucker et al., 2008) and peptide and protein assignments followed published guidelines (Bradshaw et al., 2006). The identified spots are listed in Supplementary Data (“Identified Spots” tab).
2.8 Statistical analysis: prot2D
Direct DiGE comparison was only possible for disomic and trisomic samples, but we were interested in also comparing samples by age (six-month versus twelve-month) and brain region (cerebellum versus hippocampus). We therefore extended our analysis to include 2DGE inter-gel (cerebellum versus hippocampus and six-month versus twelve-month) peak volume comparison, using the R-package prot2D (Artigaud et al., 2013). The number of samples in each comparison is shown in Supplementary Tables S4 (“Analysis Matrix”). The Norm.qt module (quantiles normalization) in the prot2D package was used to normalize the peak volumes, the data was coerced into an expression set with ES.prot, and modT.prot [the moderate t-test (Smyth, 2004)] was used to identify differentially expressed protein spots in the normalized data using method.fdr = “BH” [Classical Benjamini-Hochberg false discovery rate (Benjamini and Hochberg, 1995)] with adjusted p value < 0.05.
2.9 Principal Component Analysis
PCA values were calculated with normalized peak volumes using the prcomp() function in R (R Core Team, 2015).
2.10 Functional enrichment analysis
Eight lists of unique UniProt Accessions representing spots with highly significant differences (FDR < 0.005 for increased confidence in validity of spot differences) by age or brain region were prepared [Supplementary Tables S5 (“Spot Lists” tab). Databases were queried with these accession lists via ClueGO (version 2.3.3) + CluePedia (version 1.3.3) (Bindea et al., 2013; 2009), which are biological network plugins for Cytoscape (version 3.5.1), an open source software platform for visualizing complex networks (Shannon et al., 2003). The following databases were queried: GO 26.06.2017 (BiologicalProcess, CellularComponent, and MolecularFunction) (Ashburner et al., 2000), and Reactome 26.06.2017 (Croft et al., 2014). Results are shown in Supplementary Fig. S1 and S2, and Supplementary Tables S5.
3. Results
We used DiGE to compare the proteome of the Ts65Dn hippocampus and cerebellum at both six months and twelve months to that of diploid age-matched (control) mice. In this study, we identified 272 spots representing 170 unique proteins [Supplementary Data (“Identified Spots” tab)]. In addition to differences in protein levels between disomic and trisomic animals, we employed 2DGE to investigate differences in protein levels due to age, and differences between brain regions (i.e., cerebellum versus hippocampus). To accomplish this, we used the peak volumes obtained from DiGE analysis of gel images, and evaluated differences in abundance using prot2D (Artigaud et al., 2013), a module for R (R Core Team, 2015).
3.1 Differential expression of proteins in trisomic (Ts65Dn) versus disomic (control) samples
Six spots representing five proteins were significant comparing trisomic (Ts65Dn) versus disomic (control) samples (Table 2). Four of the identified proteins are encoded by genes that are trisomic in Ts65Dn mice. Three (Cbr1, Cryzl1, and Sod1) are Hsa21 homologs, and the fourth (Ezr) maps to a region of Mmu17 that is trisomic in Ts65Dn mice (Duchon et al., 2011). Of these, three proteins (CRYZL1, EZR, and SOD1) show increased abundance (p value < 0.05) in Ts65Dn, while the level of one (CBR1) is unchanged (p > 0.05). We also identified two proteins that are significantly reduced in trisomic samples (p value < 0.05); prohibitin (PHB; P67778) and microtubule-associated protein RP/EB family member 3 (MAPRE3; Q6PER3). These spots were not significant after applying false discovery rate (q value < 0.05, see Methods 2.8). However, we believe the expression differences in spots representing proteins from trisomic genes are likely real [they are the only trisomic protein spots identified (except CBR1), and are significant by P-value]. The only HSA21 homologs identified in this study were CBR1, CRYZL1, and SOD1.
Table 2.
Differences in Identified Proteins by Ploidy (DiGE)
| Spot Number |
Protein ID | UniProt Accession |
Gene Symbol |
p value | Ratio (trisomic: disomic) |
|---|---|---|---|---|---|
| v1567 | Microtubule-associated protein RP/EB family member 3 | Q6PER3 | Mapre3 | 1.89E-02 | 0.83 |
|
| |||||
| v3328 | Prohibitin | P67778 | Phb | 3.96E-02 | 0.89 |
|
| |||||
| v1394 | Carbonyl reductase [NADPH] 1 | P48758 | Cbr1* | 5.37E-01 | 1.00 |
|
| |||||
| v794 | Ezrin | P26040 | Ezr | 2.26E-02 | 1.12 |
|
| |||||
| v3052 | Quinone oxidoreductase-like 1 | Q921W4 | Cryzl1* | 4.80E-02 | 1.13 |
|
| |||||
| v2279 | Ezrin | P26040 | Ezr | 2.93E-02 | 1.18 |
| v1901 | Superoxide dismutase | P08228 | Sod1* | 3.14E-04 | 1.30 |
Bold gene symbols indicate genes that are trisomic in Ts65Dn. Gene symbols with asterisks (*) are Hsa21 homologs. Cbr1, which has a p value > 0.05, was included here because it is an Hsa21 homolog and trisomic in Ts65Dn.
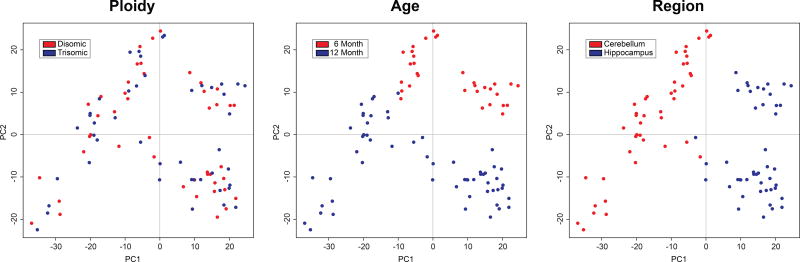
PCA indicated essentially no partitioning of samples by ploidy, but very clear partitioning by age and brain region (Figure 1).
Figure 1. Principal Component Analysis.
Spots indicate individual tissue samples, with group indicated by color. PCA values were calculated using the prcomp() function in R (R Core Team, 2015).
We therefore extended our analysis to include inter-gel (2DGE) comparisons by age and brain region (Tables 3 and 4) because changes due to age may be relevant to the loss of learning and memory, and degeneration of brain regions, observed in DS and in Ts65Dn mice.
Table 3.
Twelve-Month versus Six-Month: Significant Spots All Spots
| All Spots | |||
| Comparison | Significant Spots | ↑ Twelve-Month | ↑ Six-Month |
|
| |||
| Age in Hippocampus | 654 (372) | 319 (190) | 335 (182) |
|
| |||
| Age in Cerebellum | 766 (455) | 355 (208) | 411 (247) |
|
| |||
| Age in Trisomic | 398 (104) | 176 (43) | 222 (61) |
|
| |||
| Age in Disomic | 308 (91) | 134 (37) | 174 (54) |
| Identified Spots | |||
| Comparison | Significant Spots | ↑ Twelve-Month | ↑ Six-Month |
|
| |||
| Age in Hippocampus | 119 (87) | 56 (45) | 63 (42) |
|
| |||
| Age in Cerebellum | 156 (105) | 73 (48) | 83 (57) |
|
| |||
| Age in Trisomic | 106 (35) | 52 (17) | 54 (18) |
|
| |||
| Age in Disomic | 83 (33) | 41 (13) | 42 (20) |
Values in black are FDR < 0.05, values in red are FDR < 0.005.
Table 4.
Hippocampus versus Cerebellum: Significant Spots
| All Spots | |||
| Comparison | Significant Spots | ↑ Hippocampus | ↑ Cerebellum |
|
| |||
| Region in Twelve-Month | 1135 (825) | 570 (422) | 565 (403) |
|
| |||
| Region in Six-Month | 793 (440) | 364 (210) | 429 (230) |
|
| |||
| Region in Trisomic | 804 (489) | 388 (244) | 416 (245) |
|
| |||
| Region in Disomic | 778 (448) | 380 (217) | 398 (231) |
| Identified Spots | |||
| Comparison | Significant Spots | ↑ Hippocampus | ↑ Cerebellum |
|
| |||
| Region in Twelve-Month | 162 (127) | 84 (67) | 78 (60) |
|
| |||
| Region in Six-Month | 112 (67) | 53 (30) | 59 (37) |
|
| |||
| Region in Trisomic | 103 (70) | 50 (35) | 53 (35) |
|
| |||
| Region in Disomic | 100 (54) | 53 (30) | 47 (24) |
Values in black are FDR < 0.05, values in red are FDR < 0.005.
3.2 Functional enrichment analysis: differential expression by age
We performed functional enrichment analysis on identified proteins with significantly different levels in six versus twelve-month-old in cerebellum, hippocampus, disomic, and trisomic sample groups. Lists of unique UniProt accessions were used to query gene ontology (GO) and pathway databases using ClueGO and CluePedia (Bindea et al., 2013; 2009) which are biological network plugins for Cytoscape (Shannon et al., 2003; Smoot et al., 2011), a platform for visualizing complex networks. The following databases were queried: GO 26.06.2017 [BiologicalProcess, CellularComponent, and MolecularFunction (Ashburner et al., 2000)] and Reactome 26.06.2017 [Pathway and Reaction (Croft et al., 2014)]. Collectively querying the GO and Reactome databases returned overview term functional groups (Tables 5A – C), which include various subgroups (Supplementary Tables S1).
Tables 5.
A – C: ClueGO Age Tables
| A) Age in Cerebellum | ||||||
| GOID | Ontology Source | GOTerm | Nr. Genes | % Assoc. Genes | Term P- value | Associated Genes Found |
|
| ||||||
| Reactome:R-MMU-1428517 | Pathways | The citric acid (TCA) cycle and respiratory electron transport | 9 | 6.2 | 6.15E-10 | Atp5a1, Atp5b, Dld, Dlst, Ldhb, Mdh2, Ndufa12, Ndufs4, Ogdh |
|
| ||||||
| Reactome:R-MMU-5663213 | Pathways | RHO GTPases Activate WASPs and WAVEs | 4 | 12.5 | 2.75E-06 | Actb, Arpc5, Grb2, Mapk1 |
|
| ||||||
| Reactome:R-MMU-70895 | Pathways | Branched-chain amino acid catabolism | 10 | 4.1 | 3.52E-09 | Aspa, Dld, Dlst, Glud1, Glul, Ivd, Ogdh, Psma7, Psmb2, Psmb5 |
|
| ||||||
| Reactome:R-MMU-71291 | Pathways | Metabolism of amino acids and derivatives | 10 | 4.1 | 3.52E-09 | Aspa, Dld, Dlst, Glud1, Glul, Ivd, Ogdh, Psma7, Psmb2, Psmb5 |
|
| ||||||
| GO:0006091 | Biological Process | generation of precursor metabolites and energy | 15 | 4.3 | 1.49E-13 | Dld, Dlst, Eno1b, Eno2, Hk1, Mdh1, Mdh2, Ndufa12, Ndufs4, Ogdh, Pfkp, Pgam1, Pgk1, Pkm, Ppp1ca |
|
| ||||||
| GO:0008064 | Biological Process | regulation of actin polymerization or depolymerization | 7 | 4.3 | 6.87E-07 | Arpc5, Cfl1, Grb2, Pfn1, Sh3bgrl3, Twf2, Wdr1 |
|
| ||||||
| GO:0016836 | Molecular Function | hydro-lyase activity | 3 | 6.1 | 4.62E-04 | Car2, Eno1b, Eno2 |
|
| ||||||
| GO:0021762 | Biological Process | substantia nigra development | 5 | 13.2 | 1.09E-07 | Actb, Cnp, Cox6b1, Hspa5, Mbp |
|
| ||||||
| GO:0046164 | Biological Process | alcohol catabolic process | 3 | 6.5 | 3.83E-04 | Aldh2, Apoe, Gpd2 |
|
| ||||||
| GO:0046496 | Biological Process | nicotinamide nucleotide metabolic process | 13 | 9.6 | 2.33E-16 | Dld, Eno1b, Eno2, Hk1, Ldhb, Mdh1, Mdh2, Me1, Ogdh, Pfkp, Pgam1, Pgk1, Pkm |
|
| ||||||
| GO:1990204 | CellularComponent | oxidoreductase complex | 6 | 6.3 | 5.01E-07 | Dld, Dlst, Gpd2, Ndufa12, Ndufs4, Ogdh |
| B) Age in Hippocampus | ||||||
| GOID | Ontology Source | GOTerm | Nr. Genes | % Assoc. Genes | Term P-value | Associated Genes Found |
|
| ||||||
| Reactome:R-MMU-111885 | Pathways | Opioid Signalling | 4 | 4.4 | 9.17E-05 | Gnao1, Gnb1, Marcks, Ppp1ca |
|
| ||||||
| Reactome:R-MMU-399956 | Pathways | CRMPs in Sema3A signaling | 3 | 18.8 | 9.34E-06 | Crmp1, Crmp2, Crmp3 |
|
| ||||||
| GO:0008064 | Biological Process | regulation of actin polymerization or depolymerization | 7 | 4.3 | 2.27E-07 | Arpc5, Capzb, Cttn, Dstn, Grb2, Pfn2, Wdr1 |
|
| ||||||
| GO:0009167 | Biological Process | purine ribonucleoside monophosphate metabolic process | 11 | 4.2 | 6.39E-11 | Aprt, Atp5b, Cox5b, Eno1b, Ndufa10, Ndufs1, Ogdh, Park7, Pgam1, Pkm, Tpi1 |
|
| ||||||
| GO:0009168 | Biological Process | purine ribonucleoside monophosphate biosynthetic process | 3 | 4.3 | 7.92E-04 | Aprt, Atp5b, Cox5b |
|
| ||||||
| GO:0016836 | Molecular Function | hydro-lyase activity | 4 | 8.2 | 8.24E-06 | Aco2, Car2, Eno1b, Park7 |
|
| ||||||
| GO:0044724 | Biological Process | single-organism carbohydrate catabolic process | 7 | 5.8 | 2.76E-08 | Eno1b, Gpd2, Ogdh, Pgam1, Pkm, Ppp1ca, Tpi1 |
|
| ||||||
| GO:0098798 | Cellular Component | mitochondrial protein complex | 6 | 4.4 | 1.57E-06 | Atp5b, Cox5b, Etfa, Ndufa10, Ndufs1, Park7 |
| C) Age in Disomic | ||||||
| GOID | Ontology Source | GOTerm | Nr. Genes | % Assoc. Genes | Term P-value | Associated Genes Found |
|
| ||||||
| Reactome:R-MMU-70263 | Pathways | Gluconeogenesis | 3 | 8.3 | 1.00E-05 | Eno1b, Eno2, Tpi1 |
|
| ||||||
| GO:0016836 | Molecular Function | hydro-lyase activity | 3 | 6.1 | 2.56E-05 | Car2, Eno1b, Eno2 |
|
| ||||||
| GO:0046164 | Biological | alcohol catabolic process | 3 | 6.5 | 2.11E-05 | Aldh2, Apoe, Tpi1 |
% Associated Genes indicates the percent of the genes defining a given GO term that were listed (as accessions) in the ClueGO spot list. The “Age in Trisomic” protein accessions did not map to any networks. See Supplementary Tables S5 for more details and subnetworks.
3.3 Functional enrichment analysis: differential expression by brain region
We performed functional enrichment analysis on the identified proteins with significantly different levels in hippocampus versus cerebellum in six-month, twelve-month, disomic, and trisomic subsets. Lists of unique UniProt accessions were used to query GO and pathway databases using ClueGO and CluePedia. Collectively querying the GO and Reactome databases returned overview term functional groups (Tables 6A – D), which include various subgroups (Supplementary Tables S2).
Tables 6.
A – D: ClueGO Region Tables
| A) Region in Six-Month | ||||||
| GOID | Ontology Source | GOTerm | Nr. Genes | % Assoc. Genes | Term P-value | Associated Genes Found |
|
| ||||||
| Reactome:R-MMU-210991 | Pathways | Basigin interactions | 4 | 8.0 | 7.22E-06 | Actb, Dnm1, Crmp2, Ezr |
|
| ||||||
| GO:0008064 | Biological Process | regulation of actin polymerization or depolymerization | 7 | 4.3 | 1.56E-07 | Arpc2, Arpc5, Capzb, Cttn, Pfn2, Twf2, Wdr1 |
|
| ||||||
| GO:0016836 | Molecular Function | hydro-lyase activity | 3 | 6.1 | 2.47E-04 | Aco2, Car2, Eno1b |
|
| ||||||
| GO:0021762 | Biological Process | substantia nigra development | 3 | 7.9 | 1.15E-04 | Actb, Ckb, Cox6b1 |
|
| ||||||
| GO:0030516 | Biological Process | regulation of axon extension | 4 | 4.8 | 5.42E-05 | Apoe, Cttn, Crmp2, Twf2 |
|
| ||||||
| GO:0044724 | Biological Process | single-organism carbohydrate catabolic process | 7 | 5.8 | 1.89E-08 | Eno1b, Gpd2, Hmgb1, Pfkp, Pgam1, Pkm, Pygm |
|
| ||||||
| GO:0046164 | Biological Process | alcohol catabolic process | 3 | 6.5 | 2.04E-04 | Aldh2, Apoe, Gpd2 |
|
| ||||||
| GO:0048489 | Biological Process | synaptic vesicle transport | 6 | 4.7 | 7.65E-07 | Dnm1, Crmp2, Napb, Pfn2, Snap25, Stxbp1 |
| B) Region in Twelve-Month | ||||||
| GOID | Ontology Source | GOTerm | Nr. Genes | % Assoc. Genes | Term P-value | Associated Genes Found |
|
| ||||||
| Reactome:R-MMU-1428517 | Pathways | The citric acid (TCA) cycle and respiratory electron transport | 10 | 6.9 | 4.86E-10 | Atp5a1, Etfa, Glo1, Mdh2, Ndufa10, Ndufa12, Ndufa8, Ndufs3, Ndufs4, Ndufs8 |
|
| ||||||
| GO:0000502 | Cellular Component | proteasome complex | 3 | 4.3 | 3.11E-03 | Psmb2, Psmb5, Txnl1 |
|
| ||||||
| Reactome:R-MMU-399956 | Pathways | CRMPs in Sema3A signaling | 4 | 25.0 | 4.90E-07 | Crmp1, Crmp2, Crmp3, Crmp4 |
|
| ||||||
| GO:0008064 | Biological Process | regulation of actin polymerization or depolymerization | 8 | 4.9 | 4.01E-07 | Arpc2, Arpc5, Capzb, Cfl1, Pfn1, Pfn2, Sh3bgrl3, Wdr1 |
|
| ||||||
| GO:0009636 | Biological Process | response to toxic substance | 12 | 5.1 | 2.93E-10 | Cnp, Gsta4, Hbb-b1, Mapk1, Mbp, Nefl, Pebp1, Prdx1, Prdx3, Sod1, Txnl1, Uba52 |
|
| ||||||
| GO:0015036 | Molecular Function | disulfide oxidoreductase activity | 3 | 7.3 | 6.61E-04 | Pgk1, Sh3bgrl3, Txnl1 |
|
| ||||||
| GO:0021761 | Biological Process | limbic system development | 6 | 4.7 | 1.51E-05 | Arpc5, Cfl1, Nefl, Pebp1, Uba52, Ywhae |
|
| ||||||
| GO:0021762 | Biological Process | substantia nigra development | 6 | 15.8 | 1.08E-08 | Actb, Ckb, Cnp, Mbp, Ndufs3, Ywhae |
|
| ||||||
| GO:0023026 | Molecular Function | MHC class II protein complex binding | 3 | 18.8 | 3.74E-05 | Hsp90aa1, Pkm, Ywhae |
| GO:0033267 | Cellular Component | axon part | 9 | 4.6 | 1.18E-07 | Cplx1, Crmp2, Mbp, Nefl, Pfn2, Sod1, Stxbp1, Syn1, Uchl1 |
|
| ||||||
| GO:0042470 | Cellular Component | melanosome | 7 | 6.7 | 2.78E-07 | Atp6v1b2, Cnp, Erp29, Hsp90aa1, Hsp90b1, Prdx1, Ywhae |
|
| ||||||
| GO:0044275 | Biological Process | cellular carbohydrate catabolic process | 5 | 14.7 | 2.81E-07 | Gpd2, Hmgb1, Pgam1, Ppp1ca, Pygm |
|
| ||||||
| GO:0044306 | Cellular Component | neuron projection terminus | 8 | 5.9 | 9.97E-08 | Cplx1, Crmp2, Glul, Pebp1, Pfn2, Stxbp1, Syn1, Uchl1 |
|
| ||||||
| GO:0044724 | Biological Process | single-organism carbohydrate catabolic process | 7 | 5.8 | 6.91E-07 | Gpd2, Hmgb1, Pgam1, Pgk1, Pkm, Ppp1ca, Pygm |
| C) Region in Disomic | ||||||
| GOID | Ontology Source | GOTerm | Nr. Genes | % Assoc. Genes | Term P-value | Associated Genes Found |
|
| ||||||
| Reactome:R-MMU-210991 | Pathways | Basigin interactions | 4 | 8.0 | 3.51E-06 | Actb, Dnm1, Crmp2, Ezr |
|
| ||||||
| Reactome:R-MMU-399956 | Pathways | CRMPs in Sema3A signaling | 3 | 18.8 | 4.62E-06 | Crmp1, Crmp2, Crmp4 |
|
| ||||||
| GO:0031333 | Biological Process | negative regulation of protein complex assembly | 5 | 4.1 | 5.58E-06 | Capzb, Hmgb1, Pfn2, Stxbp1, Twf2 |
|
| ||||||
| GO:0048489 | Biological | synaptic vesicle transport | 6 | 4.7 | 2.58E-07 | Dnm1, Crmp2, Napb, Pfn2, Snap25, Stxbp1 |
| D) Region in Trisomic | ||||||
| GOID | Ontology Source | GOTerm | Nr. Genes | % Assoc. Genes | Term P-value | Associated Genes Found |
|
| ||||||
| Reactome:R-MMU-1428517 | Pathways | The citric acid (TCA) cycle and respiratory electron transport | 6 | 4.1 | 2.41E-06 | Aco2, Glo1, Mdh2, Ndufa10, Ndufa12, Ndufs8 |
|
| ||||||
| Reactome:R-MMU-210991 | Pathways | Basigin interactions | 5 | 10.0 | 2.19E-07 | Actb, Dnm1, Crmp2, Ezr, Mapk1 |
|
| ||||||
| Reactome:R-MMU-399956 | Pathways | CRMPs in Sema3A signaling | 4 | 25.0 | 8.09E-08 | Crmp1, Crmp2, Crmp3, Crmp4 |
|
| ||||||
| Reactome:R-MMU-5663213 | Pathways | RHO GTPases Activate WASPs and WAVEs | 4 | 12.5 | 1.55E-06 | Actb, Arpc2, Arpc5, Mapk1 |
|
| ||||||
| GO:0021762 | Biological Process | substantia nigra development | 5 | 13.2 | 5.31E-08 | Actb, Ckb, Cnp, Mbp, Ywhae |
|
| ||||||
| GO:0042470 | Cellular Component | melanosome | 5 | 4.8 | 8.92E-06 | Atp6v1b2, Cnp, Erp29, Hsp90b1, Ywhae |
|
| ||||||
| GO:0060052 | Biological Process | neurofilament cytoskeleton organization | 3 | 30.0 | 2.13E-06 | Nefl, Nefm, Sod1 |
% Associated Genes indicates the percent of the genes defining a given GO term that were listed (as accessions) in the ClueGO spot list. See Supplementary Tables S5 for more details and subnetworks.
3.4 Differences in CRMP proteins by age and brain region
It was clear that a large number of spots (7) had been identified as CRMP2 peptides, and that the expression of the CRMP2 spots by age and region was unusual. Given the importance of CRMP2 (and other CRMPs) in semaphorin 3A (SEMA3A) signaling and cytoskeleton mediated processes such as axon elongation and synaptic vesicle transport, we decided to investigate the CRMP levels more closely. In addition, seven more spots had been identified as other members of the CRMP family of proteins. Spots representing CRMP proteins were clearly significantly different by age and by brain region (Supplementary Table S3). 12 spots (of the 14 spots representing all the CRMP proteins) were elevated in hippocampus versus cerebellum at twelve-months. However, at six-months, there were no significant differences between CRMP spot levels, except for one CRMP2 spot (975) which was elevated in cerebellum samples. With age, 8 of the 14 CRMP spots were elevated in six-month versus twelve-month cerebellum samples, while 10 of 14 spots were elevated in twelve-month versus six-month hippocampus samples. These comparisons include both the disomic and trisomic samples. In addition, the elevated levels of spots 991 and 992 (CRMP1) in twelve-month samples were also observed only in the disomic sample set. Similarly, the elevated levels in hippocampus of spots 965 and 2413 (CRMP2) were observed only in the disomic sample set.
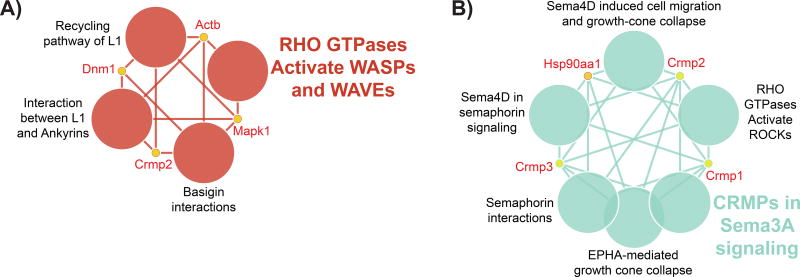
Figure 2 shows networks that are different by age in cerebellum (Figure 2 A) and in hippocampus (Figure 2 B). The only protein in common is CRMP2, which is elevated at six-months in cerebellum (spots v963, v975, v2323, and v2413), but is elevated at twelve-months in hippocampus (spots v965, v975, v2335, and v2337), which strongly suggests that CRMP2 and pathways including CRMP2 are regulated very differently in cerebellum and hippocampus.
Figure 2. CRMP2 in Aging.
A) CRMP2 containing networks that are different by age (six versus twelve-month) in cerebellum, B) networks that are different by age in hippocampus.
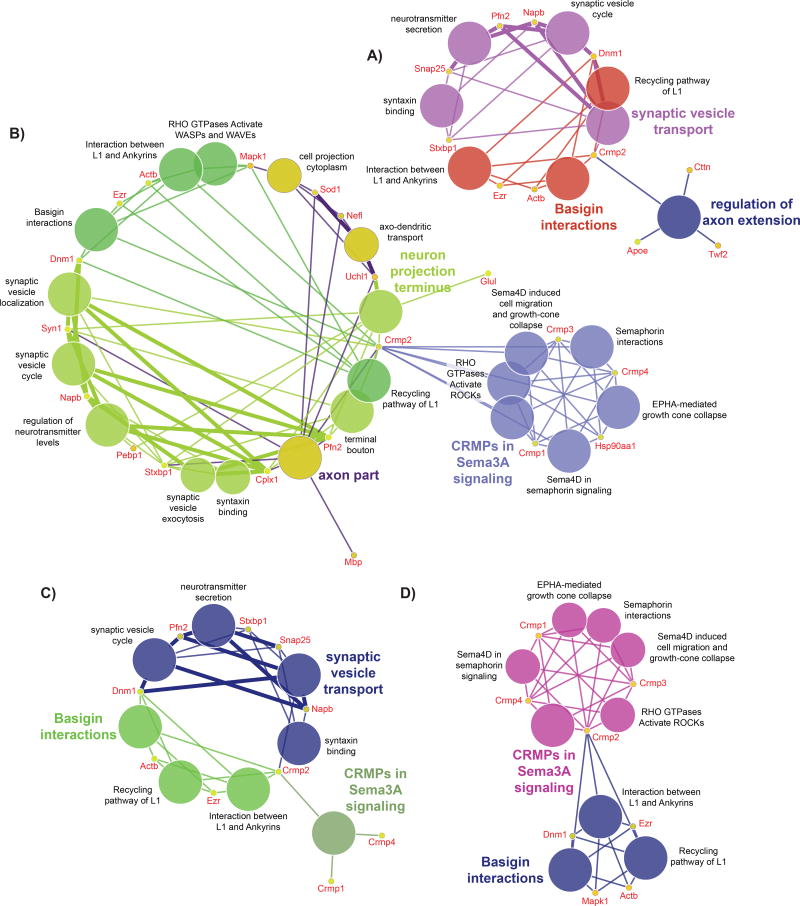
Figure 3 shows networks that are different by region in six-month (Figure 3 A) and in twelve-month (Figure 3 B). The two sets of networks are again very different, but a few of the spot levels are similar. For example, one (of four) dynamin-1 spot, v651, is elevated in hippocampus at both six and twelve-months of age. Ezr (v794 and v2279), Napb (v3312), and Uchl1 are also elevated in hippocampus at both ages. Figure 3 C) shows networks in comparison of disomic cerebellum with disomic hippocampus, and D) shows trisomic cerebellum versus trisomic hippocampus. The levels of these spots are largely the same in the two networks, except for Mapk1 (v1296), which is elevated in cerebellum in trisomic samples, and Snap25 (v3467) which is elevated in cerebellum in disomic samples. The log2 values for all the spots in the figures are reported in Supplementary Tables S1 and S2 (“log2” tabs).
Figure 3. CRMP2 in Region.
A) CRMP2 containing networks for cerebellum versus hippocampus comparison at six-months of age, and B) networks for the same comparison at twelve-months of age.
4. Discussion
4.1 Overview
In this study, we used the Ts65Dn mouse model of DS and sacrificed mice via head-focused high-energy microwave irradiation. We demonstrated previously that this method fixes brain chemistry at the time of death and preserves the in situ state of proteins (Hunsucker et al., 2008). No proteomic method is ideal and in practice, all existing approaches have (serious) limitations. For example, 1) coverage of the proteome is limited (i.e., only a subset of all proteins can be detected and quantified), 2) precision can be poor [i.e., errors associated with quantitative measurements may be larger than biological differences (Földi et al., 2011)] and 3) information on variants is often lost. The approach used in this study, 2D DiGE, is limited in its ability to comprehensively cover the proteome - only about 2,000 protein spots are seen; however, there are two important strengths of this approach. First, 2D DiGE is precise (i.e., even subtle quantitative differences can be determined) and second, because proteins are intact during the quantitative analysis, variants can be detected and independently quantified. Both these attributes are important in this study (Westermeier, 2016). When combined with peptide mass fingerprinting by MALDI TOF-MS, protein IDs can be obtained. Although alternative strategies are available (for example LC-MS/MS of each digested protein), mass fingerprinting has been demonstrated to be a reliable and expedient approach to protein identification following 2D gel separation.
Cerebellum and hippocampus were harvested at six months of age because the Ts65Dn mice show relatively good maximal learning and memory ability at this stage, and at twelve months of age, at which time the Ts65Dn mice exhibit a marked reduction in these abilities and hippocampal and cerebellar degeneration (Granholm et al., 2000). Additionally, there is a marked decrease in size and number of basal forebrain cholinergic neurons between 6 and 12 months in Ts65Dn mice (Cooper et al., 2001; Granholm et al., 2000). Recent work has demonstrated that cognitive decline can be detected by 11 months of age in euploid mice (Olmos-Serrano et al., 2016b). In addition, as discussed below, other investigators have examined Ts65Dn brain protein expression at 6 and 12 months of age. Thus, our examination of the Ts65Dn proteome at six and twelve months of age should be relevant to cognitive decline and to other studies characterizing the mice at these ages. Since differences between the disomic and trisomic proteomes are subtle in Ts65Dn mice, extra care is necessary in experimental design and interpretation of results to avoid artifacts due to age, tissue type, PMI, etc. It is possible that proteome differences in hippocampus and cerebellum samples from individuals with DS versus controls have a similar degree of subtlety. Certain variables cannot be properly controlled in studies on human tissue, illustrating the importance of mouse models.
A caveat with the current study is that it employed only male mice. This was largely due to practical considerations (see Methods 2.1). Using RPP, Block et al. measured levels of about 100 proteins/protein modifications in hippocampus, cerebellum, and cortex of male and female Dp(10)1Yey (DS model) and control (euploid) mice. Their results show that about half of the proteins differ significantly by sex in at least one brain region (40% in hippocampus) (Block et al., 2015). It’s quite likely that similar differences are present in Ts65Dn mice.
When all three variables examined in this study are considered - age, brain region, and ploidy - changes in protein expression were much more dependent on age and brain region than ploidy. This conclusion was apparent when the data were examined by PCA (Figure 1). The Ts65Dn phenotype does not involve marked changes in protein expression in the relatively abundant proteins detected here in cerebellum and hippocampus, even though both regions are significantly affected.
In contrast, we observed numerous regional (i.e., cerebellum versus hippocampus) and age-related (i.e., six-month versus twelve-month) differences in brain protein levels. This is consistent with transcriptome analysis by Olmos-Serrano et al. of three brain regions (cerebral neocortex, hippocampus, and cerebellar cortex) from DS and euploid individuals over the course of development. They observed altered expression of many genes, predominantly due to differences in brain region or developmental stage rather than ploidy (Olmos-Serrano et al., 2016a). These potentially have important effects on phenotype at birth and as the mice age. For example, changes in proteins important for synaptic plasticity, especially decreases in levels of some of these proteins in the hippocampus with age, may relate to declines in learning and memory dependent on hippocampal function. In a separate study, Olmos-Serrano et al. observed cognitive decline in euploid mice as well as a more pronounced decline in Ts65Dn mice by 11 months of age (Olmos-Serrano et al., 2016b), and we hypothesize that the alterations we observed in the proteome with age are relevant to cognitive decline associated with aging in both euploid and trisomic mice. It is reasonable to expect that abundant proteins in hippocampus are important in hippocampal function, and proteins abundant in cerebellum are important in cerebellar function. We discuss specific examples relevant to Ts65Dn mice and individuals with DS and AD below, recognizing that other alterations may also be important.
Other studies of changes in the Ts65Dn brain proteome compared to euploid controls have been reported and it is useful to compare those results with ours. Until now we know of only one study on the Ts65Dn mouse model proteome using 2DGE (Wisniewski et al., 2006). That study employed 3-month-old Ts65Dn mice and used 2DGE/silver stain and MS/MS for identification of brain proteins. The authors demonstrated increased levels of 17 proteins and decreased levels of 4 proteins. Only one of the elevated proteins, carbonyl reductase [NADPH] 3 (CBR3), is trisomic in Ts65Dn mice. We identified 8/17 of these proteins in our study: i.e., carbonic anhydrase 2 (CA2; P00920), nucleoside diphosphate kinase A (NME1; P15532), dual specificity protein phosphatase 3 (DUSP3; Q9D7X3) ubiquitin-conjugating enzyme E2 N (UBE2N; P61089), NADH dehydrogenase [ubiquinone] iron-sulfur protein 8 (NDUFS8; Q8K3J1), histidine triad nucleotide-binding protein 1 (HINT1; P70349), glutamine synthetase (GLUL; P15105), and glutathione S-transferase Mu 1 (GSTM1; P10649). None showed a significant quantitative change.
We detected only two proteins referred to by Ahmed et al. (Ahmed et al., 2017; 2015) [superoxide dismutase (SOD1; P08228) and glial fibrillary acidic protein, astrocyte (GFAP; P03995)], and two of the proteins identified by Fernandez et al. (Fernandez et al., 2009) [alpha enolase (ENO1; P17182) and elongation factor 1-alpha (EEF1A1; P10126)]. We did not detect quantitative changes in hippocampal or cerebellar levels of several other proteins reported by other investigators, including phospholipase C-beta (Ruiz de Azúa et al., 2001), G protein-activated inward rectifier potassium channel 2 (Harashima et al., 2006), or kinesin (Roberson et al., 2008). Each of these was reported to have altered levels in Ts65Dn mice. Thus, our study adds significantly to the number of proteins in cerebellum and hippocampus that do not change in level due to trisomy in Ts65Dn mice. Our results do not address changes in less abundant proteins (e.g., transcription factors) that may play a significant role in the phenotype of Ts65Dn mice and individuals with DS. Indeed, such changes have been observed.
4.2 Ploidy
Our results show that only five proteins are significantly different comparing Ts65Dn samples versus controls. Three proteins (CRYZL1, EZR, and SOD1) show increased abundance in Ts65Dn, while two proteins (PHB and MAPRE3) are significantly reduced in trisomic samples. CRYZL1, EZR, and SOD1 are all trisomic in Ts65Dn mice, but only CRYZL1 and SOD1 are Hsa21 homologs. One other Hsa21 homolog, CBR1, was identified, but it is not significantly different in disomic versus trisomic samples. EZR is located on Mmu17, and became trisomic due to the nature of the translocation which defines the Ts65Dn mouse.
Ezrin (EZR), along with Radixin and Moesin, is a member of the ERM protein family. Our results show that it is elevated by ~1.5 to 2-fold in hippocampus, and mildly elevated in trisomic samples. Active ERMs are concentrated asymmetrically in neocortical growth cones. They are rapidly (and transiently) inactivated by SEMA3A, and they are required for SEMA3A mediated growth cone collapse and guidance (Mintz et al., 2008). Ezrin is a cytoplasmic peripheral membrane protein which can interact with the actin cytoskeleton. The FERM domain of Ezrin can bind to the juxtamembrane region of L1, and this binding may be responsible for retrograde movement of L1 which generates traction forces necessary for axon growth (Sakurai et al., 2008). Given the importance of Ezrin in these processes, its overexpression in Ts65Dn may meaningfully contribute to the Ts65Dn phenotype, which may have implications concerning its validity as a model of DS. Alternatively, given the high homology of the ERM proteins, it is possible that elevated levels of Ezrin might be compensated for by alteration in levels of Moesin and/or Radixin (or vice versa). Lubec et al. report reduction of moesin (but not ezrin or radixin) in fetal DS brain (B. Lubec et al., 2001). If moesin is reduced in fetal Ts65Dn, it is possible that phenotypic effects are masked by elevated Ezrin. Barão et al. report that BACE1 (a major drug target for treating AD) is critically involved in semaphorin 3A axonal guidance in hippocampal and thalamic neurons. In response to Sema3A binding, an active membrane-bound proteolytic CHL1 fragment is generated by BACE1, which relays the Sema3A signal via ezrin-radixin-moesin (ERM) proteins to the cytoskeleton (Barão et al., 2015).
4.3 Age
We observed numerous differences in spot abundances when we compared samples from six-month-old mice to those from twelve-month-old mice. The functional networks and their component proteins are listed in Tables 5 A – C, and the ClueGO network diagrams are available in the Supplementary Material (Supplementary Fig. S1). In disomic samples, only 27 proteins showed different levels due to age (p < 0.005), and they mapped to three overview term functional groups: gluconeogenesis, hydro-lyase activity, and alcohol catabolic process. In trisomic samples, the 26 proteins that differed by age did not map to any functional networks. In contrast, the 69 proteins that differed by age in cerebellum mapped to twelve functional groups. Many of these are important in energy and metabolism. Two groups, “RHO GTPases Activate WASPs and WAVEs” and “regulation of actin polymerization or depolymerization”, indicate that regulation of actin changes dramatically during aging in cerebellum. In hippocampus, 59 proteins were significantly different by age, and these mapped to eight functional groups. Several of these are important in neurobiology, including “opioid signalling”, “regulation of actin polymerization or depolymerization”, “mitochondrial protein complex”, and “CRMPs in Sema3A signaling”.
Other studies have indicated similar changes due to aging. Ahmed et al. observed numerous changes in specific proteins involved in MAP kinase signaling, NMDA receptors, NUMB protein interactions, and the apoptosis pathway, and these changes were greater at 12 months than 6 months of age (Ahmed et al., 2017). Similarly, Dekker et al. recently demonstrated that aging affects monoamine neurotransmitters in Ts65Dn mice more than ploidy when mice aged 3 to 5.5 months were compared to 12 month old mice (Dekker et al., 2017).
Differences in some protein families suggest that these alterations may be particularly significant for alterations seen in age. One example is the collapsin response mediator proteins [CRMPs, also known as the dihydropyrimidinase-related (DPYSL) proteins] a family of cytosolic proteins originally identified via their involvement in SEMA3A signaling. The CRMPs are discussed in more detail below.
4.4 Brain region
As with age, we observed numerous differences in spot abundance when comparing cerebellum to hippocampus samples. The functional networks and their component proteins are listed in Tables 6A – D, and the ClueGO network diagrams are available in the Supplementary Material (Supplementary Fig. S2). In disomic samples, the 47 proteins that differ in abundance by region map to 7 functional groups, including “basigin interactions”, “RHO GTPases Activate WASPs and WAVEs”, “substantia nigra development”, “neurofilament cytoskeleton organization”, and “CRMPs in Sema3A signaling”. In trisomic samples, the 60 proteins that differ in abundance by region map to four functional groups, including “basigin interactions”, “synaptic vesicle transport”, and “CRMPs in Sema3A signaling”. In six-month samples, the 57 proteins that differ in abundance by region map to eight functional groups, including “Basigin interactions”, “regulation of actin polymerization or depolymerization”, “substantia nigra development”, “regulation of axon extension”, and “synaptic vesicle transport”. In twelve-month samples, the 93 proteins that differ in abundance by region map to fourteen functional groups, including “proteasome complex”, “CRMPs in Sema3A signaling”, “regulation of actin polymerization or depolymerization”, “substantia nigra development”, “axon part”, and “neuron projection terminus”.
4.5 CRMPs
There are 5 CRMP family members: CRMP1 (DPYSL1), CRMP2 (DPYSL2), CRMP3 (DPYSL4), CRMP4 (DPYSL3), and CRMP5 (DPYSL5). These proteins form homo- and hetero-tetramers and play important roles in neuronal migration, neuronal network formation, synapse formation, synaptic plasticity, and neuronal disease. They are extensively modified post-translationally via deamidation, oxidation, isoaspartyl conversion, O-glycosylation, and perhaps most importantly, phosphorylation. Crmp1 mutations have been associated with disruption in hippocampal development and other defects in brain development. CRMP2, CRMP3, and CRMP4 may be important for proper development of several brain regions as well as normal dendritic spine morphology, which is defective in Ts65Dn mice. All the collapsin response mediator proteins have important roles in the hippocampus and this may be reflected in their increased abundance in hippocampus versus cerebellum (at twelve-months of age, see Supplementary Table S3). The CRMPs have recently been proposed as therapeutic targets for neurodegenerative disease (Nagai et al., 2016).
CRMP2 is widely expressed in neuronal tissues, including brain, spinal cord, etc. and is involved in an ever-expanding range of functions, including semaphorin signaling, neurite growth/retraction, axon transport, protein endocytosis, vesicle recycling, microtubule dynamics, synaptic assembly, calcium channel regulation, and neurotransmitter release (Khanna et al., 2012). CRMP2 has multiple phosphorylation sites, and CRMP2 phosphorylation is essential for SEMA3A mediated growth cone collapse. Specifically, phosphorylation by GSK-3β and/or Rho kinase (ROCK) lowers the ability of CRMP2 to interact with tubulin, leading to axonal growth arrest and growth cone collapse (Arimura et al., 2005; Yoshimura et al., 2005). In addition, SEMA3A growth cone collapse is impaired in CRMP2 mouse mutants lacking Cdk5, GSK-3β, or Fyn phosphorylation sites (Uchida et al., 2005). Phosphorylation of both CRMP2 and Tau proteins is mediated by Cdk5 and GSK-3β. Β-amyloid induced oxidative stress leads to increased activity of both Cdk5 and GSK-3β, which may lead to CRMP2 and Tau hyperphosphorylation resulting in impaired neuronal communication, loss of synapses, and formation of neurofibrillary tangles. In transgenic mouse models of AD, CRMP2 binds to phosphorylated and non-phosphorylated forms of Tau, and hyperphosphorylated CRMP2 accumulates in the brains of these animals (Cole et al., 2007).
4.6 AD and DS
One of the proteins we identified, Apoe, plays a crucial role in AD pathogenesis. In our results, Apoe is elevated at 6 months in both cerebellum and hippocampus. It is also elevated at 6 months in disomic and trisomic samples, but fails significance in the latter, which may suggest that levels are more variable in trisomic samples. Although we did not identify other critical AD proteins such as App, Psen1 and Psen2, we did identify proteins involved in processes relevant to AD. Perhaps the best example is CRMP2 which, given its presence in neurofibrillary tangles, and its wide range of functions in neurons (discussed above), may have an extremely important role in AD. We also identified other proteins likely involved in AD progression and pathogenesis, such as cytoplasmic and mitochondrial antioxidant enzymes, synapse proteins, proteasome proteins, cytoskeletal proteins, etc.
4.7 Homeostasis?
Our results support the hypothesis that mechanisms exist that maintain homeostasis in trisomic Ts65Dn mice, and compensate for the increased dosage of trisomic genes. This is reasonable, given the degree of trisomy in Ts65Dn mice (~150 genes), and their viability. One such mechanism may be protein degradation via proteolysis. Others hypothesize that cells activate protein folding and proteolytic pathways to normalize protein stoichiometries (Pfau and Amon, 2012). Stingele et al. demonstrated that p62-dependent selective autophagy is activated in human trisomic and tetrasomic cells (Stingele et al., 2012). Nizetic and Groet discuss the observation that, although DS should, on the basis of various criteria, be a cancer-prone condition, solid tumors are in fact extremely rare in DS (Nizetic and Groet, 2012). It is possible that the mechanisms preventing solid tumor formation in DS may also act to correct protein imbalances due to the increased dosage of trisomic genes. Recent work has shown that aneuploid cells with additional single chromosomes (including Mmu16) exhibit reduced tumorigenicity relative to genetically matched euploid cells (Sheltzer et al., 2017).
4.8 Conclusions
We observed minimal differences in protein abundances (as measured by 2D DiGE peak volumes) when we compared disomic and trisomic groups. Contrastingly, we identified numerous quantitative differences associated with age (six versus twelve-month) and brain region (cerebellum versus hippocampus). We observe pronounced differences based on age (six-month versus twelve-month), and brain region (cerebellum versus hippocampus). Many of these differences are in proteins involved in energy and metabolism, while some are specific to neurological processes such as neurotransmitter transport and synapse function. These and similar studies should be helpful in defining differences in brain regions and metabolic changes due to the aging process. The data from this study is freely available, and we encourage additional analyses beyond the scope of this study. We welcome the development of hypotheses regarding DS, brain development and aging based on these findings.
Supplementary Material
Table 1.
Brain Samples
| Six-Month Cerebellum |
Six-Month Hippocampus |
Twelve-Month Cerebellum |
Twelve-Month Hippocampus |
|
|---|---|---|---|---|
| Disomic | 8 | 8 | 14 | 15 |
| Trisomic | 8 | 8 | 14 | 15 |
Highlights.
Proteome analysis of Ts65Dn hippocampus and cerebellum at 6 and 12 months.
Minimal differences in protein levels were found in trisomic vs. disomic.
Compensatory mechanisms may exist which ameliorate the effect of trisomy.
Many differences were found in hippocampus vs. cerebellum and in 12- vs. 6-month.
CRMP protein levels are distinct by age and region.
Acknowledgments
The authors would like to thank Jenna Boyd and Keri Newell for technical assistance, Dr. Stephen W. Hunsucker (Integrated Diagnostics, Inc.), Dr. Junguk Hur (University of North Dakota, Grand Forks, North Dakota), and Dr. Anne Poljak (University of New South Wales, Sydney, Australia) for critical comments and suggestions, and Dr. Tarik Haydar (Boston University School of Medicine, Boston, Massachusetts) for donation of Ts65Dn and disomic males.
Funding: This work was supported by the National Institutes of Health [grant numbers P30GM103329, R01MH100972 and R01MH105329 to JDG], NINDS [grant number NS51539 to DP], the Lowe Fund of the Denver Foundation [to DP], and the Itkin Family Foundation [to DP].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed MM, Block A, Tong S, Davisson MT, Gardiner KJ. Age exacerbates abnormal protein expression in a mouse model of Down syndrome. Neurobiol Aging. 2017;57:120–132. doi: 10.1016/j.neurobiolaging.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Ahmed MM, Dhanasekaran AR, Block A, Tong S, Costa ACS, Gardiner KJ. Protein profiles associated with context fear conditioning and their modulation by memantine. Mol Cell Proteomics. 2014;13:919–937. doi: 10.1074/mcp.M113.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed MM, Dhanasekaran AR, Block A, Tong S, Costa ACS, Stasko M, Gardiner KJ. Protein dynamics associated with failed and rescued learning in the Ts65Dn mouse model of Down syndrome. PLoS ONE. 2015;10:e0119491. doi: 10.1371/journal.pone.0119491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aït Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Créau N, Bléhaut H, Robin S, Delabar J-M, Potier M-C. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge K, Reeves RH, Olson LE, Richtsmeier JT. Differential effects of trisomy on brain shape and volume in related aneuploid mouse models. Am J Med Genet A. 2007;143A:1060–1070. doi: 10.1002/ajmg.a.31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N, Ménager C, Kawano Y, Yoshimura T, Kawabata S, Hattori A, Fukata Y, Amano M, Goshima Y, Inagaki M, Morone N, Usukura J, Kaibuchi K. Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol Cell Biol. 2005;25:9973–9984. doi: 10.1128/MCB.25.22.9973-9984-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artigaud S, Gauthier O, Pichereau V. Identifying differentially expressed proteins in two-dimensional electrophoresis experiments: inputs from transcriptomics statistical tools. Bioinformatics. 2013;29:2729–2734. doi: 10.1093/bioinformatics/btt464. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barão S, Gärtner A, Leyva-Díaz E, Demyanenko G. Antagonistic effects of BACE1 and APH1B-γ-secretase control axonal guidance by regulating growth cone collapse. Cell Reports. 2015;12:1367–1376. doi: 10.1016/j.celrep.2015.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter LL, Moran TH, Richtsmeier JT, Troncoso J, Reeves RH. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum Mol Genet. 2000;9:195–202. doi: 10.1093/hmg/9.2.195. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. JSTOR: Journal of the Royal Statistical Society. Series B (Methodological) 1995;57(1):289–300. (1995), Journal of the Royal Statistical Society Series B ‥‥. [Google Scholar]
- Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Bower C, Hussain R, Glasson EJ. The four ages of Down syndrome. Eur J Public Health. 2007;17:221–225. doi: 10.1093/eurpub/ckl103. [DOI] [PubMed] [Google Scholar]
- Block A, Ahmed MM, Dhanasekaran AR, Tong S, Gardiner KJ. Sex differences in protein expression in the mouse brain and their perturbations in a model of Down syndrome. Biol Sex Differ. 2015;6:24. doi: 10.1186/s13293-015-0043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bradshaw RA, Burlingame AL, Carr S, Aebersold R. Reporting protein identification data: the next generation of guidelines. Mol Cell Proteomics. 2006;5:787–788. doi: 10.1074/mcp.E600005-MCP200. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Blake JA, Kadin JA, Richardson JE, Mouse Genome Database Group Mouse genome database 2016. Nucleic Acids Res. 2016;44:D840–7. doi: 10.1093/nar/gkv1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dyakin VV, Branch CA, Ardekani B, Yang D, Guilfoyle DN, Peterson J, Peterhoff C, Ginsberg SD, Cataldo AM, Nixon RA. In vivo MRI identifies cholinergic circuitry deficits in a Down syndrome model. Neurobiol Aging. 2009;30:1453–1465. doi: 10.1016/j.neurobiolaging.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JHK, Berger JD, Mazzella MJ, Morales-Corraliza J, Cataldo AM, Nixon RA, Ginsberg SD, Levy E, Mathews PM. Age-dependent dysregulation of brain amyloid precursor protein in the Ts65Dn Down syndrome mouse model. J Neurochem. 2009;110:1818–1827. doi: 10.1111/j.1471-4159.2009.06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Scott HS, Papasavvas MP, Rossier C, Antonarakis ES, Barras C, Davisson MT, Schmidt C, Estivill X, Dierssen M, Pritchard M, Antonarakis SE. The mouse brain transcriptome by SAGE: differences in gene expression between P30 brains of the partial trisomy 16 mouse model of Down syndrome (Ts65Dn) and normals. Genome Res. 2000;10:2006–2021. doi: 10.1101/gr.10.12.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AR, Noble W, van Aalten L, Plattner F, Meimaridou R, Hogan D, Taylor M, LaFrancois J, Gunn-Moore F, Verkhratsky A, Oddo S, LaFerla F, Giese KP, Dineley KT, Duff K, Richardson JC, Yan SD, Hanger DP, Allan SM, Sutherland C. Collapsin response mediator protein-2 hyperphosphorylation is an early event in Alzheimer's disease progression. J Neurochem. 2007;103:1132–1144. doi: 10.1111/j.1471-4159.2007.04829.x. [DOI] [PubMed] [Google Scholar]
- Cooper JD, Salehi A, Delcroix JD, Howe CL, Belichenko PV, Chua-Couzens J, Kilbridge JF, Carlson EJ, Epstein CJ, Mobley WC. Failed retrograde transport of NGF in a mouse model of Down's syndrome: reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc Natl Acad Sci USA. 2001;98:10439–10444. doi: 10.1073/pnas.181219298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D'Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson MT, Schmidt C, Akeson EC. Segmental trisomy of murine chromosome 16: a new model system for studying Down syndrome. Prog. Clin. Biol. Res. 1990;360:263–280. [PubMed] [Google Scholar]
- Dekker AD, Vermeiren Y, Albac C, Lana-Elola E, Watson-Scales S, Gibbins D, Aerts T, Van Dam D, Fisher EMC, Tybulewicz VLJ, Potier M-C, De Deyn PP. Aging rather than aneuploidy affects monoamine neurotransmitters in brain regions of Down syndrome mouse models. Neurobiol Dis. 2017;105:235–244. doi: 10.1016/j.nbd.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchon A, Raveau M, Chevalier C, Nalesso V, Sharp AJ, Herault Y. Identification of the translocation breakpoints in the Ts65Dn and Ts1Cje mouse lines: relevance for modeling down syndrome. Mamm Genome. 2011 doi: 10.1007/s00335-011-9356-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euthanasia APO. AVMA Guidelines for the Euthanasia of Animals (2013 Edition) 2013 [Google Scholar]
- Fernandez F, Trinidad JC, Blank M, Feng D-D, Burlingame AL, Garner CC. Normal protein composition of synapses in Ts65Dn mice: a mouse model of Down syndrome. J Neurochem. 2009;110:157–169. doi: 10.1111/j.1471-4159.2009.06110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Földi I, Müller G, Penke B, Janáky T. Characterisation of the variation of mouse brain proteome by two-dimensional electrophoresis. Journal of Proteomics. 2011;74:894–901. doi: 10.1016/j.jprot.2011.03.006. [DOI] [PubMed] [Google Scholar]
- García-Cerro S, Rueda N, Vidal V, Lantigua S, Martínez-Cué C. Normalizing the gene dosage of Dyrk1A in a mouse model of Down syndrome rescues several Alzheimer's disease phenotypes. Neurobiol Dis. 2017;106:76–88. doi: 10.1016/j.nbd.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Gardiner KJ. Pharmacological approaches to improving cognitive function in Down syndrome: current status and considerations. Drug Des Devel Ther. 2015;103 doi: 10.2147/DDDT.S51476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner KJ. Molecular basis of pharmacotherapies for cognition in Down syndrome. Trends Pharmacol Sci. 2010;31:66–73. doi: 10.1016/j.tips.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasson EJ, Sullivan SG, Hussain R, Petterson BA, Montgomery PD, Bittles AH. The changing survival profile of people with Down's syndrome: implications for genetic counselling. Clinical Genetics. 2002;62:390–393. doi: 10.1034/j.1399-0004.2002.620506.x. [DOI] [PubMed] [Google Scholar]
- Granholm AC, Sanders LA, Crnic LS. Loss of cholinergic phenotype in basal forebrain coincides with cognitive decline in a mouse model of Down's syndrome. Experimental Neurology. 2000;161:647–663. doi: 10.1006/exnr.1999.7289. [DOI] [PubMed] [Google Scholar]
- Gupta M, Dhanasekaran AR, Gardiner KJ. Mouse models of Down syndrome: gene content and consequences. Mamm Genome. 2016;27:1–18. doi: 10.1007/s00335-016-9661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton TG, Stasko MR, Kale A, Amende I, Costa ACS. Gait dynamics in trisomic mice: quantitative neurological traits of Down syndrome. Physiol Behav. 2004;82:381–389. doi: 10.1016/j.physbeh.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Harashima C, Jacobowitz DM, Stoffel M, Chakrabarti L, Haydar TF, Siarey RJ, Galdzicki Z. Elevated Expression of the G-Protein-Activated Inwardly Rectifying Potassium Channel 2 (GIRK2) in Cerebellar Unipolar Brush Cells of a Down Syndrome Mouse Model. Cell Mol Neurobiol. 2006;26:717–732. doi: 10.1007/s10571-006-9066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L, Swatton J, Wengenroth M, Wayland M, Lockstone H, Holland A, Faull R, Lilley K, Bahn S. Differences in Protein Profiles in Schizophrenia Prefrontal Cortex Compared to Other Major Brain Disorders. Clinical Schizophrenia & Related Psychoses. 2007;1:73–91. [Google Scholar]
- Hartley D, Blumenthal T, Carrillo M, DiPaolo G, Esralew L, Gardiner K, Granholm A-C, Iqbal K, Krams M, Lemere C, Lott I, Mobley W, Ness S, Nixon R, Potter H, Reeves R, Sabbagh M, Silverman W, Tycko B, Whitten M, Wisniewski T. Down syndrome and Alzheimer's disease: Common pathways, common goals. Alzheimers Dement. 2015;11:700–709. doi: 10.1016/j.jalz.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Santucci D, Kilbridge J, Chua-Couzens J, Fontana DJ, Daniels SE, Johnson RM, Chen K, Sun Y, Carlson E, Alleva E, Epstein CJ, Mobley WC. Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc Natl Acad Sci USA. 1996;93:13333–13338. doi: 10.1073/pnas.93.23.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsucker SW, Solomon B, Gawryluk J, Geiger JD, Vacano GN, Duncan MW, Patterson D. Assessment of post-mortem-induced changes to the mouse brain proteome. J Neurochem. 2008;105:725–737. doi: 10.1111/j.1471-4159.2007.05183.x. [DOI] [PubMed] [Google Scholar]
- Hyde LA, Crnic LS. Age-related deficits in context discrimination learning in Ts65Dn mice that model Down syndrome and Alzheimer's disease. Behav Neurosci. 2001;115:1239–1246. [PubMed] [Google Scholar]
- Insausti AM, Megías M, Crespo D, Cruz-Orive LM, Dierssen M, Vallina IF, Insausti R, Flórez J, Vallina TF. Hippocampal volume and neuronal number in Ts65Dn mice: a murine model of Down syndrome. Neurosci Lett. 1998;253:175–178. doi: 10.1016/s0304-3940(98)00641-7. [DOI] [PubMed] [Google Scholar]
- Irving C, Basu A, Richmond S, Burn J, Wren C. Twenty-year trends in prevalence and survival of Down syndrome. Eur J Hum Genet. 2008;16:1336–1340. doi: 10.1038/ejhg.2008.122. [DOI] [PubMed] [Google Scholar]
- Khanna R, Wilson SM, Brittain JM, Weimer J. Opening Pandora's jar: a primer on the putative roles of CRMP2 in a panoply of neurodegenerative, sensory and motor neuron, and central disorders. Future ‥‥. 2012 doi: 10.2217/fnl.12.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsammer G, Jilani S, Liu H, Davis E, Gurbuxani S, Le Beau MM, Crispino JD. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinsky-McHale SJ, Devenny DA, Gu H, Jenkins EC, Kittler P, Murty VV, Schupf N, Scotto L, Tycko B, Urv TK, Ye L, Zigman WB, Silverman W. Successful aging in a 70-year-old man with down syndrome: a case study. Intellect Dev Disabil. 2008;46:215–228. doi: 10.1352/2008.46:215-228. [DOI] [PubMed] [Google Scholar]
- Kurt MA, Kafa MI, Dierssen M, Davies DC. Deficits of neuronal density in CA1 and synaptic density in the dentate gyrus, CA3 and CA1, in a mouse model of Down syndrome. Brain Res. 2004;1022:101–109. doi: 10.1016/j.brainres.2004.06.075. [DOI] [PubMed] [Google Scholar]
- Latash ML, Kang N, Patterson D. Finger coordination in persons with Down syndrome: atypical patterns of coordination and the effects of practice. Exp Brain Res. 2002;146:345–355. doi: 10.1007/s00221-002-1189-3. [DOI] [PubMed] [Google Scholar]
- Levine S, Saltzman A, Levy E, Ginsberg SD. Systemic pathology in aged mouse models of Down“s syndrome and Alzheimer”s disease. Exp Mol Pathol. 2009;86:18–22. doi: 10.1016/j.yexmp.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockrow JP, Fortress AM, Granholm A-CE. Age-related neurodegeneration and memory loss in down syndrome. Current Gerontology and Geriatrics Research. 2012;2012:463909–13. doi: 10.1155/2012/463909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi HA, Reeves RH. Hippocampal hypocellularity in the Ts65Dn mouse originates early in development. Brain Res. 2006;1104:153–159. doi: 10.1016/j.brainres.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Lott IT. Neurological phenotypes for Down syndrome across the life span. Prog. Brain Res. 2012;197:101–121. doi: 10.1016/B978-0-444-54299-1.00006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubec B, Weitzdoerfer R, Fountoulakis M. Manifold reduction of moesin in fetal Down syndrome brain. Biochem Biophys Res Commun. 2001;286:1191–1194. doi: 10.1006/bbrc.2001.5520. [DOI] [PubMed] [Google Scholar]
- Lubec G. Advances in Down Syndrome Research. Springer Science & Business Media; Vienna: 2013. [DOI] [Google Scholar]
- Mann DM. The pathological association between Down syndrome and Alzheimer disease. Mech Ageing Dev. 1988;43:99–136. doi: 10.1016/0047-6374(88)90041-3. [DOI] [PubMed] [Google Scholar]
- Miniño AM, Xu J, Kochanek KD. Deaths: preliminary data for 2008. Natl Vital Stat Rep. 2010;59:1–52. [PubMed] [Google Scholar]
- Mintz CD, Carcea I, McNickle DG, Dickson TC, Ge Y, Salton SRJ, Benson DL. ERM proteins regulate growth cone responses to Sema3A. Journal of Comparative Neurology. 2008;510:351–366. doi: 10.1002/cne.21799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JK, Alberman E. Trends in Down's syndrome live births and antenatal diagnoses in England and Wales from 1989 to 2008: analysis of data from the National Down Syndrome Cytogenetic Register. BMJ. 2009;339:b3794–b3794. doi: 10.1136/bmj.b3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhler H. Cognitive enhancement by pharmacological and behavioral interventions: the murine Down syndrome model. Biochem. Pharmacol. 2012;84:994–999. doi: 10.1016/j.bcp.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Nagai J, Baba R, Ohshima T. CRMPs Function in Neurons and Glial Cells: Potential Therapeutic Targets for Neurodegenerative Diseases and CNS Injury. Mol Neurobiol. 2016;34:1–14. doi: 10.1007/s12035-016-0005-1. [DOI] [PubMed] [Google Scholar]
- Nizetic D, Groet J. Tumorigenesis in Down's syndrome: big lessons from a small chromosome. Nat Rev Cancer. 2012;12:721–732. doi: 10.1038/nrc3355. [DOI] [PubMed] [Google Scholar]
- Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, Pletikos M, Jankovic-Rapan L, Cramer NP, Galdzicki Z, Goodliffe J, Peters A, Sethares C, Delalle I, Golden JA, Haydar TF, Sestan N. Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination. Neuron. 2016a;89:1208–1222. doi: 10.1016/j.neuron.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos-Serrano JL, Tyler WA, Cabral HJ. Longitudinal measures of cognition in the Ts65Dn mouse: refining windows and defining modalities for therapeutic intervention in Down syndrome. Experimental Neurology. 2016b;279:40–56. doi: 10.1016/j.expneurol.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, Anderson P, Mason CA, Collins JS, Kirby RS, Correa A, National Birth Defects Prevention Network Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defect Res A. 2010;88:1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- Patterson D, Costa ACS. Down syndrome and genetics - a case of linked histories. Nat. Rev. Genet. 2005;6:137–147. doi: 10.1038/nrg1525. [DOI] [PubMed] [Google Scholar]
- Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandini P, Deutsch S, Lyle R, Gagnebin M, Vivier C, Delorenzi M, Gehrig C, Descombes P, Sherman S, Bricarelli F. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. The American Journal of Human Genetics. 2007;81:252–263. doi: 10.1086/519248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presson AP, Partyka G, Jensen KM, Devine OJ, Rasmussen SA, McCabe LL, McCabe ERB. Current estimate of Down Syndrome population prevalence in the United States. J. Pediatr. 2013;163:1163–1168. doi: 10.1016/j.jpeds.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. 2015 www.R-project.org.
- Rabilloud T, Lelong C. Two-dimensional gel electrophoresis in proteomics: A tutorial. Journal of Proteomics. 2011;74:1829–1841. doi: 10.1016/j.jprot.2011.05.040. [DOI] [PubMed] [Google Scholar]
- Raz N, Torres IJ, Briggs SD, Spencer WD, Thornton AE, Loken WJ, Gunning FM, McQuain JD, Driesen NR, Acker JD. Selective neuroanatomic abnormalities in Down's syndrome and their cognitive correlates: evidence from MRI morphometry. Neurology. 1995;45:356–366. doi: 10.1212/wnl.45.2.356. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Reinholdt LG, Ding Y, Gilbert GT, Czechanski A, Solzak JP, Roper RJ, Johnson MT, Donahue LR, Lutz C, Davisson MT. Molecular characterization of the translocation breakpoints in the Down syndrome mouse model Ts65Dn. Mamm Genome. 2011 doi: 10.1007/s00335-011-9357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson R, Toso L, Abebe D, Spong CY. Altered expression of KIF17, a kinesin motor protein associated with NR2B trafficking, may mediate learning deficits in a Down syndrome mouse model. Am J Obstet Gynecol. 2008;198:313, e1–4. doi: 10.1016/j.ajog.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper RJ, Baxter LL, Saran NG, Klinedinst DK, Beachy PA, Reeves RH. Defective cerebellar response to mitogenic Hedgehog signaling in Down [corrected] syndrome mice. Proc Natl Acad Sci USA. 2006;103:1452–1456. doi: 10.1073/pnas.0510750103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Azúa I, Lumbreras MA, Zalduegui A, Baamonde C, Dierssen M, Flórez J, Sallés J. Reduced phospholipase C-beta activity and isoform expression in the cerebellum of TS65Dn mouse: a model of Down syndrome. J Neurosci Res. 2001;66:540–550. doi: 10.1002/jnr.10019. [DOI] [PubMed] [Google Scholar]
- Ruparelia A, Pearn ML, Mobley WC. Cognitive and pharmacological insights from the Ts65Dn mouse model of Down syndrome. Curr Opin Neurobiol. 2012;22:880–886. doi: 10.1016/j.conb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Gil OD, Whittard JD, Gazdoiu M, Joseph T, Wu J, Waksman A, Benson DL, Salton SR, Felsenfeld DP. Interactions between the L1 cell adhesion molecule and ezrin support tractionforce generation and can be regulated by tyrosine phosphorylation. J Neurosci Res. 2008;86:2602–2614. doi: 10.1002/jnr.21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saran NG. Global disruption of the cerebellar transcriptome in a Down syndrome mouse model. Hum Mol Genet. 2003;12:2013–2019. doi: 10.1093/hmg/ddg217. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Ko JH, Replogle JM, Habibe Burgos NC, Chung ES, Meehl CM, Sayles NM, Passerini V, Storchová Z, Amon A. Single-chromosome Gains Commonly Function as Tumor Suppressors. Cancer Cell. 2017;31:240–255. doi: 10.1016/j.ccell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3–25. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Henrique-Silva F. Trisomy 21 and Down syndrome: a short review. Braz J Biol. 2008;68:447–452. doi: 10.1590/S1519-69842008000200031. [DOI] [PubMed] [Google Scholar]
- Spellman C, Ahmed MM, Dubach D, Gardiner KJ. Expression of trisomic proteins in Down syndrome model systems. Gene. 2013;512:219–225. doi: 10.1016/j.gene.2012.10.051. [DOI] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchová Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Piccini I, Balzereit D, Herwig R, Saran NG, Lehrach H, Reeves RH, Yaspo M-L. Gene expression variation in Down's syndrome mice allows prioritization of candidate genes. Genome Biol. 2007;8:R91. doi: 10.1186/gb-2007-8-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dierssen M, Torán N, Pollak DD, Chen W-Q, Lubec G. A gel-based proteomic method reveals several protein pathway abnormalities in fetal Down syndrome brain. Journal of Proteomics. 2011;74:547–557. doi: 10.1016/j.jprot.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Sasaki Y, Suzuki H, Yanai S, Yamashita N, Nakamura F, Takei K, Ihara Y, Mikoshiba K, Kolattukudy P, Honnorat J, Goshima Y. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer's disease. Genes Cells. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- Vacano GN, Duval N, Patterson D. The use of mouse models for understanding the biology of down syndrome and aging. Current Gerontology and Geriatrics Research 2012. 2012 doi: 10.1155/2012/717315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qian W-J, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp Ii DG, Khan AH, Smith DJ, Smith RD. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J. Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijerman ME, van Furth AM, Vonk Noordegraaf A, van Wouwe JP, Broers CJM, Gemke RJBJ. Prevalence, neonatal characteristics, and first-year mortality of Down syndrome: a national study. J. Pediatr. 2008;152:15–19. doi: 10.1016/j.jpeds.2007.09.045. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Westermeier R. 2D gel-based Proteomics: there's life in the old dog yet. Archives of Physiology and Biochemistry. 2016;122:236–237. doi: 10.1080/13813455.2016.1179766. [DOI] [PubMed] [Google Scholar]
- Wimo A, Prince M. World Alzheimer Report 2010—Alzheimer's Disease International. The Global Economic Impact of Dementia 2010 [Google Scholar]
- Wisniewski KE, Kida E, Golabek AA, Walus M, Rabe A, Palminiello S, Albertini G. Neurobehavioural Specificity. Wiley; Chichester: 2006. Down syndrome: from pathology to pathogenesis. Down syndrome; pp. 17–33. [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3β Regulates Phosphorylation of CRMP-2 and Neuronal Polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Yu T, Li Z, Jia Z, Clapcote SJ, Liu C, Li S, Asrar S, Pao A, Chen R, Fan N, Carattini-Rivera S, Bechard AR, Spring S, Henkelman RM, Stoica G, Matsui S-I, Nowak NJ, Roder JC, Chen C, Bradley A, Yu YE. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum Mol Genet. 2010;19:2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.