Abstract
CRISPR-Cas systems, found in nature as microbial adaptive immune systems, have been repurposed into an important tool in biological engineering and genome editing, providing a programmable platform for precision gene targeting. These tools have immense promise as therapeutics that could potentially correct disease-causing mutations. However, CRISPR-Cas gene editing components must be transported directly to the nucleus of targeted cells to exert a therapeutic effect. Thus, efficient methods of delivery will be critical to the success of therapeutic genome editing applications. Here, we review current strategies available for in vivo delivery of CRISPR-Cas gene editing components and outline challenges that need to be addressed before this powerful tool can be deployed in the clinic.
Next generation genome editing
The development of genome editing tools has made great progress in the past decade. Originally limited to certain model organisms and inefficient at best, genome editing began to become more broadly applicable with the discovery of several programmable DNA binding proteins. Chief among these were zinc-finger (ZF) and transcription activator-like effectors (TALEs). By fusing these proteins to nucleases, it is possible to create a double-strand break at a specified site in the genome, which would then be repaired either through non-homologous end joining, usually leading to a small insertion or deletion and thus gene knock-out, or homologous recombination with a template, leading to insertion of a desired sequence of DNA [1, 2]. In practice, however, both ZFs and TALEs are difficult to reprogram, requiring extensive protein engineering for each new target.
In early 2013, genome editing made a huge leap forward with the report that the microbial CRISPR – Cas9 (clustered regularly spaced palindromic repeat – CRISPR associated protein 9) adaptive immune system could be repurposed for genome editing in mammalian cells [3–5]. Unlike ZFs and TALEs, Cas9 uses a complementary RNA to achieve sequence specificity [6]. Moreover, Cas9 is a nuclease, obviating the need to fuse it to an additional protein. Cas9 can be programmed to target nearly any gene in the genome by synthesizing a guide RNA (gRNA) molecule which is complementary to the target sequence (Box 1) [7]. The design flexibility of the CRISPR system has driven the rapid adoption of this method for an array of genome editing applications in the laboratory [8]. Recently, as the alternative CRISPR nuclease Cpf1 has been discovered [9]. CRISPR-Cpf1 acts on a similar principle to CRISPR-Cas9, and the two techniques share many of the same advantages and disadvantages (Box 2).
Even though CRISPR has already dramatically changed how molecular biology is conducted on the benchtop, some argue its true potential lies as a therapeutic. CRISPR, as well as ZFs and TALEs, offer the tantalizing possibility of treating or even eradicating thousands of human diseases at the level of DNA. By simple changing the gRNA and template DNA, Cas9 could go from repairing a mutation in CFTR in a patient with cystic fibrosis to knocking out an oncogene in a tumor of a cancer patient. However, like all genetic tools, Cas9-gRNA complexes must be delivered directly to the target cells’ nuclei to be effective. This challenge necessitates a particular focus on the delivery strategies and techniques in order to maximize the success rate and safety of this promising technology. Ultimately, successful delivery will allow for the treatment of a wide range of genetic disease and disorders in a clinical setting.
Keywords: CRISPR, Cas9, Delivery, Gene Editing, Therapeutics, Clinical
The right place at the right time
In a laboratory setting, scientists have many options for introducing Cas9 and gRNA into cells. For example, cell lines can be transfected with lentivirus carrying genetically encoded Cas9 and gRNA, or these components can be introduced through electroporation. Typical laboratory methods, however, are not viable options for human patients. Although some diseases can be addressed through ex vivo editing therapies, such as leukemia where edited bone marrow could be transplanted to the patient, most diseases will require in vivo delivery of Cas9 and gRNA. In order to achieve this, there is a need for robust delivery modes to ensure specific and efficient targeted delivery of both Cas9 and gRNA within the body in order to exert a therapeutic benefit. Finally, treating disease at the genetic level rather than addressing disease symptoms will require careful consideration of developmental timing and disease course progression. For example, many genetic disorders will need to be addressed early in development to alleviate their symptoms and cancers should be targeted as early as possible to prevent their spread.
There are a number of additional challenges specific to the delivery of Cas9 The Cas9 protein is a very large molecule (~160 kDa)[10], and the long phosphate backbone of the gRNA is highly negatively charged. These features make it exactly the type of entity that the cell membrane has evolved to reject from entering the cell [11]. A number of different approaches to address this problem have been explored, including viral, plasmid, mRNA, and protein delivery methods, the pros and cons of which we describe below. Regardless of format, however, each of these approaches faces the same obstacle: crossing the cell membrane (Figure 1).
Figure 1.
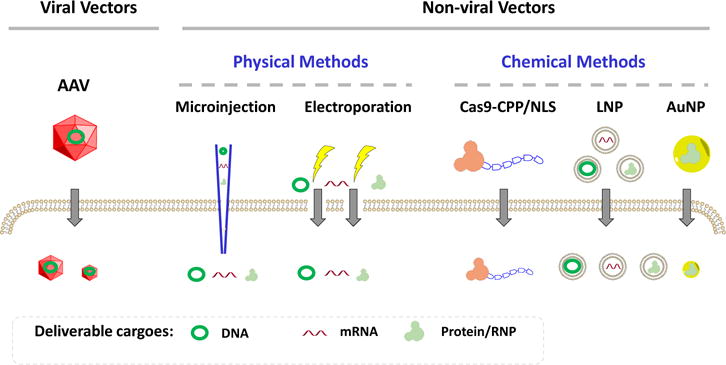
Routes of Cas9 Delivery.
A summary of the types of delivery routes discussed in this paper. Cas9 may be delivered as a DNA or mRNA molecule encoding for the cas9 gene, or it may be delivered as a functional ribonucleoprotein (RNP). Regardless of cargo format, the largest challenge lies in delivering the cargo across the cell membrane. A variety of viral and nonviral methods have been derived to achieve successful delivery across the cell membrane.
Another major challenge is that, similar to other pharmaceutical drug products, the proper dosage of CRISPR materials must be delivered to the target in the desired window of time in order to achieve therapeutic efficacy. This delivery must be specific to the desired tissues to prevent undesirable off-target gene editing events. The CRISPR complex must also evade cellular degradation mechanisms, including proteases, RNAses, and lysosomes, and must be formulated in a way to minimize eliciting an immune response. A well-designed delivery vehicle could serve to mitigate these challenges.
Cas9: DNA, mRNA or protein?
In order to achieve CRISPR-mediated gene editing, there must ultimately be a functional Cas9-gRNA ribonucleoprotein (RNP) complex present in the cell nucleus. While direct delivery of this RNP complex may sound like the most straightforward option, delivering a large protein across the cell membrane can be extremely challenging. Thus, in addition to this protein delivery approach, research has focused on delivering genetic material to instruct the target cell to produce its own Cas9 protein in situ (table 1). Delivering either DNA (in the form of either a plasmid or encoded in a viral genome) or mRNA can lead to expression of the Cas9 protein inside of a target cell, and ultimately result in Cas9-mediated gene editing [12]. Each of these delivery formats has advantages and disadvantages in overall effectiveness as well as unique delivery challenges (Figure 2). They may also exhibit different expression time courses [12], which needs to be considered when designing a therapeutic regimen.
Table 1.
Delivery Format: Examples of CRISPR Delivery as DNA, mRNA, and Protein
| Delivery format | Editing target | Model system | Study objective | Reference |
|---|---|---|---|---|
| Plasmid DNA | DMD23 | Mouse primary satellite cells | Functional gene knockdown | [14] |
| mRNA | DNMT1 | HEK293 Cell Culture | Engineering of mRNA molecules for optimal translation efficiency | [20] |
| HBV PCSK9 |
Mouse in vivo model | Fuctional viral resistance Functional gene knockdown |
[21] | |
|
GFP PCSK9 |
GFP-HEK293 Cell Culture Mouse in vivo model |
Enginering of sgRNA molecules for optimal translation efficiency | [22] | |
| HTI Fah gene | Mouse in vivo model | HDR-mediated repair of genetic mutation | [23] | |
| Protein RNP | GFP | GFP-HEK293 Cell Culture | Functional gene knockdown | [24] |
| Mdx | Mouse in vivo model | HDR-mediated repair of genetic mutation | [25] |
Figure 2.
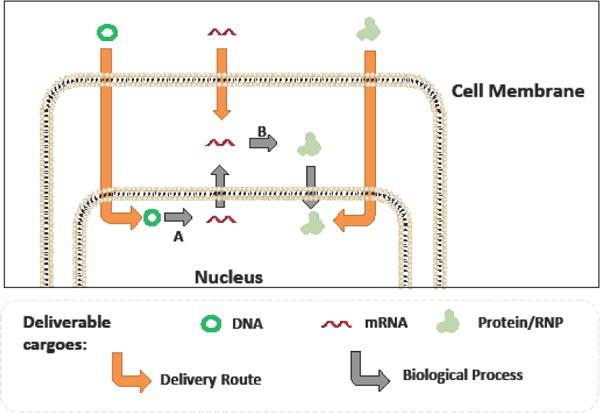
Cas9 delivery as DNA, mRNA, or Protein.
Cas9 may be successfully delivered in either a DNA, mRNA, or protein format to achieve successful gene editing. Each format carries its own particular delivery challenges. While delivery of Cas9 RNPs results in the most immediate onset of gene editing, it can also be extremely transient, and suffers the additional challenge of delivery across two cellular barriers (the cell membrane and the nuclear membrane). Delivery of DNA encoding for the cas9 gene may offer the most stable expression of the Cas9 protein, however the gene must undergo the biological processes of transcription (A) and translation (B) before the therapeutic effect can be realized, leading to a delay in the onset of gene editing. In mammalian cells, DNA generally must be delivered directly into the nucleus as well. mRNA need not be delivered into the nucleus, as the cellular translation machinery is found in the cytoplasm. However, mRNAs are extremely susceptible to enzymatic degradation, and the therapeutic effect is still somewhat delayed until the process of translation has been completed.
The cas9 gene varies from 4 – 7 kB depending on the species of origin and the inclusion of intracellular localization tags [13]. In the lab, plasmid DNA is stable and inexpensive, making a plasmid encoding Cas9 an appealing delivery route [12, 14]. In order for DNA-encoded delivery to be effective in eukaryotic cells, it must be able to penetrate both the cellular and nuclear membrane. With this delivery format care must also be taken to consider the promoter sequence or, if necessary, replication sequence encoded in the plasmid to drive the expression of Cas9. A plasmid which contains promoters for bacterial expression may not be compatible with mammalian cells. Introduction of plasmids increases the expression time compared to other modes of delivery; this may be advantageous if sustained expression is required for editing, but it could lead to increased off-target effects, raising safety concerns. Furthermore, there is some evidence that plasmid delivery may trigger an immunogenic response [15].
An alternative to encoding Cas9 in plasmid DNA is instead to encode it as messenger RNA (mRNA), which can be expressed by ribosomes found in the cytoplasm. This avoids the difficult challenge of crossing the nuclear membrane. One study comparing delivery formats showed that Cas9 mRNA delivery to cell culture resulted in quicker onset of gene editing when compared to plasmid delivery, likely due to the fact that translation of mRNA can commence immediately, while cas9 plasmid DNA must first be transcribed into mRNA [12]. The tradeoff for this quicker onset is that mRNA delivery results in transient protein expression. This transience can be leveraged to more tightly control the dose or duration of Cas9 in the cell, which may help to limit off-target editing events, but it means that sustained expression of Cas9, which may be needed to achieve beneficial levels of gene editing, will be challenging. mRNA is also less stable than DNA, and is susceptible to degradation by RNAses both in the lab and in vivo. It may therefore be beneficial to choose a delivery mechanism that protects the mRNA from enzyme-mediated degradation pathways. Recently, progress has been made in chemically modifying mRNA molecules to modulate their stability and expression inside cells, which has further improved the ability to tightly control protein expression following mRNA delivery [16–20]. Recently, mRNA delivery of Cpf1 has also been demonstrated [20]. One unique consideration for mRNA delivery is the route of delivery of the gRNA. As mRNA and gRNA are typically both single-stranded RNA molecules, often the same delivery vector is appropriate for both molecules; however, the timing of delivery may be a concern [21]. Specifically, in order to achieve genome editing, intact gRNA must be present in the cell at the same time as functional Cas9 protein, however the delivered Cas9 mRNA molecule must first be translated into protein in situ (figure 2). If the gRNA and mRNA molecules are co-delivered, the gRNA may begin to degrade by the time the mRNA has been translated. Indeed it has been demonstrated that delaying the delivery of the gRNA by up to 6 hours after the mRNA may enhance editing efficiency [21]. Alternatively, chemical modification of the gRNA molecule itself may enhance its stability after delivery [22]. Some groups have used a hybrid method in which mRNA encoding for the Cas9 protein is co-delivered with a viral genome constitutively expressing the gRNA in order to avoid this timing issue altogether [23].
The third option is to deliver Cas9 in its native protein form. This is often the most difficult delivery format due to the large size and charge of the protein, but recently successful Cas9 protein delivery has been demonstrated in vitro and in vivo [24, 25]. This format offers the most transient expression time but also has the least delay in therapeutic activity, likely due to the fact that transcription and translation are both circumvented [12]. Most frequently, both the Cas9 protein and the gRNA are produced in vitro, and then combined into a ribonucleoprotein complex (RNP) and delivered as a single unit. As with mRNA delivery, because of the presence of the gRNA in the RNP, this method also requires particular care to protect the payload from any possible degradation pathways. Obtaining pure protein is also more difficult than obtaining plasmids or mRNA, which may have drawbacks in terms of the economic viability of this approach. In particular, bacterial production of protein often results in significant endotoxin contamination, which must be removed before the protein can be used. Finally, the sudden introduction of a non-native protein of bacterial origin into a mammal might trigger immunological responses or cause toxicity if dosage levels are not carefully monitored.
Delivery vehicles
Delivery vehicles are carriers which can transport the payload to the desired target. When discussing clinical applications, transport is a particularly crucial topic. After introducing the Cas9 into the body, it must be delivered to the appropriate organ or cell type within the body, travel through the interstitial space to reach the cells of interest, and then be delivered through the cell membrane and into the nucleus, all while avoiding clearance or degradation by the body’s protective mechanisms. The selection of an appropriate vehicle may facilitate delivery at any of these stages. This review will focus predominantly on delivery through the cell membrane and protection from clearance, with some attention to targeting to a particular organ or cell type. Although delivery through the vascular wall and interstitial space is an important component to clinical delivery which may be influenced by the choice of delivery vehicles, it is not a focus of this review. This topic has been discussed in other works [26].
Viral Delivery
In the laboratory environment, one of the most popular methods for achieving Cas9 expression in cell culture has been the utilization of viruses. Viruses have evolved to become extremely efficient at invading cells and inducing the expression of non-native proteins inside that host. Viral particles have been engineered to remove the rep genes responsible for replication, and replace them with a therapeutic transgene of interest [27]. Thus, the virus is still able to enter a host cell and induce the expression of the transgene, but is no longer able to replicate itself or spread to new cells. The most common viral vector used for Cas9 delivery is the adeno-associated virus (AAV) vector [14, 28, 29]. It has been demonstrated that each AAV serotype demonstrates preferential delivery efficiency to a particular subset of cell types. This allows researchers to chose the optimal serotype to improve targeted delivery to their organ of interest [30]. Although AAV-mediated Cas9 delivery works well in the laboratory, there are concerns with its utilization in clinical settings. There is a risk that the transgene integration into the target genome could inadvertently disrupt the expression of a vital gene. There is also a physical limit to the amount of DNA that can be packaged inside a virus [27].The maximum capacity of an AAV is around 4.7kb. Thus, designing a virus to encode such a large gene as cas9 along with its promoter and gRNA is challenging. One solution to pack gRNA and Cas9 in separate viral vectors. There are also smaller Cas9 orthologs than the commonly used Streptococcus pyogenes Cas9 (SpCas9), such as Staphylococcus aureas Cas9 (SaCas9) [13]. These smaller variants are more amenable to packaging into viral particles.
Non-Viral Delivery
There are a multitude of delivery methods that do not use viral components to transfer Cas9 (whether as DNA, mRNA, or protein) past cellular barriers [11]. These include physical methods to disrupt cellular barriers, chemical modifications that improve the cargo’s delivery to evade the barriers, and physical encapsulation of the cargo within a carrier molecule [31]. Nonviral delivery has a number of significant advantages. The limiting factor of the physical size of the viral particle is avoided completely. There is a drastically reduced risk of the insertional errors that can occur with viral gene delivery. Finally, non-viral delivery vectors may allow the researcher tighter control over the dose, duration, and specificity of delivery. Nonviral delivery approaches can also often be combined in ways to reap the benefits of multiple different delivery paradigms.
Physical Delivery
A strategy for delivery often employed in vitro for Cas9 proteins is to temporarily disrupt the physical barriers impeding the cargo from reaching its intended destination. Such disruption is introduced by exposing cells to mild physical forces to temporarily open pores in the cell membranes. While care must be taken to avoid permanent damage to the membrane, when performed properly these physical disruptions are often quite successful in the laboratory environment.
Electroporation is a technique that exerts a strong electric field onto a cell’s membrane. The field temporarily reorganizes the membrane, making it more porous and non-selective. The increased permeability allows large biomolecules which would otherwise be rejected by the membrane to instead pass through. This process has had success in in vitro applications and cell lines for delivery of DNA, RNA, and even RNPs [29, 32, 33]. While this approach has been demonstrated in vivo in zygotes, it is not suitable for therapeutic use in patients. Even in cell culture, some cell types that normally react to electrical impulses such as muscle and nerve may be negatively impacted by spikes in cell surface voltage that electroporation introduces. The electric field must be finely tuned or else irreversible changes to membrane physiology might occur and threaten the long-term viability of the cell.
Hydrodynamic injection can deliver large macromolecules in vivo by injecting a liquid solution intravenously at extremely high volume and pressure. This sudden increase in volume induces the temporary generation of pores in the vasculature, allowing the large macromolecular payload to reach the target tissue. This technique is commonly paired with other delivery techniques as it excels in distribution of deliverables to in vivo tissues but does not necessarily have a method to bypass cellular membranes itself. Hydrodynamic injections via the tail vein in mice have successfully delivered plasmids carrying Cas9 and gRNA into the heart, lungs, liver, and kidney tissue [34–36]. Presently, this method is restricted to use with small animal models due to the large starting injection volume necessary (~10% body weight of the mouse). As such, it is currently not appropriate for human application, although research ongoing to optimize this technique for larger animal (and human) use.
Physical methods of delivery need not rely on manipulation of forces or fields to disrupt the cellular membrane. Microinjection, for example, uses micron-scale needles to simply pierce the cell membrane and directly inject the cargo. Development of microscopic pipettes has made microinjection a possibility for transferring a wide variety of macromolecules, including CRISPR components, to target cells. This approach has been used for DNA, RNA, or RNP delivery of Cas9, as well as direct delivery of gRNA [38–40]. Microinjections can either deliver the cargo into the cytoplasm, or in some cases into the pronucleus of fertilized eggs [41]. Microinjection offers high efficiency, precise controlled dosage, and a guarantee that the cargo is delivered exactly to the intended site. The downside is that each individual cell of the target tissue must be injected manually and it is highly impractical to perform in a high-throughput manner, let alone in a clinical or therapeutic scenario. Recently, Cas9-mediated repair of point mutations in single-celled human embryos via microinjection has been demonstrated [39].
Chemical Delivery
Although physical approaches are generally very successful in the laboratory setting, they generally do not scale well, and are thus less practical for in vivo delivery or high-throughput applications in the lab. Alternatively, instead of inducing modifications in the target cell, the deliverable itself can be modified. Chemical delivery approaches utilize the formulation of complementary molecules to assist Cas9 to bypass cellular barriers as well as protect it from degradation pathways. There are two primary forms of chemical delivery: direct chemical modification of the deliverable itself, or encapsulation of the deliverable within another chemical entity. Chemical delivery is particularly promising for in vivo applications, as there are a number of ways to use chemical moieties to target delivery to a particular tissue type (table 2).
Table 2.
Delivery Vehicles: Examples of viral, physical, and chemical CRISPR delivery
| Delivery format | Editing target | Model system | Study objective | Reference |
|---|---|---|---|---|
| AAV Virus | DMD23 | Mouse primary satellite cells | Functional gene knockdown | [14] |
| DMD | Mouse in vivo model | Functional gene Knockdown | [28] | |
| DMD | Mouse in vivo model | Functional gene knockdown | [29] | |
| Physical Delivery | Myostatin | Ovine zygotes | Genetic knockout | [38] |
|
RAG1 G6PD NEK1 HBB |
Human 3PN and 2PN zygotes | Correction of genetic mutations | [39] | |
|
Th Uhrf2 Mc3r Mc4r |
Mouse and rat embryos | Heritable gene knockdown | [41] | |
| Chemical Delivery-Lipid encapsulation | GFP | GFP-HEK293 Cell Culture | Functional gene knockdown | [24] |
| HBV PCSK9 |
Mouse in vivo model | Fuctional viral resistance Functional gene knockdown |
[21] | |
| Chemical Delivery-Gold nanoparticle | Mdx | Mouse in vivo model | HDR-mediated repair of genetic mutation | [25] |
| Chemical Delivery-CPP-modification | CCR5 | HEK293 Cell Culture | Improved delivery efficiency | [55] |
| RFP | Ai9 Mouse line primary cells Ai9 Mouse in vivo model | Improved delivery efficiency | [56] |
Delivery by encapsulation
Lipid-based carriers are a popular and efficient delivery system for CRISPR DNA, mRNA, or RNPs. Lipids are naturally occurring amphiphilic organic molecules composed of a hydrophilic head group and a long-chain hydrophobic tail group. In aqueous solution, lipids will spontaneously form nanoparticles in order to protect the hydrophobic tails from the solvent. By simple mixing, a payload may be encapsulated within a lipid nanoparticle or complexed with the lipid nanoparticle. Lipid-based drug carriers have been FDA-approved for small molecule delivery since the 1980s [42]. The commercial lipid transfection reagent Lipofectamine is a well-established delivery vehicle for nucleic acids and gene editing proteins [20, 43]. A combinatorial library of synthetic cationic lipids, which can effectively deliver a variety of payloads in vitro and in vivo, including siRNA, anticancer protein drugs, and Cas proteins, has also been developed [24, 44–47]. These cationic lipids are particularly effective at forming a complex with negatively charged payloads such as DNA or mRNA. In the case of Cas9 RNPs, the negatively charged gRNA molecule results in a net negative charge, which facilitates packaging in cationic lipids. The synthetic lipids also contain a bioreducible disulfide bond within the hydrophobic tail chain, which can be cleaved intracellularly to facilitate payload delivery and reduce lipid toxicity.
Encapsulation or complexation with a lipid carrier can protect the payload from enzymatic degradation or immunological response. This protection can be enhanced by modifying the nanoparticles with long-chain PEG molecules [48]. Lipid nanoparticles may also be modified with active targeting moieties to improve tissue-specific nanoparticle delivery. Formulation of nanoparticles with coexcipient helper molecules such as cholesterol and DOPE can tune the fluidity of the liposome and alter the pharmacokinetic distribution and uptake efficiency of nanoparticles in vivo [49, 50].
Encapsulating the Cas9 protein, DNA, or RNA within polymeric carriers is another popular approach due to the wide array of polymers available for use in biological transfection. Polymeric carriers may be synthesized from natural monomers, often sugars such as chitosan, or from synthetic chemical monomers, often poly-caprolactone (PCL) or poly-lactic/glycolic acid copolymers (PLGA) [51]. Like lipid encapsulation, polymeric encapsulation can protect and conceal the cargo from degradation pathways in vivo, and may also be functionalized with active targeting moieties that specifically bind to surface receptors of the target cell type, improving specificity of delivery. Polymeric nanoparticles may result in high toxicity, especially in the case of synthetic monomers, which can reach high concentrations upon degradation. While polymeric carriers are common for the delivery of various other proteins, very few examples of polymeric Cas9 delivery can be found in the literature to date.
Recently the use of nucleic acids themselves as a polymeric substrate for Cas9 RNP delivery has also been explored [52]. This approach should have very low toxicity, but it may need to be combined with other approaches such as polymeric coating to successfully protect the nanoparticle from degradation and facilitate cell entry.
Macromolecules can also be delivered via complexation with nanoparticles composed of inorganic materials such as gold, silicia, and carbon nanotubes. Unlike lipids or polymer carriers, nanoparticles are extremely colloidally stable, minimizing risk of early clearance. Formulation control of size, composition, and distribution is also precise and easier to handle compared to other carriers. The use of gold nanoparticles for Cas9 RNP delivery is becoming increasingly popular, and may be a promising approach for in vivo RNP delivery in the future [53]. A recent study showed that gold nanoparticle-mediated Cas9 RNP delivery was successfully able to initiate both gene editing and HDR in vivo, one of the first demonstrated instances of successful non-viral mediated HDR [25].
Delivery by Modification
Proteins and nucleic acids can be directly chemically modified to achieve improved delivery without the need for encapsulation. One common approach for this is the use of cell-penetrating peptides (CPPs) [54]. CPPs are short peptide sequences that can cross the cell membrane either through direct penetration, translocation mediated by endocytosis, or formation of micelles. Although the exact mechanisms are not fully understood, it is believed that the composition of CPPs of either highly abundant positively charged amino acids or alternating polar and nonpolar amino acids is responsible for their activity. Conjugating Cas9 protein and gRNA with CPPs was demonstrated as a means to enhance protein delivery [55, 56]. Although conjugation of Cas9 proteins with CPP may solve the problem of transport across the cell membrane, it does not provide protection from protease degradation that encapsulation can achieve. Additionally CPPs do not afford cell-type specific targeting. Thus, the successful use of CPPs may require they be combined with encapsulation or other delivery techniques.
Nuclear localization sequences (NLSs) are naturally occurring sequences that tag proteins that are synthesized in the cytoplasm for transport into the nucleus. NLSs are made up of poly-arginine or poly-lysine chains conjugated to the protein surface and stimulate nuclear import signals to allow passage through the nuclear membrane. Due to the requirement that Cas9 reach the nucleus, all Cas9 applications include an NLS. Proteins may be synthesized to contain an NLS prior to delivery, or an NLS may be encoded directly into the DNA or mRNA Cas9 construct.
Delivery vehicles and editing specificity
Editing specificity is a perpetual concern regarding the potential translational application of CRIPSR technology [57]. When it comes to in vivo editing specificity, there are two separate concerns. The first is genome target specificity. Once the Cas9 has reached the cell nucleus, it should only edit the gene proscribed by the gRNA. However, stochastically there is a risk of off-target editing an unintended gene. There are web tools available to predict the potential off-target editing locations and severity (for example, http://www.rgenome.net/cas-offinder/). To some extent, the enzyme specificity is a property of the enzyme itself—for example, an advantage of Cpf1 is improved editing specificity [58]. However, as discussed, the delivery format has been shown to improve the editing specificity [12]. This is believed to largely be a function of bioavailability—persistently expressed formats such as virus or plasmid DNA simply have more opportunity for off-target editing than transient formats such as protein delivery [59].
Delivery specificity is a separate concern. Biology is fraught with molecules which can serve entirely different purposes depending on the context or location. It is easy to imagine a therapeutic intervention which would require CRISPR activity in the liver but not in the kidneys, for example. Targeted drug delivery is not a new field, but has yet to be thoroughly applied to CRISPR research. To date, targeted CRISPR delivery has been limited to direct local injections, for example delivering gold-complexed Cas9 nanoparticles directly into the muscle in order to achieve muscle-specific editing to treat Duchenne muscular dystrophy [25]. However, drug delivery in other fields have already demonstrated a wide range of targeted delivery approaches which could be applied to CRISPR delivery. These can range from passive size-based approaches which leverage native uptake and clearance mechanisms [60] or the tumor-specific EPR effects [61], to the inclusion of a wide range of active targeting ligands to direct drug delivery to a particular organ [62, 63]. Any of the chemical delivery methods described above may be particularly amenable to these active approaches, as the chemical moieties present may be modified with active targeting ligands. The application of these techniques to CRISPR warrants further study.
Concluding Remarks and Future Perspectives
We have described many of the current approaches for delivering Cas9 and gRNA, but the CRISPR field is relatively nascent and new advances in Cas9 delivery are constantly being developed. Each form of delivery discussed here has weaknesses that must be considered before use. One of the most significant concerns with any gene editing application is the risk of off-target editing events, so delivery specificity is a pervasive concern with Cas9 delivery [57]. The CRISPR technology field is also still quickly evolving, for example, single-base editing tools based on a modified dCas9 fused to a cytidine deaminase have recently been reported [64]. Delivery approaches will need to adapt to accommodate these new and emerging technologies (see Outstanding Questions).
Outstanding Questions.
Is there a single, unified method that can be used for delivery of gRNA and Cas9 regardless of the target cell type and environment?
Are any of the currently available delivery methods reliable enough for therapeutic use of Cas9 genome editing in humans?
Aside from gene correction, are there other applications for CRISPR-mediated gene editing in vivo?
How will CRISPR gene editing be implemented as a human therapeutic?
Recently, the first human Cas9-based clinical trial commenced (clinicaltrials.gov). This study is an ex vivo CRISPR anticancer therapeutic—the patient’s T cells are isolated and edited with Cas9 in the lab, and then the engineered cells are re-introduced into the patient. In mice, an ex vivo approach has been used as a proof-of-concept approach for a therapeutic treatment of Duchenne’s muscular dystrophy [65]. With recent advances in induced pluripotent stem (iPS) cells, the concept of patient-customized therapeutics generated from ex vivo CRISPR editing of autologous iPS cells is becoming increasingly popular [66]. Ex vivo modification of autologous hematopoietic stem cells for the purpose of treating β-hemoglobinopathies is also being explored [67]. It may also become possible to adapt CRISPR technology to help combat viral and other infectious diseases in humans. Recently progress has been made towards using Cas9 to confer immunity against HIV-1 infection in human iPS cells, demonstrating that this may be an interesting and promising application of CRISPR technology [68]. In addition, Cas9 was recently used to disrupt the Hepatitis B genome in a mouse model, and was successfully able to reduce the detectable viral load [21]. In addition to direct therapeutic applications, the use of CRISPR-Cas-based tools for the generation of novel cell line models of diseases may be possible at an extremely rapid pace, driving forward the state of basic research. However this will require highly efficient delivery of these tools in a laboratory setting. Thus, advances in delivery methods are crucial for both clinical applications and laboratory research.
The power of CRISPR-Cas technology depends entirely on delivery of the Cas enzyme and guide RNA into the target cells. Care must be taken to tailor the delivery approach for the particular objectives at hand. This becomes even more important as more intricate CRISPR applications such as HDR or base editing become prominent, as these approaches require co-delivery of multiple functional entities simultaneously. Discovery and implementation of new delivery vectors and materials that can achieve continually higher efficiency, higher specificity, and lower toxicity will improve the therapeutic outcomes of CRISPR delivery.
Trends Box.
CRISPR is a novel gene editing tool that has the potential for multiple in vivo applications
Cas9 can target virtually any gene through complementarity to a synthetically produced gRNA
A major obstacle to in vivo implementation of CRISPR-mediated genome editing is an efficient, targeted delivery vehicle
Cas9 may be delivered to cells in DNA, mRNA, or protein format, and each mode has unique strengths, weaknesses, and delivery requirements.
A variety of physical and chemical delivery vectors are available for Cas9 delivery
Box 1. How CRISPR-Cas9 functions.
CRISPR-Cas9 (Figure I) functions as a pairing between a nuclease (green) and a guide RNA (gRNA) molecule (purple/red). The gRNA contains a constant structural region (red) which interacts with the Cas9 protein, and a variable targeting region (purple) which identifies a complementary region on the target DNA molecule (blue). The gRNA brings the nuclease into close proximity with the target DNA at the specified target site, where the nuclease introduces a double-stranded break (DSB). The power of CRISPR-Cas9 lies in that the Cas9 can be easily engineered to specifically target any site in the genome, just by modifying the sequence of the gRNA. The Cas9 protein itself need not be changed; by simply providing a new gRNA the same protein can now cleave a new target site. This is in stark contrast to the previous ZF and TALE nuclease technologies, where the nuclease protein itself needed to be engineered for each new target site.
Figure I.
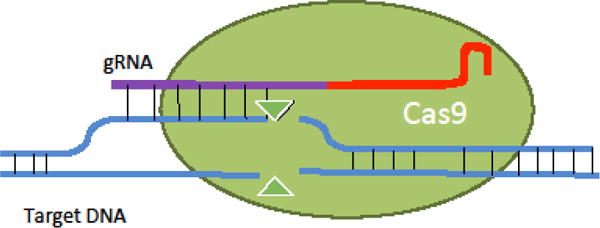
Schematic of Cas9 function.
Box 2. Cpf1: An Alternative CRISPR Nuclease.
CRISPR-Cpf1 (Figure I) functions on the same principle as Cas9. Like Cas9, Cpf1 is a nuclease paired with a gRNA composed of a variable targeting region and a constant structural region. The Cpf1 gRNA is notably shorter than the Cas9 gRNA (~45 nucleotides for Cpf1 vs ~110 nucleotides for Cas9). This length may make in vitro synthesis of gRNA cheaper and easier, or it may facilitate encoding into the limited genomic space of an AAV viral genome. Another difference between Cas9 and Cpf1 lies in the PAM sequence. This is a short sequence in the target genome which facilitates the nuclease binding to the target sequence. A common drawback of CRISPR editing is that it requires a PAM sequence to be found in the gene of interest, often in the region particularly coding for the active site of the target protein. Occasionally, there are no PAM sites conveniently located in the target gene. Cpf1 uses a different PAM than Cas9. This opens up the sample space of target genes—in case the Cas9 PAM is not available in a particular gene, the researcher could instead look for a Cpf1 PAM. Overall, this increases the flexibility of CRISPR as a whole. Research is currently ongoing to engineer both the Cas9 and Cpf1 proteins to allow for even more PAM sites. Finally, while Cas9 generates a blunt-end cut, Cpf1 generates an overhanging cut. There is some evidence that this overhanging cut may facilitate NHEJ-mediated knock-in of target genes; however more research into this idea is necessary.
Figure I.
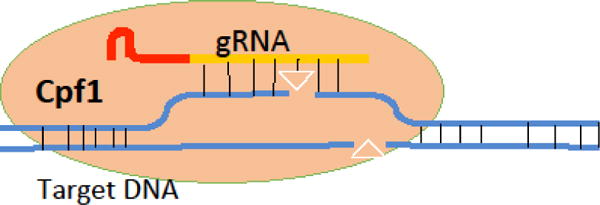
Schematic of Cpf1 function.
Acknowledgments
This work was supported by National Science Foundation Grant DMR 1452122 and NIH (1R21EB024041-01). The authors also acknowledge Dr. Rhiannon Macrae and Dr. Feng Zhang’s help in preparing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 2.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 3.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 6.Wang HX, et al. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem Rev. 2017;117(15):9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sternberg SH, Doudna JA. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell. 2015;58(4):568–74. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 9.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wujin S, Zhen G. Tailoring non-viral delivery vehicles for transporting genome-editing tools. SCIENCE CHINA Materials. 2017;60(6):511. [Google Scholar]
- 12.Liang X, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 13.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–11. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes TS, et al. Immunogenicity of intrathecal plasmid gene delivery: cytokine release and effects on transgene expression. J Gene Med. 2009;11(9):782–90. doi: 10.1002/jgm.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pardi N, et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release. 2015;217:345–51. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol Ther. 2013;21(2):358–67. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams DJ, et al. A Simple, Highly Efficient Method for Heterologous Expression in Mammalian Primary Neurons Using Cationic Lipid-mediated mRNA Transfection. Front Neurosci. 2010;4:181. doi: 10.3389/fnins.2010.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zohra FT, et al. Effective delivery with enhanced translational activity synergistically accelerates mRNA-based transfection. Biochem Biophys Res Commun. 2007;358(1):373–8. doi: 10.1016/j.bbrc.2007.04.059. [DOI] [PubMed] [Google Scholar]
- 20.Li B, et al. Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat Biomed Eng. 2017;1(5) doi: 10.1038/s41551-017-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang C, et al. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27(3):440–443. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin H, et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol. 2017 doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–33. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci U S A. 2016;113(11):2868–73. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K, et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nature Biomedical Engineering. 2017;1(11):889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Yuan F. Delivery of viral vectors to tumor cells: extracellular transport, systemic distribution, and strategies for improvement. Ann Biomed Eng. 2006;34(1):114–27. doi: 10.1007/s10439-005-9007-2. [DOI] [PubMed] [Google Scholar]
- 27.Naso MF, et al. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs. 2017 doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–3. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–7. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zincarelli C, et al. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16(6):1073–80. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 31.Wang M, et al. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017;24(3):144–150. doi: 10.1038/gt.2016.72. [DOI] [PubMed] [Google Scholar]
- 32.Kim S, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–9. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashimoto M, Takemoto T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. 2015;5:11315. doi: 10.1038/srep11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–3. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonamassa B, et al. Hydrodynamic gene delivery and its applications in pharmaceutical research. Pharm Res. 2011;28(4):694–701. doi: 10.1007/s11095-010-0338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamimura K, et al. Image-Guided Hydrodynamic Gene Delivery: Current Status and Future Directions. Pharmaceutics. 2015;7(3):213–23. doi: 10.3390/pharmaceutics7030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Astolfo DS, et al. Efficient intracellular delivery of native proteins. Cell. 2015;161(3):674–90. doi: 10.1016/j.cell.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 38.Crispo M, et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. PLoS One. 2015;10(8):e0136690. doi: 10.1371/journal.pone.0136690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang L, et al. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol Genet Genomics. 2017;292(3):525–533. doi: 10.1007/s00438-017-1299-z. [DOI] [PubMed] [Google Scholar]
- 40.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Meth. 2013;10(8):741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotech. 2013;31(8):681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 42.Allen TM, Cullis PR. Liposomal drug delivery systems: From concept to clinical applications. Advanced Drug Delivery Reviews. 2013;65(1):36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 43.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altinoglu S, et al. Combinatorial library strategies for synthesis of cationic lipid-like nanoparticles and their potential medical applications. Nanomedicine (Lond) 2015;10(4):643–57. doi: 10.2217/nnm.14.192. [DOI] [PubMed] [Google Scholar]
- 45.Altinoglu SA, et al. Intracellular delivery of the PTEN protein using cationic lipidoids for cancer therapy. Biomater Sci. 2016;4(12):1773–1780. doi: 10.1039/c6bm00580b. [DOI] [PubMed] [Google Scholar]
- 46.Wang M, et al. Reactive oxygen species-responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew Chem Int Ed Engl. 2014;53(49):13444–8. doi: 10.1002/anie.201407234. [DOI] [PubMed] [Google Scholar]
- 47.Wang M, et al. Enhanced intracellular siRNA delivery using bioreducible lipid-like nanoparticles. Adv Healthc Mater. 2014;3(9):1398–403. doi: 10.1002/adhm.201400039. [DOI] [PubMed] [Google Scholar]
- 48.Jokerst JV, et al. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond) 2011;6(4):715–28. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onuki Y, et al. Membrane Microdomain Structures of Liposomes and Their Contribution to the Cellular Uptake Efficiency into HeLa Cells. Mol Pharm. 2016;13(2):369–78. doi: 10.1021/acs.molpharmaceut.5b00601. [DOI] [PubMed] [Google Scholar]
- 50.Hattori Y, et al. The role of dioleoylphosphatidylethanolamine (DOPE) in targeted gene delivery with mannosylated cationic liposomes via intravenous route. J Control Release. 2005;108(2–3):484–95. doi: 10.1016/j.jconrel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Tong R, et al. Nanopolymeric Therapeutics. MRS Bulletin. 2011;34(6):422–431. [Google Scholar]
- 52.Sun W, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54(41):12029–33. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mout R, et al. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11(3):2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Z, et al. Cell-penetrating peptides: Possible transduction mechanisms and therapeutic applications. Biomedical Reports. 2016;4(5):528–534. doi: 10.3892/br.2016.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramakrishna S, et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24(6):1020–7. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staahl BT, et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol. 2017;35(5):431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17(5):300–12. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34(8):863–8. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 59.Glass Z, et al. Nanoparticles for CRISPR–Cas9 delivery. Nature Biomedical Engineering. 2017;1(11):854. doi: 10.1038/s41551-017-0158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Litzinger DC, et al. Effect of liposome size on the circulation time and intraorgan distribution of amphipathic poly(ethylene glycol)-containing liposomes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1994;1190(1):99–107. doi: 10.1016/0005-2736(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 61.Prabhakar U, et al. Challenges and key considerations of the enhanced permeability and retention (EPR) effect for nanomedicine drug delivery in oncology. Cancer research. 2013;73(8):2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bazak R, et al. Cancer active targeting by nanoparticles: a comprehensive review of literature. Journal of Cancer Research and Clinical Oncology. 2015;141(5):769–784. doi: 10.1007/s00432-014-1767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maruyama K, et al. Targetability of novel immunoliposomes modified with amphipathic poly(ethylene glycol) s conjugated at their distal terminals to monoclonal antibodies. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1995;1234(1):74–80. doi: 10.1016/0005-2736(94)00263-o. [DOI] [PubMed] [Google Scholar]
- 64.Komor AC, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ousterout DG, et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Translational Research. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 67.Lattanzi AM, Vasco, Pavani Giulia, Amor Fatima, Antoniani Chiara, Lee Ciaran, Porteus Matthew, Bao Gang, Amendola Mario, Mavilio Fulvio, Miccio Annarita. Viral and non-viral delivery of the CRISPR-Cas9 system in human hematopoetic stem and progenitor cells. Mol Ther. 2017;25(5):299–299. [Google Scholar]
- 68.Liao HK, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]


