Abstract
Specificity of the ubiquitin proteasome system is controlled by ubiquitin E3 ligases, including their major representatives, the multi-subunit cullin-RING ubiquitin ligases (CRLs). More than 200 different CRLs are divided into seven families according to their cullin scaffolding proteins (CUL1-7) around which they are assembled. Research over two decades has revealed that different CRL families are specialized to fulfil specific cellular functions. Whereas many CUL1-based CRLs (CRL1s) ubiquitylate cell cycle regulators, CRL4 complexes often associate with chromatin to control DNA metabolism. Based on studies about differentiation programs of mesenchymal stem cells (MSCs), including myogenesis, neurogenesis, chondrogenesis, osteogenesis and adipogenesis, we propose that CRL3 complexes evolved to fulfill a pivotal role in mammalian cell differentiation.
Keywords: Cullin 3, cullin-RING-ubiquitin ligases, BTB proteins, mesenchymal stem cells, differentiation, cytoskeleton
Cullin-RING ubiquitin ligases: Background
Cullin–RING ubiquitin (Ub) ligases (CRLs) comprise the largest family of ubiquitin E3 ligases in eukaryotic cells [1]. The ~240 different E3 enzyme complexes belonging to the CRL family control a broad array of critical biological processes [2]. CRLs consist of seven cullins designated CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5 and CUL7, which serve as scaffolds. The C-terminal part of cullins tightly associates with the RING-H2-domain proteins RBX1 or RBX2, which catalyze the transfer of Ub from the Ub-conjugating E2 to the substrate. Through N-terminal domains, cullins interact with their specific set of substrate adaptor/receptor proteins [1]. F-box proteins (FBP) function as SRs for CUL1 complexes (CRL1), VHL-box proteins for CRL2s, Bric-a-brac, Tramtrack and Broad Complex/Pox virus and Zinc finger (BTB/POZ) proteins for CRL3s [3], DXR proteins for CRL4s, and SOCS-box proteins for CRL5s [1].
Two decades after their discovery, it now appears that different CRL families evolved to fulfil specialized cellular functions. For example, CRL1 complexes control cell proliferation and cell cycle progression by targeting a wide array of cell cycle regulators [4]. CRL4 ligases, on the other hand, are widely associated with chromatin [5] and play crucial roles in DNA repair, checkpoint control and chromatin remodeling [6]. CRL3 complexes play key roles in cancer [7]. In addition, in this review, we are summarizing evidence that suggests the emerging concept that CRL3 complexes are involved in controlling mammalian cell differentiation. We will focus on multilineage differentiation of mesenchymal stem cells (MSCs).
Structure and assembly of CUL3-based CRLs
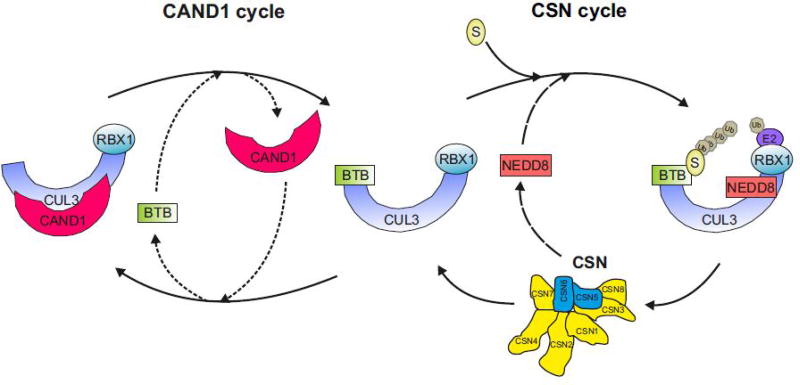
CUL3 interacts with BTB proteins, encoded by ~180 genes in the human genome [8], which act as both substrate adaptors and receptors in a single protein [3]. The BTB domain mediates CUL3 binding and an adjacent protein-interaction domain recruits substrates for ubiquitylation [8, 9]. However, it is not entirely clear how many BTB proteins actually engage CUL3 and form an ubiquitin ligase in vivo. The BTB protein family can be subdivided into five classes that include proteins with the BTB domain alone, the BTB domain with one or more zinc-fingers (BTB-ZF), the BTB domain with one or more Kelch repeats (BTB-Kelch), the BTB domain with the Pipsqueak domain (BTB-PSQ) and the BTB with another functional domain [10]. BTB proteins are involved in diverse functions including transcriptional regulation, chromatin remodeling as well as proteolysis. Specificity of function is determined by additional domains present in BTB proteins [10]. Whereas BTB-ZF proteins possessing DNA-binding zink-finger are mostly involved in transcriptional regulation and chromatin remodeling, the Kelch domain is suitable for substrate binding by forming a β-propeller structure in CRL3BTB-Kelch complexes. With few exceptions the BTB-Kelch proteins and the MATH-BTB subclass encoded by two human genes, are involved in CUL3-dependent ubiquitylation [7, 8, 10, 11]. This includes the BTB-Kelch-like proteins KEAP1 (KLHL19), an important regulator of oxidative stress defense [12], and KLHL3, a regulator of hypertension [13]. Most interestingly, there are BTB-ZF transcription factors, PLZF and BCL6, which interact with CUL3 to recruit it to the nucleus to alter the ubiquitylation pattern of chromatin for the differentiation of several B and T cell effector programs [14]. Moreover, the BTB-ZF protein BAZF acts as SR in CUL3 complex to polyubiquitylate C-promotor binding factor 1 (CBF1) for subsequent 26S proteasomal degradation, which downregulates Notch signaling [15]. RHOBTB proteins belong to yet another subclass of BTB proteins that interact with CUL3 to ubiquitylate substrates involved in vesicle trafficking or to autoubiquitylate [16]. MUF1 is a prominent substrate of RHOBTB-dependent CRL3 ubiquitylation [17], which seems to be controlled by an autoregulatory mechanism [18]. The BTB adaptors/receptors are unique among CRLs: By dimerizing, BTB proteins are able to recruit two CUL3 subunits into CRL3 complexes [9, 19, 20]. Most interestingly, CRL3s promote mono- and poly-ubiquitylation as well as ubiquitylation of non-lysine residues (see below). The different outcomes in target modification can be explained by the recruitment of specific E2s [1, 21, 22]. In addition, there is a cooperation between CRL3 and another RBR-type E3, ARIH1, which acts as a mono-ubiquitylating enzyme for the neddylated CUL3KLHL12 substrate SEC13-SEC31A [23]. Like other CRLs, CRL3s are activated by the covalent modification of cullins at a conserved lysine with the Ub-like protein NEDD8 (or Rub1 in S. cerevisiae) [24]. NEDD8 conjugation greatly stimulates CRL activity by multiple mechanisms. First, it facilitates the recruitment of ubiquitin E2s to cullins, thereby promoting their E3 activity. Second, it induces a conformational change that causes CRLs to switch from an inactive to an active conformation, a step that appears to reduce the distance between the E2 active site and the substrate target lysine [25, 26]. Neddylation is reversed by the COP9 signalosome (CSN) [27], a key regulator of CRLs, which consists of six distinct PCI (proteasome lid-CSN- initiation factor 3) proteins (CSN1–CSN4, CSN7 and CSN8) and two MPN (MPR1/PAD1 amino-terminal) domain-containing proteins (CSN5 and CSN6) of which one subunit, CSN5, is catalytic [27]. Recently, architectural and regulatory principles probably common to all CRL family members have been revealed for CRL1-CSN [28] and CRL4A-CSN complexes [29]. According to this model, the CSN is subject to autoinhibition, which is only relieved upon recruitment of a neddylated CRL [30]. Conformational changes of CSN2, CSN4 and CSN7 provide an induced fit mechanism that propagates the binding of neddylated CRL to the CSN5–CSN6 dimer and activates CSN5 [29]. The presence of CRL substrates prevents CSN-mediated cullin deneddylation, most likely due to steric hindrance [29].
The estimated total amount of all cullins in human cells is approximately 2.2 µM [28] and about 1 µM is represented by CUL3 [31]. The total amount of neddylated cullins in human cells comprises approximately 1 µM. Since the amount of CSN is about 0.5 µM [32], near saturation of the cellular CSN with neddylated cullin can be predicted. Thus, based on the estimated CRL and CSN dynamics, a highly adaptable CRL network results, that can be re-organized in response to substrate availability on a time scale of minutes [28]. CSN-mediated deneddylation of CRLs has two important regulatory consequences. First, it prevents autoubiquitylation of CRLs [33] and secondly, it is a prerequisite for the binding of Cullin-Associated and Neddylation-Dissociated 1 (CAND1) [34]. CAND1 has a pivotal role in the rapid adaptation of the CRL network to fluctuations in substrate availability [35–37]. It augments the dissociation of SRs from CRL1 by one million-fold. CAND1 is a SR exchange factor that accelerates the rate at which CUL1-RBX1 equilibrates with SKP1-FBX modules [36]. Depletion of CAND1 altered the cellular landscape of CRL1 complexes [35, 36, 38, 39]. CAND1 is also responsible for the integration of the BTB protein KEAP1 into CRL3 complexes [40]. Thus, since CAND1 binds to all cullins [32], CAND1’s SR exchange activity is presumably universally applicable to other CRLs as well. The assembly of CRL3 complexes is most likely driven by substrate binding as recently demonstrated for SCF (SKP1-Cullin-F-box) complexes [37]. In conclusion, CRL network is organized by the coordinated interplay of closely intertwined cycles of CAND1-mediated SR exchange and CSN-controlled neddylation/deneddylation [33] (Figure 1).
Figure 1. Remodeling of CRL3BTB complexes mediated by CAND1 and CSN.
Upon substrate-induced neddylation, CRL3s are activated to ubiquitylate a specific substrate (CSN cycle). Once substrate is consumed, CSN-mediated cullin deneddylation allows binding of CAND1 (CAND1 cycle). The protein exchange factor activity of CAND1 causes displacement of the BTB protein. The released CRL3 core complex can then reassemble with another BTB protein presumably driven by cognate substrate.
CRL3s in differentiation
Myogenesis
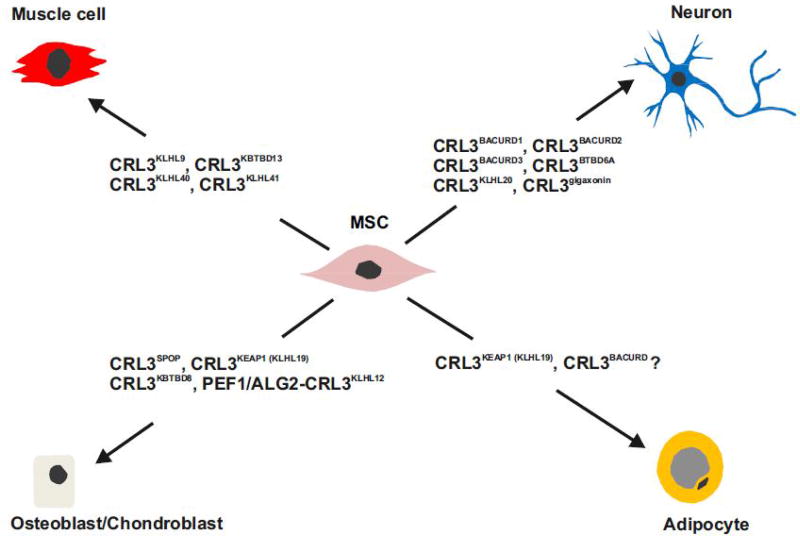
Cul3 null mice exhibit lethality by embryonic day 7.5 indicating broad and essential functions during development [41]. As shown through the use of the NEDD8 E1 inhibitor MLN4924, cullin activity is indispensable for terminal muscle cell differentiation [42]. In association with Kelch proteins, CUL3 complexes are involved in muscle cell differentiation (Figure 2, Key Figure). In particular, mutation in BTB-BACK-Kelch-like protein KLHL9 results in distal myopathy [43]. KLHL9 interacts with CUL3 and targets Aurora B kinase for ubiquitylation and subsequent degradation [44]. Since Aurora B kinase is a critical regulator of the assembly and disassembly of type III intermediate filaments, including vimentin and desmin, KLHL9 deficiency impairs skeletal muscle function [11]. Moreover, the BTB-Kelch protein KBTBD13 forms a complex with CUL3. Mutations in KBTBD13 are associated with a new type of nemaline myopathy [45]. Similarly, mutations in KLHL40 as well as KLHL41 result in an autosomal recessive form of nemaline myopathy [11]. Knockdown of KLHL41 in C2C12 myoblasts inhibited myotube formation [46]. Although CRL3KLHL40 and CRL3KLHL41 complexes were identified in muscle cells as regulators of differentiation, their exact substrates remain unknown. It was suggested that KLHL41, KLHL40, KBTBD13, and KLHL9 might regulate the stability of nebulin, actin, and other important skeletal muscle proteins that are required for normal differentiation and functioning of skeletal muscle cells [11]. Recently, Gong et al. demonstrated that KBTBD5 (KLHL40) promotes the ubiquitylation and degradation of DP1 [47]. The transcription factor E2F1 forms heterodimers with DP1 and the E2F1-DP1 complex regulates the expression of essential cell division genes involved in G1-S transition [48]. By targeting DP1 for degradation, CRL3KBTBD5 inhibits the transcription driven by E2F1-DP1, a prerequisite for skeletal muscle myogenesis [47].
Figure 2. Multilineage differentiation programs of mesenchymal stem cells (MSCs) require specific CUL3-based CRLs.
Multipotent MSCs derive from numerous vascularized tissue sources, including bone marrow, adipose and skeletal muscle. MSCs function as precursors to a variety of mature mesenchymal cell types like muscle, neuron, osteoblast and adipocyte. The differentiation of MSCs occurs in different phases. For example, adipogenesis, the differentiation of adipocytes, is characterized by two phases: the determination phase and the terminal differentiation phase. In the determination phase MSCs commit to the adipocyte lineage. Preadipocytes are morphologically not yet distinguishable from their MSC precursors. During terminal differentiation, adipocytes differentiate from preadipocytes producing lipid droplets and adipocyte-specific proteins. Whereas CRL3BACURDs are needed for neuron as well as adipocyte differentiation and CRL3KEAP1 is necessary for adipocyte and osteoblast differentiation, most so far identified CRL3s involved in multileanage differentiation programs are specific.
Neurogenesis
During development, neurons undergo directional cell migration to specific positions in the embryonic cerebral cortex and subsequently establish dendritic spine connections. Members of the family of RHO GTPases, including RND2 and RND3, play critical roles in the generation of neurons [49, 50]. In addition, it is now recognized that BTB-BACURD proteins such as BACURD1/KCTD13 are involved in neurogenesis [51] (see Figure 2). BTB-containing adaptor for CUL3-mediated RHOA degradation (BACURD) and CRL3BACURD complexes were originally discovered in Drosophila [52] where they mediate the degradation of RHOA, which controls actin cytoskeleton structure and cell movement [52, 53]. Actin cytoskeleton dynamics depending on RHO GTPases determine cell morphology, establish cell polarity, and provide driving forces for cell migration [54]. RHOA activation leads to assembly of the contractile actin and myosin filaments structure called stress fibers. Cells lacking CUL3 show impaired ubiquitylation and degradation of RHOA and hence exhibit accumulation of actin stress fibers. Interestingly, BACURDs have a clear preference for RHOA·GDP over RHOA·GTP [52].
All three human BACURDs bind members of the RHO family and CUL3. BACURD1 and BACURD2 recruit RND proteins, RND2 and RND3, to CRL3 complexes, which is essential for the development of cortical neurons [55]. BACURD2 was identified as an interaction partner of RND2, which promotes actin dynamics, neurite outgrowth and radial migration within the developing cerebral cortex [56]. Thus, CRL3BACURDs coordinate early steps of cortical neurogenesis by reorganizing the actin cytoskeleton via RND2 and RND3 [56, 57]. Two important BTB proteins in neurogenesis are the BTB-Kelch proteins gigaxonin and KLHL20, which are also related to cytoskeleton remodeling. Mutations of gigaxonin cause the human neurodegenerative disorder giant axonal neuropathy, which is associated with massive disorganization of the intermediate filaments [58]. Interestingly, CRL3KLHL20 ubiquitylates the PDZ-RHO guanine nucleotide exchange factor (RHOGEF) targeting it for degradation and thereby facilitating neurite outgrowth [59].
In addition, BTBD6A, another CRL3s substrate receptor, binds the transcriptional repressor promyelocytic leukemia zinc finger (PLZF, also known as ZBTB16, ZNF145). BTBD6A promotes the relocation of PLZF from the nucleus to the cytoplasm and targets PLZF for ubiquitylation and degradation [60]. PLZF is a negative regulator of differentiation, and degradation via CRL3BTBD6A initiates neurogenesis in zebrafish [60]. Interestingly, the highly conserved PLZF protein, which is itself a BTB domain protein, is a multifunctional transcription factor involved in a number of differentiation processes [61].
Chondrogenesis and Osteogenesis
The MATH-BTB protein SPOP is necessary for chondroblast hypertrophy and osteoblast differentiation [62] (Figure 2). CRL3SPOP is an ubiquitin ligase that targets GLI family proteins for ubiquitylation and degradation [63]. The GLI2 and GLI3 transcriptional regulators are controlled by Indian Hedgehog (IHH) signaling, which is essential for osteoblast differentiation and bone development. SPOP has a positive function in IHH signaling by downregulating the transcriptional repressor GLI3R [62]. This data provides evidence for essential roles of SPOP-mediated ubiquitylation in chondrocyte and osteoblast differentiation during skeletal development [62]. Moreover, nuclear factor-E2-related factor 2 (NRF2) is an inhibitor of osteoblast differentiation [64]. It binds to the Kelch-like BTB protein KLHL19, also known as KEAP1 (Kelch-like ECH-associated protein 1), forming complexes with CUL3. Under oxidative or electrophilic stress, specific cysteinyl residues of KEAP1 are modified and KEAP1 loses its ability to ubiquitylate NRF2, a transcription factor with oxidative stress protective activity [12]. Interestingly, CRL3KEAP1 mediated destruction of NRF2 is a prerequisite of proper osteoblast and osteoclast differentiation [64].
In complex with the SR KLHL12, CUL3 also mono-ubiquitylates the COPII coat protein SEC31, which accelerates the traffic of collagen from the endoplasmic reticulum thus enhancing chondrogenesis [65]. The latter process is Ca2+-dependent and mediated by target-specific CUL3 co-substrate receptors, PEF1 and ALG2 [66] (see Figure 4B). Moreover, when CUL3 is associated with the SR KBTBD8, it mono-ubiquitylates the nucleolar proteins TCOF1 and NOLC1, which then dimerize to generate a platform for ribosome remodeling [67]. This results in the reprogramming of ribosome translational output such as to promote the differentiation of hESC-derived neural crest cells into chondrocytes.
Figure 4. CRL3BTB-dependent remodeling of the cytoskeleton and vesicle transport adaptation during differentiation.
(A) RHO GTPases change between an active GTP bound state and an inactive GDP bound state, a process influenced by several signaling pathways as indicated by red arrows. Guanine-nucleotide exchange factor (GEF) triggers the release of GDP and the formation of active GTP-RHOA. GTPase activating protein (GAP) activates hydrolysis of GTP producing inactive GDP-RHOA, which is a substrate of CRL3BACURD ubiquitylation and subsequent degradation by the 26S proteasome. RHO-associated coiled-coil kinase (ROCK) is activated by GTP-RHOA leading to an increase of myosin light chain (MLC) phosphorylation (MLC-P). This induces actomyosin-based contractility. Furthermore, ROCK directly activates LIM kinase (LIMK), which leads to filament stabilization via cofilin phosphorylation (Cofilin-P). Degradation of RHOA via CRL3BACURD results in actin depolymerization and destabilization (red).
(B) Differentiation dependent secretion of collagen by chondrocytes is mediated by CRL3KLHL12 and its Ca2+-binding coregulators, PEF1 and ALG2. During differentiation, chondrocytes secrete extracellular matrix type II and type X collagen. This requires CRL3KLHL12 dependent mono-ubiquitylation of SEC31. In this process, CRL3KLHL12 utilizes two Ca2+-binding co-substrate receptors, PEF1 and ALG2. SEC1 mono-ubiquitylation triggers the formation of large COPII coats and collagen secretion. By this mechanism, Ca2+ from the endoplasmatic reticulum stimulates chondrocyte differentiation via CRL3KLHL12 and its coregulators, PEF1 and ALG2.
Adipogenesis
White adipocytes are derived from mesenchymal stem cells (MSCs) (see Figure 2) in a two-step differentiation program resulting in the formation of mature adipocytes. Adipogenesis coincides with considerable transcriptional reprogramming and morphological cytoskeleton remodeling. It is induced by hormones such as insulin and maintained by the interplay of critical transcription factors, including peroxisome proliferator-activated receptor γ (PPAR-γ), CCAAT/enhancer binding protein α (C/EBPα) and β (C/EBPβ) as well as their antagonist CCAAT-enhancer binding homologous protein (CHOP) [68]. CUL3 ligases are essential for initiating and completing the differentiation program. During adipogenesis of human LiSa-2 preadipocytes, the CRL regulators CAND1 and CSN increase ~2-fold, allowing a rapid remodeling of CRL1 and CRL3 complexes [39, 40]. While the F-box protein SKP2 is released from CRL1 complexes and p27 accumulates [39], CUL3 becomes almost completely neddylated. An initial adipogenesis-dependent remodeling of CRL3s includes the CAND1-mediated incorporation of KEAP1 into CRL3 [40]. CRL3KEAP1 targets CHOP, a negative regulator of adipogenesis, for degradation [69], thereby initiating the process of adipogenesis (Figure 2). CRL3s escape from CSN-mediated deneddylation by binding to membranes of lipid droplets together with RHOA. These data suggest a role for CRL3 in the degradation of RHOA, possibly by recruiting the BTB protein BACURD3 [70]. Again, CRL3-dependent ubiquitylation seems to be involved in the reorganization of the actin cytoskeleton during differentiation.
Mechanisms of CRL3 action in differentiation
From studies over the past years, the common theme has emerged that CRL3s promote cell differentiation through transcriptional/translational reprogramming and cytoskeleton remodeling. While the participation of BTB domain proteins in differentiation processes has been well established [10, 61], there is now sufficient support that many of these BTB proteins function as SRs of CRL3 complexes.
Transcriptional and translational reprogramming
Accumulating evidence confirms that CRL3s impinge on multiple signaling pathways such as VEGF/VEGFR, WNT/β-catenin and RA/RAR, which transcriptionally reprogram MSCs to promote differentiation into myogenic, neurogenic, osteoblastic, chondrogenic, adipogenic, and smooth muscle cell lineages [15, 71, 72] (see Figure 2). Lineage-dependent transcriptional reprogramming of MSCs is induced by specific hormones or cytokines and controlled by master regulators such as peroxisome proliferator-activated receptor γ (PPARγ) and Runt-related transcription factor 2 (Runx2) [71]. In an initial phase, cell lineage differentiation is put into motion by specific CRL3BTB complexes that target negative transcriptional regulators of differentiation for degradation (see Figure 3A). In addition, CUL3-based E3 ligases participate in signaling pathways promoting cell differentiation.
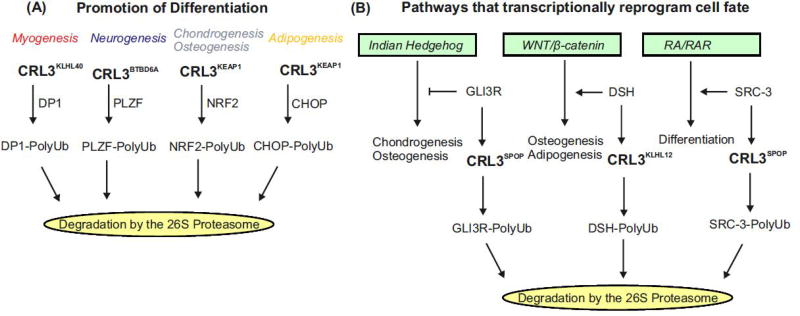
Figure 3. CRL3BTB complexes promote differentiation by targeting negative regulators of differentiation for degradation and by impinging on signaling pathways that trigger differentiation.
(A) Various differentiation programs are initiated by the ubiquitylation and degradation of transcriptions factors that are targeted by the indicated CUL3-based ubiquitin ligases. Myogenic differentiation is initiated by CRL3KLHL40 mediated degradation of DP1, neurogenic differentiation by the CRL3BTBD6A dependent ubiquitylation of PLZF, osteoblast differentiation by CRL3KEAP1 mediated degradation of NRF2 and of adipogenic differentiation by the CRL3KEAP1 ubiquitylation of CHOP and its subsequent degradation by the 26S proteasome.
(B) The commitment and differentiation of MSCs towards a myogenic, neurogenic, osteogenic or adipogenic cell fate depend on a variety of signaling and transcription factor pathways controlled by CUL3-based ubiquitin ligases. Indian Hedgehog (IHH) pathway is essential for bone development. It regulates gene expression necessary for chondrogenesis and osteogenesis via the GLI family of transcription factors. GLI3R is a repressor of IHH signaling, which is ubiquitylated by CRL3SPOP and subsequently degraded by the 26S proteasome. WNT signaling plays an essential role in cell fate determination, proliferation, and differentiation. CRL3KLHL12 mediated degradation of DSH blocks the WNT pathway inhibiting osteogenesis. Retinoic acid (RA) influences cell differentiation, proliferation and apoptosis via expression of specific target genes. The transcription of target genes is a complex process requiring RA, nuclear receptors (RARs) as well as several coregulators such as SRC-3. In response to RA, SRC-3 is degraded in a CRL3SPOP dependent manner.
Indian Hedgehog (IHH) signaling is essential for chondrogenesis and osteogenesis. IHH regulates gene expression in chondrocytes and osteoblasts through the GLI family of transcription factors, which also act as transcription repressors (Figure 3B). The repressor GLI3R is degraded by CRL3SPOP, which promotes IHH signaling required for chondrocyte and osteoblast differentiation [62].
Notch signaling is a conserved cell-fate-determination pathway involved in stem cell differentiation and neurogenesis. The pathway comprises five Notch ligands (Delta-like ligand 1 (DLL1) DLL3 and DLL4, and Jagged1 and Jagged2) and four receptors (Notch1-4) [73]. The BTB protein KCDT10 interacts with CUL3 and with Notch1, which leads to ubiquitylation and subsequent degradation of the Notch1 receptor [74]. There is a crosstalk between Notch and VEGF signaling. Both KCDT10 as well as BAZF [15] downregulate Notch signaling and thereby promoting endothelial cell differentiation and angiogenesis.
The WNT/β-catenin signaling pathway determines cell fate, deciding whether cells proliferate or differentiate. When WNT binds to the extracellular frizzled receptor, the intracellular disheveled (DSH) protein is activated, which blocks the β-catenin destruction complex. Interestingly, the canonical WNT pathway stimulates osteogenesis, but blocks adipogenesis [71]. It has been reported that CRL3KLHL12 inhibits WNT/β-catenin signaling by targeting DSH for degradation by the UPS [75] (Figure 3B).
The dopamine D4 receptor (D4R), which belongs to the superfamily of G protein-coupled receptors, is a positive regulator of osteogenesis [76] and another ubiquitylation target of CRL3KLHL12 [77]. However, this ubiquitylation does not destine D4R for degradation. In addition, CRL3KLHL12 ubiquitylates D4R on non-lysine residues, although the function of this modification remains unknown [77].
Retinoic acid (RA) controls differentiation through modulation of target gene expression, a highly coordinated process, which requires RA nuclear receptors (RARs) and transcriptional coactivators such as the p160 family member, SRC-3. SRC-3 is essential for RAR target gene expression. It is recruited to target promoters together with RAR but then becomes phosphorylated, evicted from chromatin, and degraded in a CUL3-dependent fashion [78]. Phosphorylated SRC-3 is ubiquitylated by CRL3SPOP and targeted for proteolysis [79]. SRC-3 degradation thus controls the transcriptional output of RAR and other nuclear receptors [78] (Figure 3B). The interplay between nuclear receptor signaling and CUL3-based ligases is further underscored by the finding that the estrogen-related nuclear receptor α promotes the degradation of RHOA by inducing the expression of the SR, BACURD2 [80].
Cytoskeleton remodeling and vesicle transport
Actin filaments and microtubules determine cell shape, are the basis for intracellular transport, and mediate all forms of cell migration. During differentiation, cells switch from motility to a more sessile phenotype, and re-organize intracellular trafficking. Remodeling of the actin cytoskeleton is controlled by members of the RHO family of GTPases [81]. Activated RHO family members, including RHOA, drive the formation of actin filaments known as stress fibers through a signaling cascade involving activation of the RHO-associated coiled-coiled kinases (ROCK), followed by inhibition of the actin depolarization factor cofilin [82] (see Figure 4A). These pathways control differentiation in myofibroblasts via actin and microtubule cytoskeletons [83]. RHOA/ROCK signaling plays a key role in signal transduction and cell differentiation [84]. It has a negative effect on adipogenesis [85]. In neuronal and most likely in adipogenic differentiation, the stability of RHOA is determined by the CUL3-based E3 ligases associated with the SRs, BACURD1-3 [52, 55, 70]. RHOA, RND2 and RND3 are degraded during differentiation in a CRL3-dependent manner, which is an essential event for the reorganization of the cytoskeleton, leading to a more sessile phenotype of cells (Figure 4A). In human LiSa-2 pre-adipocytes, the CUL3-mediated degradation of RHOA parallels increases in the small GTPase, RAB18 [70], a positive regulator of autophagy [86] and of CAV1, a protein involved in endocytosis [87]. The interdependence of RHOA, RAB18 and CAV1 and their role in adipogenesis is currently unknown.
Cell migration and intracellular transport require the coordinated action of the actin as well as the microtubule cytoskeletons. As with the actin cytoskeleton, CUL3-based E3 ligases play critical roles in the remodeling of the microtubule cytoskeleton. Microtubule dynamics are controlled by specific proteins including the end binding 1 (EB1) protein, which binds to the microtubule plus ends [88]. EB1 is degraded by the UPS and associated with the CSN, the latter interaction determining its stability [89]. Recently, CRL3KLHL21 was found to mediate EB1 mono-ubiquitylation [90]. Data obtained by Peter and co-workers support the hypothesis that EB1 mono-ubiquitylation promotes microtubule disassembly, and cells expressing mutant EB1 that cannot be ubiquitylated at Lys 100 are defective in cell migration [90].
The cytoskeleton facilitates vesicle transport connected with endocytosis as well as exocytosis [91, 92]. During differentiation, specific intracellular trafficking takes place, which includes, for example, CUL3-dependent collagen secretion in osteogenesis [66, 67] (see Figure 4B) and endocytosis of lipids and their transport to lipid droplets in adipogenesis [93, 94]. CUL3-based CRLs regulate trafficking of proteins via the endosomal pathway [95]. CUL3 in association with the substrate receptor SPOPL targets endosome-bound EPS15 for degradation and thereby regulates the fate of EPS15-dependent cargos: recycling versus lysosomal degradation [96]. In addition, CUL3-based ubiquitylation promotes post-Golgi trafficking. CUL3KLHL20 targets coronin 7 (CRN7) for Lys 33-linked poly-ubiquitylation, a modification that facilitates CRN7 targeting to the trans-Golgi network via Ub-dependent interaction with EPS15 [97].
Interestingly, an early endosomal BTB protein called ANKFY1 associates with CUL3 and CRL3ANKFY1 regulates endosomal trafficking of integrin β1 in endothelial cells [98]. In addition, Drosophila insomniac (INC), orthologous to mammalian KCTD2, KCTD5 and KCTD17, acts as SR and CRL3INC traffics to synapsis linking synaptic function and sleep [99]. RHOBTB proteins are also related to vesicle trafficking. They are involved in the regulation of transport from endosomes to the Golgi apparatus [16].
It is important to note that many aspects of CRL3-based cell fate specification and differentiation described here for mammalian cells are conserved in other species. This is particularly true for CRL3-mediated organization of the cytoskeleton in C. elegans [3] as well as in Drosophila [52, 100].
Concluding Remarks
A large variety of CRL3 complexes is necessary to accomplish multilineage differentiation of MSCs. The repertory of CRL3BTB complexes required for differentiation toward a particular cell lineage can be provided within minutes by CAND1- and CSN-dependent remodeling. CRL3 ligases ubiquitylate their targets for subsequent 26S proteasome-dependent proteolysis. On the other hand, they also support mono-ubiquitylation, Lys33-linked ubiquitylation and ubiquitylation at non-Lys residues. CUL3-based ligases are engaged in two major tasks crucial for differentiation: the reprogramming of transcription/translation and the remodeling of the cytoskeleton. The differentiation-dependent transcriptional reprogramming requires the CRL3-controlled degradation of negative transcriptional regulators of differentiation. CRL3-dependent and differentiation-specific cytoskeleton remodeling establishes a more sessile phenotype of cells and supports intracellular trafficking typical of differentiated cells.
In the future, it will be interesting to see whether specific BTB proteins can drive a particular differentiation process, which would have consequences for programming of stem cells and for diagnosis and treatment of diseases associated with impaired cell differentiation. Understanding the function of CRL3s in MSC differentiation has implications in diverse areas of human health including obesity, neurodegenerative diseases, osteoporosis and regenerative medicine.
Trends Box.
Whereas CRL1 complexes often target for degradation cell cycle regulators such as cyclins and the cell cycle inhibitors p21 and p27, most members of the CRL4 family are essential for chromatin stability and DNA repair. These findings support the emerging concept that different CRL families evolved to fulfil specialized cellular functions.
In contrast to CUL1 and CUL4, which are predominantly in the nucleus, CUL3 is mostly localized to and functional in the cytoplasm.
There are approximately 180 BTB proteins encoded in the human genome. However, so far only about 50 BTB proteins have been confirmed as substrate receptors (SR) in CRL3 complexes.
Inhibition of RHOA signaling by targeting RHOA for degradation via CRL3-dependent ubiquitylation transfers mesenchymal stem cells (MSCs) to a more sessile phenotype and accelerates cell differentiation.
There is increasing interest in MSCs in regenerative medicine with the potential of finding novel cures for a large variety of diseases. Understanding the exact roles of CRL3s in MSC differentiation may catalyze leaps in mesenchymal stem cell research and regenerative medicine.
Outstanding Questions.
What is the mechanism of the functional and spatial specialization of cullin-RING ubiquitin ligases (CRLs)?
Besides confirmed data, are there additional CRL3BTB complexes required for the specific differentiation programs of mesenchymal stem cells (MSCs)? Does the expression of BTB proteins of unknown function during cell differentiation reflect the existence of additional CRL3BTB complexes involved in the process?
Which signaling pathways are responsible for the timely expression of appropriate BTB proteins during specific differentiation phases? How are the signaling pathways coordinated?
What are the mechanisms of the activation of CRL3BTB complexes during cell differentiation? Are master regulators of differentiation such as peroxisome proliferator-activated receptor γ (PPARγ) and Runt-related transcription factor 2 (Runx2) involved in CRL3BTB activation through transcriptionally controlling CRL3 complex components and/or regulators? Is the neddylation of CRL3BTB complexes specifically controlled during differentiation? Do CRL3BTB complexes escape from deneddylation by the COP9 signalosome in certain phases of differentiation?
Are there common mechanisms of CRL3 mediated ubiquitylation in different cell-lineage differentiation programs? For example, is the CRL3 mediated degradation of RHO family proteins and the connected remodeling of the cytoskeleton common to many cell-lineage differentiation processes? Do lineage-specific CRL3BTB complexes, that target negative transcriptional regulators of differentiation for degradation, initiate the differentiation process?
Do differentiation programs of endodermal, ectodermal and mesodermal stem cells have CRL3BTB driven processes in common?
Will the application of our understanding of CRL3 mediated mechanisms in stem cell differentiation lead to the discovery of specific drugs against diseases associated with impaired differentiation? Will we be able to program adult stem cells based on our knowledge of the specific role of CRL3BTB complexes in stem cell differentiation in the future?
Acknowledgments
This work was supported by grants to W. D. by the Natural Science Foundation of China, No. 31770813, and to M.N. by the Ministry of Economy, Research and Digitalization (Funding of Science and Research in Saxony-Anhalt by budget funds of the European Structural and Investment Fund (ESIF), ZS/2016/04/78155). Work in D.A.W.’s lab is funded by grants GM105802 and GM121834 from the US National Institutes of Health, grant No. 81773771 from the Natural Science Foundation of China, the Foreign 1000 Talent Program of the Government of the People’s Republic of China, and the 100 Talent Program of the Province of Fujian, China.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, Peter M. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 4.Teixeira LK, Reed SI. Ubiquitin ligases and cell cycle control. Annu Rev Biochem. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 5.Gao S, Geng C, Song T, Lin X, Liu J, Cai Z, Cang Y. Activation of c-Abl Kinase Potentiates the Anti-myeloma Drug Lenalidomide by Promoting DDA1 Protein Recruitment to the CRL4 Ubiquitin Ligase. J Biol Chem. 2017;292:3683–3691. doi: 10.1074/jbc.M116.761551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng M, Ren L, Mizuno K, Nestoras K, Wang H, Tang Z, Guo L, Kong D, Hu Q, He Q, Du L, Carr AM, Liu C. CRL4(Wdr70) regulates H2B monoubiquitination and facilitates Exo1-dependent resection. Nat Commun. 2016;7:11364. doi: 10.1038/ncomms11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HY, Chen RH. Cullin 3 Ubiquitin Ligases in Cancer Biology: Functions and Therapeutic Implications. Front Oncol. 2016;6:113. doi: 10.3389/fonc.2016.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP, Schulman BA. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaharbakhshi E, Jemc JC. Broad-complex, tramtrack, and bric-a-brac (BTB) proteins: Critical regulators of development. Genesis. 2016;54:505–518. doi: 10.1002/dvg.22964. [DOI] [PubMed] [Google Scholar]
- 11.Gupta VA, Beggs AH. Kelch proteins: emerging roles in skeletal muscle development and diseases. Skelet Muscle. 2014;4:11. doi: 10.1186/2044-5040-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 13.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Valimaki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR, Stockigt JR, Keppler-Noreuil KM, Porter CC, Anand SK, Whiteford ML, Davis ID, Dewar SB, Bettinelli A, Fadrowski JJ, Belsha CW, Hunley TE, Nelson RD, Trachtman H, Cole TR, Pinsk M, Bockenhauer D, Shenoy M, Vaidyanathan P, Foreman JW, Rasoulpour M, Thameem F, Al-Shahrouri HZ, Radhakrishnan J, Gharavi AG, Goilav B, Lifton RP. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathew R, Seiler MP, Scanlon ST, Mao AP, Constantinides MG, Bertozzi-Villa C, Singer JD, Bendelac A. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature. 2012;491:618–621. doi: 10.1038/nature11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohnuki H, Inoue H, Takemori N, Nakayama H, Sakaue T, Fukuda S, Miwa D, Nishiwaki E, Hatano M, Tokuhisa T, Endo Y, Nose M, Higashiyama S. BAZF, a novel component of cullin3-based E3 ligase complex, mediates VEGFR and Notch cross-signaling in angiogenesis. Blood. 2012;119:2688–2698. doi: 10.1182/blood-2011-03-345306. [DOI] [PubMed] [Google Scholar]
- 16.Ji W, Rivero F. Atypical Rho GTPases of the RhoBTB Subfamily: Roles in Vesicle Trafficking and Tumorigenesis. Cells. 2016;5 doi: 10.3390/cells5020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schenkova K, Lutz J, Kopp M, Ramos S, Rivero F. MUF1/leucine-rich repeat containing 41 (LRRC41), a substrate of RhoBTB-dependent cullin 3 ubiquitin ligase complexes, is a predominantly nuclear dimeric protein. J Mol Biol. 2012;422:659–673. doi: 10.1016/j.jmb.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Berthold J, Schenkova K, Ramos S, Miura Y, Furukawa M, Aspenstrom P, Rivero F. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes--evidence for an autoregulatory mechanism. Exp Cell Res. 2008;314:3453–3465. doi: 10.1016/j.yexcr.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canning P, Cooper CD, Krojer T, Murray JW, Pike AC, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Ayinampudi V, Marsden BD, Gileadi O, Knapp S, von Delft F, Bullock AN. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem. 2013;288:7803–7814. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji AX, Prive GG. Crystal structure of KLHL3 in complex with Cullin3. PLoS One. 2013;8:e60445. doi: 10.1371/journal.pone.0060445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber A, Cohen I, Popp O, Dittmar G, Reiss Y, Sommer T, Ravid T, Jarosch E. Sequential Poly-ubiquitylation by Specialized Conjugating Enzymes Expands the Versatility of a Quality Control Ubiquitin Ligase. Mol Cell. 2016;63:827–839. doi: 10.1016/j.molcel.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 23.Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, de Jong A, Ovaa H, Alpi AF, Harper JW, Schulman BA. Two Distinct Types of E3 Ligases Work in Unison to Regulate Substrate Ubiquitylation. Cell. 2016;166:1198–1214. e1124. doi: 10.1016/j.cell.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 28.Mosadeghi R, Reichermeier KM, Winkler M, Schreiber A, Reitsma JM, Zhang Y, Stengel F, Cao J, Kim M, Sweredoski MJ, Hess S, Leitner A, Aebersold R, Peter M, Deshaies RJ, Enchev RI. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. Elife. 2016;5 doi: 10.7554/eLife.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavadini S, Fischer ES, Bunker RD, Potenza A, Lingaraju GM, Goldie KN, Mohamed WI, Faty M, Petzold G, Beckwith RE, Tichkule RB, Hassiepen U, Abdulrahman W, Pantelic RS, Matsumoto S, Sugasawa K, Stahlberg H, Thoma NH. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature. 2016;531:598–603. doi: 10.1038/nature17416. [DOI] [PubMed] [Google Scholar]
- 30.Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, Fischer ES, Thoma NH. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 31.Iso T, Suzuki T, Baird L, Yamamoto M. Absolute Amounts and Status of the Nrf2-Keap1-Cul3 Complex within Cells. Mol Cell Biol. 2016;36:3100–3112. doi: 10.1128/MCB.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siergiejuk E, Scott DC, Schulman BA, Hofmann K, Kurz T, Peter M. Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. EMBO J. 2009;28:3845–3856. doi: 10.1038/emboj.2009.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, Zhu W, Nhan T, Toth JI, Petroski MD, Wolf DA. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun. 2013;4:1642. doi: 10.1038/ncomms2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, Shan SO, Deshaies RJ. Cand1 Promotes Assembly of New SCF Complexes through Dynamic Exchange of F Box Proteins. Cell. 2013;153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitsma JM, Liu X, Reichermeier KM, Moradian A, Sweredoski MJ, Hess S, Deshaies RJ. Composition and Regulation of the Cellular Repertoire of SCF Ubiquitin Ligases. Cell. 2017 doi: 10.1016/j.cell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zemla A, Thomas Y, Kedziora S, Knebel A, Wood NT, Rabut G, Kurz T. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- 39.Dubiel D, Gierisch ME, Huang X, Dubiel W, Naumann M. CAND1-dependent control of cullin 1-RING Ub ligases is essential for adipogenesis. Biochim Biophys Acta. 2013;1833:1078–1084. doi: 10.1016/j.bbamcr.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Dubiel D, Ordemann J, Pratschke J, Dubiel W, Naumann M. CAND1 exchange factor promotes Keap1 integration into cullin 3-RING ubiquitin ligase during adipogenesis. Int J Biochem Cell Biol. 2015;66:95–100. doi: 10.1016/j.biocel.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blondelle J, Shapiro P, Domenighetti AA, Lange S. Cullin E3 Ligase Activity Is Required for Myoblast Differentiation. J Mol Biol. 2017;429:1045–1066. doi: 10.1016/j.jmb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cirak S, von Deimling F, Sachdev S, Errington WJ, Herrmann R, Bonnemann C, Brockmann K, Hinderlich S, Lindner TH, Steinbrecher A, Hoffmann K, Prive GG, Hannink M, Nurnberg P, Voit T. Kelch-like homologue 9 mutation is associated with an early onset autosomal dominant distal myopathy. Brain. 2010;133:2123–2135. doi: 10.1093/brain/awq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumara I, Quadroni M, Frei C, Olma MH, Sumara G, Ricci R, Peter M. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12:887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Sambuughin N, Swietnicki W, Techtmann S, Matrosova V, Wallace T, Goldfarb L, Maynard E. KBTBD13 interacts with Cullin 3 to form a functional ubiquitin ligase. Biochem Biophys Res Commun. 2012;421:743–749. doi: 10.1016/j.bbrc.2012.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.du Puy L, Beqqali A, van Tol HT, Monshouwer-Kloots J, Passier R, Haagsman HP, Roelen BA. Sarcosin (Krp1) in skeletal muscle differentiation: gene expression profiling and knockdown experiments. Int J Dev Biol. 2012;56:301–309. doi: 10.1387/ijdb.113327lp. [DOI] [PubMed] [Google Scholar]
- 47.Gong W, Gohla RM, Bowlin KM, Koyano-Nakagawa N, Garry DJ, Shi X. Kelch Repeat and BTB Domain Containing Protein 5 (Kbtbd5) Regulates Skeletal Muscle Myogenesis through the E2F1-DP1 Complex. J Biol Chem. 2015;290:15350–15361. doi: 10.1074/jbc.M114.629956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs. Cell. 2006;127:871–874. doi: 10.1016/j.cell.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Pacary E, Heng J, Azzarelli R, Riou P, Castro D, Lebel-Potter M, Parras C, Bell DM, Ridley AJ, Parsons M, Guillemot F. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron. 2011;69:1069–1084. doi: 10.1016/j.neuron.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 51.Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, Reymond A, Sun M, Sawa A, Gusella JF, Kamiya A, Beckmann JS, Katsanis N. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Wu S, Wolf DA. Destruction of RhoA CULtivates actin. Mol Cell. 2009;35:735–736. doi: 10.1016/j.molcel.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Gladwyn-Ng I, Huang L, Ngo L, Li SS, Qu Z, Vanyai HK, Cullen HD, Davis JM, Heng JI. Bacurd1/Kctd13 and Bacurd2/Tnfaip1 are interacting partners to Rnd proteins which influence the long-term positioning and dendritic maturation of cerebral cortical neurons. Neural Dev. 2016;11:7. doi: 10.1186/s13064-016-0062-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gladwyn-Ng IE, Li SS, Qu Z, Davis JM, Ngo L, Haas M, Singer J, Heng JI. Bacurd2 is a novel interacting partner to Rnd2 which controls radial migration within the developing mammalian cerebral cortex. Neural Dev. 2015;10:9. doi: 10.1186/s13064-015-0032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pacary E, Azzarelli R, Guillemot F. Rnd3 coordinates early steps of cortical neurogenesis through actin-dependent and -independent mechanisms. Nat Commun. 2013;4:1635. doi: 10.1038/ncomms2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cleveland DW, Yamanaka K, Bomont P. Gigaxonin controls vimentin organization through a tubulin chaperone-independent pathway. Hum Mol Genet. 2009;18:1384–1394. doi: 10.1093/hmg/ddp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin MY, Lin YM, Kao TC, Chuang HH, Chen RH. PDZ-RhoGEF ubiquitination by Cullin3-KLHL20 controls neurotrophin-induced neurite outgrowth. J Cell Biol. 2011;193:985–994. doi: 10.1083/jcb.201103015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sobieszczuk DF, Poliakov A, Xu Q, Wilkinson DG. A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes Dev. 2010;24:206–218. doi: 10.1101/gad.554510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu TM, Lee EH, Lim B, Shyh-Chang N. Concise Review: Balancing Stem Cell Self-Renewal and Differentiation with PLZF. Stem Cells. 2016;34:277–287. doi: 10.1002/stem.2270. [DOI] [PubMed] [Google Scholar]
- 62.Cai H, Liu A. Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc Natl Acad Sci U S A. 2016;113:14751–14756. doi: 10.1073/pnas.1612520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 64.Park CK, Lee Y, Kim KH, Lee ZH, Joo M, Kim HH. Nrf2 is a novel regulator of bone acquisition. Bone. 2014;63:36–46. doi: 10.1016/j.bone.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 65.Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgartel C, Schekman R, Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGourty CA, Akopian D, Walsh C, Gorur A, Werner A, Schekman R, Bautista D, Rape M. Regulation of the CUL3 Ubiquitin Ligase by a Calcium-Dependent Co-adaptor. Cell. 2016;167:525–538. e514. doi: 10.1016/j.cell.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 67.Werner A, Iwasaki S, McGourty CA, Medina-Ruiz S, Teerikorpi N, Fedrigo I, Ingolia NT, Rape M. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature. 2015;525:523–527. doi: 10.1038/nature14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol. Cell. Endocrinol. 2010;318:10–14. doi: 10.1016/j.mce.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang X, Ordemann J, Muller JM, Dubiel W. The COP9 signalosome, cullin 3 and Keap1 supercomplex regulates CHOP stability and adipogenesis. Biol Open. 2012;1:705–710. doi: 10.1242/bio.20121875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubiel D, Bintig W, Kahne T, Dubiel W, Naumann M. Cul3 neddylation is crucial for gradual lipid droplet formation during adipogenesis. Biochim Biophys Acta. 2017;1864:1405–1412. doi: 10.1016/j.bbamcr.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 71.James AW. Review of Signaling Pathways Governing MSC Osteogenic and Adipogenic Differentiation. Scientifica (Cairo) 2013;2013:684736. doi: 10.1155/2013/684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakaue T, Sakakibara I, Uesugi T, Fujisaki A, Nakashiro KI, Hamakawa H, Kubota E, Joh T, Imai YK, Izutani H, Higashiyama S. The CUL3-SPOP-DAXX axis is a novel regulator of VEGFR2 expression in vascular endothelial cells. Sci Rep. 2017;7:42845. doi: 10.1038/srep42845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren K, Yuan J, Yang M, Gao X, Ding X, Zhou J, Hu X, Cao J, Deng X, Xiang S, Zhang J. KCTD10 is involved in the cardiovascular system and Notch signaling during early embryonic development. PLoS One. 2014;9:e112275. doi: 10.1371/journal.pone.0112275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 76.Lee DJ, Tseng HC, Wong SW, Wang Z, Deng M, Ko CC. Dopaminergic effects on in vitro osteogenesis. Bone Res. 2015;3:15020. doi: 10.1038/boneres.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skieterska K, Shen A, Clarisse D, Rondou P, Borroto-Escuela DO, Lintermans B, Fuxe K, Xiang YK, Van Craenenbroeck K. Characterization of the interaction between the dopamine D4 receptor, KLHL12 and beta-arrestins. Cell Signal. 2016;28:1001–1014. doi: 10.1016/j.cellsig.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 78.Ferry C, Gaouar S, Fischer B, Boeglin M, Paul N, Samarut E, Piskunov A, Pankotai-Bodo G, Brino L, Rochette-Egly C. Cullin 3 mediates SRC-3 ubiquitination and degradation to control the retinoic acid response. Proc Natl Acad Sci U S A. 2011;108:20603–20608. doi: 10.1073/pnas.1102572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, O'Malley BW. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sailland J, Tribollet V, Forcet C, Billon C, Barenton B, Carnesecchi J, Bachmann A, Gauthier KC, Yu S, Giguere V, Chan FL, Vanacker JM. Estrogen-related receptor alpha decreases RHOA stability to induce orientated cell migration. Proc Natl Acad Sci U S A. 2014;111:15108–15113. doi: 10.1073/pnas.1402094111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 82.Ohashi K, Nagata K, Maekawa M, Ishizaki T, Narumiya S, Mizuno K. Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J Biol Chem. 2000;275:3577–3582. doi: 10.1074/jbc.275.5.3577. [DOI] [PubMed] [Google Scholar]
- 83.Sandbo N, Smolyaninova LV, Orlov SN, Dulin NO. Control of Myofibroblast Differentiation and Function by Cytoskeletal Signaling. Biochemistry (Mosc) 2016;81:1698–1708. doi: 10.1134/S0006297916130071. [DOI] [PubMed] [Google Scholar]
- 84.Xu B, Ju Y, Song G. Role of p38, ERK1/2, focal adhesion kinase, RhoA/ROCK and cytoskeleton in the adipogenesis of human mesenchymal stem cells. J Biosci Bioeng. 2014;117:624–631. doi: 10.1016/j.jbiosc.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 85.Chen K, He H, Xie Y, Zhao L, Zhao S, Wan X, Yang W, Mo Z. miR-125a-3p and miR-483-5p promote adipogenesis via suppressing the RhoA/ROCK1/ERK1/2 pathway in multiple symmetric lipomatosis. Sci Rep. 2015;5:11909. doi: 10.1038/srep11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feldmann A, Bekbulat F, Huesmann H, Ulbrich S, Tatzelt J, Behl C, Kern A. The RAB GTPase RAB18 modulates macroautophagy and proteostasis. Biochem Biophys Res Commun. 2017;486:738–743. doi: 10.1016/j.bbrc.2017.03.112. [DOI] [PubMed] [Google Scholar]
- 87.Fernandez-Rojo MA, Ramm GA. Caveolin-1 Function in Liver Physiology and Disease. Trends Mol Med. 2016;22:889–904. doi: 10.1016/j.molmed.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 88.Nehlig A, Molina A, Rodrigues-Ferreira S, Honore S, Nahmias C. Regulation of end-binding protein EB1 in the control of microtubule dynamics. Cell Mol Life Sci. 2017 doi: 10.1007/s00018-017-2476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peth A, Boettcher JP, Dubiel W. Ubiquitin-dependent Proteolysis of the Microtubule End-binding Protein 1, EB1, Is Controlled by the COP9 Signalosome: Possible Consequences for Microtubule Filament Stability. J Mol Biol. 2007;368:550–563. doi: 10.1016/j.jmb.2007.02.052. [DOI] [PubMed] [Google Scholar]
- 90.Courtheoux T, Enchev RI, Lampert F, Gerez J, Beck J, Picotti P, Sumara I, Peter M. Cortical dynamics during cell motility are regulated by CRL3(KLHL21) E3 ubiquitin ligase. Nat Commun. 2016;7:12810. doi: 10.1038/ncomms12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noordstra I, Akhmanova A. Linking cortical microtubule attachment and exocytosis. F1000Res. 2017;6:469. doi: 10.12688/f1000research.10729.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Margiotta A, Bucci C. Role of Intermediate Filaments in Vesicular Traffic. Cells. 2016;5 doi: 10.3390/cells5020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Le Lay S, Hajduch E, Lindsay MR, Le Liepvre X, Thiele C, Ferre P, Parton RG, Kurzchalia T, Simons K, Dugail I. Cholesterol-induced caveolin targeting to lipid droplets in adipocytes: a role for caveolar endocytosis. Traffic. 2006;7:549–561. doi: 10.1111/j.1600-0854.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- 94.Fridolfsson HN, Roth DM, Insel PA, Patel HH. Regulation of intracellular signaling and function by caveolin. FASEB J. 2014;28:3823–3831. doi: 10.1096/fj.14-252320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huotari J, Meyer-Schaller N, Hubner M, Stauffer S, Katheder N, Horvath P, Mancini R, Helenius A, Peter M. Cullin-3 regulates late endosome maturation. Proc Natl Acad Sci U S A. 2012;109:823–828. doi: 10.1073/pnas.1118744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gschweitl M, Ulbricht A, Barnes CA, Enchev RI, Stoffel-Studer I, Meyer-Schaller N, Huotari J, Yamauchi Y, Greber UF, Helenius A, Peter M. A SPOPL/Cullin-3 ubiquitin ligase complex regulates endocytic trafficking by targeting EPS15 at endosomes. Elife. 2016;5:e13841. doi: 10.7554/eLife.13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yuan WC, Lee YR, Lin SY, Chang LY, Tan YP, Hung CC, Kuo JC, Liu CH, Lin MY, Xu M, Chen ZJ, Chen RH. K33-Linked Polyubiquitination of Coronin 7 by Cul3-KLHL20 Ubiquitin E3 Ligase Regulates Protein Trafficking. Mol Cell. 2014;54:586–600. doi: 10.1016/j.molcel.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 98.Maekawa M, Tanigawa K, Sakaue T, Hiyoshi H, Kubota E, Joh T, Watanabe Y, Taguchi T, Higashiyama S. Cullin-3 and its adaptor protein ANKFY1 determine the surface level of integrin beta1 in endothelial cells. Biol Open. 2017;6:1707–1719. doi: 10.1242/bio.029579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Q, Kellner DA, Hatch HAM, Yumita T, Sanchez S, Machold RP, Frank CA, Stavropoulos N. Conserved properties of Drosophila Insomniac link sleep regulation and synaptic function. PLoS Genet. 2017;13:e1006815. doi: 10.1371/journal.pgen.1006815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hudson AM, Mannix KM, Cooley L. Actin Cytoskeletal Organization in Drosophila Germline Ring Canals Depends on Kelch Function in a Cullin-RING E3 Ligase. Genetics. 2015;201:1117–1131. doi: 10.1534/genetics.115.181289. [DOI] [PMC free article] [PubMed] [Google Scholar]