Abstract
Intracellular delivery of protein therapeutics by cationic polymer vehicles is an emerging technique that is, however, encountering poor stability, high cytotoxicity and non-specific cell uptake. Herein, we present a facile strategy to optimize the protein-polycation complexes by encapsulating with linear-dendritic telodendrimers. The telodendrimers with well-defined structures enable the rational design and integration of multiple functionalities for efficient encapsulation of the protein-polycation complexes by multivalent and hybrid supramolecular interactions to produce sub-20 nm nanoparticles. This strategy not only reduces the polycation-associated cytotoxicity and hemolytic activity, but also eliminates the aggregation and non-specific binding of polycations to other biomacromolecules. Moreover, the telodendrimers dissociate readily from the complexes during the cellular uptake process, which restores the capability of polycations for intracellular protein delivery. This strategy overcomes the limitations of polycationic vectors for intracellular delivery of protein therapeutics.
Keywords: polycation, telodendrimer, nanocomplex, intracellular protein delivery
Graphical Abstract
A facile strategy to optimize of the protein-polycation complexes was developed by encapsulating with rationally designed telodendrimers for efficient intracellular protein delivery.

1. Introduction
Cationic polymers have long been regarded as efficient non-viral vectors for gene delivery,[1–6] and recently have been applied in the field of intracellular delivery of protein therapeutics, which is particularly important for the cell-impermeable proteins to impart their functions intracellularly.[7–10] The representative cationic polymers such as polyethylenimine (PEI) show high delivery efficiency,[11, 12] however, are usually associated with several shortages including the large or uncontrollable aggregation size, high cytotoxicity, and non-specific phagocytosis by the reticuloendothelium systems in vivo.[2, 13] To overcome these shortages, PEGylated polycations have been developed towards safe therapeutic delivery.[14, 15] However, the covalently conjugated polyethylene glycol (PEG) or other biocompatible materials on the polycations in early studies significantly sacrificed the cellular uptake and reduced the delivery efficiency.[16, 17] Therefore, smart conjugations employing reversible covalent chemistries have been developed to endow the polycations with stimuli-responsive detachable shielding layers,[18, 19] wherein the shielding materials such as PEG can dissociate from the polycations to restore the high gene delivery efficiency upon stimuli such as pH change[2, 20], light[21], and redox reagents[22, 23]. Encapsulation of the therapeutic-polycation complexes with negatively charged shielding polymers based on completely reversible noncovalent interactions is an alternative way without complicated chemical conjugation to endow the delivery vehicles with spontaneously detachable shielding layers. In terms of gene delivery, copolymers of anionic peptides linked to PEG have been used to encapsulate the DNA-polycation complexes by electrostatic interactions to prevent salt- and serum albumin-induced aggregation, and the protective copolymer-coated vectors have been demonstrated to be compatible with the intracellular gene delivery.[24] However, to our knowledge, this noncovalent interaction-based shielding strategy has yet to be applied in the development of advanced polycation-based vehicles for intracellular protein delivery.
In addition to the electrostatic interactions, most proteins can provide valid hydrophobic grooves for efficient binding of other hydrophobic materials and even for the reinforcement of electrostatic interactions by decreasing the static dielectric constants of surroundings.[25–27] In previous studies, we have integrated multiple charged and hydrophobic functionalities in a well-defined linear-dendritic telodendrimer[28, 29] matrix to match protein surface chemistries through synergistic combination of electrostatic and hydrophobic interactions to produce protein-telodendrimer nanoparticles with excellent in vitro stability.[30] These telodendrimers have been demonstrated to be efficient to encapsulate and deliver proteins to tumor sites in vivo by the enhanced permeability and retention effects[31] and to manage the blood glucose level by sustained release of insulin in mouse models.[30, 32] These achievements testify the importance to adopt multivalent and hybrid noncovalent interactions in a well-defined telodendrimer system for stable protein encapsulation and efficient delivery. Remarkably, the polycation complexation may induce a change in the surface charge properties of the payload proteins, but the hydrophobic regions on the proteins can be generally maintained. Therefore, it is promising and technically feasible to encapsulate the protein-polycation complexes with rationally designed telodendrimers based on both electrostatic and hydrophobic interactions for efficient intracellular protein delivery.
In this study, we rationally designed a chemically well-defined telodendrimer, composed of linear PEG-blocking-dendritic oligolysine and the capping peripheral building blocks of oxalic acids and cholesterols, for encapsulation of the protein-polycation complexes. The negatively charged oxalic acid and hydrophobic cholesterol functionalities on the telodendrimer are selected to interact with the positively charged protein-polycation complexes by both electrostatic and hydrophobic interactions. We found this protein-polycation-telodendrimer nanocomplexes possessing proper serum stability showed eliminated non-specific binding capacity, significantly reduced nanocarrier-associated cytotoxicity and hemolytic activity, and maintained intracellular protein delivery efficiency when compared to the protein-polycation complexes. This study presents a simple and clean method to optimize protein-polycation complexes by modification with telodendrimer shielding materials based on multivalent and hybrid interactions for efficient intracellular protein delivery.
2. Materials and methods
2.1. Materials
Monomethylterminated poly(ethylene glycol) monoamine hydrochloride (MeO-PEG-NH2·HCl, MW: 5 kDa) was purchased from Jenkem Technology. (Fmoc)Lys(Fmoc)-OH and (Fmoc)Lys(Boc)-OH were obtained from AnaSpec Inc. Cholesteryl chlorofomate was purchased from Alfa Aesar. CellTiter 96® AQueous MTS reagent powder was purchased from Promega. Diisopropyl carbodimide (DIC), N-hydroxybenzotriazole (HOBt), trifluoroacetic acid (TFA), methyl chlorooxoacetate, fluorescein isothiocyanate isomer I (FITC), bovine serum albumin (BSA, Mw 66.5 kDa), N-hydroxysuccinimide (HOSu), rhodamine B (RB), polyethylenimine (PEI, branched, Mw 25 kDa) and other chemical reagents were purchased from Sigma-Aldrich. Dialysis membrane with 3,500 MW cut off was purchased from Spectrum Laboratories, Inc.
2.2. Telodendrimer synthesis
The telodendrimer with eight oxalic acid groups containing cholesterols, named as PEG5k(OAOA-L-CHO)4, was synthesized using a solution-phase condensation reaction starting from MeO-PEG-NH2·HCl (5 kDa) via stepwise peptide chemistry. The synthesis of the telodendrimer has been published separately[30] and, therefore, is not repeated here.
2.3. Fabrication of protein-polycation-telodendrimer nanocomplexes
Proteins and polycations in phosphate-buffered saline (PBS) were mixed at various mass ratios of protein to polycation, and different amounts of telodendrimers in PBS were added into the protein-polycation complex solution to form protein-polycation-telodendrimer nanocomplexes. The nanocomplex solution was diluted with PBS to desired concentrations.
2.4. Characterization
Dynamic light scattering (DLS) studies were performed using a Zetatrac (Microtrac Inc.) instrument, and the area-based mean particle sizes were presented. Zeta potential measurements were carried out on a Malvern Nano-ZS zetasizer at room temperature. Transmission electron microscopy (TEM) images were taken on a JEOL 2100 TEM instrument operating at a voltage of 80 kV. The samples were negatively stained by 1% uranyl acetate.
2.5. Fluorescently labeled proteins and telodendrimers
FITC-labeled BSA (named FITC-BSA) was prepared by mixing 3 mg of FITC dissolved in 0.3 mL of DMSO with 10 mL of BSA aqueous solution (10 mg/mL) in the presence of 0.1 M of NaHCO3 under stirring. The feeding molar ratio of FITC to BSA is approximately 5:1. After 24 h, the reaction mixture was dialyzed against deionized water in dark for one week to remove the unreacted FITC molecules, and dried by lyophilization.
RB-labeled BSA (named RB-BSA) was synthesized as follows: RB-OSu was first synthesized by mixing 10 mg of RB, 3 mg of HOSu and 4 μL of DIC dissolved in 0.5 mL of DMSO. RB-BSA was then prepared by mixing above RB-OSu solution with 20 mL of BSA aqueous solution (13 mg/mL) in the presence of 0.1 M of NaHCO3 under stirring. After 24 h, the reaction mixture was dialyzed against deionized water in the dark for one week to remove the unreacted RB molecules, and dried by lyophilization.
FITC-labeled PEG5k(OAOA-L-CHO)4 (named FITC-PEG5k(OAOA-L-CHO)4) was prepared as follows: 10 mg of PEG5k(OAOA-L-CHO)4, 0.12 mg of HOSu and 0.2 μL of DIC were mixed in 0.5 mL of DMF to fabricate PEG5k(OAOA-L-CHO)4-OSu. After 12 h, 0.26 mg of mono Boc protected 2,2′-(Ethylenedioxy)bis(ethylamine) and 0.2 μL of DIEA were added into the above reaction mixtures, allowing for 24 h reaction. The product was then precipitated in cold ether, followed by the removal of Boc protecting groups in 1/1 (v/v) mixture of TFA/CH2Cl2 and the precipitation in cold ether. The amino-functionalized PEG5k(OAOA-L-CHO)4 was dissolved in aqueous solution in the presence of 0.1 M of NaHCO3, and 0.4 mg of FITC in 0.2 mL of DMSO was added. After 24 h, the reaction mixture was dialyzed against deionized water in the dark for one week to remove the unreacted FITC molecules, and dried by lyophilization.
2.6. Agarose gel retention assay
Samples in loading buffer (30% glycerol aqueous solution) were loaded into agarose gel (1.5%wt) in Tris-acetate-EDTA (TAE) buffer. The gel tray was run for 2 h at a constant current of 20 mA. The gel was then stained with 1% coomassie blue (30 min) followed by overnight destaining. The gel was imaged by a Bio-Rad Universal Hood II Imager (Bio-Rad Laboratories, Inc.) under SYBR Green and coomassie blue modes.
2.7. Förster resonance energy transfer (FRET) studies
Equal volumes of 2 mg/mL of RB-BSA solution, 4 mg/mL of PEI solution and 4 mg/mL of FITC-PEG5k(OAOA-L-CHO)4 solution were mixed, following by stirring overnight. The fluorescence spectra with a range from 480 to 640 nm at different time points excited by 439 nm were recorded using a microplate reader (BioTek Synergy 2). The FRET ratio was calculated by the formula of [100%×I584/(I584 + I528)], where I584 and I528 were fluorescence intensity of RB-BSA at 584 nm and FITC-PEG5k(ArgArg-L-C17)4 at 528 nm, respectively.
2.8. Cell culture and MTS assays
The colon cancer cell line HT-29 and the breast cancer cell line MDA-231 were purchased from American Type Culture Collection (ATCC, Manassas, VA, U.S.A.). All cells were cultured in 100 U/mL penicillin G, and 100 μg/mL streptomycin at 37 °C using a humidified 5% CO2 incubator. Various formulations of proteins with different dilutions were added to the plate and then incubated in a humidified 37 °C, 5% CO2 incubator. After 4h incubation, McCoy’s 5A medium supplemented with 10% fetal bovine serum (FBS) was added into the above system. After 72 h incubation, a mixture solution composed of CellTiter 96 AQueous MTS, and an electron coupling reagent, PMS, was added to each well according to the manufacturer’s instructions. The cell viability was determined by measuring absorbance at 490 nm using a microplate reader (BioTek Synergy 2). Untreated cells served as the control. Results were shown as the average cell viability [100%×(ODtreat − OD blank)/(ODcontrol − ODblank)] of triplicate wells.
2.9. Hemolytic assays
One milliliter of fresh blood from healthy human volunteers was collected into 5 mL of PBS solution in the presence of 20 mM EDTA. Red blood cells (RBCs) were then separated by centrifugation at 1000 rpm for 10 min. The RBCs were washed three times with 10 mL of PBS and resuspended in 20 mL of PBS. Diluted RBC suspension (200 μL) was mixed with nanoparticle PBS solutions at serial concentrations (10, 100, and 500 μg/mL) by gentle vortex and incubated at 37 °C. After 0.5 h, 4 h, and overnight, the mixtures were centrifuged at 1,000 rpm for 5 min. The supernatant free of hemoglobin was determined by measuring the absorbance at 540 nm using a UV-vis spectrometer. Incubations of RBCs with Triton-100 (2%) and PBS were used as the positive and negative controls, respectively. The percent hemolysis of RBCs was calculated using the following formula:
| (1) |
2.10. Cellular uptake
The cellular uptake and intracellular trafficking of the protein-incorporated nanoparticles were determined by fluorescence microscopy. FITC-BSA and RB-BSA were used as model proteins. HT-29 and MDA-231 cells were seeded in chamber slide with a density of 5 ×104 cells per well in 350 μL of McCoy’s 5A and cultured for 24 h. The original medium was replaced with FITC-BSA, FITC-BSA:PEI (1:2, w/w), FITC-BSA:PEI:telodendrimer (1:2:2, w/w), BSA:PEI:FITC-telodendrimer (1:2:2, w/w) or RB-BSA at a final dye concentration of approximately 1.5 μg/mL at 37 °C. After a 3 h incubation, the cells were washed three times with cold PBS and fixed with 4% formaldehyde for 10 min at room temperature, and the cell nuclei and lysosome were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue) and Lysotracker (green), respectively. The slides were mounted with cover slips and cells were imaged with a NiKON FV1000 laser scanning confocal fluorescence microscope or a Leica fluorescence microscope.
2.11. Statistical analysis
Data are presented as means ± standard deviation (SD) unless otherwise stated. Statistical analysis was performed using two-tailed Student’s t-test for comparison of two groups. The level of significance in all statistical analyses was set at a probability of P < 0.05.
3. Results and discussion
3.1. Fabrication of protein-polycation complexes
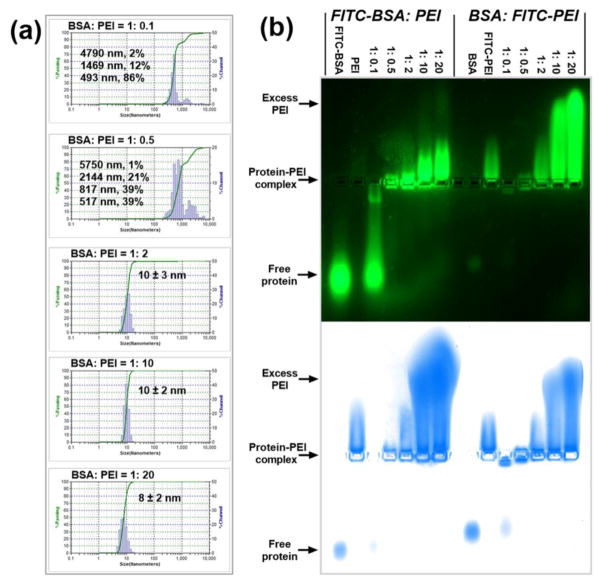
Bovine serum albumin (BSA) with a molecular weight of 66.5 kDa and an isoelectric point of 5.4, and branched polyethylenimine (PEI) with a molecular weight of 25 kDa were selected as the model protein and the model polycation, respectively, for the fabrication of protein-polycation complexes. BSA and PEI can form complex mainly based on electrostatic interactions (Fig. 1). The particle sizes and distributions of the BSA-PEI complexes at different BSA:PEI mass ratios were detected by dynamic light scattering (DLS), and the results are shown in Fig. 2a. Interestingly, large aggregates of the complexes were observed with multiple distribution peaks when less PEI molecules were used at BSA:PEI mass ratios of 1:0.1 and 1:0.5. It may be ascribed to the crosslinking effects of PEI chains in interacting with negatively charged BSA. In comparison, with the increasing amount of PEI added, small and well-defined nanoparticles with hydrodynamic diameters of ~10 nm were observed at BSA:PEI mass ratios of 1:2, 1:10 and 1:20. It reveals that BSA can be efficiently coated by PEI into individual and well-dispersed nanoparticles without further aggregation. Native agarose gel electrophoresis was used to investigate the formation of BSA-PEI complex, and fluorescently labeled BSA and PEI (noted as FITC-BSA and FITC-PEI, respectively) were used to track the migrations of nanocomplex in the gel. The gel was also stained by coomassie blue, in which case both BSA and PEI can be stained based on electrostatic and/or hydrophobic interactions. The images taken under fluorescence (upper row) and coomassie blue (lower row) modes in Fig. 2b gave a consistent result: BSA and FITC-BSA migrated towards the anode (positive electrode) while PEI and FITC-PEI migrated towards the cathode (negative electrode), and the BSA-PEI complexes were trapped in the loading well and migrated towards the cathode with the increasing proportion of PEI, because of the increased particle sizes and charge reversal.[30] When the BSA:PEI mass ratio was high (1:0.1), there were not sufficient PEI molecules for the formation of BSA-PEI complexes and free proteins could be observed. When the BSA:PEI mass ratios were as low as 1:10 and 1:20, extensive migrations towards the cathode were observed, due to the increased positive charges and reduced particle sizes as shown in DLS studies (Fig. 2a). Large amounts of excess PEI molecules in the systems may lead to high cytotoxicity. Therefore, a BSA:PEI mass ratio of 1:2 is determined to be the optimal one based on both electrophoresis and DLS analysis and will be used in the following experiments.
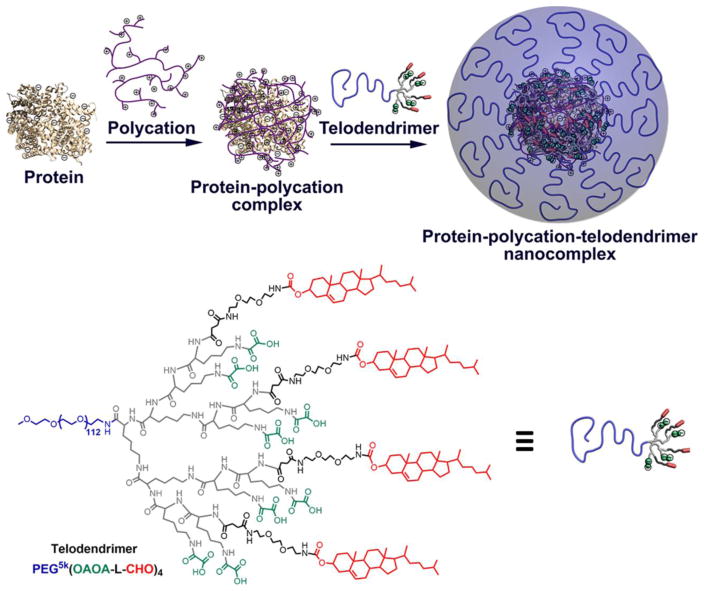
Fig. 1.
Schematic illustration of the procedure to fabricate protein-polycation-telodendrimer nanocomplexes and the chemical structure of PEG5k(OAOA-L-CHO)4 telodendrimer.
Fig. 2.
Characterization of protein-polycation complex systems. (a) DLS particle size analysis for BSA:PEI complexes at different BSA:PEI mass ratios. The value behind ‘±’ describes the half-peak width of the size distribution. (b) Agarose gel retention assays for FITC-BSA:PEI (the 7 lanes on the left) and BSA:FITC-PEI (the 7 lanes on the right) complexes at different mass ratios. The images were taken before (upper row) and after (lower row) staining by coomassie blue.
3.2. Fabrication of protein-polycation-telodendrimer nanocomplexes
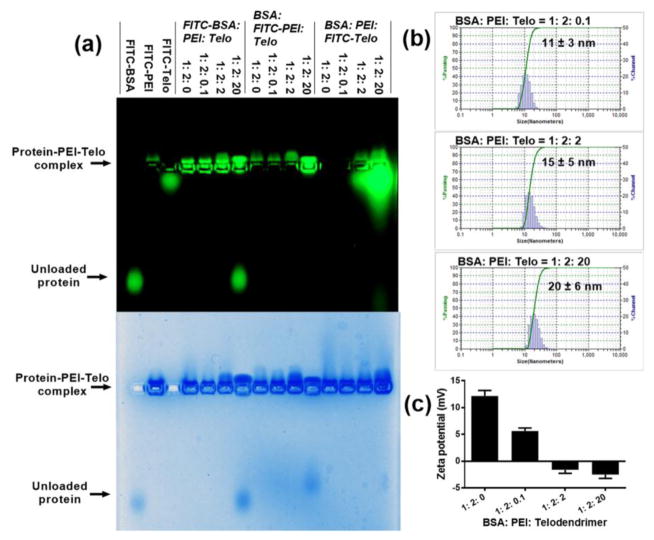
Based on the properties of the BSA-PEI complexes with positive surface charges (zeta potential: +12 mV) and hydrophobic regions, the rationally designed telodendrimer composed of a linear PEG and a dendritic oligolysine-based scaffold containing eight negatively charged oxalic acid groups and four hydrophobic cholesterols, named PEG5k(OAOA-L-CHO)4 (chemical structure, Fig. 1), was synthesized via stepwise peptide chemistry. This telodendrimer has shown availability for the delivery of positively charged proteins, e.g. lysosome, in our previous study.[30] The synthesis and characterization of this telodendrimer have been previously reported.[30] Here, we rationally apply this negatively charged telodendrimer to coat the protein-PEI nanocomplex by both electrostatic and hydrophobic interactions (Fig. 1) to reduce the cytotoxicity and non-specific cellular uptake. The PEG5k(OAOA-L-CHO)4 telodendrimers can form nanoparticles with a hydrodynamic diameter of ~16 nm and zeta potential of ~−5 mV in an aqueous solution (Fig. S1). Even though the telodendrimers form micelles above the critical micelle concentration (CMC, 1.52 μM), they can readily reconstruct and encapsulate proteins to form protein-loaded nanoparticles undergoing an absorption-reassembly process, which has been demonstrated in our previous study.[30] Agarose gel retention assays were used to study the ternary complex of BSA, PEI and PEG5k(OAOA-L-CHO)4. Notably, the negatively charged telodendrimers cannot be stained by coomassie blue. A relatively high BSA:PEI mass ratio of 1:2 was selected also in consideration of the excesses of the hydrophobic regions on protein to telodendrimer for stable nanocomplex formation. The BSA:telodendrimer (or PEI:telodendrimer) mass ratio was varied as a means to control the property of the nanocomplexes. In this study, we labeled three components, i.e. BSA, PEI and PEG5k(OAOA-L-CHO)4, respectively, with FITC to visualize and confirm the nanocomplex formation on agarose gel electrophoresis. As shown in Fig. 3a, the ternary complex was trapped in the loading well in the presence of telodendrimer. Interestingly, BSA leaked out when a large amount of telodendrimers were added in the complex system (BSA:PEI:PEG5k(OAOA-L-CHO)4 = 1:2:20, w/w). This is because large amounts of negatively charged telodendrimers can neutralize the polycation and therefore fail to stably encapsulate the negatively charged protein, e.g. BSA, as shown in our previous study.[30] This fact further demonstrates that it is critical to synergistically combine both electrostatic and hydrophobic interactions for the fabrication of stable protein-polycation-telodendrimer nanocomplexes. In comparison, these three components can form stable complexes when having other mass ratios with lower telodendrimer contents (1:2:0.1, and 1:2:2), as demonstrated by the co-localization of three components in the agarose gel. In these cases, both charge and hydrophobic interactions contribute to the formation of stable ternary nanocomplexes. The DLS results in Fig. 3b indicate that the BSA-PEI-PEG5k(OAOA-L-CHO)4 complexes at different mass ratios show similar particle sizes. However, positive zeta potential was detected for the BSA-PEI-PEG5k(OAOA-L-CHO)4 complexes at mass ratios of 1:2:0 and 1:2:0.1 (Fig. 3c), which may lead to non-specific cellular uptake. Therefore, a BSA:PEI:PEG5k(OAOA-L-CHO)4 complex at a 1:2:2 mass ratio with slightly negative zeta potential is considered to be the optimal candidate for further studies.
Fig. 3.
Characterization of protein-polycation-telodendrimer complex systems. (a) Agarose gel retention assays for FITC-BSA:PEI:PEG5k(OAOA-L-CHO)4, BSA:FITC-PEI:PEG5k(OAOA-L-CHO)4, and BSA:PEI:FITC-PEG5k(OAOA-L-CHO)4 complexes at different mass ratios. The images were taken before (upper row) and after (lower row) staining by coomassie blue. (b,c) DLS particle size (b) and zeta potential (c) analysis for BSA:PEI:PEG5k(OAOA-L-CHO)4 complexes at different mass ratios. For the DLS data in b, the value behind ‘±’ describes the half-peak width of the size distribution. The error in c is for standard deviation (n = 3).
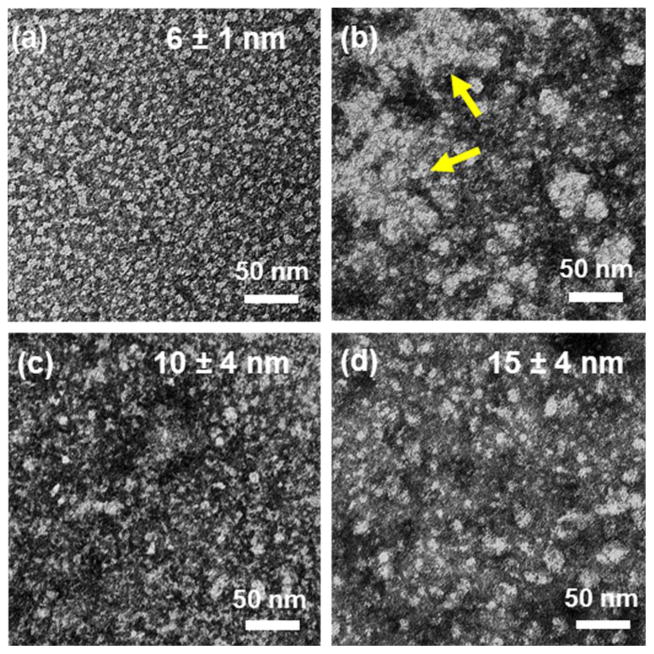
Transmission electron microscopy (TEM) was used to study the morphology of BSA-PEI complexes (1:0.5 and 1:2, w/w) and BSA-PEI-PEG5k(OAOA-L-CHO)4 complex (1:2:2, w/w) in contrast to free BSA. Free BSA shows isolated spherical particles with an average size of 6 nm (Fig. 4a). The addition of PEI to BSA to reach to a BSA:PEI mass ratio of 1:0.5 leads to a significant increase in particle size and formation of large aggregates (Fig. 4b), which is consistent with the DLS analysis (Fig. 2a). Further increasing the PEI content to a BSA:PEI mass ratio of 1:2 breaks the aggregation and decreases the complex particle size to 10 nm (Fig. 4c). The addition of PEG5k(OAOA-L-CHO)4 to the BSA-PEI complex to a mass ratio of 1:2:2 (BSA:PEI:PEG5k(OAOA-L-CHO)4) increases the particle size to 15 nm (Fig. 4d), indicating formation of the ternary complex of BSA, PEI and PEG5k(OAOA-L-CHO)4. These results obtained from the TEM images acquire good agreement with the DLS results (Fig. 2a and 3b).
Fig. 4.
TEM images of (a) free BSA, (b) BSA-PEI complex at a mass ratio of 1:0.5, (c) BSA-PEI complex at a mass ratio of 1:2, and (d) BSA-PEI-PEG5k(OAOA-L-CHO)4 complex at a mass ratio of 1:2:2 with negative staining. The average diameters of the particles are indicated in the graphs. The yellow arrows in b indicate the aggregates.
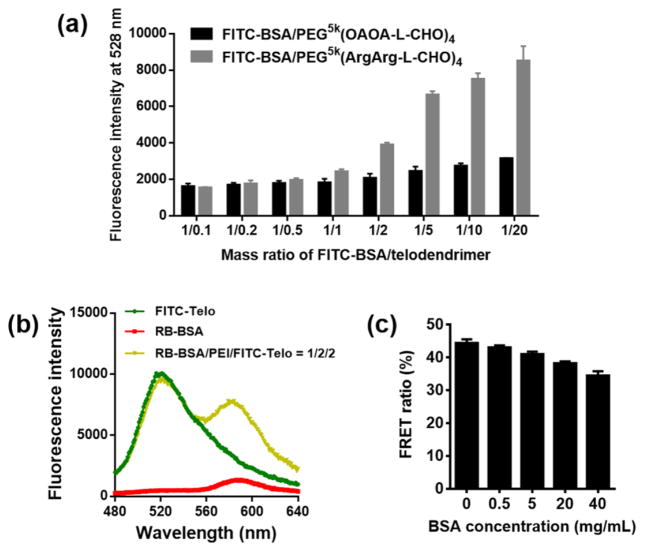
In a previous study, we have demonstrated that positively charged telodendrimers such as PEG5k(ArgArg-L-CHO)4 can encapsulate negatively charged protein such as BSA based on both electrostatic and hydrophobic interactions.[30] By addition of PEG5k(ArgArg-L-CHO)4 to FITC-BSA aqueous solutions, the fluorescence signal of FITC-BSA was observed to gradually increase, indicating the increased hydrophobic microenvironment of FITC-BSA after being coated with telodendrimer. In contrast, the fluorescence signal of FITC-BSA generally kept constant when PEG5k(OAOA-L-CHO)4 was gradually added, indicating the weak interactions between BSA and the negatively charged telodendrimer (Fig. 5a). This is consistent with our previous observation that BSA cannot form stable complex with PEG5k(OAOA-L-CHO)4 as shown in the agarose gel electrophoresis assay.[30] The addition of positively charged PEI can mediate BSA and PEG5k(OAOA-L-CHO)4 to form stable ternary complex (Fig. 3a), indicating the importance of PEI for stabilizing this nanocomplex. Förster resonance energy transfer (FRET) experiments were used to probe the molecular proximity[33] during the complex formation. Rhodamine B-labeled BSA (RB-BSA), PEI and FITC-PEG5k(OAOA-L-CHO)4 were added sequentially into an aqueous solution to prepare the ternary complex. When the ternary complex was excited at the excitation wavelength of FITC at 439 nm, a significant FRET signal was recorded at the emission wavelength of RB at 584 nm (Fig. 5b), which can be attributed to the close molecular proximity of proteins and telodendrimers. The FRET ratio (the calculation method can be found in the Experimental Section) of RB-BSA/PEI/FITC-PEG5k(OAOA-L-CHO)4 ternary complexes slightly decreased by 9.9% with the increasing serum protein concentrations up to 40 mg/mL (Fig. 5c), implying the dynamic feature of the nanocomplexes and the partial dissociation due to serum protein exchange.
Fig. 5.
(a) Fluorescent intensity of FITC-BSA incubation with either positively (PEG5k(ArgArg-L-CHO)4) or negatively (PEG5k(OAOA-L-CHO)4) charged telodendrimers at different mass ratios of FITC-BSA/telodendrimer. (b) Representative FRET spectra of RB-BSA, FITC-PEG5k(OAOA-L-CHO)4, and RB-BSA/PEI/FITC-PEG5k(OAOA-L-CHO)4 (1:2:2, w/w) complex. (c) FRET ratios of RB-BSA/PEI/FITC-PEG5k(OAOA-L-CHO)4 (1:2:2, w/w) complexes after incubation with non-fluorescent BSA solutions at different concentrations. The errors in a and c are for standard deviation (n = 3).
3.3. Cytotoxicity and hemolysis
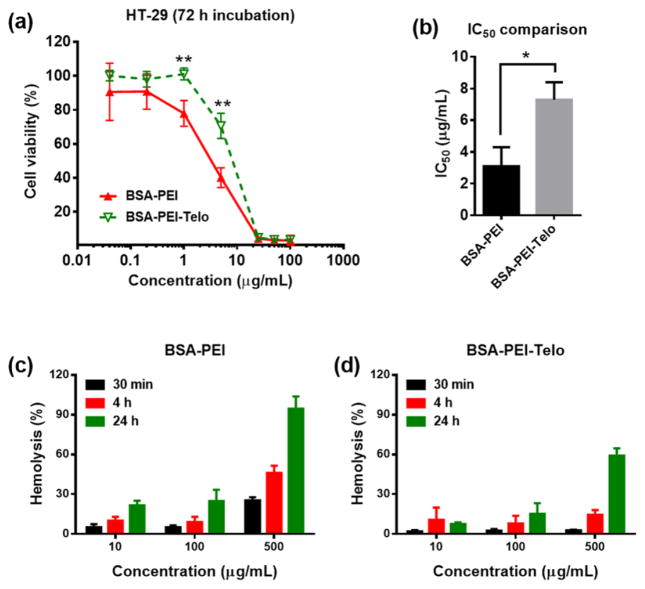
The polycationic molecules or complexes were reported to be toxic for many cell lines.[2, 4, 34] HT-29 colon cancer cell line was selected as a model cell line to investigate the cytotoxicity of protein-polycation and protein-polycation-PEG5k(OAOA-L-CHO)4 nanocomplexes. The cell viability results after a 72-h continuous incubation at 37 °C in Fig. 6 indicate that the encapsulation of the protein-polycation complexes within PEG5k(OAOA-L-CHO)4 can significantly reduce polycation-associated cytotoxicity. The half maximal inhibitory concentration (IC50) value for the BSA-PEI-PEG5k(OAOA-L-CHO)4 complexes is significantly higher than that for the BSA-PEI complexes (Fig. 6b). Moreover, the BSA-PEI-PEG5k(OAOA-L-CHO)4 complexes show significantly lower hemolytic activity when compared to the BSA-PEI complexes (Fig. 6c and d). The reduced cytotoxicity and hemolytic activity by PEG5k(OAOA-L-CHO)4 telodendrimer encapsulation are presumably because of the PEG shells in the telodendrimers that can shelter positive charges efficiently (Fig. 3c) to reduce non-specific binding to cell membranes or other biomacromolecules. In addition, the telodendrimer consists of all biocompatible components constructed by peptide bonds, which can be hydrolyzed by peptidase in vivo. The telodendrimer has a low molecular weight of ~9.7 kDa, which can be eliminated by renal clearance once dissociated from the nanocomplex. All these features of telodendrimer favor the in vivo applications with great biocompatibility.
Fig. 6.
(a) Cell viability assays on HT-29 cells after a 72-h continuous incubation at 37 °C for BSA-PEI (1:2, w/w) and BSA-PEI-PEG5k(OAOA-L-CHO)4 (1:2:2, w/w) complexes. (b) IC50 values calculated from the cell viability results in a. The errors in a and b are for standard deviation (n = 3, *P < 0.05, **P < 0.01). (c,d) Hemolytic assays for (c) BSA-PEI (1:2, w/w) and (d) BSA-PEI-PEG5k(OAOA-L-CHO)4 (1:2:2, w/w) complexes. The error is for standard deviation (n = 3).
3.4. Intracellular protein delivery
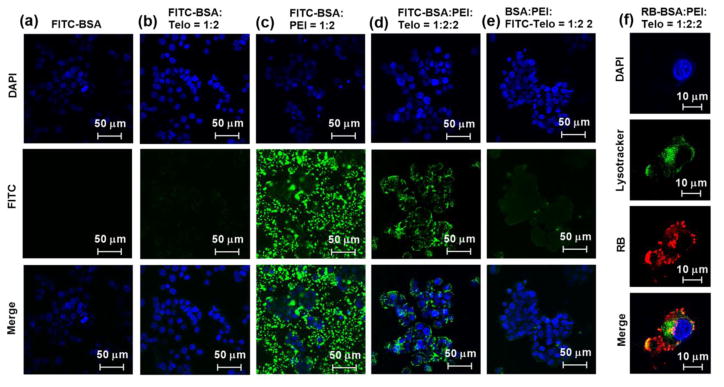
Many proteins are possible to be therapeutics if they can be delivered across plasma membrane into intracellular spaces, such as antibodies against intracellular proteins used in biochemistry assays or pathology detections, as well as truncated diphtheria toxins used in brain tumor treatment.[34, 35] However, such exogenous proteins, even some endogenous proteins are cell-impermeable by themselves due to their surface charge distributions, large molecular weights and vulnerable tertiary structures.[7, 36] In addition, the lack of receptors to mediate their cellular uptake renders these proteins inactive. Therefore, the ability to create efficient vehicles for intracellular protein delivery will expand the horizon dramatically in the development and application of therapeutic proteins in disease treatments. Cationic polymers have been widely studied over the past few decades for gene delivery and recently have been used for intracellular delivery of proteins while maintaining the bioactivity.[1, 7–9, 37] However, the advancement of these vehicles is mainly hindered by their positive surface charges, that usually cause high cytotoxicity and non-specific interactions with negatively charged biomacromolecules and cell membranes during the transport process.[2] The encapsulation of protein-polycation complexes by rationally designed telodendrimers has shown great promise to reduce the polycation-induced cytotoxicity and hemolytic activity (Fig. 6). We hypothesize that the telodendrimer encapsulation can also avoid the non-specific binding of protein-polycation complexes to other biomacromolecules during the intracellular protein delivery process. To test this hypothesis, FITC-BSA was used to probe the intracellular trafficking of proteins delivered by polycation-PEG5k(OAOA-L-CHO)4 nanocarriers in HT-29 colon cancer cell cultures. We kept the dye concentrations and the dye:BSA:PEI:PEG5k(OAOA-L-CHO)4 ratios consistent for all the groups in Fig. 7a–e. FITC-BSA could hardly get into cells (Fig. 7a) and the negatively charged group-containing telodendrimer, PEG5k(OAOA-L-CHO)4, could not intracellularly deliver FITC-BSA (Fig. 7b). FITC-BSA-PEI complexes further aggregated significantly and precipitated in the cell cultures due to the positive surface charges (Fig. 7c), and majority of the green spots (precipitates) were observed in the extracellular space in the culture (Fig. S2). It indicates that the protein-polycation complexes without shielding layers can interact with cell culture medium and form large aggregates, resulting in precipitation and inefficient intracellular protein delivery. When the FITC-BSA-PEI complexes were encapsulated by PEG5k(OAOA-L-CHO)4 telodendrimers, FITC-BSA was efficiently translocated into the cytoplasm, and no aggregation was found outside the cells (Fig. 7d and S2). This fact indicates that the telodendrimer encapsulation can significantly prevent the non-specific binding of cationic polymers to other biomacromolecules in the cell cultures. Furthermore, the PEG5k(OAOA-L-CHO)4 telodendrimer was labeled by FITC instead of BSA in the ternary nanocomplex and tested for cell uptake properties. Interestingly, as shown in Fig. 7e, no obvious FITC-PEG5k(OAOA-L-CHO)4 signal can be detected after the intracellular protein delivery with the polycation/FITC-PEG5k(OAOA-L-CHO)4 complex, which is correlated with the binary system in Fig. 7b. It indicated that the telodendrimer consisting of PEG and negative charges is not ready for cellular uptake. It serves as a shielding layer in the ternary system to reduce the cytotoxicity, hemolytic activity and prevent non-specific aggregation of PEI. The telodendrimer shielding is, however, dynamic to dissociate readily from the protein-polycation complexes for efficient intracellular protein delivery. RB-BSA and MDA-231 breast cancer cell were used as the model protein and the model cell line, respectively, to further study the intracellular protein delivery by polycation-PEG5k(OAOA-L-CHO)4 complexes. As shown in Fig. 7f, during a confocal fluorescence microscopy imaging study by counterstaining the endosome/lysosome (green) of a MDA-231 cell, the co-localization of RB-BSA (red) within the endosome/lysosome after a 3-h incubation was revealed. Moreover, significant amounts of proteins had already escaped from the endosomal compartments, as indicated by the red fluorescence signals throughout the cell, which is essential for therapeutic proteins to be functional intracellularly.
Fig. 7.
Confocal fluorescence microscopy images of (a–e) HT-29 and (f) MDA-231 cells incubated with (a) free FITC-BSA, (b) FITC-BSA:PEG5k(OAOA-L-CHO)4 (1:2, w/w), (c) FITC-BSA:PEI (1:2, w/w), (d) FITC-BSA:PEI:PEG5k(OAOA-L-CHO)4 (1:2:2, w/w), (e) BSA:PEI:FITC-PEG5k(OAOA-L-CHO)4 (1:2:2, w/w), and (f) RB-BSA:PEI:PEG5k(OAOA-L-CHO)4 (1:2:2, w/w) at 37 °C for 3 h. The cell nuclear was stained with DAPI (blue). The lysosome in f was stained with Lysotracker (green).
4. Conclusion
In this study, we presented a facile strategy of encapsulating protein-polycation complexes with rationally designed telodendrimers to minimize the polycation-induced cytotoxicity and non-specific binding capacity for efficient intracellular protein delivery. The telodendrimers possessing both negative charges and hydrophobic groups could form stable complexes with proteins and polycations to reduce the nanocarrier-associated cytotoxicity and hemolytic activity, as well as to prevent the aggregation in cell cultures. Moreover, the telodendrimer shielding layers could detach themselves from the nanocomplexes during the intracellular protein delivery, evidenced by the inefficient cellular internalization of telodendrimers, which suggested the possibility to completely restore the intracellular protein delivery efficiency of the cationic polymers. This strategy of synergistic combination of electrostatic and hydrophobic interactions for stable and detachable telodendrimer modification on protein-polycation complexes could have extensive in vitro and in vivo application prospects in intracellular delivery of proteins for disease treatments.
Supplementary Material
Highlights.
Telodendrimers encapsulate protein-polycation complexes by hybrid interactions
The encapsulation reduces the cytotoxicity and hemolytic activity
The encapsulation eliminates the extracellular aggregation
The polycation-telodendrimer complexes enable intracellular protein delivery
Acknowledgments
Prof. Mathew M. Maye of Department of Chemistry at Syracuse University gratefully acknowledged for assistance with zeta potential studies. The financial supports from the Program of Qilu Young Scholars of Shandong University (Wang), National Natural Science Foundation of China (NSFC grant 21704057) (Wang), NIH/NIBIB 1R21EB019607 (Luo), Napi Family Research Awards (Luo), and New York State Department of Health/PETER T. ROWLEY breast cancer research award (Luo) are greatly acknowledged.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at xxx.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.De Smedt SC, Demeester J, Hennink WE. Pharm Res. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 2.Guan X, Guo Z, Wang T, Lin L, Chen J, Tian H, Chen X. Biomacromolecules. 2017;18:1342–1349. doi: 10.1021/acs.biomac.7b00080. [DOI] [PubMed] [Google Scholar]
- 3.Barnard A, Posocco P, Pricl S, Calderon M, Haag R, Hwang ME, Shum VWT, Pack DW, Smith DK. J Am Chem Soc. 2011;133:20288–20300. doi: 10.1021/ja2070736. [DOI] [PubMed] [Google Scholar]
- 4.Tripathi SK, Ahmadi Z, Gupta KC, Kumar P. Colloids Surf B Biointerfaces. 2016;140:117–120. doi: 10.1016/j.colsurfb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L, Bader A, Bogorad RL, Yin H, Racie T, Dong Y, Jiang S, Seedorf D, Dave A, Singh Sandhu K, Webber MJ, Novobrantseva T, Ruda VM, Lytton-JeanAbigail KR, Levins CG, Kalish B, Mudge DK, Perez M, Abezgauz L, Dutta P, Smith L, Charisse K, Kieran MW, Fitzgerald K, Nahrendorf M, Danino D, Tuder RM, von Andrian UH, Akinc A, Panigrahy D, Schroeder A, Koteliansky V, Langer R, Anderson DG. Nat Nanotechnol. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinnapireddy SR, Duse L, Strehlow B, Schäfer J, Bakowsky U. Colloids Surf B Biointerfaces. 2017;158:93–101. doi: 10.1016/j.colsurfb.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Kamaly N, Shi J, Zhao L, Xiao Z, Hollett G, John R, Ray S, Xu X, Zhang X. Angew Chem Int Ed. 2014;53:8975–8979. doi: 10.1002/anie.201404766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AL, Wang Y, Ye WH, Yoon HS, Chan SY, Yang YY. Biomaterials. 2008;29:1224–1232. doi: 10.1016/j.biomaterials.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 9.Tamura A, Ikeda G, Nishida K, Yui N. Macromol Biosci. 2015;15:1134–1145. doi: 10.1002/mabi.201500083. [DOI] [PubMed] [Google Scholar]
- 10.Maier K, Martin I, Wagner E. Mol Pharm. 2012;9:3560–3568. doi: 10.1021/mp300404d. [DOI] [PubMed] [Google Scholar]
- 11.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akinc A, Thomas M, Klibanov AM, Langer R. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 13.Guan X, Li Y, Jiao Z, Lin L, Chen J, Guo Z, Tian H, Chen X. ACS Appl Mater Interfaces. 2015;7:3207–3215. doi: 10.1021/am5078123. [DOI] [PubMed] [Google Scholar]
- 14.Petersen H, Fechner PM, Fischer D, Kissel T. Macromolecules. 2002;35:6867–6874. [Google Scholar]
- 15.Petersen H, Fechner PM, Martin AL, Kunath K, Stolnik S, Roberts CJ, Fischer D, Davies MC, Kissel T. Bioconjug Chem. 2002;13:845–854. doi: 10.1021/bc025529v. [DOI] [PubMed] [Google Scholar]
- 16.Hatakeyama H, Akita H, Harashima H. Adv Drug Deliv Rev. 2011;63:152–160. doi: 10.1016/j.addr.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Mishra S, Webster P, Davis ME. Eur J Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 18.Ganta S, Devalapally H, Shahiwala A, Amiji M. J Control Release. 2008;126:187–204. doi: 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Shim MS, Kwon YJ. Adv Drug Deliv Rev. 2012;64:1046–1059. doi: 10.1016/j.addr.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Xiong MP, Bae Y, Fukushima S, Forrest ML, Nishiyama N, Kataoka K, Kwon GS. ChemMedChem. 2007;2:1321–1327. doi: 10.1002/cmdc.200700093. [DOI] [PubMed] [Google Scholar]
- 21.Park S-j, Park W, Na K. Adv Funct Mater. 2015;25:3472–3482. [Google Scholar]
- 22.Cai X, Dong C, Dong H, Wang G, Pauletti GM, Pan X, Wen H, Mehl I, Li Y, Shi D. Biomacromolecules. 2012;13:1024–1034. doi: 10.1021/bm2017355. [DOI] [PubMed] [Google Scholar]
- 23.Ping Y, Wu D, Kumar JN, Cheng W, Lay CL, Liu Y. Biomacromolecules. 2013;14:2083–2094. doi: 10.1021/bm400460r. [DOI] [PubMed] [Google Scholar]
- 24.Finsinger D, Remy JS, Erbacher P, Koch C, Plank C. Gene Ther. 2000;7:1183–1192. doi: 10.1038/sj.gt.3301227. [DOI] [PubMed] [Google Scholar]
- 25.Dao-Pin S, Anderson D, Baase W, Dahlquist F, Matthews BW. Biochemistry. 1991;30:11521–11529. doi: 10.1021/bi00113a006. [DOI] [PubMed] [Google Scholar]
- 26.Robinson AC, Castañeda CA, Schlessman JL. Proc Natl Acad Sci USA. 2014;111:11685–11690. doi: 10.1073/pnas.1402900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walther TH, Ulrich AS. Curr Opin Struct Biol. 2014;27:63–68. doi: 10.1016/j.sbi.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Luo J, Xiao K, Li Y, Lee JS, Shi L, Tan Y-H, Xing L, Holland Cheng R, Liu G-Y, Lam KS. Bioconjug Chem. 2010;21:1216–1224. doi: 10.1021/bc1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi C, Guo D, Xiao K, Wang X, Wang L, Luo J. Nat Commun. 2015;6:7449. doi: 10.1038/ncomms8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Shi C, Zhang L, Bodman A, Guo D, Wang L, Hall WA, Wilkens S, Luo J. Biomaterials. 2016;101:258–271. doi: 10.1016/j.biomaterials.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabral H, Matsumoto Y, Mizuno K, Chen Q, Murakami M, Kimura M, Terada Y, Kano M, Miyazono K, Uesaka M, Nishiyama N, Kataoka K. Nat Nanotechnol. 2011;6:815–823. doi: 10.1038/nnano.2011.166. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Shi C, Zhang L, Lin MY, Guo D, Wang L, Yang Y, Duncan TM, Luo J. ACS Macro Lett. 2017;6:267–271. doi: 10.1021/acsmacrolett.6b00982. [DOI] [PubMed] [Google Scholar]
- 33.Cho H, LaMarca R, Chan C. Biochem Biophys Res Commun. 2012;427:764–767. doi: 10.1016/j.bbrc.2012.09.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Bodman A, Shi C, Guo D, Wang L, Luo J, Hall WA. Small. 2016;12:4185–4192. doi: 10.1002/smll.201601234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canton I, Massignani M, Patikarnmonthon N, Chierico L, Robertson J, Renshaw SA, Warren NJ, Madsen JP, Armes SP, Lewis AL, Battaglia G. FASEB J. 2013;27:98–108. doi: 10.1096/fj.12-212183. [DOI] [PubMed] [Google Scholar]
- 36.Gu Z, Biswas A, Zhao M, Tang Y. Chem Soc Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 37.Memanishvili T, Zavradashvili N, Kupatadze N, Tugushi D, Gverdtsiteli M, Torchilin VP, Wandrey C, Baldi L, Manoli SS, Katsarava R. Biomacromolecules. 2014;15:2839–2848. doi: 10.1021/bm5005977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.