Highlights
-
•
Virus metagenomics has expanded known RNA virus diversity and emphasized connections between evolution of viruses and hosts.
-
•
Genome comparisons revealed that the extent of gene module shuffling among RNA viruses is greater than previously thought.
-
•
Expanded phylogenetic analysis implied pervasive horizontal virus transport between diverse hosts of RNA viruses.
-
•
Evolution of plant and vertebrate RNA viromes appears to be driven by HVT from vast RNA viromeof invertebrates.
Keywords: RNA viruses, Virus evolution, Horizontal virus transfer, Virus metagenomics, Metaviromics
Abstract
Virus metagenomics is a young research filed but it has already transformed our understanding of virus diversity and evolution, and illuminated at a new level the connections between virus evolution and the evolution and ecology of the hosts. In this review article, we examine the new picture of the evolution of RNA viruses, the dominant component of the eukaryotic virome, that is emerging from metagenomic data analysis. The major expansion of many groups of RNA viruses through metagenomics allowed the construction of substantially improved phylogenetic trees for the conserved virus genes, primarily, the RNA-dependent RNA polymerases (RdRp). In particular, a new superfamily of widespread, small positive-strand RNA viruses was delineated that unites tombus-like and noda-like viruses. Comparison of the genome architectures of RNA viruses discovered by metagenomics and by traditional methods reveals an extent of gene module shuffling among diverse virus genomes that far exceeds the previous appreciation of this evolutionary phenomenon. Most dramatically, inclusion of the metagenomic data in phylogenetic analyses of the RdRp resulted in the identification of numerous, strongly supported groups that encompass RNA viruses from diverse hosts including different groups of protists, animals and plants. Notwithstanding potential caveats, in particular, incomplete and uneven sampling of eukaryotic taxa, these highly unexpected findings reveal horizontal virus transfer (HVT) between diverse hosts as the central aspect of RNA virus evolution. The vast and diverse virome of invertebrates, particularly nematodes and arthropods, appears to be the reservoir, from which the viromes of plants and vertebrates evolved via multiple HVT events.
1. Introduction
With the advent of the powerful and inexpensive next generation technologies for DNA sequencing, virus metagenomics has been rapidly expanding and transforming our understanding of virus diversity, ecology, evolution and taxonomy (Brum and Sullivan, 2015, Chow and Suttle, 2015, Lecuit and Eloit, 2013, Mokili et al., 2012, Paez-Espino et al., 2016, Rosario and Breitbart, 2011, Simmonds et al., 2017). Although RNA viruses lag behind DNA viruses relative to the volume of the generated metagenomic data, substantial recent progress in this field challenges the existing concepts of RNA virus origins and evolutionary pathways. Thus, an attempt to re-evaluate these concepts appears to be timely.
In addition to metagenomics proper that focuses on ‘sequencing distinct environments’, irrespective of the organisms that inhabit these environments, virus research benefits greatly from ‘holobiont metagenomics’, which targets a particular organism complete with its co-habitants, from viruses to symbionts to parasites (Coetzee et al., 2010, Fauver et al., 2016, Lachnit et al., 2015, Li et al., 2015, Marzano et al., 2016, Roossinck et al., 2015, Shi et al., 2016a). Ideally, it would be enormously useful to know the exact host organism(s) for each virus found in both environmental and holobiont metagenomes. Such knowledge would contribute to tracing evolution of the virus host ranges and empower development of evidence-based scenarios for the origins and evolution of each virus class. In the real world, we are limited to educated guesses on the likely hosts of ‘metagenomic viruses’ based on the known host ranges of their closest relatives. In some instances, such inferences come with high confidence: e.g., when a typical bacteriophage is found upon sequencing a spider, chances are that this bacteriophage infects spider-associated bacteria. Furthermore, for some viruses, metagenomics could provide ways for definitive assignment of the host(s), such as identification of multiple CRISPR spacers for a bacterial or archaeal virus genome (Edwards et al., 2016, Willner and Hugenholtz, 2013).
Additional considerations for assigning putative viral hosts are based on the size selectivity for the biological entities present in environmental samples. For instance, aquatic samples are typically run through filters that could enrich either for viruses or for bacteria and pico-eukaryotes, while leaving multicellular eukaryotes and large protists behind (Brum and Sullivan, 2015, Chow and Suttle, 2015, Hayes et al., 2017). Another layer of selectivity is provided by the abundance bias. For aquatic environments, it could be expected that most of the isolated viruses would come from the most abundant organisms, such as prokaryotes and eukaryotic plankton, rather than from large animals, such as sharks, walruses or whales, the global abundance of which in the oceans is low (even taking into account that larger hosts could yield more viruses than small ones).
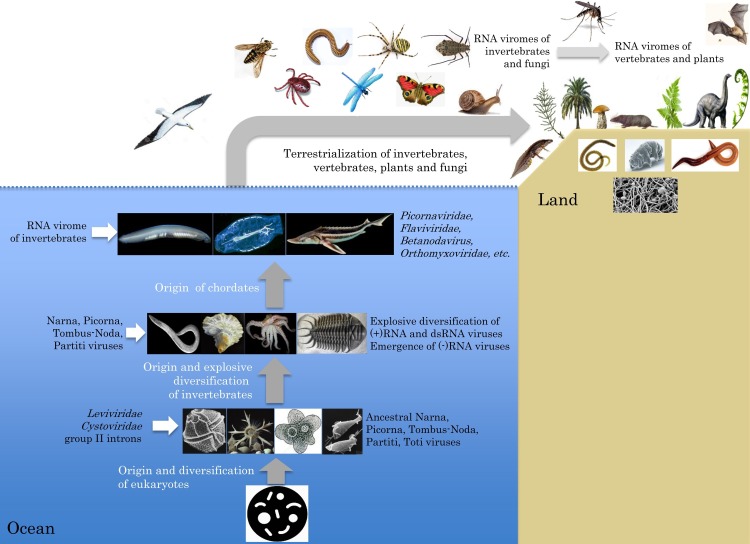
Due to the limited known diversity of prokaryotic RNA viruses that include a single family each of bacterial (+)RNA and dsRNA viruses, and only one putative (+)RNA virus of archaea (Bolduc et al., 2012), most of this review deals with the RNA viruses of eukaryotes that represent the majority of the eukaryotic virome. By any measure, the emergence of eukaryotes (eukaryogenesis) was one of the principal evolutionary transitions in the history of life (Booth and Doolittle, 2015, Koonin, 2015, Martijn and Ettema, 2013). Cohabitation of two types of cells, the archaeal host and the bacterial “guest” that became an endosymbiont and eventually evolved into the mitochondrion, as well as the emergence of the endomembrane system and the nucleus, dramatically changed cell biology and resulted in a tectonic shift in host-parasite interactions. This shift presented enormous challenges and opportunities for then extant viruses and other selfish genetic elements. One major outcome of this process was the massive proliferation of Group II introns, bacterial retroelements that are thought to have reshaped eukaryotic genome by spawning spliceosomal introns and numerous types of retrotransposons (Irimia and Roy, 2014, Rogozin et al., 2012). Another striking outcome was a cardinal change in the virome composition: whereas the viromes of archaea and bacteria are heavily dominated by DNA viruses, the eukaryotic virome exhibits a substantial excess of RNA viruses, albeit with a healthy proportion of DNA ones (Koonin et al., 2015).
To account for the composition of eukaryotic virome, we have previously proposed a scenario, in which the main groups of eukaryotic viruses originated in the crucible of eukaryogenesis via mixing and matching of gene modules derived from viruses and selfish elements of prokaryotes, as well as from the emerging eukaryotic genome (Koonin et al., 2015, Koonin et al., 2006). This turbulent process was followed by rapid diversification of the newborn ancestors of the major virus lineages along with the radiation of the supergroups of eukaryotes. It has been further proposed that the representation of DNA viruses in the eukaryotic virome was limited due to the separation of the nucleus, the ‘DNA-RNA compartment’, from the cytosol, the endomembrane-rich ‘RNA compartment’. Due to this compartmentalization of eukaryotic cells, DNA viruses that evolved to utilize at least some elements of DNA replication and transcription machineries of the prokaryotic cell were challenged with finding ways to enter the nucleus through the membrane barrier or else evolving autonomous replication and transcription systems, which hampered their spread in eukaryotes. Concurrently, eukaryotes evolved antiviral innate immunity mechanisms that identify and inactivate ‘illegitimate’ cytosolic viral DNA, further compromising reproduction of DNA viruses (Goubau et al., 2013, Mansur et al., 2014, Paludan and Bowie, 2013, Sparrer and Gack, 2015).
To explain the greatly increased share of RNA viruses in the eukaryotic virome, it has been proposed that the cytosol provided an environment conducive to the survival and replication of this type of viruses. Unlike the short-lived, co-transcriptionally translated mRNAs of prokaryotes, mature mRNAs of eukaryotes, typically show much lower turnover rates (Chen and Shyu, 2017, Mohanty and Kushner, 2016). Perhaps even more importantly, the cytosol is highly compartmentalized by the endomembrane system that consists of the expansive endoplasmic reticulum network, various vesicles and larger organelles, such as Golgi stacks, peroxisomes and mitochondria. Therefore, the cytosol provides a hospitable habitat for RNA viruses, most of which recruit various endomembranes, especially, the ER, to form ‘viral factories’, distinct membranous formations within the cytosol in which viral genomes are replicated and expressed, and virions are formed (Ahlquist, 2006, Diaz and Ahlquist, 2012). These factories also provide a level of protection against antiviral responses that target ‘alien’ RNAs.
This model also implied that, both at the time of the explosive emergence and diversification of the viral lineages and during the following virus co-evolution with the hosts, vertical transmission and evolutionary continuity of these lineages was the principal trend in virus evolution. This modality of virus evolution was generally supported by in-depth studies of several virus lineages including the vast picornavirus-like superfamily of small (+)RNA viruses although vertical evolution clearly was confounded by numerous exchanges of gene modules among viruses (Koonin et al., 2008). In addition, we have considered another route of evolution, namely, horizontal virus transfer (HVT) between diverse hosts. The HVT scenario has found support in phylogenomic analyses of plant viruses with (−)RNA genomes which appear to be a derived subset of related animal viruses, a likely result of HVT facilitated by herbivorous arthropods (Dolja and Koonin, 2011). However, at the time, the relative contribution of HVT appeared to be limited compared to the ancient origin and evolutionary continuity scenario.
The recent surge in viral metagenomics has shown that many lineages of RNA viruses whose host ranges have been thought to be restricted to plants and/or vertebrates have numerous relatives that infect marine and terrestrial invertebrates, fungi and other eukaryotic hosts (Bekal et al., 2011, Felix et al., 2011, Li et al., 2015, Marzano et al., 2016, Shi et al., 2016a, Shi et al., 2016b). Although invertebrates are not a taxonomically or phylogenetically valid group, we use this term hereinafter to denote all metazoan lineages other than the chordates. The groups that have been sampled for viruses both by traditional approaches and by metagenomics are arthropods and, more recently and to a lesser extent, nematodes and mollusks, which all represent the Protostomia, one of the two major branches of bilaterian animals, the second one being Deuterostomia including chordates (Adoutte et al., 2000, Dunn et al., 2008, Philippe et al., 2005). Thus, when it comes to reconstruction of virus evolution from the current genomic data, the invertebrate-chordate divide largely reflects phylogenetic relationships.
Notably, the newly discovered viruses of the diverse eukaryotes have been found to exhibit a greater diversity in the gene repertoires and genome architectures than their plant and vertebrate counterparts. Here we review the main results of RNA virus metagenomics and propose that pervasive HVT played a paramount role in the evolution of eukaryotic (+)RNA viruses. We further compare the RNA viromes of different types of organisms and present evidence that distinct RNA virus lineages originated from prokaryotic RNA viruses, prokaryotic reverse-transcribing elements, or from pre-existing groups of eukaryotic RNA viruses. The resulting new evolutionary scenario for RNA viruses signifies a major shift in our understanding of virus evolution and implies that the extant (+)RNA virome of eukaryotes was shaped through a complex process whereby evolutionary continuity of major viral lineages was complemented by rampant HVT.
2. The RNA virome of prokaryotes and related groups of eukaryotic viruses
The numerous and enormously diverse groups of prokaryotes host an even greater diversity of double-strand and single-strand DNA bacteriophages and archaeal viruses (Iranzo et al., 2016, Krupovic et al., 2017, Paez-Espino et al., 2016, Shulman and Davidson, 2017), but only one family of (+)RNA viruses, Leviviridae, and one family of dsRNA viruses, Cystoviridae. Although metagenomic studies of distinct environments and holobionts have resulted in the identification of many new leviviruses, the genome diversity of this family has not increased substantially (Friedman et al., 2012, Greninger and DeRisi, 2015c, Rumnieks and Tars, 2012, Shi et al., 2016a). However, this status quo was challenged by a recent, broad survey of (+)RNA phages in diverse environments that dramatically expanded their known host range and spectrum of genome architectures (Krishnamurthy et al., 2016). For the first time, (+)RNA phages have been detected in viral marine metagenomes and in a gram-positive bacterium. In addition to the four proteins encoded by most members of the Leviviridae (maturation, coat, RdRp, and lysis), several proteins with no recognizable homologs in databases were identified.
As mentioned above, the very existence of archaeal RNA viruses remains unproven (Bolduc et al., 2012). Given that bacteria are far more ancient, ancestral organisms relative to eukaryotes, the most parsimonious, intuitive scenario for the origin of eukaryotic (+)RNA viruses would involve direct descent from Leviviridae-like ancestors. The problem with this scenario is that, although not outright false, it seems to be substantially incomplete because there is no clear indication of a direct evolutionary relationship between leviviruses and the bulk of eukaryotic RNA viruses. Actually, the RdRps of the eukaryotic viruses appear to be more closely related to reverse transcriptases (RT) of prokaryotic mobile elements, namely Group II self-splicing introns, than to the levivirus RdRps (Koonin et al., 2008). Phylogenetic trees combining the RdRps of eukaryotic and prokaryotic (+)RNA viruses with RTs are inherently unreliable due to the large evolutionary distances. Nevertheless, the higher sequence similarity between eukaryotic viral RdRp and RTs of group II introns along with the wide spread of these retroelements in bacteria, which contrasts the relatively narrow host range of leviviruses, seems to make the RT origin plausible for the RdRps of the eukaryotic (+)RNA viruses. It is unclear whether the RdRp of leviviruses (Kidmose et al., 2010) itself also originates from a bacterial RT or from a primordial RdRp that is generally thought to have been one of the central players in the transition from an ancient RNA world to the RNA-protein world. The coat protein that forms the icosahedral levivirus capsids has a unique structural fold (Golmohammadi et al., 1993), and its origin remains obscure (Krupovic and Koonin, 2017).
Although (+)RNA phages may not have sired most of the eukaryotic (+)RNA viruses, there is a direct line of descent from the leviviruses to several eukaryotic virus families. The first such family to be described was Narnaviridae, which includes capsid-less, non-infectious, vertically transmitted RNA elements that reproduce either inside the mitochondria (genus Mitovirus) or in the cytosol (genus Narnavirus) of the host cells (Hillman and Cai, 2013). Recent metatranscriptomics analysis of the viromes of five plant-pathogenic fungi has revealed substantial prevalence of mitoviruses (Marzano et al., 2016).
The Narnaviridae hosted by fungi remained the only known descendants of (+)RNA phages until the unexpected discovery of the genus Ourmiavirus that includes several plant viruses related to Narnaviridae (Rastgou et al., 2009). It took another 7 years and the latest advances of metagenomics to find out that the ‘eukaryotic progeny’ of (+)RNA phages is not limited to two somewhat exotic groups of fungal and plant viruses. The recent massive metagenomics study has identified numerous viruses related to Ourmiavirus and narna-like viruses in holobionts of diverse invertebrates (Shi et al., 2016a). Although some of these viruses could originate from host-associated fungi, the sheer genomic diversity of these capsid-less or encapsidated viruses implies a broad host range, most likely, among invertebrates.
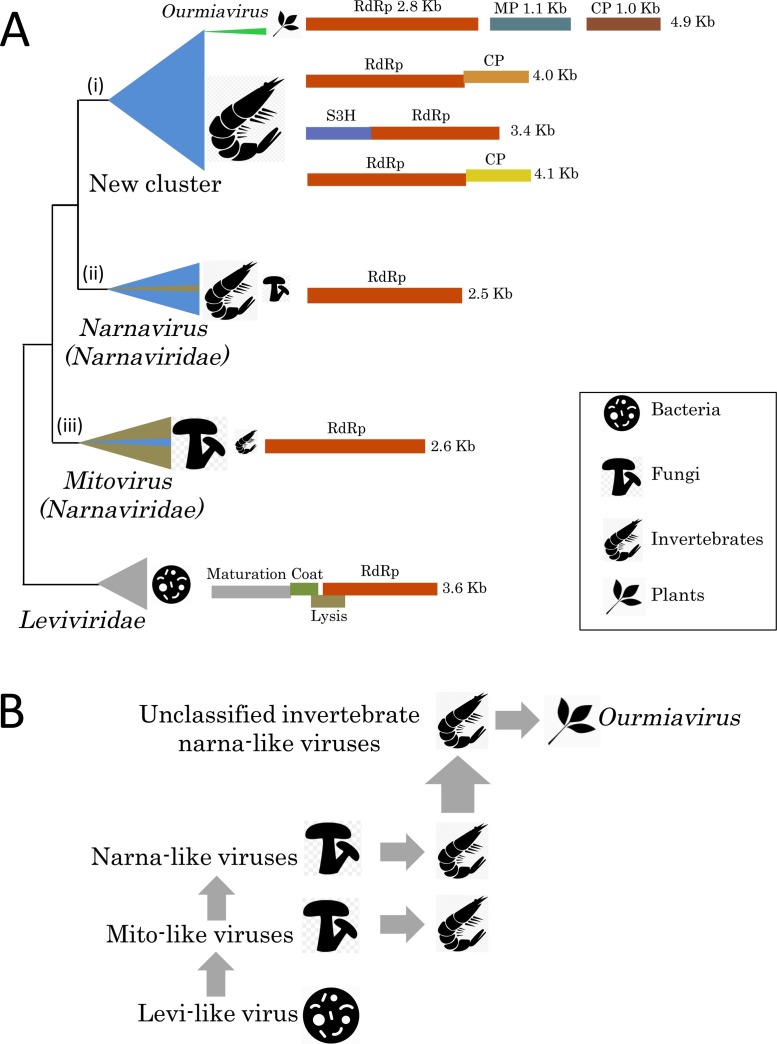
Phylogenetic analysis has identified three lineages of phage-related (+)RNA viruses of eukaryotes: i) small genus Ourmiavirus (plant hosts) and the diverse related invertebrate viruses; ii) small genus Narnavirus (fungal hosts) and numerous related invertebrate viruses; iii) large genus Mitovirus (fungal hosts) and a few viruses from invertebrate holobionts (Shi et al., 2016a) (Fig. 1 A). The ourmia-like group shows remarkable diversity of genome architectures. In particular, plant ourmiaviruses possess three monocistronic genomic segments, whereas the genomes of related viruses of invertebrates consist of a single, mono- or dicistronic RNA segment. The ourmia-like viruses also possess capsid proteins distinct from each other and from those of plant ourmiaviruses (Fig. 1A). Furthermore, several of the invertebrate ourmia-like viruses encode a superfamily 3 helicase (S3H) that is typical of picorna-like viruses, but is not found in other levivirus-derived viruses of eukaryotes. In contrast to the ourmia-like viruses, all narna-like and mito-like viruses have minimal genomes that encode RdRp alone.
Fig. 1.
Evolution of the narnavirus-like (+)RNA viruses. (A) Schematic dendrogram based on the phylogenetic tree for RNA-dependent RNA polymerases (RdRp) of Narna-Levi clade from Shi et al. (2016a). Major clusters of related viruses are shown as triangles colored in accord with virus host ranges: grey, bacteria; olive, fungi; blue, invertebrates; green, plants. Rough diagrams of typical virus genomes for each cluster showing encoded proteins (rectangles; homologous proteins are in the same color), their functions and the genome size in kilobases (Kb), are at the right. CP, capsid protein; MP, movement protein; S3H, superfamily 3 helicase. (B) Hypothetical scenario for the evolution of narnavirus-like viruses. Vertical arrows denote virus transmission that accompanies host evolution, whereas horizontal arrows show presumed horizontal virus transfer (HVT) between distinct host organisms.
The currently defined, limited host range of the narna-like viruses that includes fungi, invertebrates and plants might seem to be poorly compatible with the apparent origin of the RdRps of these viruses from (+)RNA bacteriophages. Given the bacterial origin of mitochondria, it can be assumed that a levivirus-like virus of the α-proteobacterial ancestor of the mitochondrion was brought along during eukaryogenesis, and subsequently, upon the transition of the endosymbiont to the organelle life style, lost its coat and other non-replicational genes, becoming a selfish RNA element of the genus Mitovirus (Koonin and Dolja, 2014). The Narnavirus elements could originate from either a Mitovirus that escaped to the cytosol or (less parsimoniously) from another levi-like virus that adopted to reproduction in the cytosol. This scenario implies that the mitoviruses are ancestral in eukaryotes even though these viruses so far have not been discovered in any hosts other than the unikont and archaeplastida supergroups (Fig. 1B). Although the descendants of (+)RNA bacteriophages might have been lost in the rest of the eukaryotic lineages, we suspect that the major reason for their apparently narrow spread is the limited scope of protist metagenomics. Once such studies advance, mito-like viruses can be expected to be identified in diverse protists. Indeed, narna-like viruses have been recently detected in parasitic trypanosomatid protists (Akopyants et al., 2016a, Lye et al., 2016), as well as in Baltic sea meta-transcriptomes, although the hosts of these marine viruses remain known (Zeigler Allen et al., 2017).
Whereas mitoviruses and narnaviruses are most likely to be of ancestral, protoeukaryotic origin, a different evolutionary scenario appears likely for the ourmia-like virus branch. Given that the plant ourmiaviruses represent but a small twig within a huge and diverse lineage (i) of the levi-like viruses (Fig. 1A), most of which so far have been detected in invertebrates, HVT from herbivorous invertebrates (e.g., nematodes or arthropods) to plants emerges as the preferred evolutionary scenario (Fig. 1B). To switch to a plant host, an ancestral ourmia-like virus had to acquire a movement protein (a must for all non-defective plant viruses) from a pre-existing plant virus (Rastgou et al., 2009).
From an evolutionary taxonomy standpoint, the three major lineages of bacteriophage-derived (+)RNA viruses of eukaryotes, the mito-like, narna-like and ourmia-like viruses, each can be expected to eventually attain the family status. Furthermore, although the criteria for higher taxa of viruses are not currently well established, it appears likely that an umbrella taxon for these viruses, probably, at the order level, eventually would be created.
Similar to the (+)RNA virome, the dsRNA virome of bacteria is represented by a single, very small, family Cystoviridae (Krishnamurthy et al., 2016, Makeyev and Grimes, 2004, Poranen and Bamford, 2012), whereas no dsRNA viruses of archaea have been identified so far. The three-partite cystovirus genomes encode 14 proteins, which form a complex capsid that consists of two icosahedral shells. An internal shell has a T = 1 symmetry (often referred to as T = 2, because unlike true T = 1 capsids, a capsomer is a CP dimer), whereas an external T = 13 shell is formed from a different capsid protein (Makeyev and Grimes, 2004). The outer-most virion layer is formed by a lipoprotein envelope containing receptor-binding spikes. The crystal structure of the cystovirus RdRp exhibits a ‘palm-and-fingers’ fold typical of the viral RdRps and RTs, which supports a common origin for all of these polymerases. Structural comparisons have shown significant similarity between the cystovirus RdRp and that of the Hepatitis C flavivirus (Butcher et al., 2001). More recent comparisons, however, indicate a closer structural similarity between the RdRps of cystoviruses and picobirnaviruses, one of the eukaryotic dsRNA virus families (Krupovic, personal communication). The picobirnaviruses also share a T = 2 capsid symmetry with that of the internal shell of cystoviruses. Thus, the possibility seems to exist that picobirnaviruses are direct descendants of cystoviruses in eukaryotes, in parallel to the origin of nano-, mito- and ourmiaviruses from leviviruses (see the preceding section). Interestingly, the dsRNA reoviruses of diverse eukaryotes possess a T = 2 and T = 13 double-shelled capsid (Poranen and Bamford, 2012, Trask et al., 2012) similar to that of cystoviruses but not seen in any other viruses, suggesting an evolutionary connection between these virus families. Thus, the dsRNA bacteriophages and their apparent eukaryotic descendants exhibit complex evolutionary relationships that appear to involve both the replicational and the structural gene modules. Clarifying these relationships is in important problem on the way to a general concept of RNA virus evolution.
3. The RNA virome of unicellular eukaryotes is dominated by picornavirus-like and tombusvirus-like viruses
Unicellular eukaryotes (aka protists) is an umbrella name that covers extremely diverse organisms from all major eukaryotic lineages (Katz, 2012, Parfrey et al., 2006). Many protists are free-living organisms, both heterotrophs like amoebae and autotrophs like many photosynthesizing classes of phytoplankton. Other diverse protists, such as Leishmania or Plasmodium, are parasites. Although the extant protists are no less evolved than more complex, multicellular eukaryotes, they play a special role in discerning the origins of present-day viruses. One reason for this is that protists represent all major eukaryotic lineages (supergroups) that apparently radiated from the Last Common Eukaryotic Ancestor (LECA) over a relatively short time interval following the initial events of eukaryogenesis (Katz and Grant, 2015, Keeling et al., 2005, Koonin, 2010, Yoon et al., 2008). Therefore, direct ancestors of modern protists presumably were already around at the time when the ancestors of eukaryotic viruses have emerged. In contrast, the multicellular eukaryotes, such as some brown and green algae, oomycetes and fungi, as well as the more complex animals and vascular plants, form relatively small twigs in the phylogenic tree of eukaryotes that have emerged much later than protists (Berbee et al., 2017, Hinchliff et al., 2015, Katz and Grant, 2015). Furthermore, most protists are aquatic organisms that have never left the environment in which both the prokaryotic and eukaryotic life forms have evolved accompanied by their respective viromes. This primeval environment also provided a medium conducive to virus dispersal through diffusion, as well as to virus survival thanks to the protection from UV irradiation and desiccation.
Unlike protists, the majority of vascular plants and vertebrates, as well as a good share of invertebrates, adopted terrestrial life styles, which emerged much later than the aquatic ones and challenged viruses with developing distinct transmission pathways including aerosols or vector organisms. Taken together, all these evolutionary considerations imply that protists were the hosts to the most ancient groups of viruses and that viruses found in diverse protists evolved earlier that those whose host ranges are limited to multicellular eukaryotes.
The current knowledge on RNA viruses of protists comes both from metagenomic studies and from more traditional but much more limited in scope studies of viruses from known hosts. This traditional virology research initially uncovered only a few protist RNA viruses including yeast (+) RNA narnaviruses (Hillman and Cai, 2013), small dsRNA viruses (totiviruses from yeast and excavate parasites, and a single partitivirus from an apicomplexan parasite) (Ghabrial et al., 2015, Nibert et al., 2014) and a reovirus of a prasinophyte (a green alga) (Brussaard et al., 2004). More recently, several viruses of eukaryotic marine plankton of the alveolate and stramenopile groups have been discovered; intriguingly, all these plankton viruses appear to belong to the picorna-like superfamily (Kimura and Tomaru, 2015, Koonin et al., 2008, Lang et al., 2004, Nagasaki et al., 2005, Shirai et al., 2008, Takao et al., 2006, Tomaru et al., 2009).
The metagenomic studies have changed this barren landscape dramatically. Strikingly, the very first investigation of marine eukaryotic (+)RNA viruses revealed a community that appeared to be dominated by picorna-like viruses most similar to previously described viruses of eukaryotic plankton (Culley et al., 2006) implying that these viruses infect marine protists. A few tombus-like viruses also have been identified; although these viruses encoded RdRps related to those of plant tombusviruses, they lacked movement proteins typical of plant viruses indicating distinct, most likely protist hosts (Culley et al., 2006, Culley et al., 2007). Two other tombus-like viruses have been identified in holobionts of freshwater shrimp; because these viruses use ciliate genetic code, they likely infect these alveolate protists associated with shrimp (Shi et al., 2016a). Several subsequent studies on marine and freshwater RNA viromes have greatly expanded the geographic coverage from tropical to Antarctic seas to Antarctic inland lakes (Culley et al., 2014, Lopez-Bueno et al., 2015, Miranda et al., 2016, Moniruzzaman et al., 2017). However, the general trend in the RNA virome structure remained the same: the majority of new viruses were picorna-like, with a handful of tombus-like viruses. Likewise, a large variety of picorna-like viruses most similar to those found in marine metagenomes and various unicellular alveolates and stramenopiles have been identified in reclaimed water (Rosario et al., 2009).
Somewhat unexpectedly, several novel viruses that likely infect protists have been discovered in holobionts of aquatic invertebrates, such as mollusks, octopi, crustaceans and sipunculid worms (Shi et al., 2016a). These viruses dubbed Zhaoviruses represent a separate clade of (+)RNA viruses and use a variant genetic code typical of ciliates. The RdRps of several zhaoviruses show highly significant similarity to the RdRps of two previously described viruses, Ciliovirus and Brinovirus, that were isolated from wastewater and also use the ciliate code (Greninger and DeRisi, 2015b).
Although assigning hosts to viruses identified by metagenomics is not entirely straightforward and the sampling of diverse protists for viruses is quite patchy, the existing data provide for some potentially important, even if preliminary, generalizations. Because the lion share of RNA viruses discovered in a variety of diverse protists are (+)RNA viruses from the picorna-like lineage, this group most likely emerged and diversified very early in the evolution of eukaryotes (Koonin et al., 2008). Other lineages of potential ancient origin include (+)RNA tombus-like viruses and dsRNA totiviruses. The apparent absence of (−)RNA viruses in prokaryotes and unicellular eukaryotes implies their more recent origin, presumably associated with the emergence of multicellularity. Clearly, this evolutionary scenario would be falsified if (−)RNA viruses were discovered in unicellular eukaryotes. This latter possibility was emphasized by the detection of Mononegavirales-like viruses in the Baltic sea RNA virome although it is not clear if the hosts of these viruses are protists because some fish and mammalian viruses were also present in the samples (Zeigler Allen et al., 2017). Furthermore, a Bunya-like virus has been identified in a trypanosomatid protist parasite of insects (Akopyants et al., 2016b); in this case, an HVT from the host to the parasite is a suspect.
4. Overlapping RNA viromes of fungi, plants, vertebrates and invertebrates
The data on RNA viruses of primitive multicellular eukaryotes including oomycetes as well as brown and red algae are extremely scarce. In plant-pathogenic oomycetes, divergent viruses with picorna-like and noda-like RdRps have been identified (Heller-Dohmen et al., 2011, Yokoi et al., 1999, Yokoi et al., 2003). Intriguingly, the noda-like viruses encode capsid proteins that are most closely related to those of plant tombusviruses. Recently, a related virus has been identified in wastewater metagenomic samples and dubbed ‘tombunodavirus’ (Greninger and DeRisi, 2015a). The only RNA viruses isolated from red algae are dsRNA toti-like viruses and a picorna-like virus related to viruses of diatoms (Lachnit et al., 2015, Rousvoal et al., 2016).
Considerable progress has been made over the last decade towards characterization of the fungal RNA virome using both traditional and metagenomic approaches to virus identification (Deakin et al., 2017, Ghabrial et al., 2015, Marzano et al., 2016, Xie and Jiang, 2014). The fungal virome, as explored so far, is dominated by RNA viruses in general and dsRNA viruses in particular. It should be recognized, however, that sampling of the fungal virome remains limited, primarily, to yeast and plant-pathogenic fungi, whereas the enormous diversity of the fungal branch of eukaryotes, with estimated several million species (Berbee et al., 2017), remains a virtual terra incognita.
Among (+)RNA viruses, fungi are hosts to the capsid-less Narnaviridae discussed above, as well as diverse viruses from the alpha-like superfamily. Many of these fungal viruses are also capsid-less, including the fungal members of the families Endornaviridae and Gammaflexiviridae (genus Mycoflexivirus) (Koonin and Dolja, 2014). The RNA replication modules encoded by these viruses are most closely related to those of distinct lineages of plant viruses (Ghabrial et al., 2015). In the case of another group of capsid-less fungal viruses, the Hypoviridae, the replicative module clearly was acquired from an ancestor that belonged to Potyviridae, a large family of plant (+)RNA viruses (Dawe and Nuss, 2013, Koonin et al., 1991).
However, not all (+)RNA viruses of fungi are capsid-less. The fungal members of the family Alphaflexiviridae (genera Botrexvirus and Sclerodarnavirus) possess RNA replication modules and filamentous capsids typical of plant viruses of the same family, but lack homologs of plant virus movement proteins (Ghabrial et al., 2015). A recent metatranscriptome analyses recovered several fungal (+)RNA virus genomes related to Benyviridae, Betaflexiviridae, Virgaviridae, Tombusviridae, and Tymoviridae, large families of plant viruses for which no fungal members have been identified previously (Deakin et al., 2017, Marzano et al., 2016). Given the tight association between plants and plant-parasitic- and symbiotic fungi, an evolutionary scenario in which the fungal (+)RNA viruses have originated via HVT from plants appears to be plausible. An additional argument in support of this scenario is that plants host a greater diversity of viruses from each of the families mentioned above, implying a longer co-evolution of these virus families with plants than with the fungi. The inter-kingdom HVT to fungi might not be limited to plants: a capsid-less Sclerotinia sclerotiorum virus L exhibits the closest evolutionary affinity to animal viruses of the alpha-like superfamily (Liu et al., 2009). In contrast to the fungal (+)RNA viruses discussed above, Mushroom bacilliform virus and other recently discovered members of the family Barnaviridae (Marzano et al., 2016, Revill et al., 1994), possess distinct bacilliform capsids and appear to be the ‘original’ mycoviruses, with only very remote relatives within the picornavirus-like superfamily (Koonin et al., 2008).
As mentioned above, the fungal virome is enriched in dsRNA viruses, some families of which are thought to be unique to fungi (Ghabrial et al., 2015). These families include Chrysoviridae with four-component genomes and ‘true’ T = 1 icosahedral capsids built of a single capsid protein, Quadriviridae, whose four genome segments are encapsidated into isometric virions built of two capsid proteins, and Megabirnaviridae with icosahedral capsids and two genome segments.
Other families of dsRNA viruses that are common in fungi, but also found in other types of hosts include Totiviridae with monopartite genomes and Partitiviridae with bipartite genomes (Ghabrial et al., 2015). Partitiviruses are prevalent in both fungi and plants, and a partitivirus infecting a parasitic alveolate also has been characterized (Nibert et al., 2014). Two of the partitivirus genera, Alphaparititivirus and Betapartitivirus, harbor closely related viruses that infect either fungi or plants, again suggestive of extensive HVT between plant-parasitic fungi and their hosts. A less expansive dsRNA virus family isolated from both fungi and plants is Amalgaviridae, with monopartite genomes (Depierreux et al., 2016). Virions of amalgaviruses remain to be identified but protein sequence analysis has unexpectedly shown that these viruses encode a homolog of the nucleocapsid proteins of (−)RNA viruses of the genera Phlebovirus and Tenuivirus, suggesting evolution via unprecedented recombination between a dsRNA virus and a (−)RNA virus (Krupovic et al., 2015a). Notably, an amalgavirus genome has been recently identified also in the transcriptome of a microsporidium, a highly derived parasitic eukaryote distantly related to fungi; this is the first virus ever reported for Microsporidia (Pyle et al., 2017).
Recent metatranscriptomic analyses of marine fungi associated with a sea grass, as well as of an entomopathogenic fungus, also revealed prevalence of dsRNA viruses from families Totiviridae, Partitiviridae, Chrysoviridae, and Megabirnaviridae and several divergent dsRNA viruses that are yet to be formally classified (Kotta-Loizou and Coutts, 2017, Nerva et al., 2016). In addition, a handful of isosahedral dsRNA viruses of fungi appear to be related to Reoviridae (genus Mycoreovirus), a vast family of dsRNA viruses with the two-layered icosahedral capsids and 11–12 monocistronic genome segments (Ghabrial et al., 2015).
An unexpected addition to the fungal virome are (−)RNA viruses (Liu et al., 2014, Marzano et al., 2016, Yu et al., 2010). Among several recently discovered fungal (−)RNA viruses, Sclerotinia sclerotiorum negative-stranded RNA virus-1 (SsNSRV-1) has a monopartite genome, an enveloped helical nucleocapsid and belongs to a newly established family Mymonaviridae (genus Sclerotimonavirus) with an evolutionary affinity to family Nyamiviridae, also within the order Mononegavirales. Nyamiviruses infect vertebrates, arthropods and plant-parasitic soil nematodes that thrive in the same habitat as many soil fungi. Other (−)RNA viruses identified in fungal metagenomes possess segmented genomes related to either Bunyaviridae or Ophioviridae families (Marzano et al., 2016). The latter family, along with the genus Tenuivirus, is unusual among (−)RNA viruses in lacking the lipoprotein envelope (Kormelink et al., 2011). Ophioviruses infect plants, and at least some of them are vectored by soil-born fungi, once again suggesting HVT between plants and fungi.
In summary, the fungal RNA virome is rich in dsRNA and (+)RNA viruses with a dash of (−)RNA viruses. A notable trend in fungal virus evolution is the substantial representation of minimalist genomes that encode a single protein, the RdRp (Narnaviridae), or two proteins, RdRp and the capsid protein (Partitiviridae, Totiviridae). Another conspicuous tendency is the apparent extensive, two-way HVT between plant-parasitic fungi and their hosts. Such HVT is likely facilitated by the tight fungus-plant association that involves penetration of fungal appressoria through the plant cell walls (Ryder and Talbot, 2015), as well as by recruitment of fungal zoospores as transmission vectors by some plant viruses (Rochon et al., 2004). As discussed below, this form of HVT likely operates also between fungi and arthropods.
Although the plant RNA virome is heavily dominated by a diverse repertoire of (+)RNA viruses, it also includes a substantial variety of dsRNA and (−)RNA viruses (Dolja and Koonin, 2011). The plant virome had been relatively well sampled over the decades, mostly thanks to the traditional studies on viral diseases of crop plants. More recent metagenomics studies, also called ‘ecological genomics’, because viruses have been identified in individual plants collected from particular environments, such as prairie or tropical forest, have revealed prevalence of persistent viruses that cause no obvious disease in their wild plant hosts (Roossinck et al., 2015). These studies have also identified many new viruses that belonged to already known plant virus families. Although the obtained data sets have been claimed to include additional sequences of highly divergent, novel viruses, to our knowledge, none of these have been released so far. Likewise, metagenomics approaches to virus identification in crop plants, e.g., grapevine, yielded mostly RNA viruses of known families (Coetzee et al., 2010).
Until a few years ago, most of the known animal viruses have been identified in vertebrates. Similar to the plant virome, the vertebrate virome includes a rich variety of (+) RNA viruses and dsRNA viruses. The (−)RNA viruses are more broadly represented and more diverse in vertebrates than in plants. Comparison of the plant RNA virome to that of vertebrates has shown that many groups of (+)RNA, (−)RNA and dsRNA viruses contained related families or genera of both plant and animal viruses (Dolja and Koonin, 2011). The evolutionary relationships between animal and plant viruses were inferred primarily from shared modules of homologous genes involved in genome replication and expression, such as RdRps, RNA helicases, capping enzymes and proteases (Goldbach and Wellink, 1988, Koonin and Dolja, 1993, Koonin et al., 2015). However, apart from these genes involved in basic replication and expression functions, plant and animal viruses typically encode unrelated capsid proteins (with the exception of some picorna-like viruses) and proteins involved in virus-host interactions.
The evolutionary trajectories that resulted in these complex relationships among plant and animal viruses are not easy to reconstruct. One illuminating example is the alpha-like superfamily of (+)RNA viruses that is defined by the replication module comprised of a distinct capping enzyme, superfamily 1 RNA helicase and a specific lineage of RdRp (Koonin et al., 2015). The presence of this conserved module in both vertebrate and plant viruses clearly indicates monophyly of the alpha-like superfamily. Given the extreme evolutionary divergence and relatively recent origins of these ‘crown groups’ of multicellular, most complex eukaryotes, it could be proposed that the ancestral alpha-like virus emerged in a unicellular eukaryote prior to the divergence of the plant and vertebrate lineages. However, such a scenario is poorly compatible with the fact that so far no alpha-like viruses have been identified in unicellular eukaryotes or in marine viromes.
Furthermore, many alpha-like viruses of plants (Benyviridae, Virgaviridae, Alphaflexiviridae, Betaflexiviridae and Closteroviridae) possess simple, non-enveloped helical capsids (Dolja et al., 1991), in contrast to icosahedral capsids of the animal alphavirus-like viruses (Togaviridae with enveloped virions and Hepeviridae with non-enveloped ones). The capsid protein folds of helical plant viruses (Namba et al., 1989, Yang et al., 2012) are unrelated to those of icosahedral togaviruses and hepeviruses. So, what evolutionary scenario could reconcile the undeniable monophyly of the replication modules of plant and vertebrate alpha-like viruses with the equally obvious polyphyly of their capsids?
This problem could not be solved until an enormous variety of diverse RNA viruses including (+)RNA, dsRNA and (−)RNA viruses have been identified by holobiont metagenomics of invertebrates (Li et al., 2015, Shi et al., 2016a, Shi et al., 2016b). These recent studies show that the vast invertebrate RNA virome provides a pool of diverse genes and gene modules from which both plant and vertebrate RNA viruses could borrow repeatedly during the evolution of their respective viromes. Below we consider the major lineages of RNA viruses that are common to invertebrates, vertebrates, plants and fungi and propose that the evolution of RNA viruses in each of these lineages was dominated by extensive inter-kingdom HVT.
5. The origins and evolution of (+)RNA viruses
As discussed above, RNA phages of the family Leviviridae apparently gave rise to a distinct lineage of eukaryotic (+)RNA viruses (mitoviruses, narnaviruses, ourmiaviruses and their numerous invertebrate relatives) via HVT spanning two domains of life by way of mitochondrial symbiosis (Fig. 1). Following the original HVT event, presumably, from the proto-mitochondrion to the proto-eukaryote, the Levi-Narna clade was firmly established in fungi, being represented by the Mitovirus and Narnavirus genera. Intriguingly, a handful of crustacean viruses are lodged within the mitovirus cluster in the phylogenetic tree of the RdRp. Moreover, the yeast and other fungal viruses of the narnavirus cluster are sandwiched between branches filled with viruses of crustaceans and other invertebrates (Shi et al., 2016a). Assuming that fungi, which are omnipresent in aquatic environments, are older than invertebrates (Berbee et al., 2017), it appears likely that the latter acquired their narna-like viruses from the former via HVT. The opposite HVT direction is also possible, e.g., via fungi that are invertebrate parasites. As to plant Ourmiavirus, which clusters with the invertebrate narna-like viruses, there is little doubt about their origin through HVT from invertebrates (Fig. 1).
Because the remaining lineages of the eukaryotic RNA viruses have no immediate counterparts in prokaryotic RNA virome, it has been proposed that most of the eukaryotic RNA (and DNA) virus ancestors were conceived via mixing and matching of genes derived from viruses of prokaryotes, from the bacterial symbiont and from the emerging eukaryotic genome (Koonin et al., 2006). This hypothesis has found substantial support in the case of the picorna-like superfamily, where the conserved signature gene array appears to have been drawn from bacteriophages, bacterial selfish elements and the mitochondrial endosymbiont (Koonin et al., 2008). The global ecology of picorna-like viruses that includes diverse unicellular and multicellular eukaryotes is also in agreement with the early origin of these viruses followed by rapid diversification and continuous evolution along the emerging branches of eukaryotes. Moreover, picorna-like viruses appear to dominate extant marine viromes that consist mostly of viruses of unicellular eukaryotes, which persisted in this environment since the dawn of the eukaryotic life.
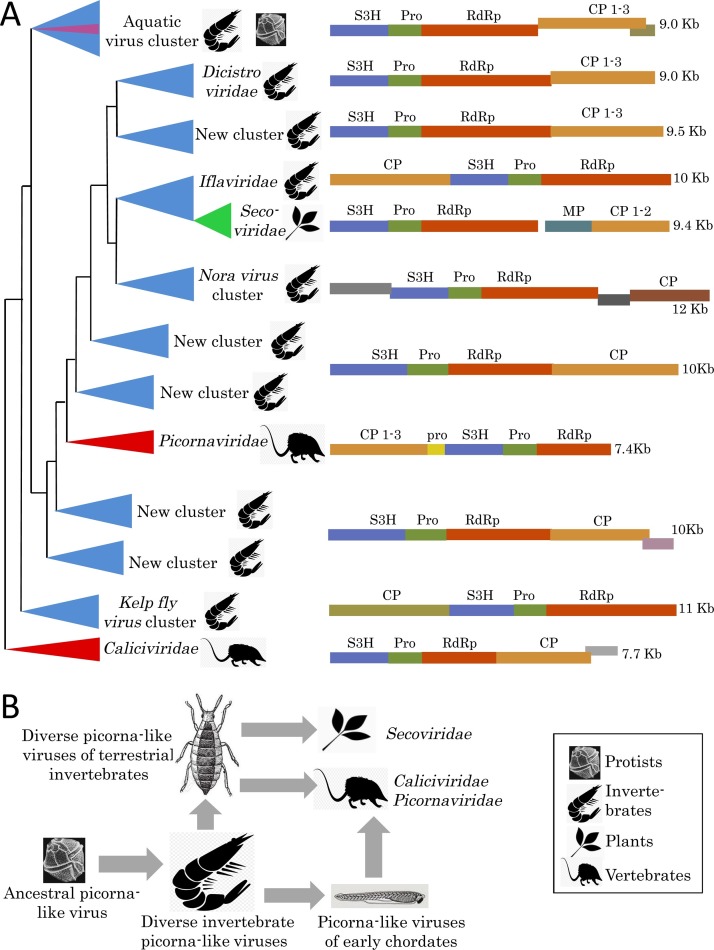
By and large, this ‘evolutionary continuity’ scenario for picorna-like viruses has survived the onslaught of metagenomics, but was substantially amended by the new data (Fig. 2 ). Among the staggering variety of picorna-like viruses, a vast ‘Aquatic picorna-like cluster’ has been identified as a monophyletic group (Shi et al., 2016a). This lineage combines viruses of diverse aquatic invertebrates including crustaceans, chelicerates and lophotrochozoans. Remarkably, different branches of the aquatic lineage also include viruses of diverse marine unicellular hosts such as diatoms, raphidophytes and thraustrochytrids, organisms that are phylogenetically far removed from invertebrates, but share the marine environment with many of them. Such tight evolutionary affinity between viruses infecting unrelated hosts but sharing the same habitat seems best compatible with extensive HVT. Should this be the case, it can be further hypothesized that the contiguous lineage of the ancient picornavirus-like viruses present in marine plankton seeded the emerging invertebrate virome early in the evolution of invertebrates (Fig. 2B). It stands to reason that this insemination occurred in the shared marine environment, during the Cambrian explosion, the rapid animal diversification that spawned most of the extant invertebrate lineages (Janvier, 2015, Knoll and Nowak, 2017).
Fig. 2.
Evolution of the picorna-like (+)RNA viruses. (A) Schematic dendrogram based on phylogenetic tree for RNA-dependent RNA polymerases (RdRp) of the Picorna-Calici clade from Shi et al. (2016a). Major clusters of the related viruses are shown as triangles colored in accord with virus host ranges: red, vertebrates; blue, invertebrates; green, plants; plum, protists. Approximate diagrams of typical virus genomes for each cluster showing encoded proteins and their functions (rectangles; homologous proteins are in the same color) and the genome size in kilobases (Kb) are at the right. CP, capsid protein; MP, movement protein; Pro, protease; S3H, superfamily 3 helicase. (B) Hypothetical scenario for the evolution of picornavirus-like viruses. Vertical arrows denote virus transmission that follows host evolution, whereas horizontal arrows show presumed horizontal virus transfer (HVT) events between distinct host organisms.
This hypothesis is also compatible with the clustering of the dinoflagellate picorna-like viruses with viruses of marine and freshwater mollusks, octopi, crabs, crayfish and millipedes within one branch of the ‘Luteo-sobemo clade’ of divergent picorna-like viruses (Shi et al., 2016a). Within this clade, plant Luteoviridae and Sobemovirus are deeply lodged among diverse viruses of invertebrates, suggesting HVT as the founding event for the origin of plant viruses. There are also fungal Badnaviridae in this mix, hinting at extreme promiscuity of HVT among highly diverse eukaryotic hosts.
The theme of rampant HVT keeps reappearing in other clades of the picorna-like viruses. Thus, a large family of plant viruses, Secoviridae, falls within a wider variety of the Iflaviridae and other viruses found in insects, spiders, ticks and other invertebrates (Fig. 2A), again suggesting that the ancestral secovirus arrived to the plant kingdom via HVT from arthropods followed by host- specific evolution in flowering plants (Shi et al., 2016a). The ‘Iflaviridae-Secoviridae cluster’ itself is but one branch within the vast ‘Picorna-Calici clade’, an expanded version of the recognized order Picornavirales. This clade also includes the ‘Dicistroviridae cluster’ packed with insect, crustacean and other invertebrate viruses, but also containing Antarctic picorna-like virus 1 from a lake, likely infecting a eukaryotic protist host (Lopez-Bueno et al., 2015).
Deep within the ‘Picorna-Calici clade’ is the ‘Picornaviridae’ branch, the thoroughly studied group of viruses that gave the name to the rest of the picorna-like viruses. The position of this branch leaves little doubt of its later emergence compared with the clusters containing viruses of unicellular eukaryotes and invertebrates (Fig. 2A) (Shi et al., 2016a). However, the exact ancestry of picornaviruses remains somewhat enigmatic because there is no compact sister group closely related to this family of vertebrate viruses. Such a sister group might be still lurking undetected in some unexpected habitat and would come up in subsequent metagenomics studies. Alternatively, it could be the case that, when the ancestor of Picornaviridae, most likely, an invertebrate virus, was transferred to a primitive chordate, rapid evolution required to adapt to a new type of host all but obliterated the ancestral evolutionary signal. Similar reasoning could apply to the ‘Caliciviridae’ branch that consists of a family of vertebrate viruses at the root of the expansive ‘Picorna-Calici clade’ (Fig. 2). In this case, however, a lone virus closely related to the caliciviruses has been identified in a freshwater mollusk. Ditto the ‘Astro’ clade, a lineage remotely related to other picorna-like viruses. This clade includes a family of vertebrate viruses Astroviridae and so far, only four viruses of diverse invertebrates (Shi et al., 2016a).
The evolutionary patterns among the picorna-like viruses of plants and vertebrates are notably contrasting. The families of picorna-like viruses of angiosperms are positioned within clusters of invertebrate viruses, apparently, indicative of relatively recent origins of the former from contiguous lineages of the latter via HVT (Fig. 2). In contrast, vertebrate picornaviruses, caliciviruses and astroviruses appear far diverged from their presumable invertebrate ancestors, implying longer and/or faster host-dependent evolution of the vertebrate viruses compared to that of their plant counterparts. This implication is compatible with the currently accepted timeline of evolution, according to which Chordates emerged in the marine environments, perhaps, as early as early Cambrian, ∼540 Mya (Janvier, 2015, Knoll and Nowak, 2017). The flowering plants’ line of decent comes from Zygnematophycea, filamentous or single-celled, freshwater algae of the Characean lineage (Umen, 2014). Upon terrestrialization ∼400 Mya, evolution of land plants included emergence of liverworts, mosses, lycophytes, horsetails and gymnosperms. A recent transcriptome analysis of the pre-flowering plant lineages has revealed only a marginal presence of (+)RNA viruses (Mushegian et al., 2016) implying that the enormous extant RNA virome of flowering plants emerged later, i.e. at the time when these plants conquered the land as late as early to mid-Cretaceous, ∼100 Mya (Bowman et al., 2007, Kenrick and Crane, 1997). Thus, the chordates would enjoy extra ∼400 M years for evolving their viromes after the presumed HVT from ancient marine invertebrates, which explains why these viromes appear farther diverged from invertebrate ones than those of plants (Fig. 2B). All the indications of HVT notwithstanding, the possibility remains that the early chordates received their pro-virome via vertical inheritance from their ancestors. Investigation of the extant viromes of basal branches of metazoa, such as sponges and Cnidaria, will help to choose between these possibilities.
A ‘picorna-like scenario’ of ancient origin and continuous evolution punctuated by bouts of HVT that coincide with explosive diversification of the hosts appears to be also applicable to the newly established Tombu-Noda clade of (+)RNA viruses (Shi et al., 2016a). Previous attempts at discerning the evolutionary relationships between (+)RNA viruses have suggested affinity between plant Tombusviridae and animal Flaviviridae that, however, relied solely on clustering of these two groups in the phylogenetic trees of the RdRp (Koonin and Dolja, 1993). The drastic increase in the number and diversity of the available virus genomes resulted in an apparent flavi-tombus breakup. An even more feeble relationship between nodaviruses and picorna-like viruses has also ended in separation that was buttressed by the identification of a putative capping enzyme encoded by nodaviruses, but never found in picorna-like viruses (Ahola and Karlin, 2015). Furthermore, when the enormous ecological and evolutionary diversity of both tombus-like viruses and noda-like viruses has been revealed, these two major lineages showed mutual attraction in the amended phylogenetic trees of the RdRps, resulting in the ‘Tombus-noda clade’ of small icosahedral (+)RNA viruses (Shi et al., 2016a).
Phylogenetic analysis of the RdRp shows that the tombus-like branch of the Tombus-noda clade encompasses an enormous variety of viruses found in highly diverse aquatic and terrestrial invertebrates, from nematodes to mollusks to crustaceans to insects. Somewhat shockingly, the genera of the plant Tombusviridae and the related genera Umbravius and Luteovirus are buried deep within distinct, well-supported branches that consist primarily of viruses of aquatic invertebrates (Shi et al., 2016a). This phylogenetic pattern clearly implies a relatively recent, polyphyletic origin of plant viruses via multiple events of HVT from separate invertebrate viruses.
The ‘classical’ Nodaviridae were sharply split between two genera of insect Alphanodavirus and fish Betanodavirus, a disconnected host range, given the evolutionary and ecological distance between the insects and fish. This clean split started to deteriorate upon identification of divergent noda-like viruses in oomycetes (Heller-Dohmen et al., 2011, Yokoi et al., 2003) and nematodes (Felix et al., 2011, Franz et al., 2012). The massive transcriptome analysis of various invertebrates further bloated this virus lineage with new viruses that possessed either alphanodavirus-like or tombusvirus-like capsid protein (also present in betanodaviruses), as well as with some viruses with a capsid protein that appeared related to those of marine picorna-like viruses (Shi et al., 2016a). These new findings have blurred the boundaries between the two nodavirus genera while closing the gulf in the host range of the Nodaviridae by expanding it across a wide range of animals. Furthermore, the large group of viruses closely related to fish Betanodavirus turned to be viruses of crustaceans, an older animal group than fish, but sharing the same environment. Thus, the same, now familiar evolutionary scenario would involve diversification of the noda-like lineage in invertebrates followed by HVT from crustaceans to fish. Recently, the Tombus-noda clade has received another reinforcement in Statoviruses, a novel group of viruses found in mammalian stool samples (Janowski et al., 2017). It remains to be established if statoviruses reproduce in mammalian hosts or in organisms present in their alimentary tracts. The extremely broad host range of the Tombus-noda group that includes diverse unicellular and multicellular eukaryotes, together with the existence of all possible combinations of the noda-like and tombus-like RdRps with noda-like and tombus-like capsid proteins, imply an ancient origin, co-habitation and notable host promiscuity of these small viruses.
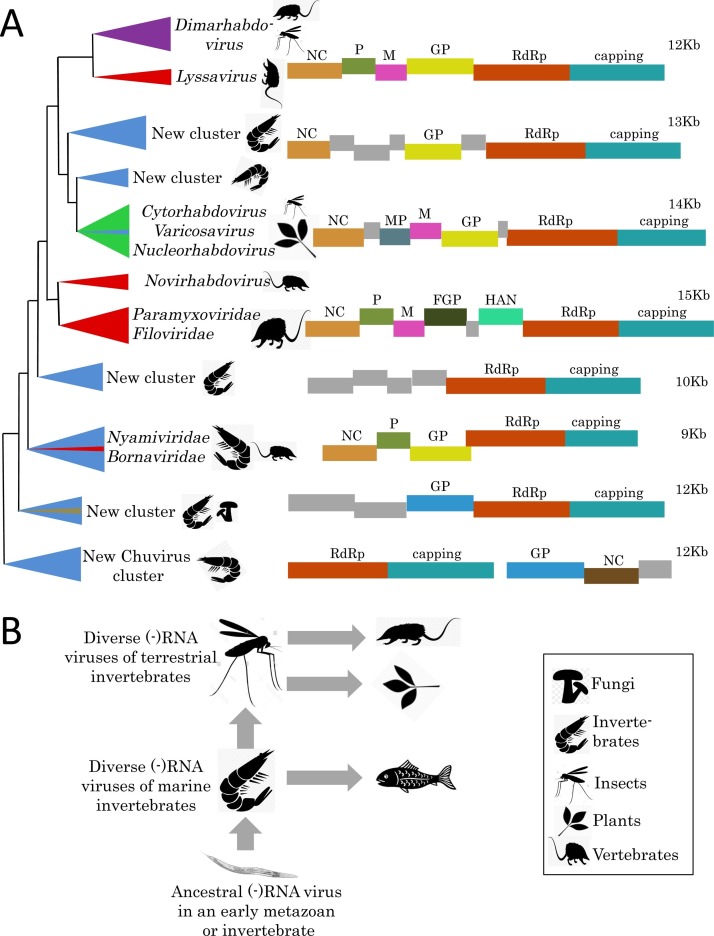
The phylogenetic divorce between tombus-like viruses and Flaviviridae-like viruses was accompanied by a radical makeover of the latter clade, thanks to the flood of metagenomic data (Bekal et al., 2014, Fauver et al., 2016, Hall et al., 2016, Kobayashi et al., 2013, Qin et al., 2014, Shi et al., 2016b, Teixeira et al., 2016, Webster et al., 2016). Only a few years ago, this clade had encompassed the single family Flaviviridae, primarily mammalian viruses, some of which also infect arthropods that serve as vectors. These flaviviruses have relatively uniform, monopartite, 10–12 kb genomes with a conserved gene module that encodes protease, superfamily 2 helicase, capping enzyme and RdRp (Koonin et al., 2015). Metagenomic analysis resulted in the discovery of flavi-like viruses that are specific to arthropods (rather than shuttling between mammals and arthropods as the classical flaviviruses do), nematodes and plants, with monopartite genomes up to 26 kb in length, twice the size limit of the ‘orthodox’ Flaviviridae. In addition, several novel branches of flavi-like viruses, such as arthropod Jingmenvirus were discovered (Qin et al., 2014, Shi et al., 2016b); in contrast to other viruses of this clade, the genomes of jingmenviruses are tetrapartite.
The phylogenetic trees that reflect the evolution of the most conserved parts of the flavivirus-like polyproteins (including the RdRp) clearly show that vertebrate viruses comprise a subset of the diversity of viruses identified in invertebrates (Shi et al., 2016b). These findings imply that vertebrates have acquired flavi-like viruses via HVT that was probably facilitated by tight associations of blood-sucking arthropods with their vertebrate prey. Given the existence of closely related viruses of plant-parasitic nematodes (Bekal et al., 2014) and aphids (Teixeira et al., 2016), the origin of the plant-infecting flavi-like viruses so far discovered (Kobayashi et al., 2013) similarly could be explained by HVT from invertebrates feeding on plant roots or leaves. So far, no flavi-like viruses have been found in unicellular eukaryotes, implying a later origin of this group, perhaps in ancestral metazoa, although discovery of related viruses infecting protists could refute this hypothesis.
Similar considerations apply to the alpha-like superfamily of (+)RNA viruses (named Hepe-Virga clade by Shi et al., 2016a), which have not yet been identified in protist or aquatic viromes, but are highly abundant in invertebrates and plants, although much less so in vertebrates and fungi. A peculiar ‘TMV-like’ virus has been identified in Chara australis, a representative of the algal branch that is ancestral to the flowering plants (Gibbs et al., 2011). Because the CP of this virus is similar to those of invertebrate viruses (Shi et al., 2016a) and the MP is apparently absent, this virus could be a result of HVT from aquatic invertebrates (e.g., snails).
The latest phylogenetic tree of the RdRps of alpha-like viruses (the ‘Hepe-Virga clade’) splits into two major clusters, each including viruses of invertebrates, plants and vertebrates, with a pinch of fungal viruses here and there (Shi et al., 2016a). Deep within one of these clusters, are three families of plant viruses (Virgaviridae, Bromoviridae and Closteroviridae) and the Alphavirus genus of the vertebrate virus family Togaviridae, interspersed with invertebrate viruses. This tree topology clearly indicates that the plant and vertebrate viruses of this cluster evolved from diverse groups of the older invertebrate viruses.
Comparison of the genome architectures of the alpha-like viruses further supports their origin in invertebrates. The most striking relevant finding is the discovery of homologs of the Tobacco mosaic virus (TMV)-like capsid proteins in several novel crustacean and insect viruses. Previously, the TMV-like CPs have been thought to be specific to an array of plant viruses with rigid, rod-shaped capsids (Dolja et al., 1991). The discovery of TMV-like capsids in invertebrate viruses, although not revealing the ultimate ancestry of their unique α-helical fold, implies an origin outside the plant virome. It should be emphasized, however, that the diverse proteins that drive cell-to-cell movement of plant viruses, as well as those that suppress plant RNAi defense, remain unique to the plant RNA virome (Csorba et al., 2015, Heinlein, 2015). Notably, one of the insect viruses that encodes a TMV-like capsid protein also encompasses an unusual, large RNA replication module similar in size to those of Closteroviridae, suggesting that this family of the largest plant viruses with ∼15–20 kb genomes (Dolja et al., 2006) also emerged from a particular lineage of invertebrate viruses.
Strikingly, inside the first cluster in the Hepe-Virga clade (Shi et al., 2016a), there are a few plant viruses of the recently established genus Cilevirus, surrounded by the diverse invertebrate viruses (Locali-Fabris et al., 2006, Melzer et al., 2013, Roy et al., 2013). These viruses, similarly to many vertebrate and invertebrate viruses of the alpha-like virus superfamily, possess enveloped particles, a feature that is so far unique among plant alpha-like viruses, which typically form simple proteinaceous capsids. Given the position of the cilevirus RdRps in the phylogenetic tree and their transmission by arthropod vectors, it seems most likely that cileviruses are a result of a relatively recent HVT from an arthropod to a plant. Apparently, all it takes for a virus that normally infects a plant-parasitic arthropod to expand its host range to plants, is to acquire a movement protein and, for good measure, an RNAi suppressor.
The family of capsid-less, persistent viruses Endornaviridae is the most divergent branch of the first cluster of the Hepe-Virga clade. The host range of Endornaviridae, which are common in plants and fungi (Koonin and Dolja, 2014, Roossinck et al., 2011), has been recently expanded to include oomycetes and invertebrates (Kozlakidis et al., 2010, Shi et al., 2016a). The basal position of this branch suggests either that endornaviruses are the oldest in this cluster or that their switch to persistent life style with no extracellular phase triggered rapid divergence from their infectious relatives.
The second major cluster of alpha-like viruses encompasses the Rubellavirus genus of the vertebrate Togaviridae and the Benyviridae family of plant viruses with closely related replication modules, the vertebrate family Hepeviridae, the Alphatetraviridae family of insect viruses within variety of invertebrate viruses, and the most basal branch comprised of the plant Tymovirales (Shi et al., 2016a). One surprising feature in this cluster is the discovery of invertebrate viruses that fall within icosahedral Tymoviridae and helical Alphaflexiviridae, two families of plant viruses that each also include a few fungal members. Although the evolutionary trajectory of the Tymovirales is difficult to reconstruct with the existing data, HVT clearly played a major role in shaping the diverse host range of this virus order. Another notable feature of this cluster is the clear split of the Togaviridae genera into two distinct lineages of alpha-like viruses, suggesting reclassification of this family into two separate taxa.
Collectively, the new data on the alpha-like virus superfamily put us in a better position to solve the conundrum of virus evolution formulated above: the uniform conservation of the replication module in this superfamily versus the seemingly erratic distribution of the host-specific structural modules (or lack thereof, as in endornaviruses) among vertebrate- and plant-infecting viruses. It is not yet certain whether the alpha-like replication module emerged early in unicellular eukaryotes, or later in metazoa. Obviously, however, the phylogenomic diversity of invertebrate alpha-like viruses encompasses prototypes of the structural modules found in both vertebrate and plant viruses. Moreover, some of the invertebrate viruses of this superfamily possess structural modules that include glycoproteins closely related to those of (−)RNA bunyaviruses (Shi et al., 2016a). This unexpected connection between (+)RNA and (−)RNA viruses becomes even more intriguing given the structural similarity between the capsid proteins of the Alphaflexiviridae and the nucleocapsid proteins of Phlebovirus (Bunyaviridae) (Agirrezabala et al., 2015). If borne out by additional observations, this similarity would suggest a common origin of the structural proteins that form flexuous helical capsids of numerous plant viruses and the similarly flexuous, helical nucleocapsids of many animal viruses.
The primary conclusion from the expanded analysis of the alpha-like virus superfamily is the same as that for the other (+)RNA viruses discussed above: the phylogenomic diversity of the invertebrate virome far exceeds those of plant and vertebrate viruses. Because the origin of invertebrates antedates that of vertebrates and flowering plants, it seems all but certain that the invertebrate virome served as a pool from which evolving plants and vertebrates have drawn the ancestors of their respective viromes.
The order of animal viruses Nidovirales is something of an evolutionary puzzle. It cannot be readily placed into the currently recognized superfamilies of the (+)RNA viruses although affinity with picorna-like viruses has been proposed based on the RdRp comparison and the presence of a 3C-like protease typical of picorna-like viruses in the Coronaviridae family of Nidovirales (Koonin et al., 2015). So far, no nido-like viruses have been found in unicellular eukaryotes or any branches of the eukaryotic tree other than animals. The phylogenetic tree of the ‘Nido clade’ is currently split into four major lineages: i) the deepest-rooted branch with two insect and one spider virus; ii) vertebrate Coronaviridae (Corovavirinae); iii) insect Mesoniviridae and crustacean Roniviridae intermixed with divergent crustacean and mollusk viruses; iv) vertebrate Arteriviridae and Coronaviridae (Torovirinae) (Shi et al., 2016a). Surprisingly, the latter subfamily also contains two viruses associated with the snake nematodes, both closely related to a Ball python nidovirus, which causes a severe respiratory disease in this snake (Stenglein et al., 2014). This could be a case of a recent HVT although a nematode contamination with virus-containing snake material cannot be ruled out. The overall topology of the RdRp tree suggests that the nido-like clade of animal viruses emerged in early invertebrates and then invaded the vertebrate virome via HVT. The relatively small genome size of the deep-rooted arthropod viruses (roughly half of that of vertebrate coronaviruses) agrees with this interpretation, further suggesting that the largest known RNA genomes of Coronaviridae (∼30 kb) have evolved via accretion of genes during co-evolution with the vertebrate hosts.
As follows from the discussion above, the accelerating expansion of the RNA virosphere brought about by the progress of metagenomics has already reshaped the evolutionary tree of (+)RNA viruses. The flavi-like superfamily, with its dubious unification of flaviviruses and tombusviruses, is gone but the departure of the tombus-like viruses is compensated by the dramatic expansion in the diversity and host range of new flavi-like viruses. In parallel, the vast picorna-like superfamily has lost its purported peripheral members, the noda-like viruses, and yet, became even vaster by absorbing new large clusters of undeniable picorna-like viruses, such as the ‘aquatic cluster’. The third of the previously established (+)RNA virus superfamilies, the alpha-like one, while absorbing numerous invertebrate viruses, retained all the previously included families of the plant and vertebrate viruses. A major new addition to the evolutionary layout of the (+)RNA virome is the expansive, widespread tombus-noda supergroup that includes the smallest and simplest among the known RNA viruses of eukaryotes. This simplicity of genome architecture, along with the broad host range, suggests that tombus-noda viruses, together with the picorna-like viruses, are the most ancient (+)RNA viruses of eukaryotes. However, caution is still due, given that the positive evidence in support of the tombusvirus-nodavirus affinity stems solely from the topology of the RdRp tree. Although the current genome collection vastly exceeds that of two decades ago, lending extra credence to the phylogeny, it is difficult to rule out that, with further accumulation of diverse genomes, this supergroup goes the way of the flavi-tombus assemblage.
6. The dsRNA virome
A leading concept of the origins of dsRNA viruses is the polyphyly of this virus class with distinct lineages of dsRNA viruses evolving from separate (+)RNA virus ancestors (Koonin, 1992, Koonin et al., 2015). This interpretation is beyond reasonable doubt for at least two families of viruses that have been traditionally included in the dsRNA class, Endornaviridae and Hypoviridae. However, it is now realized that members of these two families are not bona fide dsRNA viruses which, by definition, possess encapsidated dsRNA genomes. Although dsRNAs of endorna- and hypoviruses could be isolated from their persistently infected hosts, both families represent capsid-less RNA replicons with clear relationships with distinct groups of (+)RNA viruses, the alpha-like superfamily in the case of the endornaviruses and potyviruses in the case of the hypoviruses (Koonin and Dolja, 2014).
Among the dsRNA viruses proper, several families appear to be distantly related to the picorna-like (+)RNA viruses (Koonin et al., 2008), whereas bacteriophages of the Cystoviridae family could be direct ancestors of the eukaryotic virus family Picobirnaviridae (see above) and have been also tentatively linked to the Reoviridae, an expansive family of eukaryotic dsRNA viruses (Koonin et al., 2015). It remains to be seen whether these proposed evolutionary associations withstand future, more comprehensive phylogenetic analyses of the dsRNA viruses. Nevertheless, the substantially expanded knowledge of the dsRNA virome of invertebrates, fungi, plants and other eukaryotes, is already changing the scenarios developed previously for a much smaller dsRNA virus diversity.
The small and simple toti-like viruses, many of which encode only a RdRp and a capsid protein in their monopartite dsRNA genomes, show little congruence between the virus phylogeny and host range. The ‘Toti-Chryso clade’ of viral RdRps combines the families Totiviridae, Chrysoviridae and Quadriviridae with a panoply of newly discovered viruses of invertebrates, fungi, plants, vertebrates and likely diatoms, and splits into two major clusters (Shi et al., 2016a). In one cluster, there is a well-supported branch that encompasses Victorivirus, Leishmaniavirus and Trichomonasavirus genera of Totiviridae. A notable feature of this branch is that the former genus includes fungal viruses, whereas the other two consist of viruses of parasitic excavate protists. The enormous evolutionary distance that separates the two host taxa implies that the close phylogenetic relationship between viruses infecting parasites of vertebrates (Leishmania and Trichomonas) and of plants (many of the fungi hosting victoriviruses are plant-parasitic) is due to HVT. It is difficult to imagine an ecological niche where such HVT could have occurred but missing links are likely to exist. Notably, this branch of the Toti-Chryso clade also includes a Diatom colony-associated virus (Urayama et al., 2016). The diatoms belong to stramenopiles, a eukaryotic lineage that is equally distant from fungi and excavates. This diatom virus is most closely related to a virus identified in a razor clam holobiont; this relationship could be due either to HVT from a diatom to a mollusk that feeds by filtrating water often containing diatoms, or to contamination of the mollusk holobiont with totivirus-infected diatoms.
The remaining branches in the first cluster contain two families of quadripartite fungal viruses, Chrysoviridae and Quadriviridae, and the genus Totivirus, each now intermixed with viruses of insects and some other invertebrates (Shi et al., 2016a). Once again, it is not immediately clear how HVT between insects and mostly plant-pathogenic fungi could occur, although involvement of mycophagous insects is a possibility. This cluster also contains another virus from a diatom colony related to one found in barnacles and a plant virus related to arthropod virus. These latter cases could result from more transparent HVT events between a marine crustacean and a diatom, and a terrestrial insect and a plant, respectively.
The second major cluster of the Toti-Chryso clade is populated by a variety of invertebrate viruses. However, in the midst of these, there is a Giardia lamblia virus (Giardiavirus, Totiviridae), another virus of an excavate, a common intestinal parasite of vertebrates. An additional oddity in this cluster is Piscine myocarditis virus that is common in Atlantic salmon (Haugland et al., 2011) and is closely related to a virus isolated from crabs in China. Given that totiviruses are common in invertebrates, whereas the salmon virus is so far the only known vertebrate totivirus, the direction of HVT in this case is probably salmon-eats-crab, not the other way around, even though the latter happens no less frequently than the former.
What have we learned from the much expanded host range and diversity of the toti-like viruses? The one clear conclusion is the ultimate host promiscuity and propensity of toti-like viruses to HVT although the exact routes of HVT often appear puzzling. Obviously, deeper sampling of excavates and fungi including non-parasitic ones, as well as a better understanding of ecological contacts between excavates, fungi and invertebrates is needed to get a better grasp of the HVT pathways taken by toti-like viruses.
‘The Partiti-Picobirna clade’ of dsRNA viruses in many ways resembles the toti-like viruses. This clade is split into four major clusters (Shi et al., 2016a). The first cluster includes Alpha- and Betapartitivirus genera of Partitiviridae; each of these genera contains closely related viruses of fungi, plants and a few invertebrate viruses, clearly implying HVT between their hosts. The sister branch in this cluster contains a multitude of invertebrate (mainly insect) viruses. Remarkably, whereas the typical partitiviruses possess small bipartite genomes encoding only RdRp and capsid protein, their arthropod relatives have mono-, bi-, quadri- or even hexapartite genomes.
The second cluster in this clade consists largely of crustacean viruses together with viruses of other invertebrates and a few unclassified fungal viruses. The third cluster combines plant Delta- and fungal Gammapartitivirus genera of Partitiviridae and invertebrate viruses. Finally, the fourth, most basal cluster includes primarily mammalian Picobirnaviridae, many invertebrate viruses that infect crustaceans and other marine invertebrates and a likely diatom virus.
Thus, the Partiti-Picobirna clade, similarly to toti-like viruses, gives indications of rampant HVT among viruses infecting fungi, plants, invertebrates, vertebrates and other divergent hosts including the parasitic alveolate Cryptosporidium parvum (Khramtsov and Upton, 2000), likely diatoms (Urayama et al., 2016), and green algae (Mushegian et al., 2016). Intriguingly, a subset of partiti-like invertebrate viruses use the mitochondrial genetic code suggesting either their ancient origin during eukaryogenesis or a later switch to intra-mitochondrial reproduction (Shi et al., 2016a). The former possibility seems to be better compatible with the existence of partitivirus-like RNA replicons in mitochondria and chloroplasts of green algae (Koga et al., 1998, Koga et al., 2003). However, no convincing phylogenetic affinity between partitiviruses and the only known group of prokaryotic dsRNA viruses, the cystoviruses, has been established so far. To solve this conundrum, search for dsRNA bacteriophages demonstrably related to partitiviruses, as well as a more detailed comparison of the amino acid sequences and 3D structures of the cystovirus and partitivirus RdRps and capsid proteins will be helpful.
Compared to the haphazard phylogenetic patterns of toti-like and partiti-like viruses, the ‘Reo-like clade’ that includes a well-established, 15 genera-strong family Reoviridae, with the addition of new viruses of arthropods, is quite orderly with respect to the apparent virus-host co-evolution (Shi et al., 2016a). The reovirus genomes encompass from 9 to 12 segments of dsRNA reaching the total capacity of 32 kb, which seems to come close to the upper size limit for RNA genomes; in addition to the segmented genomes of reoviruses, this limit is reached only by some Nidovirales and flavi-like viruses, both possessing monopartite (+)RNA genomes.
The dominant evolutionary pattern in reoviruses is that most of the known genera include viruses with homogenous host ranges. Thus, the genus Cypovirus consists of insect viruses, the genera Orthoreovirus, Rotavirus and Aquareovirus are vertebrate-specific, whereas viruses in the genus Mycoreovirus infect fungi. Several reovirus genera exhibit double loyalty to vector arthropods and either vertebrates (Orbivirus, Coltivirus and Seadornavirus) or plants (Phytoreovirus, Fijivirus and Oryzavirus). The new reoviruses discovered in invertebrates also tend to form compact clusters of viruses that infect insects or crustaceans. The only known outliers among reoviruses in terms of unusual hosts are two divergent viruses that infect a picoeukaryote green alga Micromonas pusilla (Brussaard et al., 2004) and a blood fluke parasitic flatworm (Platyhelmintes) (Shi et al., 2016a).
The overall topology of the phylogenetic tree of the reovirus RdRp resembles those for several large taxa of (+)RNA and (−)RNA viruses (see below): i) diversity of invertebrate viruses supersedes that of both vertebrate and plant viruses, which appears to be indicative of their evolutionary primacy; ii) plant virus branches are deep inside the diversity of arthropod viruses suggesting HVT, perhaps during the plant radiation in the Cretaceous; iii) reoviruses that infect either only vertebrates or vertebrates and arthropods have more complex genomic layouts (more segments and accordingly more encoded proteins) than the plant reoviruses suggestive of longer co-evolution with the vertebrate hosts, perhaps starting in the Cambrian. Although the uncanny similarity between double-shelled cystovirus and reovirus capsids implies potential ancestor-descendant relationship (see above), a verdict on the ultimate origin of the reoviruses has to await a more detailed understanding of the phylogeny of the reovirus RdRp and the provenance of the other genes of these viruses.
To summarize, the overview of the three largest clades of dsRNA viruses identifies two general evolutionary patterns. One is the rampant HVT that is apparently driven by the host promiscuity among the small dsRNA viruses (toti-like and partiti-like), many of which lead non-infectious, persistent life style. The other pattern is the much greater degree of host specificity in the expansive family Reoviridae than among the rest of the dsRNA viruses, apparently due to a greater extent of host adaptation via acquisition of genes involved in virus-host interactions.
7. The (−)RNA virome
The origin of the (−)RNA viruses was and remains a major puzzle of evolutionary virology. Until relatively recently, these viruses have been identified only in vertebrates, plants and arthropods that vectored some (−)RNA viruses between their vertebrate or plant hosts. Phylogenomic comparisons of the viruses infecting vertebrates and plants, two distant lineages of the multicellular eukaryotes, indicated that the plant (−)RNA virome is a subset of that of the vertebrates (Kormelink et al., 2011). For instance, the family Tospoviridae of plant (−)RNA viruses placed within the much larger order of the primarily vertebrate viruses, Bunyavirales. Moreover, comparison of genome architectures shows that the genomes of plant (−)RNA viruses are closely similar to those of the vertebrate relatives, with the additions of plant virus-specific movement proteins and RNAi suppressors that were likely acquired from plant (+)RNA viruses (Dolja and Koonin, 2011, Kormelink et al., 2011). To explain this evolutionary pattern, it has been proposed that the plant (−)RNA virome evolved via HVT from vertebrates mediated by arthropod vectors (Dolja and Koonin, 2011).
The amino acid sequences of the (−)RNA virus RdRps, although conserved within this virus class, are so divergent from the other known virus RdRps that sequence comparison failed even to clearly show that they possess the prototypical palm-and-fingers structure, let alone to allow inferences of their origin. Analysis of the first solved 3D structure of a (−)RNA virus RdRp, that of Bat influenza virus A, has unequivocally shown that not only this RdRp shared the fold with other virus RdRps and RTs, but that it appeared to be most similar to the flavivirus RdRps (Pflug et al., 2014). This outcome implied a monophyletic origin of virus RdRps and suggested the possibility that (−)RNA viruses have evolved from the more ancient (+)RNA viruses, plausibly, in primitive metazoa. This possibility is well compatible with the apparent absence of (−)RNA viruses in protists.
The recent forays into RNA virus metagenomics of invertebrates revealed an unexpected diversity of (−)RNA viruses including novel virus taxa for which the names Chuvirus, Quinvirus and Yuevirus have been proposed (Bekal et al., 2011, Li et al., 2015, Shi et al., 2016a). In addition, several diverse (−)RNA viruses have been identified in fungi (Liu et al., 2014, Marzano et al., 2016). Investigation of the expanded (−)RNA virome has shown that previously established taxa are nested within a much greater phylogenomic diversity of invertebrate (−)RNA viruses, suggesting that invertebrates represent a natural reservoir from which the ancestors of the related plant and vertebrate (−)RNA viruses were recruited via HVT. This notion is further supported by the evolutionary pattern whereby diverse invertebrates in the same ecological niche often share similar viruses, with incongruent virus and host phylogenies, again pointing to pervasive HVT and host promiscuity of invertebrate viruses (Li et al., 2015).
According to the recent, large-scale phylogenetic analysis, RdRps of (−)RNA viruses are divided into at least 6 major clades (Shi et al., 2016a). One of the two largest ones is the ‘Mono-Chu clade’ combining Mononegavirales with monopartite genomes and the Chuviruses, which possess monopartite or bipartite, apparently, circular genomes (Fig. 3 A). In the phylogenetic tree of the RdRps, the Chuvirus is the basal lineage, whereas in the rest of tree, the traditional families of vertebrate and plant mononegaviruses are buried within the dominant diversity of the invertebrate viruses, implying the ancestral status of the latter. In a remarkable instance of arthropod-transmitted plant viruses of the genus Cytorhabdovirus, such viruses as Persimmon virus A or Lettuce necrotic yellows virus have viruses of insects rather than other plant viruses as their closest kin. Together with the tendency of the plant rhabdoviruses to group according to their insect vectors rather than plant hosts (Kormelink et al., 2011), this phylogenetic outcome clearly favors a relatively recent HVT of rhabdoviruses from insects to plants.
Fig. 3.
Evolution of the mononegavirus-like (−)RNA viruses. (A) Schematic dendrogram based on the phylogenetic tree for RNA-dependent RNA polymerases (RdRp) of the Mono-Chu clade from Shi et al. (2016a). Major clusters of the related viruses are shown as triangles colored in accord with the virus host ranges: blue, invertebrates; red, vertebrates; olive, fungi; green, plants (corresponds to arthropod-transmitted plant viruses); purple, insects and vertebrates. Rough diagrams of typical virus genomes for each cluster showing encoded proteins and their functions (rectangles; homologous proteins are in the same color) and the genome size in kilobases (Kb) are shown at the right. NC, nucleocapsid protein; M, matrix protein; P, phosphoproten; GP, glycoprotein; FGP, fusion glycoprotein; MP, movement protein; HAN, hemagglutinin-neuraminidase. (B) Hypothetical scenario for the evolution of (−)RNA viruses. Vertical arrows denote virus transmission that accompanies host evolution, whereas horizontal arrows show presumed horizontal virus transfer (HVT) events between distinct host organisms.
The second large group of (−)RNA viruses is the ‘Bunya-Arena clade’, which includes the order Bunyavirales and the family Arenaviridae, again within the overwhelming diversity of the related invertebrate viruses that are typically basal relative to the plant and vertebrate virus taxa (Shi et al., 2016a). Although most of the virus genomes in this clade possess 2 or 3 minus strand or ambisense genome segments, plant Tenuivirus have 4–6 segments. The ‘Orthomyxo clade’ of (−)RNA viruses seems to show some affinity to the Bunya-Arena clade; fittingly, the genomes of orthomyxo-like viruses consist of 6–8 segments. In a phylogenetic pattern that is already too familiar from the preceding discussion of RNA virus evolution, the bona fide influenza viruses form but a twig deep within the RdRp phylogenetic tree that is full of invertebrate viruses, many of which were identified in insects. However, because the most basal of the orthomyxo-like viruses were identified in Lophotrochozoa (an earthworm and a mollusk), as well as in marine tunicates (early chordates), it seems that vertebrate influenza-like viruses might have evolved in marine invertebrates, were passed on to early chordates and then followed those during their terrestrialization.
The remaining three, relatively small clades of (−)RNA viruses are quinviruses and yueviruses that have been identified in crustaceans and other invertebrates, as well as the plant virus family Ophioviridae. These viruses possess 2–3 genome segments and occupy intermediate positions between the Bunya-Arena/Orthomyxo and Mono-Chu clades. In contrast to the vast majority of enveloped (−)RNA viruses, plant Ophyoviridae and Tenuivirus form atypical, non-enveloped helical, flexuous nucleocapsids similar to those of typical enveloped viruses (Kormelink et al., 2011). Because of the thick plant cell walls that make plasma membranes inaccessible for interaction with the virus envelopes, it seems that these viruses simply shed the genes encoding glycoproteins and switched to the naked capsid lifestyle that is characteristic of most plant viruses.
Thus, the large-scale metagenomics studies on invertebrate viruses point to these abundant and diverse eukaryotes as the hosts, in which (−)RNA viruses have diversified and from which these viruses were transmitted to other complex eukaryotes, namely, vertebrates and plants (Fig. 3B). Because the majority of the vertebrate and plant (−)RNA viruses are arthropod-transmitted, this evolutionary trajectory appears to be effectively certain. Importantly, not only these viruses lack any prokaryotic relatives, but they are also conspicuously absent from the aquatic or protist viromes that have been sampled so far. Thus, a plausible origin of (−)RNA viruses could date back to primitive metazoa or their early invertebrate descendants. A broader sampling of the most ancient diverging lineages of multicellular eukaryotes is needed to support or refute this hypothesis. What is even less certain, however, is where the gene modules that were recruited into ancestral (−)RNA viruses come from. As discussed above, the more ancient (+)RNA or dsRNA viruses are potential sources of at least the RdRp of (−)RNA viruses.
8. Concluding remarks
In a few short years, the dramatic increase in the numbers and diversity of new virus genomes discovered through the metagenomics approaches in various environments, from marine to terrestrial, from tropical forests to sewage, from hot springs to Antarctic lakes, has transformed our understanding of the global virome in general and the RNA virome in particular.
Far from being trivial extensions of the previous knowledge derived primarily from comparisons of plant and animal viruses, the new metagenomics studies change the existing picture of virus evolution. Moreover, these new discoveries seem to be revealing glimpses of the intimate and intricate connections between virus evolution and the evolution and ecology of the hosts that remained obscure before. These changes to the existing ideas on virus evolution come primarily in three areas.
First, major expansion of many groups of viruses provided for more robust phylogenetic trees of the conserved genes (primarily, the RdRps, in the case of RNA viruses). The elimination of the dubious clustering of flavi-like and tombus-like (+)RNA viruses that was replaced by the stronger supported tombus-noda branch, which reflects the similarity of the genome architectures among the smallest and simplest RNA viruses, is the prime case in point but the trend appears general. Nevertheless, the substantial changes notwithstanding, major supergroups of viruses identified previously, such as picorna-like and alpha-like viruses, although losing some peripheral members, in general, seem to stand the test of the vastly increased diversity.
Second, the extent of gene module shuffling among diverse virus genomes that has already been recognized as a major evolutionary trend in the pre-metagenomic era, is dramatically increased by metagenomic discoveries. The hybrid tombus-noda viruses and the invertebrate alpha-like viruses encoding TMV-like capsid proteins can be mentioned as some of the most notable examples. This trend is dominant in the evolution of viruses with different types of genomes as demonstrated, in particular, by the discovery of the enormous diversity of chimeric ssDNA viruses (Krupovic et al., 2015b, Stedman, 2015). The latest metagenomics studies show that RNA viruses are not far behind in this respect.
Third, and arguably, most dramatically, inclusion of the massive metagenomic data in phylogenetic analyses has led to the identification of numerous, strongly supported groups that combine RNA viruses that infect diverse hosts including different groups of protists, animals and plants. Certainly, these observations come with serious caveats, which include possible cases of contamination; phylogenetic artifacts, such as long branch attraction; and, probably, most important, incomplete and uneven sampling of viromes from different host taxa. These caveats notwithstanding, the consistent mixing of viruses from different hosts in phylogenetic tree branches across the entire range of RNA viruses appears to reflect the reality of virus evolution.
A key aspect of this reality is the broad host range of many virus groups because of which HVT is emerging as the defining factor of the RNA virome evolution. The confidence in the major contribution of HVT in virus evolution differs between virus groups and the hosts. In particular, the huge, highly diverse plant RNA virome is densely populated by picorna-like, alpha-like, tombus-like, partiti-like, reo-like, bunya-like, rhabdo-like and some other clades of RNA viruses. Such massive RNA virome could have emerged by either vertical descent from the plant ancestral lineage of Zygnematophycea algae, or by HVT from the much more expansive invertebrate virome, or via a combination of these pathways. The small ancestral algal taxon appears unlikely to host all extant components of the flowering plant virome. Moreover, the algal virome is known to be dominated by Phycodnaviridae, large DNA viruses that are completely excluded from the plant virome. Thus, although some components of the plant virome could lurk in Zygnematophycea, the hypothesis that the RNA virome of plants flourished through HVT from marine environment via freshwater, soil and air routes seems to be preferable (Fig. 4 ). Such HVT was likely mediated by nematodes, aquatic and soil inhabitants that are the most abundant animals on earth (Telford et al., 2008), arthropods, and, perhaps to a smaller degree, mollusks and fungi.
Fig. 4.
Evolutionary relationships between viruses and their host organisms. The vertical and bended arrows denote major transitions in the evolution of the cellular organisms, whereas horizontal arrows show hypothetical events of horizontal virus transfer (HVT) from the viromes of the ancestral organisms to the newly evolving groups of organisms.
Similar reasoning could apply to vertebrates albeit with less confidence: there are currently no data on viruses of Deuterostomes other than vertebrates, whereas the viromes of Protostomes, in particular, arthropods and nematodes, have been shown to be enormously diverse. Combined with the known vector properties of many arthropods and nematodes, this makes HVT from these animals the most plausible route of evolution of the vertebrate virome, given the current data (Fig. 4). However, proper sampling of the viromes of echinoderms, the sister group of vertebrates, as well as those of protists related to animals, such as Choanoflagellates, is essential to support or refute this line of reasoning.
A role for HVT in the evolution of some RNA viruses has been suggested previously, e.g. for the case of plant (−)RNA viruses whose ancestors appear to have been acquired from animals, but the pervasive character of this phenomenon revealed through metagenomics signifies a sea change in our understanding of RNA virus evolution. This realization comes from the sum total of many important studies, but the tour de force investigation of the invertebrate RNA virome stands apart in its scale, quality and impact, and represent a tipping point in our thinking on RNA virus evolution (Li et al., 2015, Shi et al., 2016a, Shi et al., 2016b, Shi et al., 2017).
Some ‘highways’ of HVT as well as connections with the host ecology are becoming apparent. In particular, the strikingly diverse RNA virome of invertebrates that temporally antedate both vertebrates and angiosperm plants clearly was the reservoir that gave rise to both the vertebrate and plant viromes (Fig. 4). This role of invertebrates in virus evolution is compatible with the vector life style of many arthropods and nematodes. Apart from the vector transmission, HVT is clearly facilitated in aquatic, compared to terrestrial, environments, where viruses seem to be shuttled between highly diverse hosts such as various protists and animals. Accurate dating of the likely HVT events is problematic because of the likely changes in the evolutionary rates following a host switch. On many occasions, however, it appears possible to plausibly link HVT to well characterized events in host evolution and global ecology, such as the Cambrian explosion of animals or terrestrialization of animals and vascular plants, thus obtaining rough dates for the HVT.
The new metaviromic data appear to support the three major routes through which the RNA viruses of eukaryotes have emerged and evolved. The first one, a continuous line of descent from (+)RNA bacteriophages, was taken by narna-like viruses whose RdRp is related to that of Leviviridae and some of which still reproduce in the mitochondria (Fig. 1B). The second route, exemplified by the picorna-like viruses, involves assembly of the ancestral viral genome from genes derived from bacteria, their viruses and selfish elements (Fig. 2B). Although the ultimate evolutionary provenance of picorna-like RdRp is not firmly established, it appears plausible that it evolved from the RT of bacterial Group II introns. The third route involves more recent origin of major groups of viruses; given the current lack of evidence of presence of (−)RNA viruses, flavi-like viruses and alpha-like viruses in eukaryotes predating invertebrates, it appears likely that these groups of viruses emerged late in evolution, perhaps, at the root of the Metazoa (Fig. 3B). As transpired upon deep sampling of invertebrate RNA viruses, all three routes seem to have converged during the extensive diversification of aquatic invertebrates yielding the vast invertebrate RNA pan-virome (Fig. 4).
It remains to be established why did invertebrates grow into such a fertile niche for incubating an enormous variety of the RNA viruses that includes all currently known major virus lineages. As pointed out by Shi et al. (2017), certain invertebrate taxa, in particular nematodes and arthropods, themselves represent major parts of the animal diversity, while being globally abundant and often living in extremely large and dense populations. Furthermore, many invertebrates are excellent HVT agents because they form tight biological associations with such organisms as vertebrates and plants, often serving as vectors that shuttle viruses between these organisms (Blanc and Gutierrez, 2015).
With all the importance of invertebrates, they could not have been alone in seeding the extant aquatic and terrestrial viromes. The first glimpses of the fungal virome show that it is richer than previously expected. Given that fungi are ubiquitous organisms whose diversity and population sizes could be comparable to or exceed those of invertebrates, future deep sampling of fungi for viruses potentially could reveal major contributions of fungi to the formation of the eukaryotic RNA pan-virome.
We have to admit that, despite the transformative impact of metaviromics on our understanding of RNA virus evolution, a unifying scenario that would explain the relationships between (+)RNA, (−)RNA, dsRNA and retroelements is still lacking. To develop such a grand scheme, major improvements are required in at least two directions: first, large-scale phylogenetic analysis of highly diverged proteins, such as the RdRps and RTs, and second, ‘evolutionary viromics’, a systematic study of the viromes of all major host taxa. Furthermore, the possibility remains that further trawling of metagenomes, especially with most powerful methods for sequence similarity detection, results in the discovery of completely new groups of viruses within what currently looks like “dark matter” of RNA metagenomes. Such findings would have the potential to dramatically change the picture of RNA virus evolution.
Acknowledgements
The authors are grateful to Mart Krupovic (Institut Pasteur) for critical reading of the manuscript and thoughtful comments, as well as for sharing unpublished observations. The VVD work on this article was in part supported by NIH, NCBI Scientific Visitor Program of which he was a Fellow. EVK is supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
References
- Adoutte A., Balavoine G., Lartillot N., Lespinet O., Prud'homme B., de Rosa R. The new animal phylogeny: reliability and implications. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X., Mendez-Lopez E., Lasso G., Sanchez-Pina M.A., Aranda M., Valle M. The near-atomic cryoEM structure of a flexible filamentous plant virus shows homology of its coat protein with nucleoproteins of animal viruses. Elife. 2015;4:e11795. doi: 10.7554/eLife.11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T., Karlin D.G. Sequence analysis reveals a conserved extension in the capping enzyme of the alphavirus supergroup, and a homologous domain in nodaviruses. Biol. Direct. 2015;10:16. doi: 10.1186/s13062-015-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyants N.S., Lye L.F., Dobson D.E., Lukes J., Beverley S.M. A narnavirus in the trypanosomatid protist plant pathogen phytomonas serpens. Genome Announc. 2016;4 doi: 10.1128/genomeA.00711-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyants N.S., Lye L.F., Dobson D.E., Lukes J., Beverley S.M. A novel bunyavirus-like virus of trypanosomatid protist parasites. Genome Announc. 2016;4 doi: 10.1128/genomeA.00715-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal S., Domier L.L., Niblack T.L., Lambert K.N. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J. Gen. Virol. 2011;92:1870–1879. doi: 10.1099/vir.0.030585-0. [DOI] [PubMed] [Google Scholar]
- Bekal S., Domier L.L., Gonfa B., McCoppin N.K., Lambert K.N., Bhalerao K. A novel flavivirus in the soybean cyst nematode. J. Gen. Virol. 2014;95:1272–1280. doi: 10.1099/vir.0.060889-0. [DOI] [PubMed] [Google Scholar]
- Berbee M.L., James T.Y., Strullu-Derrien C. Early diverging fungi: diversity and impact at the dawn of terrestrial life. Annu. Rev. Microbiol. 2017;71:41–60. doi: 10.1146/annurev-micro-030117-020324. [DOI] [PubMed] [Google Scholar]
- Blanc S., Gutierrez S. The specifics of vector transmission of arboviruses of vertebrates and plants. Curr. Opin. Virol. 2015;15:27–33. doi: 10.1016/j.coviro.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Bolduc B., Shaughnessy D.P., Wolf Y.I., Koonin E.V., Roberto F.F., Young M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J. Virol. 2012;86:5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth A., Doolittle W.F. Eukaryogenesis, how special really? Proc. Natl. Acad. Sci. U. S. A. 2015;112:10278–10285. doi: 10.1073/pnas.1421376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Floyd S.K., Sakakibara K. Green genes-comparative genomics of the green branch of life. Cell. 2007;129:229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Brum J.R., Sullivan M.B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015;13:147–159. doi: 10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- Brussaard C.P., Noordeloos A.A., Sandaa R.A., Heldal M., Bratbak G. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology. 2004;319:280–291. doi: 10.1016/j.virol.2003.10.033. [DOI] [PubMed] [Google Scholar]
- Butcher S.J., Grimes J.M., Makeyev E.V., Bamford D.H., Stuart D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Chen C.A., Shyu A.B. Emerging themes in regulation of global mRNA turnover in cis. Trends Biochem. Sci. 2017;42:16–27. doi: 10.1016/j.tibs.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.E., Suttle C.A. Biogeography of viruses in the sea. Annu. Rev. Virol. 2015;2:41–66. doi: 10.1146/annurev-virology-031413-085540. [DOI] [PubMed] [Google Scholar]
- Coetzee B., Freeborough M.J., Maree H.J., Celton J.M., Rees D.J., Burger J.T. Deep sequencing analysis of viruses infecting grapevines: virome of a vineyard. Virology. 2010;400:157–163. doi: 10.1016/j.virol.2010.01.023. [DOI] [PubMed] [Google Scholar]
- Csorba T., Kontra L., Burgyan J. Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology. 2015;479–480:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- Culley A.I., Lang A.S., Suttle C.A. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Culley A.I., Lang A.S., Suttle C.A. The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol. J. 2007;4:69. doi: 10.1186/1743-422X-4-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A.I., Mueller J.A., Belcaid M., Wood-Charlson E.M., Poisson G., Steward G.F. The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. MBio. 2014;5:e01210–e01214. doi: 10.1128/mBio.01210-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe A.L., Nuss D.L. Hypovirus molecular biology: from Koch's postulates to host self-recognition genes that restrict virus transmission. Adv. Virus Res. 2013;86:109–147. doi: 10.1016/B978-0-12-394315-6.00005-2. [DOI] [PubMed] [Google Scholar]
- Deakin G., Dobbs E., Bennett J.M., Jones I.M., Grogan H.M., Burton K.S. Multiple viral infections in Agaricus bisporus—characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci. Rep. 2017;7:2469. doi: 10.1038/s41598-017-01592-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depierreux D., Vong M., Nibert M.L. Nucleotide sequence of Zygosaccharomyces bailii virus Z: evidence for +1 programmed ribosomal frameshifting and for assignment to family Amalgaviridae. Virus Res. 2016;217:115–124. doi: 10.1016/j.virusres.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Ahlquist P. Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication. Curr. Opin. Microbiol. 2012;15:519–524. doi: 10.1016/j.mib.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V.V., Koonin E.V. Common origins and host-dependent diversity of plant and animal viromes. Curr. Opin. Virol. 2011;1:322–331. doi: 10.1016/j.coviro.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V.V., Boyko V.P., Agranovsky A.A., Koonin E.V. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology. 1991;184:79–86. doi: 10.1016/0042-6822(91)90823-t. [DOI] [PubMed] [Google Scholar]
- Dolja V.V., Kreuze J.F., Valkonen J.P. Comparative and functional genomics of closteroviruses. Virus Res. 2006;117:38–51. doi: 10.1016/j.virusres.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C.W., Hejnol A., Matus D.Q., Pang K., Browne W.E., Smith S.A., Seaver E., Rouse G.W., Obst M., Edgecombe G.D., Sorensen M.V., Haddock S.H., Schmidt-Rhaesa A., Okusu A., Kristensen R.M., Wheeler W.C., Martindale M.Q., Giribet G. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Edwards R.A., McNair K., Faust K., Raes J., Dutilh B.E. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol. Rev. 2016;40:258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver J.R., Grubaugh N.D., Krajacich B.J., Weger-Lucarelli J., Lakin S.M., Fakoli L.S., 3rd, Bolay F.K., Diclaro J.W., 2nd, Dabire K.R., Foy B.D., Brackney D.E., Ebel G.D., Stenglein M.D. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology. 2016;498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- Felix M.A., Ashe A., Piffaretti J., Wu G., Nuez I., Belicard T., Jiang Y., Zhao G., Franz C.J., Goldstein L.D., Sanroman M., Miska E.A., Wang D. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C.J., Zhao G., Felix M.A., Wang D. Complete genome sequence of Le Blanc virus, a third Caenorhabditis nematode-infecting virus. J. Virol. 2012;86:11940. doi: 10.1128/JVI.02025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S.D., Snellgrove W.C., Genthner F.J. Genomic sequences of two novel levivirus single-stranded RNA coliphages (family Leviviridae): evidence for recombinationin environmental strains. Viruses. 2012;4:1548–1568. doi: 10.3390/v4091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial S.A., Caston J.R., Jiang D., Nibert M.L., Suzuki N. 50-plus years of fungal viruses. Virology. 2015;479–480:356–368. doi: 10.1016/j.virol.2015.02.034. [DOI] [PubMed] [Google Scholar]
- Gibbs A.J., Torronen M., Mackenzie A.M., Wood J.T., 2nd, Armstrong J.S., Kondo H., Tamada T., Keese P.L. The enigmatic genome of Chara australis virus. J. Gen. Virol. 2011;92:2679–2690. doi: 10.1099/vir.0.033852-0. [DOI] [PubMed] [Google Scholar]
- Goldbach R., Wellink J. Evolution of plus-strand RNA viruses. Intervirology. 1988;29:260–267. doi: 10.1159/000150054. [DOI] [PubMed] [Google Scholar]
- Golmohammadi R., Valegard K., Fridborg K., Liljas L. The refined structure of bacteriophage MS2 at 2.8 A resolution. J. Mol. Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- Goubau D., Deddouche S., Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft genome sequence of tombunodavirus UC1. Genome Announc. 2015;3 doi: 10.1128/genomeA.00655-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft genome sequences of Ciliovirus and Brinovirus from San Francisco wastewater. Genome Announc. 2015;3 doi: 10.1128/genomeA.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft genome sequences of Leviviridae RNA phages EC and MB recovered from San Francisco wastewater. Genome Announc. 2015;3 doi: 10.1128/genomeA.00652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Bielefeldt-Ohmann H., McLean B.J., O'Brien C.A., Colmant A.M., Piyasena T.B., Harrison J.J., Newton N.D., Barnard R.T., Prow N.A., Deerain J.M., Mah M.G., Hobson-Peters J. Commensal viruses of mosquitoes: host restriction, transmission, and interaction with arboviral pathogens. Evol. Bioinform. Online. 2016;12:35–44. doi: 10.4137/EBO.S40740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland O., Mikalsen A.B., Nilsen P., Lindmo K., Thu B.J., Eliassen T.M., Roos N., Rode M., Evensen O. Cardiomyopathy syndrome of atlantic salmon (Salmo salar L.) is caused by a double-stranded RNA virus of the Totiviridae family. J. Virol. 2011;85:5275–5286. doi: 10.1128/JVI.02154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S., Mahony J., Nauta A., van Sinderen D. Metagenomic approaches to assess bacteriophages in various environmental niches. Viruses. 2017;9 doi: 10.3390/v9060127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M. Plant virus replication and movement. Virology. 2015;479–480:657–671. doi: 10.1016/j.virol.2015.01.025. [DOI] [PubMed] [Google Scholar]
- Heller-Dohmen M., Gopfert J.C., Pfannstiel J., Spring O. The nucleotide sequence and genome organization of Plasmopara halstedii virus. Virol. J. 2011;8:123. doi: 10.1186/1743-422X-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman B.I., Cai G. The family narnaviridae: simplest of RNA viruses. Adv. Virus Res. 2013;86:149–176. doi: 10.1016/B978-0-12-394315-6.00006-4. [DOI] [PubMed] [Google Scholar]
- Hinchliff C.E., Smith S.A., Allman J.F., Burleigh J.G., Chaudhary R., Coghill L.M., Crandall K.A., Deng J., Drew B.T., Gazis R., Gude K., Hibbett D.S., Katz L.A., Laughinghouse E.J., McTavish H.D., Midford P.E., Owen C.L., Ree R.H., Rees J.A., Soltis D.E., Williams T., Cranston K.A. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12764–12769. doi: 10.1073/pnas.1423041112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J., Krupovic M., Koonin E.V. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. MBio. 2016;7 doi: 10.1128/mBio.00978-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M., Roy S.W. Origin of spliceosomal introns and alternative splicing. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski A.B., Krishnamurthy S.R., Lim E.S., Zhao G., Brenchley J.M., Barouch D.H., Thakwalakwa C., Manary M.J., Holtz L.R., Wang D. Statoviruses, a novel taxon of RNA viruses present in the gastrointestinal tracts of diverse mammals. Virology. 2017;504:36–44. doi: 10.1016/j.virol.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier P. Facts and fancies about early fossil chordates and vertebrates. Nature. 2015;520:483–489. doi: 10.1038/nature14437. [DOI] [PubMed] [Google Scholar]
- Katz L.A., Grant J.R. Taxon-rich phylogenomic analyses resolve the eukaryotic tree of life and reveal the power of subsampling by sites. Syst. Biol. 2015;64:406–415. doi: 10.1093/sysbio/syu126. [DOI] [PubMed] [Google Scholar]
- Katz L.A. Origin and diversification of eukaryotes. Annu. Rev. Microbiol. 2012;66:411–427. doi: 10.1146/annurev-micro-090110-102808. [DOI] [PubMed] [Google Scholar]
- Keeling P.J., Burger G., Durnford D.G., Lang B.F., Lee R.W., Pearlman R.E., Roger A.J., Gray M.W. The tree of eukaryotes. Trends Ecol. Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kenrick P., Crane P.R. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]
- Khramtsov N.V., Upton S.J. Association of RNA polymerase complexes of the parasitic protozoan Cryptosporidium parvum with virus-like particles: heterogeneous system. J. Virol. 2000;74:5788–5795. doi: 10.1128/jvi.74.13.5788-5795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidmose R.T., Vasiliev N.N., Chetverin A.B., Andersen G.R., Knudsen C.R. Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10884–10889. doi: 10.1073/pnas.1003015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Tomaru Y. Discovery of two novel viruses expands the diversity of single-stranded DNA and single-stranded RNA viruses infecting a cosmopolitan marine diatom. Appl. Environ. Microbiol. 2015;81:1120–1131. doi: 10.1128/AEM.02380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A.H., Nowak M.A. The timetable of evolution. Sci. Adv. 2017;3:e1603076. doi: 10.1126/sciadv.1603076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Atsumi G., Iwadate Y., Tomita R., Chiba K., Akasaka S., Nishihara M., Takahashi H., Yamaoka N. Gentian Kobu-sho-associated virus: a tentative, novel double-stranded RNA virus that is relevant to gentian Kobu-sho syndrome. J. Gen. Plant Pathol. 2013;79:56–63. [Google Scholar]
- Koga R., Fukuhara T., Nitta T. Molecular characterization of a single mitochondria-associated double-stranded RNA in the green alga Bryopsis. Plant Mol. Biol. 1998;36:717–724. doi: 10.1023/a:1005907310553. [DOI] [PubMed] [Google Scholar]
- Koga R., Horiuchi H., Fukuhara T. Double-stranded RNA replicons associated with chloroplasts of a green alga, Bryopsis cinicola. Plant Mol. Biol. 2003;51:991–999. doi: 10.1023/a:1023003412859. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 2014;78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Choi G.H., Nuss D.L., Shapira R., Carrington J.C. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. U. S. A. 1991;88:10647–10651. doi: 10.1073/pnas.88.23.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Senkevich T.G., Dolja V.V. The ancient Virus World and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Wolf Y.I., Nagasaki K., Dolja V.V. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V., Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. Evolution of double-stranded RNAviruses: a case forpolyphyletic origin from different groups of positive-stranded RNA viruses. Semin. Virol. 1992;3:327–339. [Google Scholar]
- Koonin E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20140333. doi: 10.1098/rstb.2014.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormelink R., Garcia M.L., Goodin M., Sasaya T., Haenni A.L. Negative-strand RNA viruses: the plant-infecting counterparts. Virus Res. 2011;162:184–202. doi: 10.1016/j.virusres.2011.09.028. [DOI] [PubMed] [Google Scholar]
- Kotta-Loizou I., Coutts R.H. Studies on the virome of the entomopathogenic fungus beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog. 2017;13:e1006183. doi: 10.1371/journal.ppat.1006183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlakidis Z., Brown N.A., Jamal A., Phoon X., Coutts R.H. Incidence of endornaviruses in Phytophthora taxon douglasfir and Phytophthora ramorum. Virus Genes. 2010;40:130–134. doi: 10.1007/s11262-009-0421-7. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S.R., Janowski A.B., Zhao G., Barouch D., Wang D. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol. 2016;14:e1002409. doi: 10.1371/journal.pbio.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Koonin E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2401–E2410. doi: 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Dolja V.V., Koonin E.V. Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol. Direct. 2015;10:12. doi: 10.1186/s13062-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Zhi N., Li J., Hu G., Koonin E.V., Wong S., Shevchenko S., Zhao K., Young N.S. Multiple layers of chimerism in a single-stranded DNA virus discovered by deep sequencing. Genome Biol. Evol. 2015;7:993–1001. doi: 10.1093/gbe/evv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Cvirkaite-Krupovic V., Iranzo J., Prangishvili D., Koonin E.V. Viruses of archaea: structural, functional, environmental and evolutionary genomics. Virus Res. 2017 doi: 10.1016/j.virusres.2017.11.025. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit T., Thomas T., Steinberg P. Expanding our understanding of the seaweed holobiont: RNA viruses of the red alga delisea pulchra. Front. Microbiol. 2015;6:1489. doi: 10.3389/fmicb.2015.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A.S., Culley A.I., Suttle C.A. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology. 2004;320:206–217. doi: 10.1016/j.virol.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Lecuit M., Eloit M. The human virome: new tools and concepts. Trends Microbiol. 2013;21:510–515. doi: 10.1016/j.tim.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.X., Shi M., Tian J.H., Lin X.D., Kang Y.J., Chen L.J., Qin X.C., Xu J., Holmes E.C., Zhang Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4 doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fu Y., Jiang D., Li G., Xie J., Peng Y., Yi X., Ghabrial S.A. A novel mycovirus that is related to the human pathogen hepatitis E virus and rubi-like viruses. J. Virol. 2009;83:1981–1991. doi: 10.1128/JVI.01897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Xie J., Cheng J., Fu Y., Li G., Yi X., Jiang D. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12205–12210. doi: 10.1073/pnas.1401786111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locali-Fabris E.C., Freitas-Astua J., Souza A.A., Takita M.A., Astua-Monge G., Antonioli-Luizon R., Rodrigues V., Targon M.L., Machado M.A. Complete nucleotide sequence, genomic organization and phylogenetic analysis of Citrus leprosis virus cytoplasmic type. J. Gen. Virol. 2006;87:2721–2729. doi: 10.1099/vir.0.82038-0. [DOI] [PubMed] [Google Scholar]
- Lopez-Bueno A., Rastrojo A., Peiro R., Arenas M., Alcami A. Ecological connectivity shapes quasispecies structure of RNA viruses in an Antarctic lake. Mol. Ecol. 2015;24:4812–4825. doi: 10.1111/mec.13321. [DOI] [PubMed] [Google Scholar]
- Lye L.F., Akopyants N.S., Dobson D.E., Beverley S.M. A narnavirus-like element from the trypanosomatid protozoan parasite leptomonas seymouri. Genome Announc. 2016;4 doi: 10.1128/genomeA.00713-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E.V., Grimes J.M. RNA-dependent RNA polymerases of dsRNA bacteriophages. Virus Res. 2004;101:45–55. doi: 10.1016/j.virusres.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Mansur D.S., Smith G.L., Ferguson B.J. Intracellular sensing of viral DNA by the innate immune system. Microbes Infect. 2014;16:1002–1012. doi: 10.1016/j.micinf.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Martijn J., Ettema T.J. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem. Soc. Trans. 2013;41:451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- Marzano S.Y., Nelson B.D., Ajayi-Oyetunde O., Bradley C.A., Hughes T.J., Hartman G.L., Eastburn D.M., Domier L.L. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J. Virol. 2016;90:6846–6863. doi: 10.1128/JVI.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer M.J., Simbajon N., Carillo J., Borth W.B., Freitas-Astua J., Kitajima E.W., Neupane K.R., Hu J.S. A cilevirus infects ornamental hibiscus in Hawaii. Arch. Virol. 2013;158:2421–2424. doi: 10.1007/s00705-013-1745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.A., Culley A.I., Schvarcz C.R., Steward G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016;18:3714–3727. doi: 10.1111/1462-2920.13291. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Regulation of mRNA decay in bacteria. Annu. Rev. Microbiol. 2016;70:25–44. doi: 10.1146/annurev-micro-091014-104515. [DOI] [PubMed] [Google Scholar]
- Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniruzzaman M., Wurch L.L., Alexander H., Dyhrman S.T., Gobler C.J., Wilhelm S.W. Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics. Nat. Commun. 2017;8:16054. doi: 10.1038/ncomms16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian A., Shipunov A., Elena S.F. Changes in the composition of the RNA virome mark evolutionary transitions in green plants. BMC Biol. 2016;14:68. doi: 10.1186/s12915-016-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K., Shirai Y., Takao Y., Mizumoto H., Nishida K., Tomaru Y. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 2005;71:8888–8894. doi: 10.1128/AEM.71.12.8888-8894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K., Pattanayek R., Stubbs G. Visualization of protein-nucleic acid interactions in a virus: refined structure of intact tobacco mosaic virus at 2.9 Å resolution by X-ray fiber diffraction. J. Mol. Biol. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- Nerva L., Ciuffo M., Vallino M., Margaria P., Varese G.C., Gnavi G., Turina M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016;219:22–38. doi: 10.1016/j.virusres.2015.10.028. [DOI] [PubMed] [Google Scholar]
- Nibert M.L., Ghabrial S.A., Maiss E., Lesker T., Vainio E.J., Jiang D., Suzuki N. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014;188:128–141. doi: 10.1016/j.virusres.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C. Uncovering earth's virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- Paludan S.R., Bowie A.G. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey L.W., Barbero E., Lasser E., Dunthorn M., Bhattacharya D., Patterson D.J., Katz L.A. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflug A., Guilligay D., Reich S., Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- Philippe H., Lartillot N., Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol. Biol. Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Poranen M.M., Bamford D.H. Assembly of large icosahedral double-stranded RNA viruses. Adv. Exp. Med. Biol. 2012;726:379–402. doi: 10.1007/978-1-4614-0980-9_17. [DOI] [PubMed] [Google Scholar]
- Pyle J.D., Keeling P.J., Nibert M.L. Amalga-like virus infecting Antonospora locustae, a microsporidian pathogen of grasshoppers, plus related viruses associated with other arthropods. Virus Res. 2017;233:95–104. doi: 10.1016/j.virusres.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.C., Shi M., Tian J.H., Lin X.D., Gao D.Y., He J.R., Wang J.B., Li C.X., Kang Y.J., Yu B., Zhou D.J., Xu J., Plyusnin A., Holmes E.C., Zhang Y.Z. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6744–6749. doi: 10.1073/pnas.1324194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastgou M., Habibi M.K., Izadpanah K., Masenga V., Milne R.G., Wolf Y.I., Koonin E.V., Turina M. Molecular characterization of the plant virus genus Ourmiavirus and evidence of inter-kingdom reassortment of viral genome segments as its possible route of origin. J. Gen. Virol. 2009;90:2525–2535. doi: 10.1099/vir.0.013086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill P.A., Davidson A.D., Wright P.J. The nucleotide sequence and genome organization of mushroom bacilliform virus: a single-stranded RNA virus of Agaricus bisporus (Lange) Imbach. Virology. 1994;202:904–911. doi: 10.1006/viro.1994.1412. [DOI] [PubMed] [Google Scholar]
- Rochon D., Kakani K., Robbins M., Reade R. Molecular aspects of plant virus transmission by olpidium and plasmodiophorid vectors. Annu. Rev. Phytopathol. 2004;42:211–241. doi: 10.1146/annurev.phyto.42.040803.140317. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Carmel L., Csuros M., Koonin E.V. Origin and evolution of spliceosomal introns. Biol. Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck M.J., Sabanadzovic S., Okada R., Valverde R.A. The remarkable evolutionary history of endornaviruses. J. Gen. Virol. 2011;92:2674–2678. doi: 10.1099/vir.0.034702-0. [DOI] [PubMed] [Google Scholar]
- Roossinck M.J., Martin D.P., Roumagnac P. Plant virus metagenomics: advances in virus discovery. Phytopathology. 2015;105:716–727. doi: 10.1094/PHYTO-12-14-0356-RVW. [DOI] [PubMed] [Google Scholar]
- Rosario K., Breitbart M. Exploring the viral world through metagenomics. Curr. Opin. Virol. 2011;1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Rosario K., Nilsson C., Lim Y.W., Ruan Y., Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Rousvoal S., Bouyer B., Lopez-Cristoffanini C., Boyen C., Collen J. Mutant swarms of a totivirus-like entities are present in the red macroalga Chondrus crispus and have been partially transferred to the nuclear genome. J. Phycol. 2016;52:493–504. doi: 10.1111/jpy.12427. [DOI] [PubMed] [Google Scholar]
- Roy A., Choudhary N., Guillermo L.M., Shao J., Govindarajulu A., Achor D., Wei G., Picton D.D., Levy L., Nakhla M.K., Hartung J.S., Brlansky R.H. A novel virus of the genus Cilevirus causing symptoms similar to citrus leprosis. Phytopathology. 2013;103:488–500. doi: 10.1094/PHYTO-07-12-0177-R. [DOI] [PubMed] [Google Scholar]
- Rumnieks J., Tars K. Diversity of pili-specific bacteriophages: genome sequence of IncM plasmid-dependent RNA phage M. BMC Microbiol. 2012;12:277. doi: 10.1186/1471-2180-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder L.S., Talbot N.J. Regulation of appressorium development in pathogenic fungi. Curr. Opin. Plant Biol. 2015;26:8–13. doi: 10.1016/j.pbi.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Lin X.D., Tian J.H., Chen L.J., Chen X., Li C.X., Qin X.C., Li J., Cao J.P., Eden J.S., Buchmann J., Wang W., Xu J., Holmes E.C., Zhang Y.Z. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- Shi M., Lin X.D., Vasilakis N., Tian J.H., Li C.X., Chen L.J., Eastwood G., Diao X.N., Chen M.H., Chen X., Qin X.C., Widen S.G., Wood T.G., Tesh R.B., Xu J., Holmes E.C., Zhang Y.Z. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the flaviviridae and related viruses. J. Virol. 2016;90:659–669. doi: 10.1128/JVI.02036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Zhang Y.-Z., Holmes E.C. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 2017 doi: 10.1016/j.virusres.2017.10.016. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai Y., Tomaru Y., Takao Y., Suzuki H., Nagumo T., Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 2008;74:4022–4027. doi: 10.1128/AEM.00509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman L.M., Davidson I. Viruses with circular single-stranded DNA genomes are everywhere! Annu. Rev. Virol. 2017;4:159–180. doi: 10.1146/annurev-virology-101416-041953. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Adams M.J., Benko M., Breitbart M., Brister J.R., Carstens E.B., Davison A.J., Delwart E., Gorbalenya A.E., Harrach B., Hull R., King A.M., Koonin E.V., Krupovic M., Kuhn J.H., Lefkowitz E.J., Nibert M.L., Orton R., Roossinck M.J., Sabanadzovic S., Sullivan M.B., Suttle C.A., Tesh R.B., van der Vlugt R.A., Varsani A., Zerbini F.M. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- Sparrer K.M., Gack M.U. Intracellular detection of viral nucleic acids. Curr. Opin. Microbiol. 2015;26:1–9. doi: 10.1016/j.mib.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman K.M. Deep recombination: RNA and ssDNA virus genes in DNA virus and host genomes. Annu. Rev. Virol. 2015;2:203–217. doi: 10.1146/annurev-virology-100114-055127. [DOI] [PubMed] [Google Scholar]
- Stenglein M.D., Jacobson E.R., Wozniak E.J., Wellehan J.F., Kincaid A., Gordon M., Porter B.F., Baumgartner W., Stahl S., Kelley K., Towner J.S., DeRisi J.L. Ball python nidovirus: a candidate etiologic agent for severe respiratory disease in Python regius. MBio. 2014;5:e01484–01414. doi: 10.1128/mBio.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao Y., Mise K., Nagasaki K., Okuno T., Honda D. Complete nucleotide sequence and genome organization of a single-stranded RNA virus infecting the marine fungoid protist Schizochytrium sp. J. Gen. Virol. 2006;87:723–733. doi: 10.1099/vir.0.81204-0. [DOI] [PubMed] [Google Scholar]
- Teixeira M., Sela N., Ng J., Casteel C.L., Peng H.C., Bekal S., Girke T., Ghanim M., Kaloshian I. A novel virus from Macrosiphum euphorbiae with similarities to members of the family Flaviviridae. J. Gen. Virol. 2016;97:1261–1271. doi: 10.1099/jgv.0.000414. [DOI] [PubMed] [Google Scholar]
- Telford M.J., Bourlat S.J., Economou A., Papillon D., Rota-Stabelli O. The evolution of the Ecdysozoa. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:1529–1537. doi: 10.1098/rstb.2007.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y., Takao Y., Suzuki H., Nagumo T., Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 2009;75:2375–2381. doi: 10.1128/AEM.02580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask S.D., McDonald S.M., Patton J.T. Structural insights into the coupling of virion assembly and rotavirus replication. Nat. Rev. Microbiol. 2012;10:165–177. doi: 10.1038/nrmicro2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G. Green algae and the origins of multicellularity in the plant kingdom. Cold Spring Harb. Perspect. Biol. 2014;6:a016170. doi: 10.1101/cshperspect.a016170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urayama S., Takaki Y., Nunoura T. FLDS: a comprehensive dsRNA sequencing method for intracellular RNA virus surveillance. Microbes Environ. 2016;31:33–40. doi: 10.1264/jsme2.ME15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C.L., Longdon B., Lewis S.H., Obbard D.J. Twenty-five new viruses associated with the drosophilidae (Diptera) Evol. Bioinform. Online. 2016;12:13–25. doi: 10.4137/EBO.S39454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D., Hugenholtz P. From deep sequencing to viral tagging: recent advances in viral metagenomics. Bioessays. 2013;35:436–442. doi: 10.1002/bies.201200174. [DOI] [PubMed] [Google Scholar]
- Xie J., Jiang D. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 2014;52:45–68. doi: 10.1146/annurev-phyto-102313-050222. [DOI] [PubMed] [Google Scholar]
- Yang S., Wang T., Bohon J., Gagne M.E., Bolduc M., Leclerc D., Li H. Crystal structure of the coat protein of the flexible filamentous papaya mosaic virus. J. Mol. Biol. 2012;422:263–273. doi: 10.1016/j.jmb.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi T., Takemoto Y., Suzuki M., Yamashita S., Hibi T. The nucleotide sequence and genome organization of Sclerophthora macrospora virus B. Virology. 1999;264:344–349. doi: 10.1006/viro.1999.0018. [DOI] [PubMed] [Google Scholar]
- Yokoi T., Yamashita S., Hibi T. The nucleotide sequence and genome organization of Sclerophthora macrospora virus A. Virology. 2003;311:394–399. doi: 10.1016/s0042-6822(03)00183-1. [DOI] [PubMed] [Google Scholar]
- Yoon H.S., Grant J., Tekle Y.I., Wu M., Chaon B.C., Cole J.C., Logsdon J.M., Jr., Patterson D.J., Bhattacharya D., Katz L.A. Broadly sampled multigene trees of eukaryotes. BMC Evol. Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li B., Fu Y., Jiang D., Ghabrial S.A., Li G., Peng Y., Xie J., Cheng J., Huang J., Yi X. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8387–8392. doi: 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler Allen L., McCrow J.P., Ininbergs K., Dupont C.L., Badger J.H., Hoffman J.M., Ekman M., Allen A.E., Bergman B., Venter J.C. The baltic sea virome: diversity and transcriptional activity of DNA and RNA viruses. mSystems. 2017:2. doi: 10.1128/mSystems.00125-16. [DOI] [PMC free article] [PubMed] [Google Scholar]