Summary
The jaw is central to the extensive variety of feeding and predatory behaviors across vertebrates. The bones of the lower but not upper jaw form around an early-developing cartilage template. Whereas Endothelin1 patterns the lower jaw, the factors that specify upper jaw morphology remain elusive. Here, we identify Nuclear Receptor 2f genes (Nr2fs) as enriched in and required for upper jaw formation in zebrafish. Combinatorial loss of Nr2fs transforms maxillary components of the upper jaw into lower jaw-like structures. Conversely, nr2f5 misexpression disrupts lower jaw development. Genome-wide analyses reveal that Nr2fs repress mandibular gene expression and early chondrogenesis in maxillary precursors. Rescue of lower jaw defects in endothelin1 mutants by reducing Nr2f dosage further demonstrates that Nr2f expression must be suppressed for normal lower jaw development. We propose that Nr2fs shape the upper jaw by protecting maxillary progenitors from early chondrogenesis, thus preserving cells for later osteogenesis.
Keywords: Nr2f1, Nr2f2, Nr2f5, Coup-TF, nuclear receptor, jaw, skeletal differentiation, cranial neural crest, Endothelin1, pharyngeal arches, cartilage, bone
eTOC Blurb
Barske et al. use combinatorial genetics in zebrafish to identify the Nr2f nuclear receptors as critical factors for specifying upper jaw identity. Nr2fs selectively repress early cartilage formation in upper jaw skeletal progenitors, thus creating morphological asymmetry with the lower but not upper jaw forming around a prominent cartilage template.
Introduction
The upper and lower jaws have evolved to be complementary yet morphologically asymmetrical, shaped by distinct selective pressures on their more symmetrical ancestral forms (Mallatt, 1996). Unlike the mobile lower jawbone, which ossifies around an early-differentiating and extensive Meckel’s cartilage, the stationary upper jaw is formed primarily of dermal bones that directly ossify later in development. In zebrafish, prior to ossification of the premaxilla and maxilla bones, the upper jaw function of securing the pharyngeal skeleton to the neurocranium is fulfilled by the small cartilaginous pterygoid process (Ptp), a late-forming extension of the palatoquadrate cartilage (Cubbage and Mabee, 1996). Previous fate-mapping studies have shown that the zebrafish Ptp is derived from cells located in the maxillary prominence of the first pharyngeal arch, in contrast to the main body of the palatoquadrate that derives from a common condensation with the lower jaw Meckel’s cartilage (Fig. 1A) (Eberhart et al., 2006). In amphibia and birds, the palatoquadrate also derives from a proximal region of the mandibular arch yet lacks a prominent maxillary-derived Ptp process; the maxillary prominence instead gives rise to dermal bones of the upper jaw as well as elements of the anterior neurocranium (Cerny et al., 2004, Lee et al., 2004). As mammals evolved, the palatoquadrate was transformed into the incus and embedded in the middle ear (reviewed by Maier and Ruf, 2016), with the upper jaw consisting exclusively of maxillary-derived dermal bones (Lee et al., 2004). A common theme in all these species is that the maxillary-derived portion of the upper jaw contains a much higher proportion of late-forming dermal bone to early-forming cartilage than the mandibular-derived lower jaw.
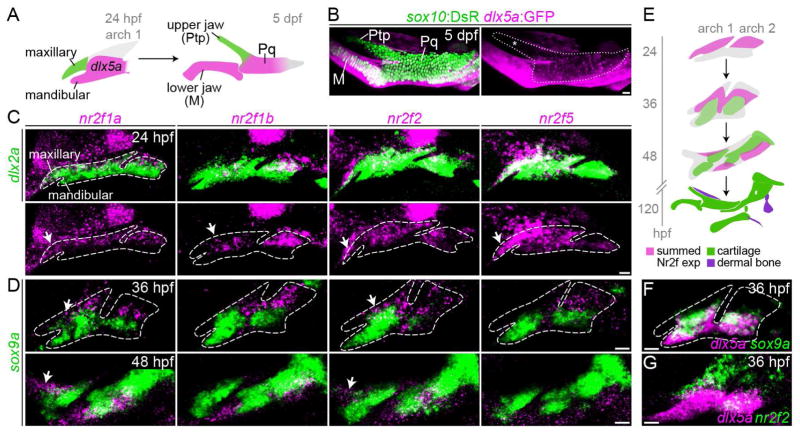
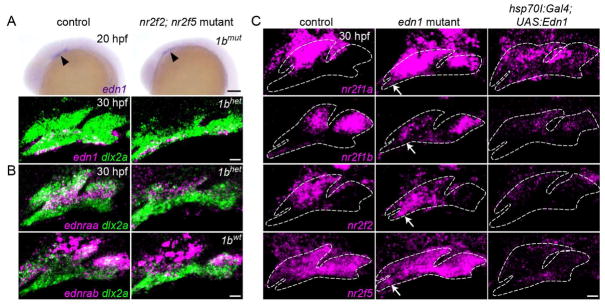
Fig. 1. Enrichment of Nr2f genes in the maxillary domain.
(A) The pterygoid process (Ptp) of the larval upper jaw derives from cells in the maxillary prominence (green), whereas the body of the palatoquadrate cartilage (Pq) derives from a large dlx5a+ precartilaginous condensation (magenta) that also gives rise to the lower jaw Meckel’s cartilage (M). (B) dlx5a:GFP staining (magenta) in the body of the Pq but not the Ptp (asterisk) confirms their distinct origins. Chondrocytes are labeled by sox10:DsRed (green). (C) Double fluorescent in situ hybridizations for four Nr2f genes (magenta) at 24 hpf, with dlx2a (green) marking arch NCCs. Dashed lines represent arch boundaries; white arrows indicate maxillary expression. Images are representative maximum intensity projections. Also see Fig. S1 for quantitative expression data and Table S3 for in situ probe information. (D) Nr2f expression (magenta) is largely excluded from nascent sox9a+ prechondrocytes (green) at 36 and 48 hpf. Dashed lines reflect approximate arch boundaries. Top images are single z-slices; bottom images are maximum intensity projections. (E) Summed Nr2f expression domains in the first and second arches and relative to developing cartilage elements. (F) dlx5a co-localizes with sox9a expression (single z-slice). (G) Co-staining shows a sharp boundary between maxillary nr2f2 and mandibular dlx5a expression (maximum intensity projection). Scale bars = 20 μm.
A major signal driving asymmetric development of the jaw is Endothelin1 (Edn1), which acts on the Ednra receptor in mandibular neural crest-derived cells (NCCs) of the first arch to promote the expression of an array of downstream genes, including the transcription factors Dlx5, Dlx6, and Hand2 (reviewed by Clouthier et al., 2010). Mandibular gene expression is lost and the lower jaw transforms into a mirror image of the upper jaw in zebrafish and mice with loss-of-function mutations in Edn1 (Kurihara et al., 1994, Miller et al., 2000), Ednra (Clouthier et al., 1998), Dlx5/6 (Depew et al., 2002, Beverdam et al., 2002), or Hand2 (Miller et al., 2003, Yanagisawa et al., 2003). Conversely, forced misexpression of Edn1 (Kimmel et al., 2007, Sato et al., 2008) or Hand2 (Sato et al., 2008) activates mandibular genes in the maxillary region, transforming the maxillary portions of the upper jaw into a reflection of the lower jaw, complete with an ectopic Meckel’s cartilage. Bmp signaling also promotes lower jaw development through direct regulation of mandibular genes in NCCs (e.g. Hand2) and by inducing edn1 expression in the overlying ectoderm (Alexander et al., 2011, Zuniga et al., 2011). Less is known about upper jaw regulation. Although Jagged-Notch signaling restricts mandibular gene expression in parts of the upper first and second arches, maxillary-derived portions of the upper jaw develop normally in zebrafish and mice with reduced Jagged-Notch signaling (Zuniga et al., 2010, Humphreys et al., 2012, Barske et al., 2016, Teng et al., 2017). Six1 mutant mice show a partial transformation of the posterior end of the maxilla into a mandible-like structure, but this phenotype is attributable to an upstream role for Six1 in limiting Edn1 expression within the dorsal pharyngeal endoderm (Tavares et al., 2017).
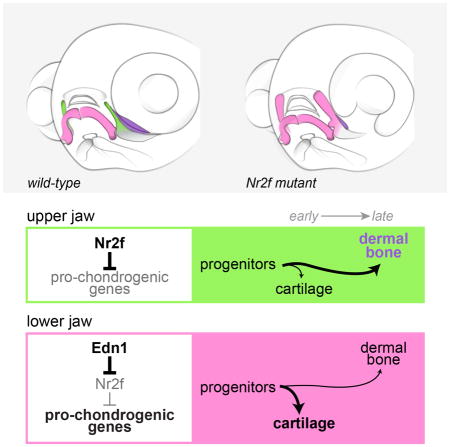
In this study, we reveal that the Nr2f nuclear receptors are necessary and sufficient for maxillary molecular identity and skeletal morphology in zebrafish. Also called chicken ovalbumin upstream promoter transcription factors (COUP-TFs), these nuclear receptors are known for their cell fate-determining functions in the heart, veins, eye, brain, and reproductive system (reviewed by Lin et al., 2011). A previous in vitro study implicated human NR2F1/2 as key drivers of neural crest-associated gene expression in induced human neural crest-like cells (Rada-Iglesias et al., 2012), yet their requirement in craniofacial development was not definitively tested in a genetic model. In zebrafish nr2f mutants, we observe broad misexpression of mandibular genes in maxillary precursors and development of a large lower jaw-like cartilage in place of the small cartilage and dermal bone of the normal upper jaw. Reduction of Nr2f gene dosage also fully restores lower jaw gene expression and skeletal development in edn1 mutants, demonstrating that the primary function of Edn1 in lower jaw development is to exclude Nr2f expression from the mandibular domain. We propose a new model of jaw patterning in which Nr2fs limit early cartilage differentiation in the maxillary precursors of the upper jaw, potentially preserving progenitors for later dermal bone formation. The absence of extensive cartilage in the developing upper jaw could therefore promote a distinct arrangement of dermal bones compared to those of the lower jaw that are organized around a prominent Meckel’s cartilage template.
Results
Nr2f genes are enriched in maxillary precursors of the upper jaw
To identify novel genes enriched in the maxillary prominence at 36 hours post-fertilization (hpf), we queried our transcriptome data (Askary et al., 2017) for candidates depleted in hand2:GFP+ cells (which include mandibular precursors) relative to all arch NCCs. Four members of the Nr2f orphan nuclear receptor family, nr2f1a, nr2f1b, nr2f2, and nr2f5, were −2.36, −2.58, −1.79, and −1.48-fold under-enriched in hand2:GFP+ cells relative to all arch NCCs. With the exception of nr2f5, which is highly expressed by 20 hpf, Nr2f gene expression generally increases as the arches develop, consistent with a post-migratory role for these factors in arch patterning (Fig. S1). All four genes were enriched in dlx2a+ NCCs of the maxillary relative to the mandibular domain at 24 hpf, as well as in dorsal relative to ventral NCCs of the hyoid arch, with nr2f1a and nr2f2 persisting in maxillary NCCs at 36 and 48 hpf (Fig. 1C–E). In zebrafish, dlx5a:GFP expression marks mandibular precursors of Meckel’s cartilage and the body of the palatoquadrate, but not the maxillary-derived Ptp (Fig. 1B). Consistent with nr2f2 enrichment in maxillary NCCs, we observe a sharp boundary between nr2f2 and dlx5a expression in the first arch at 36 hpf (Fig. 1G). Further, expression of all four Nr2f genes was generally excluded from sox9a+ pre-chondrogenic cells, a trend which continues through 48 hpf when cartilage differentiation is more extensive (Fig. 1D–E). Nr2f genes are therefore present at high levels within the maxillary domain during key patterning stages, with their later exclusion from differentiating cartilage consistent with a potential role in repressing chondrogenesis in the upper jaw. By contrast, most dlx5a+ cells in the mandibular territory co-express sox9a (Fig. 1F), suggestive of a possible pro-chondrogenic function for the dlx5a/6a-driven molecular program.
Transformation of the upper jaw in Nr2f mutants
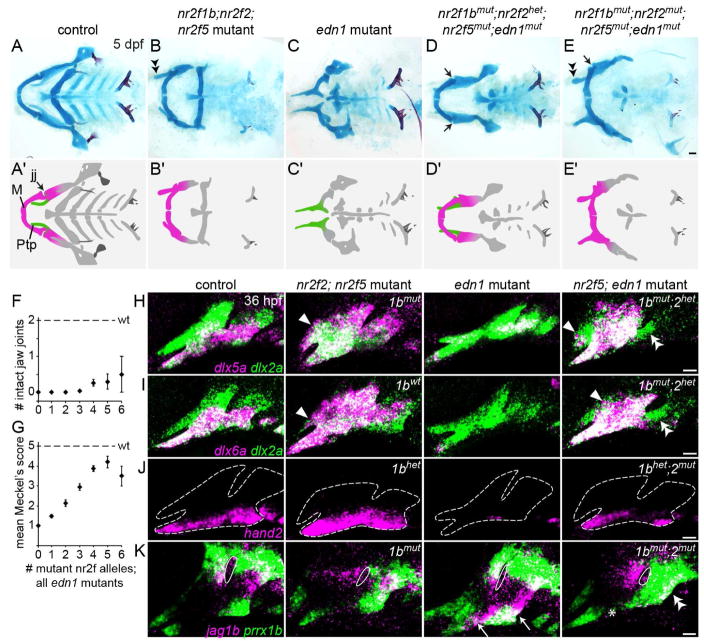
To test requirements for Nr2f genes in maxillary development, we designed TALE nucleases to make loss-of-function alleles, with care taken to introduce early frame-shift mutations likely abrogating all protein function (Fig. S2A). Single nr2f1ael512, nr2f1bel523, and nr2f5el611 mutant zebrafish are viable and fertile, and nr2f2el506 mutants die of undetermined causes in the second week of life. As no craniofacial defects were observed in single mutant larvae (Fig. S2B), we examined combinatorial mutants. The upper jaw Ptp cartilage was conspicuously enlarged in ~45% of double nr2f2; nr2f5 mutants, with the penetrance enhanced by further loss of nr2f1b alleles (Fig. 2A–D). The enlarged Ptp was reminiscent of the lower jaw Meckel’s cartilage, similar to what is observed upon misexpression of Edn1 or Bmp4 (Kimmel et al., 2007, Alexander et al., 2011). Unlike Meckel’s, the enlarged upper jaw cartilages did not meet at the midline and remained articulated with the anterior neurocranium (Fig. 2B). In wild-type fish, the proximal end of Meckel’s cartilage develops an endochondral process called the retroarticular. In contrast to chicken, where the retroarticular is built from second arch cells (Kontges and Lumsden, 1996), fate mapping by photoconversion of sox10:kikGR protein shows that the retroarticular process is first arch-derived in zebrafish (Fig. S2C). Intriguingly, we occasionally observed an ectopic protrusion resembling the retroarticular on the enlarged Ptp of nr2f2; nr2f5 mutants (Fig. 2E), further supporting transformation of the Ptp into an ectopic Meckel’s.
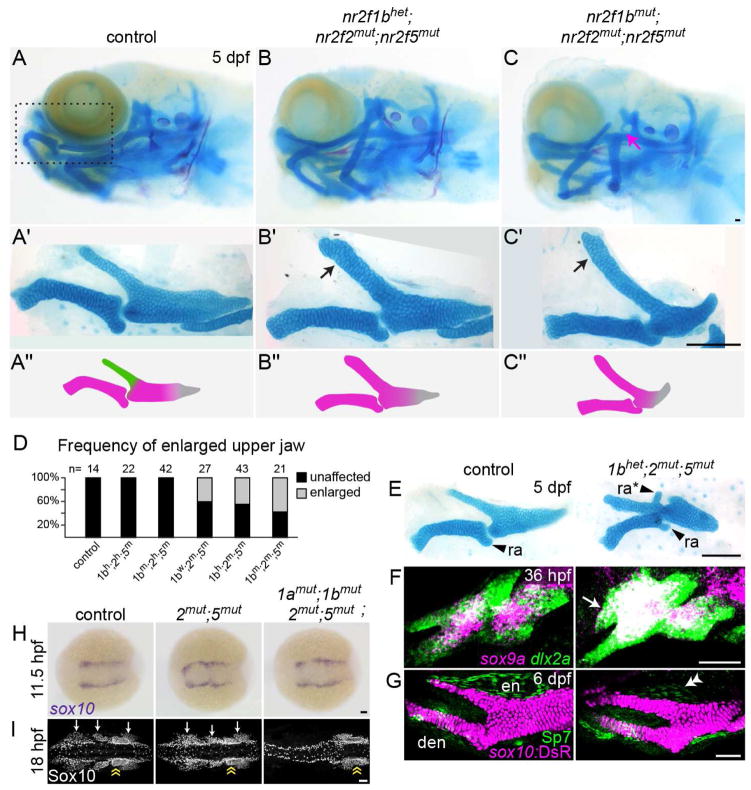
Fig. 2. Transformation of the upper jaw in Nr2f mutants.
(A–C) Larval heads stained for cartilage (Alcian blue) and bone (Alizarin red). Boxed area in A represents the approximate region of the dissected upper and lower jaw cartilages in A′–C′. In mutants (B′–C′), the pterygoid process (Ptp, green in A″) of the palatoquadrate (Pq) is thickened (black arrows) to resemble Meckel’s (M, magenta in A″). Skeletal structures derived from the dorsal hyoid arch are reduced in the triple mutant (magenta arrow in C). (D) Frequency of the thickened Ptp phenotype across different nr2f1b; nr2f2; nr2f5 genotypes. (E) Some nr2f2; nr2f5 mutants form an ectopic process (ra*) that resembles the retroarticular process (ra) of Meckel’s. (F) Fluorescent in situs show expansion of sox9a expression into the mutant maxillary prominence (white arrow; dlx2a labels all arch NCCs). (G) Formation of a larger cartilage (sox10:DsRed+, magenta) in the mutant is accompanied by a reduction in the number of Sp7+ osteoblasts (green) in the neighboring dermal entopterygoid bone (en; white double arrowhead). Mutants have normal numbers of Sp7+ osteoblasts in the lower jaw dentary bone (den). (H) sox10+ NCCs are specified at the neural plate border in all Nr2f mutants (n = 18 nr2f1aany; nr2f1bany; nr2f2−/−; nr2f5−/−; n = 2 quadruple). The nr2f2; nr2f5 double mutant shown is also nr2f1a+/−; nr2f1b−/−. (I) Three streams of Sox10+ NCCs (white arrows) are evident in all mutant combinations (n = 13 nr2f1aany; nr2f1bany; nr2f2−/−; nr2f5−/−) except the quadruple mutant (n = 1). Sox10+ otic cells (yellow double arrow) are indicated for reference. Scale bars: C, E = 100 μm; F–I = 50 μm. Also see Fig. S2 and Table S4 for mutation details and skeletal phenotypes of single and other combinatorial mutants.
The increase in upper jaw cartilage in nr2f2; nr2f5 mutants was prefigured by elevated sox9a expression and an increase in the size of the maxillary domain as early as 36 hpf (Fig. 2F). The dermal bone that forms next to Ptp was also reduced in size, containing fewer Sp7+ osteoblasts (Fig. 2G). Lethality of nr2f2; nr2f5 mutants in the second week of life prevented us from assessing whether later-forming lower jaw features, such as the intramembranous dentary bone, are duplicated in the mutant upper jaw. The second arch-derived hyoid joint, dorsal hyomandibula cartilage, and opercle bone were also progressively reduced in nr2f1b; nr2f2; nr2f5 combinatorial mutants, and further loss of nr2f1a resulted in fusions between the upper and lower jaws and cardiac edema, with quadruple mutants showing severe reductions of the entire facial skeleton (Fig. 2C, Fig. S2B).
Given the previous report that human NR2F1/2 drive early NCC gene expression in vitro (Rada-Iglesias et al., 2012), we examined NCC specification and migration in combinatorial mutants. At 11.5 hpf, the sox10+ domain marking premigratory NCCs was unchanged in all mutant combinations, including quadruple mutants (Fig. 2H). By 18 hpf, three streams of Sox10+ NCCs had migrated to the arches in all double and triple mutants, including the nr2f2; nr2f5 mutants with upper jaw phenotypes (Fig. 2I). However, the streams were absent in a quadruple knockout, consistent with large-scale loss of facial cartilage in these mutants (Fig. S2B) and frogs with nr2f1 knockdown (Rada-Iglesias et al., 2012). These results argue against the upper jaw phenotypes of nr2f2; nr2f5 mutants being due to NCC specification or migration defects, consistent with a post-migratory role for Nr2fs in maxillary NCCs.
Nr2f5 misexpression disrupts lower jaw development
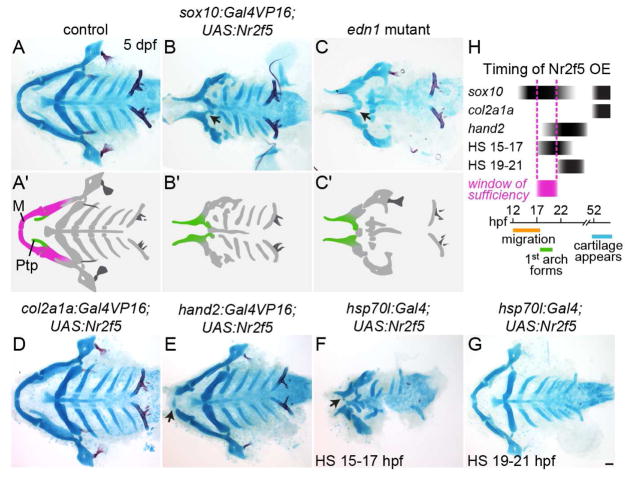
We next asked whether misexpression of Nr2f genes is sufficient to transform the lower jaw. In zebrafish, cranial NCCs begin migration at ~12 hpf, colonize the arches and are first exposed to signals such as Edn1 by ~18 hpf, and then differentiate into cartilage starting ~48 hpf. When nr2f5 was misexpressed in NCCs by the sox10 promoter (sox10:Gal4VP16; UAS:nr2f5) from ~13 hpf onwards (Das and Crump, 2012), we observed that the lower jaw Meckel’s cartilage was very reduced and fused to the palatoquadrate, a phenotype closely resembling that of edn1 mutants (Fig. 3A–C) (Miller et al., 2000). We did not observe ectopic dermal bone in the lower jaw, again similar to edn1 mutants, a finding we confirmed by analyzing the osteoblast marker Sp7 (Fig. S3A). The lack of ectopic bone upon nr2f5 misexpression suggests that Nr2fs function to inhibit cartilage formation rather than to directly promote bone differentiation. Although the sox10:Gal4VP16 driver is also re-activated in differentiating cartilage (Das and Crump, 2012), misexpression of nr2f5 in chondrocytes using col2a1a-R2:Gal4VP16 did not affect the jaw skeleton (Fig. 3D), suggesting that it is the earlier NCC misexpression that disrupts mandibular development. Further, nr2f5 misexpression in mandibular NCCs shortly after arch colonization, using a hand2:Gal4VP16 driver, resulted in distal truncation of Meckel’s cartilage (Fig. 3E). We also employed a heat-shock-inducible hsp70l:Gal4 driver to precisely define when Nr2f5 is sufficient to alter lower jaw development. Whereas a heat shock from 15–17 hpf disrupted lower jaw development, similar to pan-NCC expression of nr2f5, a 19–21 hpf heat shock had no effect (Fig. 3F–G). Given the approximate 2-h lag in UAS-driven protein expression after the start of the heat shock (Scheer et al., 2002), these results indicate that, to alter lower jaw development, Nr2f5 needs to be present at high levels in post-migratory NCCs when they are first exposed to Edn1 (Fig. 3H).
Fig. 3. nr2f5 misexpression reduces and transforms the lower jaw.
(A–G) Alcian blue and Alizarin red staining of dissected facial skeletons shown from the ventral perspective. The upper jaw (Ptp, green in A′–C′) remains, but the lower jaw (M, magenta in A′) is reduced and transformed (black arrows) in sox10:Gal4VP16; UAS:Nr2f5el662 transgenic fish (B), similar to edn1 mutants (C). Putative transformations are schematized in A′–C′. Late-onset nr2f5 misexpression in chondrocytes (col2a1a:Gal4VP16) did not affect the facial skeleton (D). Misexpression in post-migratory mandibular NCCs (hand2:Gal4VP16) reduced the distal tips of Meckel’s (black arrow, E). Heat-shock-induced embryo-wide misexpression (hsp70l:Gal4) reduced the lower jaw (black arrow) when applied at 15–17 (F) but not 19–21 hpf (G). (H) Approximate timing of Nr2f5 protein overexpression (OE) in the different driver lines, with the approximate window of sufficiency indicated in magenta. Scale bar = 50 μm. Also see Fig. S3 for additional analyses of the sox10:Gal4VP16; UAS:Nr2f5 phenotype.
Nr2fs broadly inhibit mandibular gene expression in the maxillary domain
To understand how Nr2fs globally regulate jaw molecular identity, we performed RNA sequencing of FACS-purified arch NCCs (36 hpf) from nr2f2; nr2f5 double mutant and sox10:Gal4VP16; UAS:nr2f5 misexpression embryos. Double mutant embryos were selected based on their ocular coloboma phenotype (Fig. S2D), similar to conditional Rx:Cre; Nr2f1flox/flox; Nr2f2flox/flox mouse mutants (Tang et al., 2010). Genes 2-fold higher or lower in the misexpression versus mutant line were respectively classified as ‘Nr2f-activated’ and ‘Nr2f-inhibited’ (see Methods). Among the 350 genes with arch-enriched expression in wild-type embryos, 112 were strongly inhibited by Nr2f2/5 but only 5 were activated (Tables 1, S1–S2). Nr2f-inhibited genes are associated with lower jaw development (e.g. dlx3b/4a/4b/5a, hand2, msx1a (formerly msxe), nkx3.2) and cartilage differentiation (barx1, sox9a, sim2). Consistent with the reciprocal mutant skeletal phenotypes, many genes were oppositely regulated by Nr2f and Edn1, including 15 of the top 20 Nr2f-inhibited genes and 3 of the 5 Nr2f-activated genes (Fig. 4A, Tables 1, S2). Hierarchical clustering of transcriptome data grouped the two independent nr2f2; nr2f5 mutant samples with a previously published Edn1-misexpressing sample, whereas the Nr2f5-misexpressing sample clustered with two edn1 mutants (Fig. 4B) (Barske et al., 2016, Askary et al., 2017). Thus, Nr2fs and Edn1 act oppositely to regulate both jaw molecular identity and skeletal composition/morphology.
Table 1.
Top 20 Nr2f-inhibited genes.
| Gene | full name | nr2f2/5 mutant vs. control | nr2f5 OE vs. control | nr2f5 OE vs. nr2f2/5 mutant | Edn1 regulation |
|---|---|---|---|---|---|
| dlx4b | distal-less homeobox 4b | +1.15 | n.d. in OE | n.d. in OE | activated |
| fgfbp2a | fibroblast growth factor binding protein 2a | +1.86 | n.d. in OE | n.d. in OE | activatedd |
| six5 | sine oculis-related homeobox 5 | +1.51 | −25.67 | −38.69 | - |
| dlx5a | distal-less homeobox 5a | +1.68 | −8.57 | −14.40 | activateda |
| Nkx3.2 | NK3 homeobox 2 | +1.80 | −6.75 | −12.18 | activatedb |
| dlx4a | distal-less homeobox 4a | +1.25 | −9.32 | −11.64 | activated |
| Sim2 | single-minded homolog 2 | +1.25 | −9.10 | −11.34 | - |
| TWSG1 | twisted gastrulation BMP signaling modulator 1 | +1.40 | −8.01 | −11.19 | activated |
| f3b | coagulation factor IIIb | +1.75 | −5.33 | −9.32 | activated |
| hand2 | heart and neural crest derivatives expressed transcript 2 | +1.91 | −4.72 | −9.04 | activatedc |
| zgc:110626 | protein-coding gene | +2.17 | −3.84 | −8.12 | activated |
| barx1 | BARX homeobox 1 | +1.57 | −5.03 | −7.90 | activateda |
| dlx3b | distal-less homeobox | +1.27 | −5.48 | −6.97 | activatedc |
| Lrrc8c | leucine rich repeat containing 8 family, member C | +1.60 | −3.76 | −6.03 | activated |
| sema3bl | semaphorin 3bl | +1.47 | −3.93 | −5.77 | activatedd |
| Msx1a | muscle segment homeobox 1a | +1.80 | −2.91 | −5.24 | activatedc |
| Nog2 | noggin 2 | +1.60 | −3.02 | −4.84 | activated |
| cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | +5.50 | +1.16 | −4.73 | - |
| Hsd11b2 | hydroxysteroid (11-beta) dehydrogenase 2 | +2.36 | −1.95 | −4.60 | - |
| Foxf2a | forkhead box F2a | +1.74 | −2.62 | −4.56 | - |
n.d., not detected. Unless otherwise referenced, Edn1 regulation was determined using published RNAseq data (Askary et al., 2017), with Edn1 activated and inhibited genes showing overexpression/control fold-change: mutant/control fold-change TPM ratios > 1.5 and < 0.667, respectively. A dash indicates no clear activation or repression. Predicted regulation confirmed by in situ:
Askary et al. 2017. Also see Table S2 for Nr2f-activated genes.
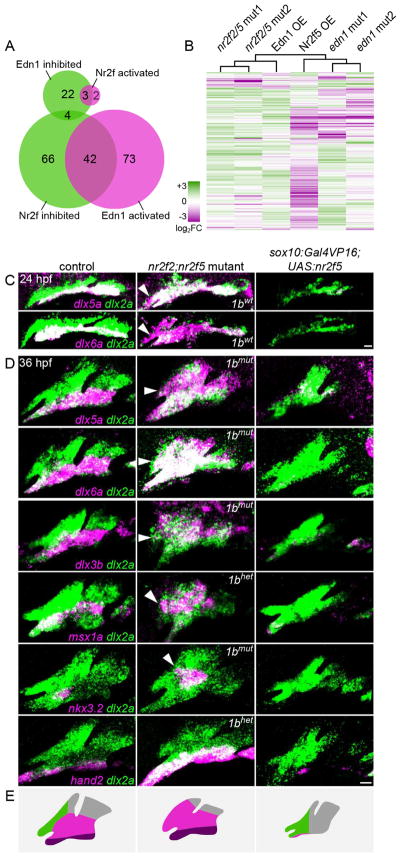
Fig. 4. Nr2f2/5 broadly inhibit mandibular gene expression.
(A) Venn diagrams depicting the overlap between genes inhibited and activated by Nr2fs and Edn1. (B) Hierarchical clustering of log2 fold-change values relative to controls for 350 arch-enriched genes in nr2f2; nr2f5 mutants, sox10:Gal4VP16; UAS:Nr2f5 transgenics (Nr2f5 OE), edn1 mutants, and hsp70l:Gal4; UAS:Edn1 (Edn1 OE) transgenics. Reference data are from Askary et al. (2017). See Table S1 for TPM values of all arch-enriched genes. (C–D) Fluorescent in situs for dysregulated genes (magenta) relative to all arch NCCs (dlx2a+, green) in nr2f2; nr2f5 mutants (nr2f1b genotype indicated) and sox10:Gal4VP16; UAS:Nr2f5 transgenics at 24 (C) or 36 hpf (D). Arrowheads indicate ectopic maxillary expression. Images are representative maximum intensity projections. Scale bars = 20 μm. (E) Schematized shifts in arch gene expression territories. See Fig. S4 for expression patterns of non-mandibular genes.
In line with the RNAseq data, in situ hybridizations confirmed expansion of dlx3b, dlx5a, dlx6a, and msx1a expression into the maxillary domain of nr2f2; nr2f5 mutants, and reduction upon nr2f5 misexpression (Fig. 4C–D). The shifts in dlx5a/6a expression were evident as early as 24 hpf (Fig. 4C), consistent with the early sufficiency window for nr2f5 affecting lower jaw morphology. We also found that nkx3.2, which is specifically expressed in the first arch and required for formation of the lower jaw retroarticular process (Miller et al., 2003), was expanded in mutants and lost in misexpression embryos, consistent with the ectopic retroarticular-like process observed in some nr2f mutants. Distal mandibular expression of hand2 was also lost upon nr2f5 misexpression and elevated in mutants, although not into the maxillary domain. Thus, loss of Nr2fs triggers an extensive yet incomplete molecular transformation of the maxillary domain.
In addition to the gene expression changes observed, the first arch of nr2f5 misexpression embryos was smaller. This reduction in arch size correlated with a modest decrease in proliferation yet no change in apoptosis of arch NCCs (Fig. S3B–C), again similar to what has been reported for edn1 mutants (Sasaki et al., 2013). As many dlx2a+ NCCs are still present in the mandibular domain of nr2f5 misexpression and edn1 mutant embryos, the loss of mandibular gene expression cannot solely be due to reduced numbers of NCCs. In addition, the second arch of nr2f2; nr2f5 mutants was also smaller compared with controls, and changes in gene expression were less straightforward than in the first arch, with dlx5a and hand2 expanded yet dlx3b, dlx6a, and msx1a reduced. Given the reduction of the upper hyoid skeleton in the mutants (Fig. 2C), these molecular differences suggest distinct roles for Nr2f genes in the maxillary domain versus the second arch.
In contrast to the extensive inhibition of the mandibular program by Nr2fs, RNAseq identified only 5 putative Nr2f-activated genes. One of these, pitx2, was expressed in cells adjacent to but not co-localizing with dlx2a+ maxillary NCCs, whereas meis2b was mostly confined to the hyoid arch at 36 hpf, with expression of both largely unaltered in nr2f2; nr2f5 mutants (Fig. S4A). Expression of jag1b, which marks dorsal mandibular and hyoid but not maxillary NCCs (Zuniga et al., 2010), was also largely unaffected in mutants (Fig. S4B), as were markers of the frontonasal NCC population (alx1, gata3) (Fig. S4C). Consistently, the anterior neurocranium, derived in part from frontonasal NCCs, formed normally in mutants (Fig. S4D). Nr2fs therefore function primarily to silence the mandibular expression program in the maxillary domain rather than activate maxillary-specific genes (Fig. 4E).
Reducing Nr2f dosage fully rescues lower jaw development in edn1 mutants
Given the opposite effects of Nr2fs and Edn1 on mandibular gene expression, we investigated their epistatic relationship in lower jaw patterning. Expression of edn1 was unaltered in nr2f2; nr2f5 mutants at 20 and 30 hpf (Fig. 5A), and the Edn1 receptors were either unchanged (ednrab) or reduced (ednraa) (Fig. 5B), arguing against expansion of mandibular gene expression being due to increased Edn1 signaling. Conversely, we observed varying degrees of ectopic expression of all four Nr2f genes in the mandibular domain of edn1 mutants at 30 hpf, and loss of their maxillary expression upon Edn1 misexpression (Fig. 5C, S5).
Fig. 5. Edn1 inhibits Nr2f expression in the mandibular prominence.
(A) edn1 is expressed comparably in the ventral ectoderm of wild-type controls and mutants at 20 hpf (arrowheads) and 30 hpf (magenta). (B) Expression of the Edn1 receptors is reduced (ednraa) or largely unaffected (ednrab) in Nr2f mutants. Arch NCCs are labeled with dlx2a (green), and nr2f1b genotypes are indicated. (C) In edn1 mutants, expression of all four Nr2f genes (magenta) ectopically increases to varying degrees in the mandibular domain (white arrows). Misexpression of edn1 induced by a heat shock of hsp70l:Gal4; UAS:Edn1 embryos from 20–24 hpf reduced Nr2f expression throughout the arches. Images are maximum intensity projections. Merged images with dlx2a arch NCC staining are presented in Fig. S5. Scale bars: A (top panel) = 100 μm; A (bottom panel), B, C = 20 μm.
We therefore tested whether ectopic mandibular expression of Nr2fs might underlie the lower jaw defects of edn1 mutants. Strikingly, removal of 3–6 alleles of nr2f1b, nr2f2, and nr2f5 completely rescued Meckel’s cartilage formation in many edn1 mutants (Fig. 6A–E, G; S6). We also infrequently observed restoration of the jaw joint, the element most sensitive to Edn1 loss (Fig. 6D, F, 12/106 mutants) (Kimmel et al., 2007). In contrast, the hyoid arch-derived ceratohyal and posterior arch-derived ceratobranchial defects of edn1 mutants were not rescued. Some nr2f2; nr2f5; edn1 mutants also developed an enlarged upper jaw (Fig. 6E), further indicating that the upper jaw transformations of Nr2f mutants are not due to inappropriate Edn1 signaling. The rescue of lower jaw cartilage defects in nr2f; edn1 mutants was preceded by restoration of dlx5a, dlx6a, and hand2 expression to relatively normal patterns in mandibular arch NCCs (Fig. 6H–J). Furthermore, progenitor markers jag1b and prrx1b, which are upregulated in ventral NCCs in edn1 mutants (Zuniga et al., 2010, Barske et al., 2016), were also restored to more typical patterns in the first arch of nr2f; edn1 mutants (Fig. 6K). However, hyoid arch expression of all five genes was not restored, correlating with the failure of hyoid cartilage rescue in compound mutants. In summary, the complete rescue of mandibular gene expression and lower jaw skeletal morphology in some nr2f; edn1 mutants demonstrate that the primary function of Edn1 is to prevent the Nr2f-dependent repression of mandibular gene expression.
Fig. 6. Reducing Nr2f gene dosage can fully rescue the lower jaw in edn1 mutants.
(A–E) Facial skeletons of fish carrying nr2f and/or edn1 mutations, schematized in A′–E′. Whereas nr2f2; nr2f5 mutants have a thickened upper jaw (double arrowhead), edn1 mutants have a much reduced lower jaw. Reducing Nr2f alleles in edn1 mutants can rescue the jaw joint (black arrows in D, E) and Meckel’s cartilage (compare C to D–E). The upper jaw is still enlarged in nr2f2; nr2f5; edn1 combinatorial mutants (double arrowhead). M, Meckel’s cartilage; Ptp, pterygoid process; jj, jaw joint. Scale bar in E = 50 μm. (F, G) Quantification of jaw joint and Meckel’s rescue across different summed nr2f; edn1 genotypes. One or both jaw joints were rescued in 12/106 mutants lacking 3–6 Nr2f alleles. See Fig. S6 for Meckel’s scoring system. n = 16, 29, 30, 53, 42, 10, and 2 for edn1 mutants with 0–6 mutant Nr2f alleles, respectively. Error bars are the standard error of the mean. Scores in G were non-evenly distributed across genotype classes (p < 0.0001, Chi-squared). (H–J) dlx5a, dlx6a, and hand2 (magenta) are expanded in nr2f2; nr2f5 mutants (white arrowheads), reduced in edn1 mutants, and restored in the first arch of nr2f5; edn1 mutants (nr2f1b/nr2f2 genotypes indicated). Some ectopic expansion into maxillary cells was still evident in the combinatorial mutants (white arrowheads), yet second arch expression was not restored (double arrowheads). dlx2a (green) labels arch NCCs, and dashed lines in J indicate arch boundaries. (K) Ectopic ventral expression of jag1b (magenta) and prrx1b (green) in edn1 mutants (white arrows) is reversed in the mandibular (asterisk) but not hyoid (double arrowheads) domains of nr2f1b; nr2f2; nr2f5; edn1 mutants. White outlines depict the first endodermal pouch. Scale bars in E = 50 μm; H–K = 20 μm.
Discussion
Here we demonstrate that Nr2f nuclear receptors function in post-migratory NCCs to confer a distinct identity to maxillary precursors of the upper jaw. The broad expression of mandibular genes throughout nr2f mutant arches, even in the absence of Edn1 signaling, highlights a key role for Nr2fs in restricting mandibular gene expression. An extensive body of work in cell culture and mammalian models has shown that Nr2fs can function as both transcriptional activators and repressors, dependent on the cellular context (reviewed in Pereira et al., 2000)). A previous study uncovered an activator function for human NR2F1/2 in migratory neural crest-like cells, with NR2F1/2 detected at more than 3000 neural crest gene-associated enhancers with active chromatin signatures (Rada-Iglesias et al., 2012). Whether Nr2fs bind to similar enhancers in post-migratory NCCs of the maxillary domain remains to be determined. As our RNA sequencing was performed at 36 hpf, ~16 hours after the critical window of Nr2f function, the observed mandibular upregulation in mutants might be indirect. It therefore remains possible that genes transiently activated by Nr2fs just after NCC migration might instead account for the observed mandibular upregulation at later stages, which could be addressed in the future through Nr2f chromatin binding assays.
The robust expression of mandibular genes (e.g. dlx5a/6a) in the combined absence of Edn1 and Nr2fs was also surprising given the large body of work showing a central role for Edn1 in their expression (reviewed in Clouthier et al., 2010). Although we had previously shown that Bmp4 misexpression also partially rescues the skeletal defects of edn1 mutants, Bmp4 was unable to restore Dlx expression (Zuniga et al., 2011), in contrast to what we show here for Nr2f reduction. It is therefore unlikely that the lower jaw rescue in nr2f; edn1 mutants is due to increased Bmp signaling. Data from mice suggest that Edn1 drives lower jaw development through a Calmodulin-CamKII-histone deacetylase cascade that de-represses Mef2c, which then transactivates Dlx5/6 expression (Verzi et al., 2007, Hu et al., 2015). One possibility is that Mef2c functions to reverse Nr2f repression of dlx5a/6a, and potentially other mandibular genes, such that, if Nr2f repression is reduced, Edn1-mediated Mef2c transactivation is no longer required. Alternately, reduced Nr2f levels could allow Mef2c to be activated in an Edn1-independent manner. Future identification and analysis of direct targets of Nr2fs in post-migratory NCCs will be required to address these questions.
A marked asymmetry in jaw development is that cartilage differentiation initiates earlier and to a greater extent in the lower jaw (Schilling and Kimmel, 1997). This disparity first manifests as an early mandibular bias in barx1 and sox9a expression (Barske et al., 2016), which mark prechondrogenic condensations. A number of mandibular genes, including Dlx5 and Msx1, have also been implicated in skeletal differentiation (e.g. Orestes-Cardoso et al., 2001, Ferrari and Kosher, 2002) and co-localize with sox9a expression in pre-chondrocytes. One possibility then is that the mandibular program represents an early pro-differentiation network, with chondrocytes being the first differentiated cell type arising in the arches. As such, Nr2fs might function to block this early differentiation network in the maxillary domain, thus preserving a pool of progenitors for later dermal bone formation. Consistently, nr2f2; nr2f5 mutants show elevated cartilage and reduced bone formation in the maxillary domain, and Nr2f expression continues to be excluded from the chondrogenic sox9a+ domain as arch development proceeds. Future lineage studies will be required to test if this represents a direct fate shift of cells from bone to cartilage. In the mouse limb, NR2F2 protein is similarly enriched in mesenchymal precursors relative to differentiating chondrocytes. However, in contrast to our findings in the skull, Nr2f2 knockdown in mesenchymal cell culture leads to increased osteogenesis, and conditional ablation of Nr2f2 in the limb results in delayed cartilage maturation (Xie et al., 2011). Further genetic studies are therefore necessary to determine whether Nr2f genes have multiple and/or discrete roles in in the NCC- and mesoderm-derived skeletons.
Our work suggests a simple model of jaw asymmetry, in which the large Meckel’s cartilage of the lower jaw serves as a template that distinctly organizes later dermal bone development relative to the upper jaw. In developing mammalian embryos, differences in skeletal composition are even more striking, with the upper jaw forming without any cartilage template. Although mammalian upper jawbones derive from a homologous population of maxillary NCCs, whether mammalian upper jaw identity also depends on Nr2f function remains to be determined. Single Nr2f1 mutants die perinatally without overt craniofacial abnormalities (Qiu et al., 1997), while Nr2f2 mouse mutants die with angiogenic and cardiac defects by E10.5 (Pereira et al., 1999). A recent study has shown that conditional mutation of Nr2f2 within NCCs results in ectopic Sox9 expression and abnormalities in the tympanic ring (Hsu et al., 2017), a proximal first arch-derived dermal bone (Mallo, 2001, Hsu et al., 2017), consistent with our finding that Nr2fs inhibit maxillary sox9a expression in fish. Deletions spanning the NR2F1 and NR2F2 genomic loci have also been identified in isolated human patients with craniofacial anomalies (Poot et al., 2007, Brown et al., 2009). Our finding of widespread redundancy between Nr2f family members means that a conditional double mouse mutant will likely be needed to determine whether Nr2f genes are similarly required to establish morphological asymmetry of the mammalian jaws.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gage Crump (gcrump@usc.edu).
EXPERIMENTAL MODEL DETAILS
Animals
The Institutional Animal Care and Use Committee of the University of Southern California approved all animal experiments performed in this study (No. 10885, 20540). Zebrafish (Danio rerio) embryos were raised in defined Embryo Medium (Westerfield, 2007) at 28.5°C and staged as described (Kimmel et al., 1995). Mutant and transgenic lines were maintained on the Tuebingen wild-type background (Haffter et al., 1996) and housed in groups of 10–15 fish. Published mutant and transgenic lines include sucker/edn1tf216 (RRID: ZDB-GENO-070209-215) (Miller et al., 2000), nr2f1ael512 (Duong et al., 2017), Tg(fli1a:EGFP)y1 (RRID: ZDB-ALT-011017-8) (Lawson and Weinstein, 2002), Tg(sox10:DsRed-Express)el10 and Tg(sox10:Gal4VP16)el159 (Das and Crump, 2012), Tg(-1.5hsp70l:Gal4)kca4 (RRID: ZDB-GENO-070209-168) (Scheer et al., 2001), Tg(~4.9sox10:kikGR)el2 (Balczerski et al., 2012), dlx5aj1073Et (RRID: ZDB-ALT-100209-11) (referred to as dlx5a:GFP; Talbot et al., 2010), Tg(hand2:Gal4VP16)b1228 (Nichols et al., 2013) and Tg(UAS:Edn1;α-crystallin:Cerulean)el249 (Zuniga et al., 2011). edn1tf216 and Gal4 and UAS transgenes were genotyped as described (Miller et al., 2000, Barske et al., 2016; also see Table S4). Two new transgenic lines (Tg(UAS:Nr2f5;α-crystallin:Cerulean) and Tg(col2a1a-R2:Gal4VP16)el647) and three targeted mutant lines (nr2f1bel523, nr2f2el506, and nr2f5el611) were generated for this study (see below). Transgenic lines were propagated by selecting larvae expressing fluorescent protein markers or by outcrossing putative adult Gal4 carriers to UAS reporter lines. For mutant lines, fin biopsies were collected at 14 dpf or 3 mpf, lysed with proteinase K, genotyped by PCR using GoTaq DNA polymerase (Promega, Madison, WI), and digested with the appropriate restriction enzymes where necessary. Genotyping details for each line are provided in Table S4.
METHOD DETAILS
Generation of zebrafish transgenic and mutant lines
The UAS:Nr2f5; α-crystallin:Cerulean construct was assembled with the Gateway (Invitrogen) Tol2kit (Kwan et al., 2007) by creating a pME containing the nr2f5 coding sequence and combining it with p5E-UAS, p3E-polyA and pDestTol2AB2. col2a1a-R2:Gal4VP16 was derived from the R2-E1b:EGFP plasmid (Dale and Topczewski, 2011) via restriction cloning to replace EGFP with the Gal4VP16 sequence. Tg(col2a1a-R2:Gal4VP16)el647 and two alleles of Tg(UAS:Nr2f5;α-crystallin:Cerulean) (el662, el663) were generated by injecting these constructs with transposase RNA and selecting for germline transmission of Cerulean fluorescence in the lens or expression of a UAS reporter construct, respectively.
Zebrafish mutant lines for nr2f1a, nr2f1b, nr2f2, and nr2f5 were generated via TALEN-based targeted mutagenesis. TALEN constructs were generated using the PCR-based platform (Sanjana et al., 2012) and digested with StuI (New England Biolabs, Ipswich, MA). RNAs were synthesized off the linearized constructs using the mMessage mMachine T7 Ultra kit (Ambion/Life Technologies, Carlsbad, CA, USA). TALEN RNAs were injected at 100 ng/μl into 1-cell-stage embryos, and mosaic germline founders were identified by screening their progeny by PCR and restriction digestion. Two to five separate mutant alleles were examined for each single mutant. The nr2f1ael512, nr2f1bel523, nr2f2el506, and nr2f5el611 alleles are frameshift mutations that interrupt the coding sequence upstream of or in the first functional domain (Fig. S2). The TALEN sequences and genotyping primers used to produce and identify mutations in each gene are provided in Table S4.
Misexpression analyses
Gal4 expression was induced in hsp70l:Gal4; UAS:Edn1 and hsp70l:Gal4; UAS:Nr2f5 embryos by ramping up a programmable incubator to 40°C for the indicated duration and then allowing it to cool back to 28.5°C. Distal truncation of Meckel’s cartilage was observed upon hand2:Gal4-driven misexpression only in fish carrying the stronger Tg(UAS:nr2f5;α-crystallin:Cerulean)el663 allele. For each Gal4/UAS combination, 4–15 individual larvae carrying both transgenes were selected for dissection and imaging.
FACS of arch NCCs
To profile the 36 hpf stage of development, we moved 27 hpf embryos to a 22°C incubator to slow their development so that they acquired morphological criteria associated with the desired stage the following morning. fli1a:EGFP; sox10:DsRed double-positive embryos were selected under a fluorescent dissecting microscope. nr2f2; nr2f5; fli1a:EGFP; sox10:DsRed double mutant embryos were identified based on their fully penetrant coloboma phenotype (Fig. S2D). sox10:Gal4VP16; fli1a:EGFP; sox10:DsRed and sox10:Gal4VP16; UAS:Nr2f5; fli1a:EGFP; sox10:DsRed transgenics were identified by genotyping fin biopsies taken at 24 hpf for Gal4 and Cerulean (Table S4). Two independent replicates of the nr2f2; nr2f5 cross were assayed, and sox10:Gal4VP16+ individuals were used as intra-run controls. To prepare cell suspensions for FACS (Covassin et al., 2006), batches of 30–40 dechorionated embryos were de-yolked by gentle agitation in fresh Ringer’s solution for approximately 10 min, then transferred to a protease cocktail containing 0.25% trypsin (Life Technologies), 1 mM EDTA, and 2 mg/ml Collagenase P (Roche Life Science, Indianapolis, IN) in PBS. The samples were pipetted intermittently over a 15-min incubation at 28.5°C to promote dissociation. The enzymatic reaction was stopped by adding 1/6 volume of 6 mM CaCl2 and 30% fetal bovine serum (FBS) in PBS. Cells were centrifuged at 2000 rpm for 5 min at 4°C, resuspended in suspension medium (1% FBS, 0.8 mM CaCl2, 50 U/ml penicillin, and 0.05 mg/ml streptomycin (Sigma-Aldrich, St. Louis, MO) in Leibovitz’s L15 medium (Life Technologies)), centrifuged as above, resuspended in 500 μl suspension medium, filtered into a FACS tube, and chilled on ice. Cells were sorted for GFP and DsRed on a MoFlo Astrios (Beckman-Coulter, Brea, CA), and double-positive cells were collected into RLT lysis buffer (Qiagen). For the two nr2f2; nr2f5 mutant replicates, we dissociated 37 and 84 embryos and collected 18,858 and 30,655 double-positive cells, respectively. For the sox10:Gal4VP16; UAS:Nr2f5 and sox10:Gal4VP16 embryos, 84 and 69 embryos yielded 56,866 and 86,517 cells, respectively.
RNA sequencing
Total RNA was immediately extracted from sorted cells using the RNeasy Micro kit (Qiagen) according to the manufacturer’s instructions. The purified RNA was evaluated for quality and quantity on a BioAnalyzer Pico RNA chip (Agilent, Santa Clara, CA) and then reverse-transcribed into cDNA using the SMARTer kit (Clontech, Mountain View, CA). cDNA amplification was adjusted based on the RNA input quantity, and the size and amount of generated cDNA was measured by BioAnalyzer. The samples were sonicated on a S2 ultrasonicator (Covaris, Woburn, MA) according to Clontech’s recommendations. DNA libraries were assembled using the Kapa Hyper Prep kit (Kapa Biosystems, Wilmington, MA) with NextFlex adapters (Bioo Scientific, Austin, TX), assessed by Bioanalyzer, and quantified using qPCR (Kapa Library Quantification kit, Kapa Biosystems). Sequencing was performed on a NextSeq500, using 75-bp paired end reads (Illumina, San Diego, CA). Library construction and sequencing were performed at the USC Molecular Genomics Core.
In situ hybridization, immunostaining, and cell death assay
In situ probes include dlx2a and dlx3b (Akimenko et al., 1994), dlx6a (Walker et al., 2006), edn1 (Miller et al., 2000), dlx5a and jag1b (Zuniga et al., 2010), hand2 (Angelo et al., 2000), msx1a (Akimenko et al., 1995), nkx3.2 (Miller et al., 2003), prrx1b (Barske et al., 2016), sox9a (Yan et al., 2002), and sox10 (Dutton et al., 2001). Partial cDNAs for alx1, gata3, meis2b, nr2f1a, nr2f1b, nr2f2, nr2f5, and pitx2 were amplified with Herculase II Fusion DNA Polymerase (Agilent) and cloned into pCR™-Blunt II-TOPO™ (Thermofisher Scientific, Waltham, MA). Plasmids were linearized by restriction digestion, and antisense probes were synthesized with Sp6 or T7 polymerase using dioxygenin (DIG)- or dinitrophenol (DNP)-labeled nucleotides (Roche) (Table S3).
Fluorescent in situ hybridizations were performed as described (Talbot et al., 2010), with minor modifications. The full protocol is available online (https://wiki.zfin.org/display/prot/Triple+Fluorescent+In+Situ). We increased the proteinase K concentration to 1.25 μg/ml, applied the anti-DIG-peroxidase (#11207733910, Roche) and anti-DNP-peroxidase antibodies (#NEL747B001KT, Perkin Elmer) at 1:500 and 1:200, respectively, and diluted the TSA-fluorescein and TSA-Cy3 to 1:75 in Perkin Elmer Amplification Diluent. Colorimetric in situs were performed following the same protocol until after the stringency washes. At this point, the embryos were washed twice in phosphate-buffered saline with 0.1% Tween-20 (PBST), blocked in PBST containing 5 mg/ml BSA and 10% sheep serum, and incubated overnight at 4°C with an anti-DIG-alkaline phosphatase antibody (1:10,000; #11093274910, Roche). The next day, embryos were washed ten times in PBST, equilibrated twice in NTMT buffer (100 mM NaCl, 50 mM MgCl2, 100 mM Tris-HCl, 0.05% Tween-20), and exposed to NBT/BCIP (stock diluted 1:50 in NTMT; #11681451001, Roche) in the dark. The color reaction was stopped by replacing the solution with PBST, and embryos were post-fixed for 20 min in 4% paraformaldehyde (PFA) and moved into glycerol for imaging. For each probe combination, 3–10 mutants or misexpression embryos were evaluated alongside a similar number of controls. Representative examples are shown.
For immunostaining, embryos were fixed in 4% PFA overnight and then exposed to rabbit anti-Sox10 (1:200, Genetex GTX128374), rabbit anti-Osterix (Sp7) (1:100, Santa Cruz Biotechnology sc-22536; RRID: AB_831618), rabbit anti-phospho-Histone H3 (Ser10) (1:500, Millipore 06-570; RRID: AB_310177), rabbit anti-GFP (1:1000, Torrey Pines Biolabs TP401; RRID: AB_10013661), or chicken anti-mCherry (1:200, Novus Biologicals NBP2-25158; RRID: AB_2636881) primary antibodies, followed by Alexa 568-donkey anti-rabbit, Alexa 647-goat anti-rabbit, or Alexa 633-goat anti-chicken secondary antibodies (Thermofisher) and counterstaining with Hoechst 33342 (Life Technologies H1399).
Dying cells in 32 hpf sox10:Gal4VP16; UAS:Nr2f5 transgenic embryos were labeled using the ApopTag® Fluorescein In Situ Apoptosis Detection Kit (Millipore-Sigma S7110) according to the manufacturer’s instructions. Labeled cells that co-express the arch NCC marker sox10:DsRed within the first two arches were manually counted.
All fluorescently stained samples were imaged on a Zeiss LSM800 confocal microscope and are presented as representative optical sections or maximum intensity projections. Colorimetric in situs were imaged on a Leica S8APO. Levels were modified consistently across samples in Adobe Photoshop CS6.
Photoconversion and fate mapping
Transgenic sox10:kikGR embryos were anesthetized with Tricaine (Sigma A5040) at 35 hpf and mounted in agarose for imaging on a Zeiss LSM800 confocal. A region of interest within the first arch was defined using the ROI tool in the ZEN software and then exposed to a 405 nm laser at 65% power for approximately 3 s to irreversibly convert the green fluorescent kikGR protein to red. The embryos were then allowed to recover in fresh Embryo Medium in the dark. At 5 dpf, photoconverted red cells were located within the larval facial skeleton and reimaged. By this stage, only chondrocytes strongly express high levels of green kikGR protein (Balczerski et al., 2012).
Skeletal staining
Alcian Blue (cartilage) and Alizarin Red (bone) staining was performed at room temperature as described (Walker and Kimmel, 2007). Briefly, larvae were anesthetized with Tricaine (Sigma A5040), fixed for one hour in 2% PFA, rinsed for 10 min in 100 mM Tris (pH 7.5) with 10 mM MgCl2, then incubated overnight in Alcian Blue solution (0.04% Alcian Blue, 80% ethanol, 100 mM Tris pH 7.5, 10 mM MgCl2). Larvae were rehydrated through 10-min washes in 80%, 50%, 25% ethanol solutions containing 100 mM Tris (pH 7.5) and 10 mM MgCl2, then bleached for 10 min in 3% H2O2 with 0.5% KOH under a lamp. Samples were washed twice in 25% glycerol with 0.1% KOH, stained for 30–45 min with Alizarin Red (0.01% Alizarin Red, 25% glycerol, 100 mM Tris (pH 7.5)), and de-stained in 50% glycerol containing 0.1% KOH. Whole-mount and dissected specimens were mounted in 50% glycerol and imaged with LAS v4.4 software on Leica S8APO and DM2500 microscopes, respectively.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNAseq data analysis
In pre-alignment QC, the raw reads from all samples were typically of high quality, with average sample quality scores exceeding 33. The reads were trimmed at both ends based on the Phred quality score, with a minimum end quality level of 20 and a minimum acceptable read length of 25. Paired-end reads were then aligned to the Danio rerio GRCz10 assembly (Ensembl_v80) by TopHat2 (Kim et al., 2013) in Partek Flow® (Partek Inc., St. Louis, MO). Aligned reads were quantified with the Partek E/M algorithm and normalized to generate Transcript Per Million (TPM) values. Values obtained for the two nr2f2; nr2f5 mutant replicates were compared in Excel (Microsoft) and proved highly similar (R2 = 0.89262, or 0.95312 with ten most highly expressed genes removed).
To focus our analysis on genes with enriched expression in the pharyngeal arches, we referenced published data from wild-type fli1a:GFP; sox10:DsRed double- and single-positive cells (GEO Accession No. GSE95812) (Askary et al., 2017) and filtered the new dataset to include only those genes with an average TPM value ≥ 2 in double-positive cells of three published 36 hpf wild-type samples and with enrichment ≥ 1.5-fold in GFP+/DsRed+ arch NCCs versus single GFP+ and DsRed+ cells. Filtering for genes enriched in double-positive cells removes those genes known to be associated with the blood/endothelial (fli1a:GFP+) or ear (sox10:DsRed) lineages, the presence of which likely reflects some small degree of contamination of our double-positive population with single-positive cells (Barske et al., 2016). We applied a final filter for a TPM ≥ 0.5 in the new control sox10:Gal4VP16+ sample to remove genes with very low expression in the current batch.
This process left 350 genes with arch-enriched expression in wild-type embryos at 36 hpf. We then calculated the fold-change values of these genes in the nr2f2; nr2f5 mutant and sox10:Gal4VP16; UAS:Nr2f5 (OE) transgenic samples relative to the sox10:Gal4VP16 control, with the reciprocal presented in Table 1 for ratios < 1. Genes for which the OE/control ratio was ≥ 1 and the OE/mutant ratio ≥ 2 were classified as ‘Nr2f-activated’, and genes with a mutant/control ratio ≥ 1 and an OE/mutant ratio ≤ −2 were deemed ‘Nr2f-inhibited’.
For hierarchical clustering, we included fold-change values relative to controls for the 350 arch-enriched genes in edn1 mutants and hsp70l:Gal4; UAS:Edn1 transgenics, calculated from published TPM values (GEO Accession No. GSE95812) (Askary et al., 2017). Log-transformed fold-change values were submitted to Gene Cluster v.3.0 (de Hoon et al., 2004) for hierarchical clustering with the average linkage method and visualized in Java TreeView v.1.1.6r4 (Saldanha, 2004).
Scoring of Meckel’s cartilage rescue and analysis of cell death and proliferation
The degree of lower jaw rescue in nr2f; edn1 mutants was scored as 1 (typical edn1 mutant) through 5 (typical wild-type) (see Fig. S5). We summed these scores across individuals with the same number of mutant Nr2f alleles, which ranged from 0–6. For example, individuals with the genotype nr2f1b+/−; nr2f2−/−; nr2f5−/− were grouped with nr2f1b−/−; nr2f2+/−; nr2f5−/− and nr2f1b−/−; nr2f2−/−; nr2f5+/− individuals, all of which carried 5 mutant Nr2f alleles. Chi-squared analysis was performed in Excel to analyze the distribution of scores across genotypes, with p < 0.01 considered significant.
The numbers of dying (ApopTag+) and proliferating (pHH3+) cells in the first two arches were compared between control and sox10:Gal4VP16; UAS:Nr2f5 embryos using the non-parametric Mann Whitney U test, with p < 0.05 taken as marginally significant.
DATA AND SOFTWARE AVAILABILITY
Raw RNAseq files are accessible through NCBI’s Gene Expression Omnibus Series, Accession No. GSE101719. Filtered TPM and fold-change values for all arch-enriched genes are provided in Table S1.
Supplementary Material
Table S1, related to Fig. 4. Expression changes in 350 arch-enriched genes in nr2f2; nr2f5 mutant and nr2f5 misexpression embryos.
Highlights.
The upper jaw transforms into a lower jaw-like structure in Nr2f mutant zebrafish
Nr2fs globally inhibit lower jaw gene expression in upper jaw precursors
Reducing Nr2f dosage fully rescues lower jaw defects in endothelin1 mutants
Nr2fs drive jaw asymmetry by limiting cartilage differentiation in the upper jaw
Acknowledgments
We thank Megan Matsutani and Jennifer DeKoeyer Crump for fish care, Yang Chai, Robert Maxson, and Ruchi Bajpai for helpful discussions, Jeffrey Boyd at the USC Stem Cell Flow Cytometry Core Facility for FACS, Jason Kim at the USC Molecular Genomics Core for RNA sequencing, and the USC Norris Medical Library Bioinformatics Service for sequence analysis. Funding was provided by the National Institute of Dental and Craniofacial Research [R01 DE018405 (J.G.C) and K99 DE026239 (L.B.)] and the A.P. Giannini Foundation (L.B.). The bioinformatics software and computing resources were funded by the USC Office of Research and the Norris Medical Library. The authors declare no conflicts of interest.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Author contributions
Conceptualization, L.B. and J.G.C.; Methodology, L.B. and S.G.C.; Investigation, L.B., P.R., K.B., L.D.R.; Writing, L.B. and J.G.C.; Funding Acquisition, L.B. and J.G.C.; Supervision, L.B. and J.G.C.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–86. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimenko MA, Johnson SL, Westerfield M, Ekker M. Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development. 1995;121:347–57. doi: 10.1242/dev.121.2.347. [DOI] [PubMed] [Google Scholar]
- Alexander C, Zuniga E, Blitz IL, Wada N, Le Pabic P, Javidan Y, Zhang T, Cho KW, Crump JG, Schilling TF. Combinatorial roles for BMPs and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. 2011;138:5135–46. doi: 10.1242/dev.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo S, Lohr J, Lee KH, Ticho BS, Breitbart RE, Hill S, Yost HJ, Srivastava D. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231–7. doi: 10.1016/s0925-4773(00)00334-8. [DOI] [PubMed] [Google Scholar]
- Askary A, Xu P, Barske L, Bay M, Bump P, Balczerski B, Bonaguidi MA, Crump JG. Genome-wide analysis of facial skeletal regionalization in zebrafish. Development. 2017;144:2994–3005. doi: 10.1242/dev.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczerski B, Matsutani M, Castillo P, Osborne N, Stainier DY, Crump JG. Analysis of sphingosine-1-phosphate signaling mutants reveals endodermal requirements for the growth but not dorsoventral patterning of jaw skeletal precursors. Developmental biology. 2012;362:230–41. doi: 10.1016/j.ydbio.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske L, Askary A, Zuniga E, Balczerski B, Bump P, Nichols JT, Crump JG. Competition between Jagged-Notch and Endothelin1 signaling selectively restricts cartilage formation in the zebrafish upper face. PLOS Genet. 2016;12:e1005967. doi: 10.1371/journal.pgen.1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A, Merlo GR, Paleari L, Mantero S, Genova F, Barbieri O, Janvier P, Levi G. Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: mirror of the past? Genesis. 2002;34:221–7. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- Brown KK, Alkuraya FS, Matos M, Robertson RL, Kimonis VE, Morton CC. NR2F1 deletion in a patient with a de novo paracentric inversion, inv(5)(q15q33.2), and syndromic deafness. Am J Med Genet A. 2009;149A:931–8. doi: 10.1002/ajmg.a.32764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny R, Lwigale P, Ericsson R, Meulemans D, Epperlein HH, Bronner-Fraser M. Developmental origins and evolution of jaws: new interpretation of “maxillary” and “mandibular”. Dev Biol. 2004;276:225–36. doi: 10.1016/j.ydbio.2004.08.046. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Garcia E, Schilling TF. Regulation of facial morphogenesis by endothelin signaling: insights from mice and fish. Am J Med Genet A. 2010;152A:2962–73. doi: 10.1002/ajmg.a.33568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–24. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Covassin L, Amigo JD, Suzuki K, Teplyuk V, Straubhaar J, Lawson ND. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol. 2006;299:551–62. doi: 10.1016/j.ydbio.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubbage CC, Mabee PM. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, cyprinidae) J Morphol. 1996;229:121–160. doi: 10.1002/(SICI)1097-4687(199608)229:2<121::AID-JMOR1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Dale RM, Topczewski J. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev Biol. 2011;357:518–31. doi: 10.1016/j.ydbio.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Crump JG. Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLOS Genet. 2012;8:e1002710. doi: 10.1371/journal.pgen.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–5. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Duong TB, Ravisankar P, Song YC, Gafranek JT, Rydeen AB, Dohn TE, Barske LA, Crump JG, Waxman JS. Nr2f1a balances atrial chamber and atrioventricular canal size via BMP signaling-independent and -dependent mechanisms. Dev Biol. 2017 doi: 10.1016/j.ydbio.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–25. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Eberhart JK, Swartz ME, Crump JG, Kimmel CB. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development. 2006;133:1069–77. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- Ferrari D, Kosher RA. Dlx5 is a positive regulator of chondrocyte differentiation during endochondral ossification. Dev Biol. 2002;252:257–70. doi: 10.1006/dbio.2002.0862. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, Van Eeden FJ, Jiang YJ, Heisenberg CP, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hsu WH, Chen CM, You LR. COUP-TFII is required for morphogenesis of the neural crest-derived tympanic ring. Sci Rep. 2017;7:12386. doi: 10.1038/s41598-017-12665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Verzi MP, Robinson AS, Tang PL, Hua LL, Xu SM, Kwok PY, Black BL. Endothelin signaling activates Mef2c expression in the neural crest through a MEF2C-dependent positive-feedback transcriptional pathway. Development. 2015;142:2775–80. doi: 10.1242/dev.126391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys R, Zheng W, Prince LS, Qu X, Brown C, Loomes K, Huppert SS, Baldwin S, Goudy S. Cranial neural crest ablation of Jagged1 recapitulates the craniofacial phenotype of Alagille syndrome patients. Hum Mol Genet. 2012;21:1374–83. doi: 10.1093/hmg/ddr575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Walker MB, Miller CT. Morphing the hyomandibular skeleton in development and evolution. J Exp Zool B Mol Dev Evol. 2007;308:609–24. doi: 10.1002/jez.b.21155. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–42. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao WH, Kamada N, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–10. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–99. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–18. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lee SH, Bedard O, Buchtova M, Fu K, Richman JM. A new origin for the maxillary jaw. Dev Biol. 2004;276:207–24. doi: 10.1016/j.ydbio.2004.08.045. [DOI] [PubMed] [Google Scholar]
- Lin FJ, Qin J, Tang K, Tsai SY, Tsai MJ. Coup d’Etat: an orphan takes control. Endocr Rev. 2011;32:404–21. doi: 10.1210/er.2010-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Ruf I. Evolution of the mammalian middle ear: a historical review. J Anat. 2016;228:270–83. doi: 10.1111/joa.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallatt J. Ventilation and the origin of jawed vertebrates: A new mouth. Zool J Linn Soc-Lond. 1996;117:329–404. [Google Scholar]
- Mallo M. Formation of the middle ear: recent progress on the developmental and molecular mechanisms. Developmental biology. 2001;231:410–9. doi: 10.1006/dbio.2001.0154. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–28. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–65. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Pan L, Moens CB, Kimmel CB. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development. 2013;140:2765–75. doi: 10.1242/dev.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orestes-Cardoso SM, Nefussi JR, Hotton D, Mesbah M, Orestes-Cardoso MD, Robert B, Berdal A. Postnatal Msx1 expression pattern in craniofacial, axial, and appendicular skeleton of transgenic mice from the first week until the second year. Dev Dyn. 2001;221:1–13. doi: 10.1002/dvdy.1120. [DOI] [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–49. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira FA, Tsai MJ, Tsai SY. COUP-TF orphan nuclear receptors in development and differentiation. Cell Mol Life Sci. 2000;57:1388–98. doi: 10.1007/PL00000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Eleveld MJ, Van ‘T Slot R, Van Genderen MM, Verrijn Stuart AA, Hochstenbach R, Beemer FA. Proportional growth failure and oculocutaneous albinism in a girl with a 6.87 Mb deletion of region 15q26.2-->qter. Eur J Med Genet. 2007;50:432–40. doi: 10.1016/j.ejmg.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Pereira FA, Demayo FJ, Lydon JP, Tsai SY, Tsai MJ. Null mutation of mCOUP-TFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes Dev. 1997;11:1925–37. doi: 10.1101/gad.11.15.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–48. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–92. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki MM, Nichols JT, Kimmel CB. edn1 and hand2 interact in early regulation of pharyngeal arch outgrowth during zebrafish development. PloS one. 2013;8:e67522. doi: 10.1371/journal.pone.0067522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kurihara Y, Asai R, Kawamura Y, Tonami K, Uchijima Y, Heude E, Ekker M, Levi G, Kurihara H. An endothelin-1 switch specifies maxillomandibular identity. Proc Natl Acad Sci USA. 2008;105:18806–11. doi: 10.1073/pnas.0807345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- Scheer N, Riedl I, Warren JT, Kuwada JY, Campos-Ortega JA. A quantitative analysis of the kinetics of Gal4 activator and effector gene expression in the zebrafish. Mech Dev. 2002;112:9–14. doi: 10.1016/s0925-4773(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–60. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2507–17. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Xie X, Park JI, Jamrich M, Tsai S, Tsai MJ. COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development. 2010;137:725–34. doi: 10.1242/dev.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares ALP, Cox TC, Maxson RM, Ford HL, Clouthier DE. Negative regulation of endothelin signaling by SIX1 is required for proper maxillary development. Development. 2017;144:2021–2031. doi: 10.1242/dev.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng CS, Yen HY, Barske L, Smith B, Llamas J, Segil N, Go J, Sanchez-Lara PA, Maxson RE, Crump JG. Requirement for Jagged1-Notch2 signaling in patterning the bones of the mouse and human middle ear. Sci Rep. 2017;7:2497. doi: 10.1038/s41598-017-02574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Agarwal P, Brown C, Mcculley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Dev Cell. 2007;12:645–52. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–8. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Coffin Talbot J, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Westerfield M. THE ZEBRAFISH BOOK: A guide for the laboratory use of zebrafish (Danio rerio) University of Oregon Press; 2007. [Google Scholar]
- Xie X, Qin J, Lin SH, Tsai SY, Tsai MJ. Nuclear receptor chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) modulates mesenchymal cell commitment and differentiation. Proc Natl Acad Sci USA. 2011;108:14843–8. doi: 10.1073/pnas.1110236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, et al. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–79. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Clouthier DE, Richardson JA, Charite J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–78. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- Zuniga E, Rippen M, Alexander C, Schilling TF, Crump JG. Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development. 2011;138:5147–56. doi: 10.1242/dev.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E, Stellabotte F, Crump JG. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development. 2010;137:1843–52. doi: 10.1242/dev.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Fig. 4. Expression changes in 350 arch-enriched genes in nr2f2; nr2f5 mutant and nr2f5 misexpression embryos.