Abstract
The Mycobacterium tuberculosis genome encodes twenty cytochrome P450 enzymes, most or all of which appear to have specific physiological functions rather than being devoted to the removal of xenobiotics. However, in many cases their specific functions remain obscure. Considerable spectroscopic, biophysical, crystallographic, and catalytic information is available on nine of these cytochrome P450 enzymes, although gaps exist in our knowledge of even these enzymes. The available evidence indicates that at least three of the better-characterized enzymes are promising targets for antituberculosis drug discovery. This review summarizes the information on the nine relatively well-characterized cytochrome P450 enzymes, with a particular emphasis on CYP121, CYP125, and CYP142 from Mycobacterium tuberculosis and Mycobacterium smegmatis.
Keywords: Mycobacterium tuberculosis, cytochrome P450, cholesterol degradation, azole drugs, enzyme inhibitors
Graphical Abstract
Tuberculosis is a long-standing human scourge. Hippocrates mentioned it in 400 BCE in “Of the Epidemics”, stating “The greatest and most dangerous disease and the one that proved fatal to the greatest number, was consumption” (1). In 1680, John Bunyan termed it the “captain of all these men of death” (2). Despite a very long history, effective drugs for the treatment of tuberculosis did not become available until the 1950s, when isoniazid, pyrazinamide, and rifampicin were introduced. These drugs lowered the profile of tuberculosis in economically advanced societies, but it remains a devastating disease in many parts of the world, with 10.4 million new cases and 1.8 million deaths in 2015 (3). Although neglected for decades, the development of new drugs for tuberculosis has again become a priority due to two factors: the synergistic action of tuberculosis with AIDS, and the growing spread of M. tuberculosis strains that are multiply resistant (MDR) and extremely resistant (XDR) to current therapies, all of which date from the 1950s or early 1960s. Only recently have two new drugs, bedaquiline (in 2012) and delamanid (in 2014) been brought to the clinic.
The potential of cytochrome P450 enzymes as targets for anti-tuberculosis drugs is highlighted by the finding that antifungal drugs known to target cytochrome P450 enzymes readily inhibit the growth of both Mycobacterium tuberculosis (4) and M. smegmatis (4, 6, 7). For example, clotrimazole and econazole at a concentration of 0.125 mg mL−1 completely inhibit the growth of the H37Rv strain of M. tuberculosis in culture (8). Furthermore, reactivation of latent H37Rv in mice was only partially inhibited by isoniazid and pyrazinamide, but completely inhibited if rifampicin was included in the drug cocktail. Importantly, rifampicin in the drug cocktail could be replaced by econazole with the same result (9). Ketoconazole was also shown to improve the in vivo prognosis in mice when added to a cocktail of antituberculosis drugs (10). Other investigators reported that several azole drugs inhibit the growth of M. smegmatis (7). These results clearly suggest that one or more cytochrome P450 enzymes are suitable targets for agents against mycobacterial infections.
The M. tuberculosis genome has genes that code for twenty cytochrome P450 enzymes. The cytochrome P450 proteins produced heterologously by nine of these genes have received particular attention: the crystal structures of eight of them have been determined and the physiological substrates for five of them have been identified (Table 1). This review focuses on these cytochrome P450 enzymes. The other eleven cytochrome P450 enzymes (Table 2) are at various stages of investigation, but for none of them is there sufficient information to warrant a discussion of their biochemistry, role, or potential utility as drug discovery targets. A number of reviews are available that focus on various aspects of the cytochrome P450 complement of M. tuberculosis (22, 23, 24, 25).
Table 1.
The M. tuberculosis cytochrome P450 enzymes for which sufficient evidence is available to initiate a discussion of their potential as drug targets.
| Gene number | P450 number | Crystal structure | Substrate |
|---|---|---|---|
| Rv0764c | CYP51B1 | (11) | Lanosterol |
| Rv2276 | CYP121A1 | (12, 13) | Cyclodityrosine (cYY) |
| Rv2266 | CYP124A1 | (14) | Dimethyl chain termini: e.g., phytanic acid, farnesol |
| Rv3545c | CYP125A1 | (15, 16) | Cholesterol Cholest-4-en-3-one |
| Rv0778 | CYP126A1 | (17) | Unknown |
| Rv2268c | CYP128A1* | Menaquinone MK9(H2) | |
| Rv1256c | CYP130A1 | (18) | Unknown |
| Rv3518c | CYP142A1 | (19, 20) | Cholesterol Cholest-4-en-3-one |
| Rv1777 | CYP144A1 | (21) | Unknown |
No crystal structure is available for this enzyme
Table 2.
The M. tuberculosis cytochrome P450 enzymes for which there is relatively little information.
| Gene number | P450 number |
|---|---|
| Rv0766c | CYP123A1 |
| Rv1394c | CYP132A1 |
| Rv0568 | CYP135B1 |
| Rv0327c | CYP135A1 |
| Rv3059 | CYP136A1 |
| Rv3685c | CYP137A1 |
| Rv0136 | CYP138A1 |
| Rv1666c | CYP139A1 |
| Rv1880c | CYP140A1 |
| Rv3121 | CYP141A1 |
| Rv1785c | CYP143A1 |
CYP51
The discovery that the M. tuberculosis genome encodes a CYP51 enzyme (CYP51B1) immediately raised interest in its potential utility as a drug target. The CYP51 family of cytochrome P450 enzymes consists of substrate-specific enzymes that catalyze the 14α-demethylation of sterols such as that of lanosterol in the cholesterol biosynthetic pathway (Fig. 1) (26). Homologous enzymes catalyze the same or similar reactions in other species, including fungi and parasites. Because of the critical role of sterols with an extended hydrocarbon side-chain at position 17 in the maintenance of membrane structure, inhibition of 14α-demethylation has proven an effective target for antifungal and antiparasitic drugs (27). The 14α-methyl sterols that accumulate in the membrane when the 14α-demethylase is inhibited cause membrane disruptions that lead to cell death. Indeed, the proliferation of azole drugs such as ketoconazole and posaconazole stems from their efficacy in the treatment of fungal infections.
Fig. 1.
The three sequential reactions of the cholesterol biosynthetic pathway catalyzed by CYP51.
M. tuberculosis CYP51B1 has been heterologously expressed, purified, biophysically characterized, and shown to catalyze the conversion of lanosterol and closely related 14α-methyl sterols to the 14α-desmethyl derivatives (28, 29, 30). CYP51B1 was the first M. tuberculosis cytochrome P450 for which a crystal structure was determined (11). In addition to the binding of azole agents (7, 11, 31), small molecule inhibitors of the enzyme have been identified by high-throughput ligand screening and other approaches (32, 33, 34, 35).
The utility of M. tuberculosis CYP51B1 as a drug target, however, is compromised by the fact that M. tuberculosis does not have a functional sterol biosynthetic pathway. It was reported on the basis of a radiolabel incorporation assay that mycobacteria produce sterols (36), but it is now believed that this was the result of contamination of the cultures (37). Analysis of the M. tuberculosis genome shows that it lacks, among others, the genes coding for squalene epoxidase and oxidosqualene cyclase, two key enzymes required for construction of the sterol skeleton. Furthermore, it does not appear from genetic studies that CYP51B1 is required for mycobacterial growth (38, 39). Given that lanosterol or a close analog is not produced endogenously by the mycobacteria, and that CYP51B1 appears not to be essential for growth, the role of this enzyme in mycobacterial biology is unclear. CYP51B1 could contribute to the carbon flux by helping to degrade lanosterol in the host cell, but levels of lanosterol in mammals are so low that such a metabolic contribution is unlikely to be significant in view of the high concentrations of cholesterol that are present. In the absence of a defined role for the enzyme, it is questionable whether inhibition of CYP51B1 contributes to the growth inhibitory effect of azole drugs, and it is unclear whether it is a suitable target for drug development.
CYP121
Among the twenty M. tuberculosis cytochrome P450 enzymes, one of the most promising from the point of view of antituberculosis drug development is CYP121A1 because it is essential for viability of the mycobacteria (40). Although the relationship of mycobacterial viability to some the cytochrome P450 enzymes has not been unambiguously tested, in many cases they do not appear to be required for mycobacterial growth in culture. For example, Mycobacterium tuberculosis strains in which CYP125A1 (16) and CYP128A1 (41) were individually knocked out nevertheless grew well in normal media. Of course, an enzyme may not be required for in vitro growth, but still be essential for infection and virulence in vivo, as will be noted in our later discussion of CYP125A1. On the basis of sequence comparisons, CYP121A1 was first suspected of involvement in lipid or polyketide metabolism (42). It was expressed heterologously in E. coli and its biophysical properties, including its absorption, EPR, MCD and resonance Raman spectra, were found to be unexceptional. However, UV-visible spectroscopic screens did not confirm the binding of fatty acids, sterols, or polyketides (42), although azole drugs that are general inhibitors of cytochrome P450 enzymes were shown to bind to the enzyme (7). The crystal structures of the ligand-free enzyme (12) and of the enzyme with fluconazole bound in the active site were then determined, but unfortunately these structures did not help to define the true substrate for the enzyme.
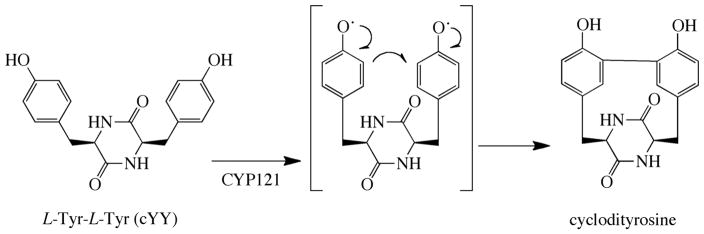
Identification of the substrate of CYP121A1 and the function of this enzyme came from unrelated studies that were not specifically intended to elucidate the biology of M. tuberculosis P450 enzymes. Gondry et al., in the course of investigating the formation of cyclodipeptides, one of which derives from two tyrosine residues, noted that the gene coding for the formation of this Tyr-Tyr cyclodipeptide was adjacent to that coding for CYP121A1 (13, 43). This led to the discovery by Belin et al. that CYP121A1 catalyzes oxidative crosslinking of the two tyrosines in the cyclodipeptide (Fig. 2) (13). This conclusion was supported by a crystal structure of CYP121A1 with the Tyr-Tyr cyclodipeptide bound in the active site, and by a subsequent study exploring the binding of substrate analogues that showed the enzyme is highly specific for the Tyr-Tyr cyclodipeptide (44). Computational (45) and experimental (46) results suggest a mechanism for the enzyme in which Compound I of CYP121A1 mediates a one-electron oxidation of each of the two tyrosine rings, which then undergo radical-radical coupling to form the crosslink (Fig. 2).
Fig. 2.
Intramolecular crosslinking of the L-Tyr-L-Tyr cyclodipeptide catalyzed by CYP121A1.
In view of the requirement of CYP121A1 for mycobacterial growth, the development of inhibitors for this enzyme is a promising avenue to novel antituberculosis drugs. Comparison of the data on the sensitivity of M. tuberculosis P450 enzymes to inhibition by azole drugs is informative in this regard. As already discussed, econazole and clotrimazole were used to establish that azole drugs can inhibit the growth of M. tuberculosis both in culture and in infected animals (8, 9, 47). Table 3, which is based on a compilation by the Munro group of the data in the literature on the inhibition of M. tuberculosis P450 enzymes by azole drugs (17), shows that CYP121A1 binds econazole, clotrimazole, and miconazole more tightly than any of the other M. tuberculosis P450 enzymes in the table. As econazole and clotrimazole were the azoles used to establish that M. tuberculosis growth could be inhibited by this class of drugs, it is likely that inhibition of CYP121A1 contributes to the reported inhibition of M. tuberculosis growth by these azole compounds.
Table 3.
Kd values for the binding of azoles to M. tuberculosis cytochrome P450 enzymes.
| Azole | 51B1 μM |
121A1 μM |
126A1 μM |
125A1 μM |
130A1 μM |
142A1 μM |
144A1 μM |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Econazole | 0.31 | 0.02 | 4.0 | 11.7 | 1.93 | 4.6 | 0.78 |
| Clotrimazole | 0.18 | 0.07 | 3.9 | 5.3 | 13.3 | 3.8 | 0.37 |
| Miconazole | 0.20 | 0.14 | 1.3 | 4.6 | 1.70 | 4.0 | 0.98 |
| Ketoconazole | 3.57 | 3.41 | 0.34 | 27.1 | 48.0 | 21 | 134 |
| Fluconazole | 5.82 | 8.61 | NBa | 43.2 | NDb | 860 | >10000 |
| Voriconazole | 2.10 | 16.3 | NB | NB | ND | NB | 6510 |
| 4-Phenyl-imidazole | 452 | 32.3 | NB | 216 | ND | 12 | 280 |
| Imidazole | 11700 | ND | 2590 | 536 | ND | ND | 2965 |
NB indicates no significant perturbation of the spectrum occurred on addition of the azole to the enzyme.
ND indicates the data is not available.
Fragment screening has been implemented in the search for specific non-azole inhibitors of CYP121A1 (48, 49). This has led to the identification of a series of phenolic compounds that bind to CYP121A1 with spectroscopically determined Kd values as low as 15 μM. The structures of some of these agents bound to CYP121A1 have been determined.
CYP126
CYP126A1 has been heterologously expressed in E. coli, its crystal structure has been determined, and its biophysical properties, including the UV-vis, EPR, and MCD spectra, and the iron redox potentials in the substrate-free and ligand bound forms, have been determined (17). High throughput and fragment screens implemented in efforts to identify the substrate of the enzyme indicated that chlorophenol derivatives and structures with three aromatic rings, particularly nitroaromatics, are viable ligands (17, 50). However, the ligands that were thus identified are, at best, poor substrates for the enzyme, so that these results shed little light on the true substrate of the enzyme. CYP126A1 is thus among the better characterized M. tuberculosis P450 enzymes in terms of biophysical properties and structure, but its role and importance remain obscure, casting a shadow on its potential as a drug target.
CYP130
CYP130A1 is readily expressed in E. coli and the expressed protein is amenable to purification and crystallization (18). A comparison of the crystal structures of ligand-free CYP130A1 and the protein with econazole bound in the active site shows that the ligand-free protein exists in a conformation with a relatively “open” active site, whereas the active site is in a more “closed” conformation in the econazole bound form. These conformational changes involve repositioning of the BC-loop and the F and G helices. Binding of econazole and clotrimazole is subject to positive cooperativity, which may stem from a tendency of CYP130A1 to associate into a dimeric structure.
Because NO is deployed by mammals in their defense against M. tuberculosis, a comparison was made of the susceptibility of several M. tuberculosis P450 enzymes to inhibition by NO (51). These studies demonstrate that CYP51B1 and CYP130A1 form ferrous-NO complexes that are resistant to subsequent exposure to oxygen, whereas CYP125A1 and CYP142A1 form complexes that, when exposed to oxygen, revert to their ferric state. It is not known, however, whether interactions of the M. tuberculosis P450 enzymes with NO is significant in terms of the pathology or therapy of mycobacterial infections.
Screening efforts with a diversity of cytochrome P450 substrates, including conventional P450 probes, fatty acids, and steroids, did not lead to identification of a substrate for CYP130A1. Furthermore, a spectroscopic screen of 20,000 compounds in a search for enzyme ligands failed to identify any compounds giving rise to a Type I spectral shift (i.e., potential substrates), but did identify a range of compounds that gave the Type II spectral shifts associated with enzyme inhibitors (52). In general these compounds were heteroaromatic amines, with the best one having a Kd of approximately 1 μM. The crystal structures of CYP130A1 with two of these ligands bound in the active site showed that in both cases the aromatic amine group coordinated to the heme iron atom. These results provide information on potential avenues for the development of non-azole inhibitors of CYP130A1. A purely computational study has claimed identification of a possible inhibitor for CYP130A1, but in the absence of experimental data, the value of this claim is uncertain (53). More importantly, as in the case of CYP126A1, the results have provided little insight into the substrate and function of this cytochrome P450 enzyme. The absence of this information limits enthusiasm for the pursuit of CYP130A1 as a drug target.
CYP128
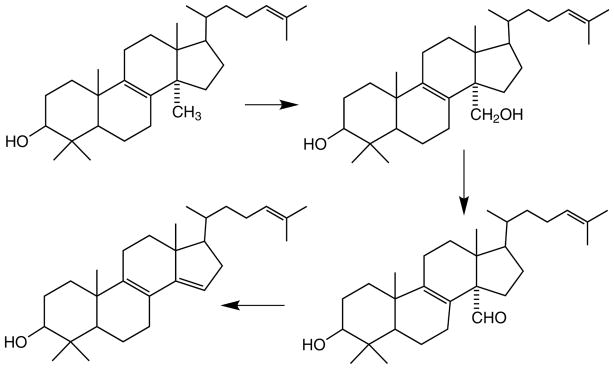
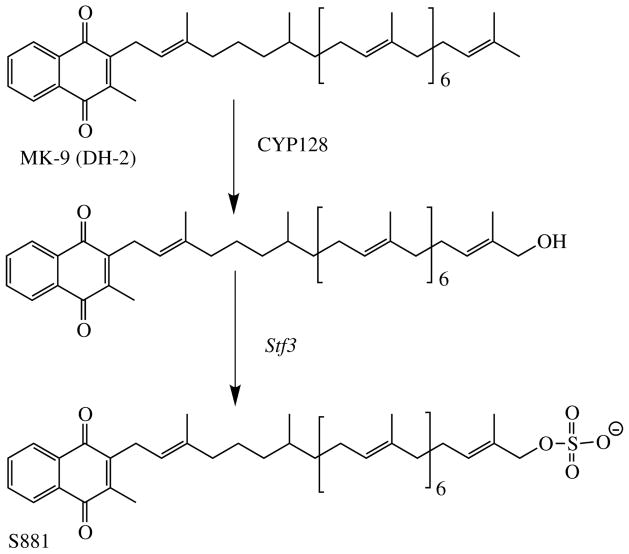
CYP128A1 is unusual among the M. tuberculosis cytochrome P450 enzymes in that the protein has not been heterologously expressed, its catalytic properties in vitro have not been determined, and its crystal structure is not known; nevertheless, its substrate and biological role have been clearly defined. In the process of determining the biogenesis of S881, a sulfated menaquinone, the sulfotransferase responsible for introducing the sulfate group was identified (Fig. 3) (54, 55). Analysis of the M. tuberculosis genome showed that the genes coding for two putative cytochrome P450 enzymes were located close to that for the sulfotransferase. One of these genes codes for CYP128A1, which has subsequently been implicated as the enzyme that introduces the hydroxyl group into menaquinone MK-9 (DH2) to which the sulfate group is attached in the synthesis of S881. This conclusion rests on genetic knockout experiments, as efforts in several laboratories to express catalytically active CYP128A1 in E. coli or other heterologous expression systems have failed.
Fig. 3.
Structure of the sulfolipid S881 and the role of CYP128A1 in its formation.
The CYP128A1-deficient M. tuberculosis strains, like those in which the sulfotransferase is suppressed, reveal that the sulfated menaquinone is a repressor of virulence (41). That is, S881 decreases the virulence of the mycobacteria. It is therefore not a suitable target for antituberculosis drug development, as inhibition of CYP128A1 (or the sulfotransferase) would lead to more, not less, virulent mycobacteria.
CYP144
CYP144A1 has been expressed and purified and many of its biophysical parameters have been determined (55). Knockout of the cyp144A1 gene in the H37Rv strain of M. tuberculosis established that CYP144A1 is not essential for growth of the mycobacteria in culture. However, azole drugs bind to the purified protein (Table 1) and inhibit the growth of the knockout strain to a greater extent than the parent H37Rv strain, suggesting a possible role for CYP144A1 in cellular biology or in modulating resistance to azole drugs. However, a similar finding was observed when CYP125A1 was knocked out (56). More detailed analysis revealed that there are two possible start codons for CYP144A1, both of which give rise to proteins that can be expressed and purified (21). The terminal region of the longer protein appears to be disordered, but the truncated, shorter protein was amenable to crystallization and its ligand-free structure was determined. The substrate and role of CYP144A1 remain unknown, however. A fragment profiling approach was pursued in efforts to obtain a clue to the native substrate, but the results did not yield a specific identity. However, they may be useful if the further development of inhibitors for the enzyme is warranted (57)
CYP124
CYP124A1, like CYP128A1, is located close to the sulfotransferase involved in the formation of S881. However, unlike CYP128A1, the catalytically active protein has been expressed, purified, crystallized, and partially characterized (14). Analysis of its substrate specificity indicates that it catalyzes the terminal hydroxylation of methyl-branched hydrocarbon chains such as those of phytanic acid and farnesol (14), cholesterol and related sterols (58, 59), and vitamin D3 (60). The methyl-branched terminus of the hydrocarbon chain is important for efficient oxidation, as fatty acids without the terminal methyl branch are either very poor or unacceptable CYP124A1 substrates. Not all methyl-branched termini are oxidized, however, as the side-chain of MK-9 (DH2), which is hydroxylated by CYP128A1, is surprisingly not oxidized in vitro by CYP124A1. All our efforts to detect this transformation with recombinant CYP124A1 have been unsuccessful.
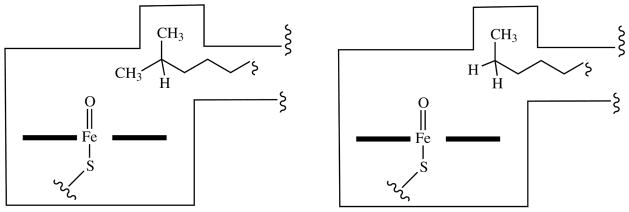
The crystal structure of CYP124A1 with phytanic acid bound in the active site rationalizes the importance of methyl branching in making a hydrocarbon chain a suitable substrate (14). As shown schematically in Fig. 4, a lipophilic cavity near the heme iron atom binds one terminal methyl, positioning the other one for oxidation. In the absence of the branching methyl, the chain terminus binds in the cavity and no carbon atom of the hydrocarbon chain is close enough to be oxidized. Of course, superimposed on this feature of the active site are additional structural constraints that determine which specific compounds can be bound and oxidized.
Fig. 4.
Schematic model of the CYP124A1 active site illustrating why a methyl-branched hydrocarbon terminus, in which one methyl is bound in the lipophilic cavity and the other is presented for oxidation, is a good substrate, whereas an unbranched chain terminus, in which binding of the methyl in the lipophilic cavity prevents its presentation for oxidation, is not.
Although we know much about the nature of CYP124A1 substrates, the identity of the specific substrate or substrates for this enzyme in M. tuberculosis remains undetermined. Strains of mycobacteria lacking functional CYP124A1, such as CDC1551, grow readily in culture (58), indicating that this enzyme is not essential for growth, although it could play a role in the more complex context of an in vivo infection. In the absence of more incriminating evidence, the viability of CYP124A1 as a potential target for antituberculosis drug development remains uncertain.
CYP125
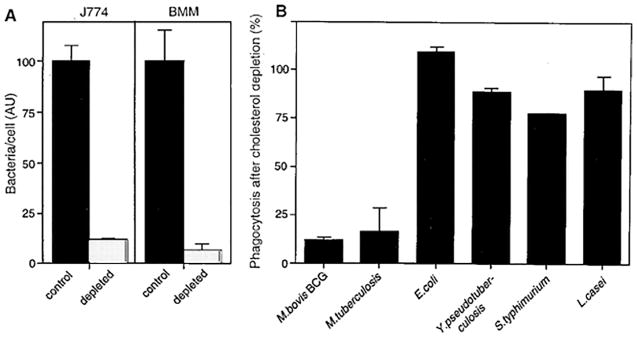
Host cholesterol plays a critical role in phagocytosis and growth of mycobacteria, as shown by the demonstration that phagocytosis of Mycobacterium bovis BCG, a model for M. tuberculosis, is severely impaired if the macrophage cell membranes are first depleted of cholesterol, a treatment that does not interfere with the phagocytosis of E. coli and other bacterial strains (Fig. 5) (61).
Fig. 5.
Phagocytosis of M. bovis BCG by two macrophage cell lines (J774, BMM) before and after cholesterol depletion of the macrophages. In panel A the phagocytosis by the control (black bars) and depleted (light bars) is compared. In panel B the bars indicate phagocytosis in cholesterol depleted cells as a percentage of that observed in control cells. Reprinted with permission from (61).
A set of genes, termed the igr (for intracellular growth) locus, was identified that is required for cholesterol metabolism and for growth and virulence of M. tuberculosis (62, 63). Interestingly, these studies also found that a toxic metabolite accumulated in igr knockout strains that prevented growth not only on cholesterol, but also on other carbon sources. One of the six genes in the igr operon (Rv3545c) codes for the cytochrome P450 enzyme CYP125A1 (63). Knockout of a highly conserved transcriptional TetR-type repressor (Rv3574) that regulates a large number of lipid degradation genes caused the up-regulation of many genes, including those that produce CYP125A1 and CYP142A1 (64). Finally, high-density mutagenesis and deep sequencing identified CYP125A1 as a monooxygenase in the H37Rv strain of M. tuberculosis required for growth on cholesterol (65).
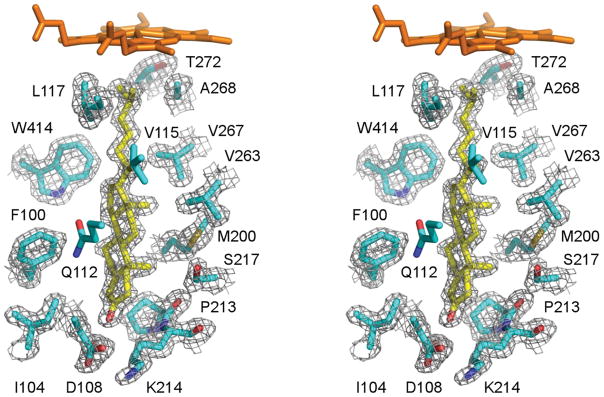
The role and function of CYP125A1 were fully decoded through the work of several laboratories. Knockout of the gene coding for the CYP125A1 in Rhodococcus jostii RHA1 revealed that the bacteria could grow on 26-carboxy cholesterol, but not on cholesterol itself (66). CYP125A1 was expressed and purified and its identity as a P450 enzyme was confirmed, although its catalytic activity could not be demonstrated in vitro. The CYP125A1 protein from M. tuberculosis heterologously expressed in Rhodococcus jostii RHA1 and then purified was shown, on reconstitution with a surrogate electron donor, to catalyze the 26-hydroxylation of cholesterol and cholest-4-en-3-one (67). M. tuberculosis CYP125A1 heterologously expressed in E. coli also catalyzed the 26-hydroxylation of cholesterol (15). These investigators obtained the crystal structures of CYP125A1 complexed with econazole and androstenedione, which unfortunately lacks the crucial 17-sidechain of cholesterol and cholest-4-en-3-one. Finally, purified CYP125A1 expressed in E. coli was shown to catalyze not only 26-hydroxylation, but also the subsequent conversion of the 26-alcohol to the 26-aldehyde and 26-carboxylic acid (58). Furthermore, the crystal structure of CYP125A1 with cholest-4-en-3-one bound in the active site demonstrated that this substrate binds in a tight-fitting active site with the side-chain methyl terminus located close to the heme iron atom (Fig. 6) (16).
Fig. 6.
Stereoscopic view of cholest-4-en-3-one bound in the active site of CYP125A1. Reprinted with permission from (16)
M. smegmatis, a non-pathogenic mycobacterium, has not one, but two enzymes that are homologous to M. tuberculosis CYP125A1 (20, 68). CYP125A3, like CYP125A1, oxidizes the 26-methyl group of cholesterol and cholest-4-en-3-one to the corresponding aldehyde and carboxylic acid (20). CYP125A4, the second enzyme, also catalyzes the 26-hydroxylation of cholesterol, but has a much higher activity for 26-hydroxylation of 7-hydroxycholesterol (68). This can be attributed to the presence of a bulky tryptophan (Trp83) in CYP125A3 instead of the smaller tyrosine in CYP125A4 at a position where it interferes with binding of the 7-hydroxy group. Replacing the tryptophan of CYP125A3 by a tyrosine by site-specific mutagenesis enhances the ability of this enzyme to oxidize 7-hydroxycholesterol. The crystal structure of the CYP125A3 W83Y mutant is consistent with this interpretation (68).
The carbon atoms of the cholesterol side-chain are incorporated into surface lipids of M. tuberculosis. Radiocarbon labeling showed that the label of [26-14C]cholesterol was incorporated into the virulence factors PDIM (phthiocerol dimycocerosate) (69) and SL-1 (sulfolipid-1) (70). This finding was more precisely demonstrated in the case of PDIM by mass spectrometric analysis of the PDIM lipid fraction after growth on cholesterol with 25,26,26,26,27,27,27-heptadeuterated cholesterol (16). As expected, the increased mass of PDIM seen when cells are grown in the presence of cholesterol is also seen when the cells are grown with propionic acid in the medium, supporting the inference that propionoyl CoA released in the degradation of the cholesterol side-chain serves as a feedstock for the synthesis of PDIM and SL-1 (16, 71)
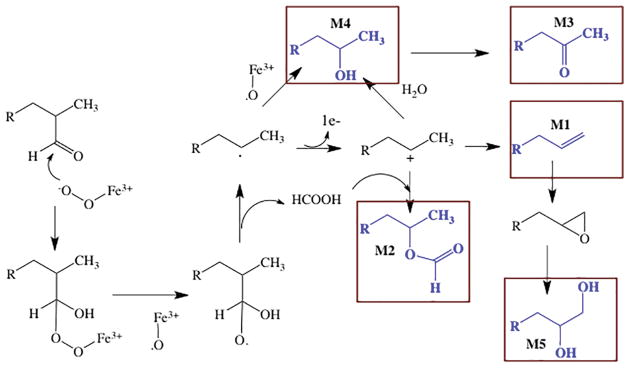
Despite the tight fit of cholest-4-en-3-one in the CYP125A1 active site seen in the crystal structure (Fig. 6), detailed studies of the oxidation of unlabeled and 25,26,26,26,27,27,27 deuterium labeled cholesterol in the presence of 16O2 versus 18O2 established that side-products are formed, at least when the oxidation of cholesterol is performed in vitro (72). Mass spectrometric and chromatographic comparisons allowed detection of the products M1 through M5 (Fig. 7), in addition to the expected 26-alcohol, 26-aldehyde, and 26-carboxylic acid. Comparison of the products formed from cholest-4-en-3-one, its 26-alcohol metabolite, and the 26-aldehyde showed that the side-products are formed from alternative reactions of the 26-aldehyde. Some support for the mechanisms proposed in Fig. 7 for the formation of these side-products comes from the finding that replacement of the cysteine thiolate of CYP125A1 by a seleno cysteine anion shifts product formation in the direction of the alternative pathways, all of which require nucleophilic attack of the iron dioxy complex on the aldehyde group. The seleno ligand is a better electron donor than the thiolate, as is illustrated by the longer wavelength absorption (~458 nm) of the ferrous carbonmonoxy complex compared to that of the normal thiolate-ligated P450 enzyme (~450 nm) (73). Further confirmation of the reactivity shift caused by selenocysteine replacement of the thiolate ligand iis provided by the more recent work of Onderko et al. (74).
Fig. 7.
Side products detected in the oxidation of cholest-4-en-3-one by M. tuberculosis CYP125A1. The side-products, shown in blue, are boxed and are denoted by the labels M1–M5. The mechanisms proposed for their formation from the aldehyde intermediate are indicated.
CYP142
The importance of cholesterol side-chain degradation is emphasized by the number of cytochrome P450 enzymes that specifically oxidize the C26 methyl group to the carboxylic acid. As discussed above, in M. tuberculosis the primary enzyme that performs the required oxidative steps is CYP125A1, although CYP124A1 is a much poorer catalyst of these reactions. In M. smegmatis, CYP125A3 and CYP125A4 are able to oxidize cholesterol, although CYP125A4 preferentially oxidizes 7α-hydroxycholesterol. However, there is an additional family of enzymes specifically evolved to catalyze terminal oxidation of the cholesterol and cholest-4-en-3-one side-chains.
CYP142A1 of M. tuberculosis was heterologously expressed, shown to be a cholesterol 26-hydroxylase, and crystallized (19). The ligand-free crystal structure had a similar structure to that of CYP125A1 in the regions that defines the ligand-binding pocket. Heterologously expressed CYP142A2 from M. smegmatis also catalyzes the oxidation of cholesterol and cholest-4-en-3-one to their 26-carboxylic acid derivatives (20). The crystal structure of this protein as a complex with cholest-4-en-3-one revealed that the sterol binds with the C17-sidechain methyl groups close to the heme iron atom, as expected given the catalytic activity of the enzyme. A difference is found in the 26-hydroxylated products, however, in that CYP142A1 (and CYP124A1) produce the 26-alcohol with a 25R configuration, whereas CYP125A1 produces the 26-alcohol with a 25S configuration (58).
CYP125-CYP142 System
The contributions of the M. tuberculosis cytochrome P450 enzymes that oxidize cholesterol and cholest-4-en-3-one in physiological situations depends on several factors, including the levels at which they are expressed, whether expression is inducible, and the relative kinetic parameters of the enzymes. In fact, the H37Rv strain of M. tuberculosis commonly used in experimental work has functional genes that encode CYP124A1, CYP125A1 and CYP142A1. However, immunoblotting measurements of the proteins found in mycobacterial cells indicate that CYP125A1 is well expressed in cells grown on glycerol as the sole carbon source, but it is expressed at much higher levels in mycobacteria grown on cholesterol (58). CYP142A1 is weakly expressed in glycerol-grown cells, but its expression is somewhat induced when the mycobacteria are grown on cholesterol. In contrast, CYP124A1 is not detected at the protein level by immunoblotting experiments in the H37Rv cells whether they are grown on glycerol or cholesterol. Furthermore, the virulent CDC1551 strain of M. tuberculosis does not have functional genes for either CYP142A1 or CYP124A1, but does have one for CYP125A1. In accord with this, CYP124A1 and CYP142A1 are not detected by immunoblotting experiments in mycobacteria grown on either glycerol or cholesterol as the carbon source (58). M. bovis BCG also only has a gene coding for a CYP125 protein and when this is knocked out, the mycobacteria do not grow on cholesterol; they just accumulate cholest-4-en-3-one (67).
A comparison of the kinetic parameters for M. tuberculosis CYP124A1, CYP125A1, and CYP142A1 clearly shows that CYP124A1 is a much poorer catalyst for the oxidation of cholesterol and cholest-4-en-3-one than the other two enzymes (Table 4) (58). Cholesterol and cholest-4-en-3-one bind more tightly to M. smegmatis CYP142A2 than CYP125A3 (20). The relative efficiency of CYP125A1 and CYP142A1 is confirmed when the cyp125 gene, which codes for the only cholesterol oxidizing enzyme in the CDC1551 strain, is knocked out and is replaced by overexpression of either CYP142A1 or CYP124A1. In the absence of complementation, the CDC1551 Δcyp125 strain does not grow on cholesterol. However, complementation of the Δcyp125A1 mycobacteria with the cyp142A1 gene produces a strain that grows just as well as the wild-type CDC1551 strain. In contrast, complementation with the cyp124A1 gene yields a strain that grows only poorly on cholesterol and accumulates cholest-4-en-3-one, albeit to a lower level than it in the uncomplemented Δcyp125 strain.
Table 4.
Kinetic constants for 26-hydroxylation of cholesterol and cholest-4-en-3-one by purified CYP124A1, CYP125A1, and CYP142A1. KD is the spectroscopically determined binding constant.
| Substrate | Enzyme | KD (nM) | Kmapp (μM) | kcatapp (min−1) | kcatapp/Kmapp (μM−1 min−1) |
|---|---|---|---|---|---|
| Cholesterol | 124A1 | - | 11.6 | 1.5 | 0.13 |
| 125A1 | 107 | 10.7 | 28 | 2.6 | |
| 142A1 | 18.4 | 7.7 | 16.7 | 2.2 | |
|
| |||||
| Cholest-4-en-3-one | 124A1 | 1056 | 20.8 | 11.7 | 0.56 |
| 125A1 | 1180 | 20.8 | 175 | 8.4 | |
| 142A1 | 114 | 11.8 | 84 | 7.1 | |
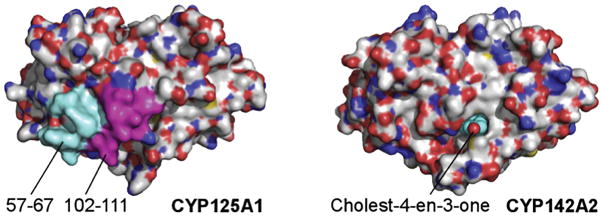
Comparison of the specificities of CYP124A1, CYP125A1, and CYP142A1 for substrates with modified C17-sidechains shows that all four enzymes tolerate the introduction of double bonds into the sidechain and replacement of a methyl group by a halide, albeit with some decrease in the catalytic rate (59). However, comparison of the crystal structures of CYP125A1 and CYP142A2 with cholest-4-en-3-one bound in the active site showed that the active site of CYP125A1 is capped by the peptides defined by amino acid residues 57–67 and 102–111, peptides that are lacking in CYP142A2 (Fig. 8) (75). Investigation of this difference led to the finding that the CYP125A1 has good activity for cholesterol and cholest-4-en-3-one, but low 26-hydroxylase activity with cholesterol sulfate and none with cholesterol propionate, substrates that would sterically clash with the capping peptides. CYP142A1 and CYP142A2 also have good activities with cholesterol and cholest-4-en-3-one, but they are better able to oxidize cholesterol sulfate and cholesterol propionate, as no cap exists to sterically interfere with binding of the esterified cholesterol derivatives. It is of interest in this context that quantitation of the sterols in macrophages shows that infection with mycobacteria elevates the concentration of esterified cholesterol (76). CYP142A1 may therefore contribute to utilizing these forms of cholesterol, in addition to its contribution to the metabolism of cholesterol itself.
Fig. 8.
Comparison of the surfaces of CYP125A1 and CYP142A2 illustrating capping of the CYP125A1, but not CYP142A2, active site channel by peptide segments in CYP125A1 not present in CYP142A2.
As mentioned earlier, knockout of the igr operon prevented growth on cholesterol and led to the accumulation of a toxic metabolite (62, 63). Subsequent work has identified this “toxic” metabolite as cholest-4-en-3-one, a metabolite that accumulates when degradation of its side-chain is prevented by knocking out the gene coding for CYP125A1 (16, 58, 67). Indeed, cholest-4-en-3-one prevents the growth of M. tuberculosis not only on cholesterol as the sole carbon source, but also on other carbon sources, including glycerol, acetate, and glucose (77). In the presence of an active 26-hydroxylase enzyme, notably CYP125A1 or CYP142A1, the cholest-4-en-3-one is metabolically removed, preventing cell growth inhibition. In the presence of a large initial excess of cholest-4-en-3-one, a lag phase is observed in the growth of the wild-type mycobacteria even if they express CYP125A1 (16). However, in the absence of such an activity, the growth inhibition is maintained.
Inhibition of CYP125 and CYP142
CYP125A1 and CYP142A1 are inhibited by azole drugs such as clotrimazole, miconazole, and econazole, although CYP121A1 is more sensitive to inhibition by these agents than CYP125A1 (Table 3). Inhibition of CYP125A1 and CYP142A1 may contribute to the inhibition of mycobacterial growth by azole drugs (47).
α-[(2-methylcyclohexyl)carbonyl amino]-N-4-pyridinyl-1H-indole-3-propanamide (LP10), a compound initially identified as an inhibitor of Trypanosoma cruzi CYP51, binds to M. tuberculosis CYP125A1 (78). Spectroscopic and crystallographic analyses indicate that this agent has an unusual binding mode that favors tighter binding of the distal water ligand to the heme iron atom rather than the usual displacement of the water ligand by a nitrogen of the compound. A similar binding mode was observed in the complex of fluconazole with CYP121A1, in which fluconazole hydrogen bonds to the distal water rather than displacing it (79).
A screen of 19 sterols identified several that inhibited the growth of M. tuberculosis, although no studies were performed to elucidate the basis for growth inhibition (80). One of sterols was 16,26-dihydroxycholesterol, here termed the “triol” (Fig. 9). The presence of a 26-hydroxyl group in this sterol led us to investigate its mechanism of action (77). Spectroscopic and catalytic studies with purified, recombinant CYP125A1 and CYP142A1 established that the triol bound to these enzymes, but was not a substrate for either one; i.e., that its side-chain was resistant to degradation. Incubation of the sterol with a 3β-hydroxysteroid dehydrogenase showed that it was readily converted to the 3-keto form. The triol is therefore also an analogue of cholest-4-en-3-one, and has an inhibitor profile similar to that sterol in its ability to inhibit the growth of mycobacteria grown on diverse media. Most importantly, as its side-chain cannot be degraded even if CYP125A1 and CYP142A1 are expressed, this molecule produces a persistent, inhibitory activity against wild-type strains of M. tuberculosis.
Fig. 9.
Oxidation of 16,26-dihydroxycholesterol (the “triol”) to its 3-keto-4-ene derivative.
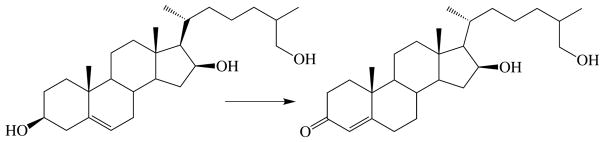
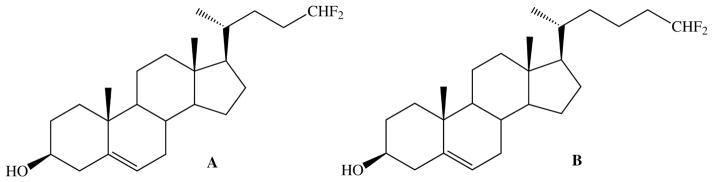
An alternative approach in which a sterol is used to inhibit CYP125A1 directly, preventing removal of endogenous cholest-4-en-3-one as it is formed, has been explored in preliminary experiments (77). Two synthetic cholesterol analogues with terminally unbranched sidechains of different lengths, each containing a difluoromethyl terminus, have been examined as potential M. tuberculosis growth inhibitors (Fig. 10). Both compounds blocked the growth of M. tuberculosis even with glycerol as the carbon source. Direct inhibition of CYP125A1 and CYP142A1 was not established, although it was shown that one of the compounds (B) is oxidized by these enzymes to the corresponding acylfluoride that can be trapped with methanol. It is therefore possible, but was not shown, that the enzyme is covalently modified.
Fig. 10.
Structures of two terminally difluorinated sterols that inhibit the growth of M. tuberculosis.
The utilization of cholesterol by M. tuberculosis can be blocked at the level of 26-hydroxylation by the cytochrome P450 enzymes, but once the 26-methyl is converted to the 26-carboxylic acid, a series of additional enzymes is required to further degrade the side-chain as well as the sterol core. Inhibition of some of these enzymes provides an additional route to the development of agents that inhibit growth at the level of cholesterol degradation (e.g., 81). These alterative approaches have been reviewed (82).
Conclusion
As summarized here, CYP121A1 is a crucial enzyme for mycobacterial growth and is a viable target for drug development efforts. CYP125A1 and CYP142A1 are closely related enzymes in terms of their catalytic function, the oxidation of cholesterol, cholest-4-en-3-one, and possibly cholesterol esters. Although the enzymes are not essential for in vitro growth on most media, they are critical for growth on cholesterol and for phagocytosis and virulence in vivo. As such, they are suitable targets for drug design efforts. The biological role of CYP128A1 in biogenesis of a sulfated menaquinone is well established even if the protein has proven difficult to characterize at the biochemical level. However, as CYP128A1 produces a molecule that attenuates virulence, it is not a drug design candidate. The precise substrates and roles of the other enzymes discussed here, CYP51B1, CYP124A1, CYP126A1, CYP130A1, and CYP144A1 are not known, so that even though crystal structures and biophysical data are available, their potential utility in drug design efforts is unclear. This applies also to the other eleven cytochrome P450 enzymes (Table 2) for which little molecular level information is available, although this may change as these enzymes are better characterized
Highlights.
Tuberculosis persists as highly lethal scourge, particularly in the developing world
M. tuberculosis encodes 20 P450 enzymes, for 8 of which crystal structures are available
Three of these eight enzymes, CYP121, CYP125, and CYP142, are potential drug targets
The development of inhibitors for these three enzymes is underway
Acknowledgments
The work from the author’s laboratory and the preparation of this manuscript were supported by National Institutes of Health grant AI074824. The author thanks the talented individuals who carried out the work in his laboratory reported in this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hippocrates, Of the Epidemics (400 B.C.E.) Adams F, translator. Book II, Section III, 13, Internet Classics Archive. http://classics.mit.edu/Hippocrates/epidemics.2.ii.html.
- 2.Bunyan J. The Life and Death of Mr. Badman. Grace Abounding & the Life and Death of Mr. Badman. 1928;Chapter XVIII(1680):282. [Google Scholar]
- 3.WHO Global Tuberculosis Report. 2016 https://reliefweb.int/report/world/global-tuberculosis-report-2016.
- 4.Sun Z, Zhang Y. Antituberculosis activity of certain antifungal and antihelminthic drugs. Tuber Lung Dis. 1999;79:319–320. doi: 10.1054/tuld.1999.0212. [DOI] [PubMed] [Google Scholar]
- 5.Jackson CJ, Lamb DC, Kelly DE, Kelly SL. Bactericidal and inhibitory effects of azole antifungal compounds on Mycobacterium smegmatis. FEMS Microbiol Lett. 2000;192:159–162. doi: 10.1111/j.1574-6968.2000.tb09375.x. [DOI] [PubMed] [Google Scholar]
- 6.Guardiola-Diaz HM, Foster L-A, Mushrush D, Vaz ADN. Azole-antifungal binding to a novel cytochrome P450 from Mycobacterium tuberculosis: implications for treatment of tuberculosis. Biochem Pharmacol. 2001;61:1463–1470. doi: 10.1016/s0006-2952(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 7.McLean KJ, Marshall KR, Richmond A, Hunter IS, Fowler K, Kieser T, Gurcha SS, Bestra GS, Munro AW. Azole antifungals are potent inhibitors of cytochrome P450 mono-oxygenases and bacterial growth in mycobacteria and streptomyces. Microbiology. 2002;148:2937–2949. doi: 10.1099/00221287-148-10-2937. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad Z, Sharma S, Khuller GK. In vitro and ex vivo antimycobacterial potential of azole drugs against Mycobacterium tuberculosis H37Rv. FEMS Microbiol Lett. 2005;251:19–22. doi: 10.1016/j.femsle.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad Z, Sharma S, Khuller GK. The potential of azole antifungals against latent/persistent tuberculosis. FEMS Microbiol Lett. 2006;258:200–203. doi: 10.1111/j.1574-6968.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 10.Byrne ST, Denkin SM, Gu P, Nuermberger E, Zhang Y. Activity of ketoconazole against Mycobacterium tuberculosis in vitro and in the mouse model. J Med Microbiol. 2007;56:1047–1051. doi: 10.1099/jmm.0.47058-0. [DOI] [PubMed] [Google Scholar]
- 11.Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc Natl Acad Sci USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leys D, Mowat CG, McLean KJ, Richmond A, Chapman SK, Walkinshaw MD, Munro AW. Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450. J Biol Chem. 2003;278:5141–5147. doi: 10.1074/jbc.M209928200. [DOI] [PubMed] [Google Scholar]
- 13.Belin P, Le Du MH, Fielding A, Lequin O, Jacquet M, Charbonnier J-B, Lecoq A, Thai R, Courçon M, Masson C, Dugave C, Genet R, Pernodet J-L, Gondry M. Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2009;106:7426–7431. doi: 10.1073/pnas.0812191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston JB, Kells PM, Podust LM, Ortiz de Montellano PR. Biochemical and structural characterization of CYP124, a methyl-branched lipid ω-hydroxylase from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2009;106:20687–20692. doi: 10.1073/pnas.0907398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLean KJ, Lafite P, Levy C, Cheesman MR, Mast N, Pikuleva IA, Leys D, Munro AW. The structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection. J Biol Chem. 2009;284:35524–35533. doi: 10.1074/jbc.M109.032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouellet H, Guan S, Johnston JB, Chow ED, Petrea PM, Burlingame AL, Cox JS, Podust LM, Ortiz de Montellano PR. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Molec Microbiol. 2010;77:730–742. doi: 10.1111/j.1365-2958.2010.07243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chenge JT, Le DV, Swami S, McLean KJ, Kavanagh ME, Coyne AG, Rigby SEJ, Cheesman MR, Girvan HM, Levy CW, Rupp B, von Kries JP, Abell C, Leys D, Munro AW. Structural characterization and ligand/inhibitor identification provide functional insights into the Mycobacterium tuberculosis cytochrome P450 CYP126A1. J Biol Chem. 2017;292:1310–1329. doi: 10.1074/jbc.M116.748822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouellet H, Podust LM, Ortiz de Montellano PR. Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs. J Biol Chem. 2008;283:5069–5080. doi: 10.1074/jbc.M708734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Driscoll MD, McLean KJ, Levy C, Mast N, Pikuleva IA, Lafite P, Rigby SEJ, Leys D, Munro AW. Structural and biochemical characterization of Mycobacterium tuberculosis CYP142: Evidence for multiple cholesterol 27-hydroxylase activities in a human pathogen. J Biol Chem. 2010;285:38270–38282. doi: 10.1074/jbc.M110.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Fernández E, Frank DJ, Galán B, Kells PM, Podust LM, García JL, Ortiz de Montellano PR. A highly conserved mycobacterial cholesterol catabolic pathway. Environmental Microbiol. 2013;15:2342–2359. doi: 10.1111/1462-2920.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chenge J, Kavanagh ME, Driscoll MD, McLean KJ, Young DB, Cortes T, Matak-Vinkovic D, Levy CW, Rigby SEJ, Leys D, Abell C, Munro AW. Structural characterization of CYP144A1 – a cytochrome P450 enzyme expressed from alternative transcripts in Mycobacterium tuberculosis. Scientif Rep. 2016;6:26628. doi: 10.1038/srep26628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean KJ, Munro AW. Structural biology and biochemistry of cytochrome P450 systems in Mycobacterium tuberculosis. Drug Metab Rev. 2008;4:427–446. doi: 10.1080/03602530802186389. [DOI] [PubMed] [Google Scholar]
- 23.McLean KJ, Belcher J, Driscoll MD, Fernandez CC, Le Van D, Bui S, Golovanova M, Munro AW. The Mycobacterium tuberculosis cytochromes P450: physiology, biochemistry & molecular intervention. Future Med Chem. 2010;2:1339–1353. doi: 10.4155/fmc.10.216. [DOI] [PubMed] [Google Scholar]
- 24.Ouellet H, Johnston JB, Ortiz de Montellano PR. The Mycobacterium tuberculosis cytochrome P450 system. Arch Biochem Biophys. 2010;493:82–95. doi: 10.1016/j.abb.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouellet H, Johnston JB, Ortiz de Montellano PR. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011;19:530–539. doi: 10.1016/j.tim.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz de Montellano PR. Substrate oxidation by cytochromes P450. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 4. Springer; New York: 2015. pp. 111–176. [Google Scholar]
- 27.Becher R, Wirtsel SGR. Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl Microbiol Biotechnol. 2012;95:825–840. doi: 10.1007/s00253-012-4195-9. [DOI] [PubMed] [Google Scholar]
- 28.Bellamine A, Mangla AT, Nes WD, Waterman MR. Characterization and catalytic properties of the sterol 14α-demethylase from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1999;96:8937–8942. doi: 10.1073/pnas.96.16.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellamine A, Mangla AT, Dennis AL, Nes WD, Waterman MR. Structural requirements for substrate recognition of Mycobacterium tuberculosis 14α-demethylase: implications for sterol biosynthesis. J Lipid Res. 2001;42:128–136. [PubMed] [Google Scholar]
- 30.McLean KJ, Warman AJ, Seward HE, Marshall KR, Girvan HM, Cheesman MR, Waterman MR, Munro AW. Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin, and their interactions. Biochemistry. 2006;45:8427–8443. doi: 10.1021/bi0601609. [DOI] [PubMed] [Google Scholar]
- 31.Matsuura K, Yoshioka S, Tosha T, Hori H, Ishimori K, Kitagawa T, Morishima I, Kagawa N, Waterman MR. Structural diversities of active site in clinical azole-bound forms between sterol 14α-demethylases (CYP51s) from human and Mycobacterium tuberculosis. J Biol Chem. 2005;280:9088–9096. doi: 10.1074/jbc.M413042200. [DOI] [PubMed] [Google Scholar]
- 32.Podust LM, von Kries JP, Eddine AN, Kim Y, Yermalitskaya LV, Kuehne R, Ouellet H, Warrier T, Alteköster M, Lee J-S, Rademann J, Oschkinat H, Kaufmann SHE, Waterman MR. Small-molecule scaffolds for CYP51 inhibitors identified by high-throughput screening and defined by X-ray crystallography. Antimicrob Agents Chemother. 2007;51:3915–3923. doi: 10.1128/AAC.00311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddine AN, von Kries JP, Podust MV, Warrier T, Kaufmann SHE, Podust LM. X-ray structure of 4,4′-dihydroxybenzophenone mimicking sterol substrate in the active site of sterol 14α-demethylase (CYP51) J Biol Chem. 2008;283:15152–15159. doi: 10.1074/jbc.M801145200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zampieri D, Mamolo MG, Vio L, Banfi E, Scialino G, Fermeglia M, Ferrone M, Pricl S. Synthesis, antifungal and antimycobacterial activities of new bis-imidazole derivatives, and prediction of their binding to P45014DM by molecular docking and MM/PBSA method. Bioorg Med Chem. 2007;15:7444–7458. doi: 10.1016/j.bmc.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 35.Zampieri D, Mamolo MG, Laurini E, Fermeglia M, Posocco P, Pricl S, Banfi E, Scialino G, Vio L. Antimycobacterial activity of new 3,5-disubstituted 1,3,4-oxadiazol-2(3H)-one derivatives, Molecular modeling investigations. Bioorg Med Chem. 2009;17:4693–4707. doi: 10.1016/j.bmc.2009.04.055. [DOI] [PubMed] [Google Scholar]
- 36.Lamb DC, Kelly DE, Manning NJ, Kelly SI. A sterol biosynthetic pathway in mycobacterium. FEBS Lett. 1998;437:142–144. doi: 10.1016/s0014-5793(98)01218-6. [DOI] [PubMed] [Google Scholar]
- 37.Jackson CJ, Lamb DC, Marczylo TH, Parker JE, Manning NL, Kelly DE, Kelly SL. Conservation and cloning of CYP51: a sterol 14α-demethylase from Mycobacterium smegmatis. Biochem Biophys Res Commun. 2003;301:558–563. doi: 10.1016/s0006-291x(02)03078-4. [DOI] [PubMed] [Google Scholar]
- 38.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Molec Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 40.McLean KJ, Carroll P, Lewis DG, Dunford AJ, Seward HE, Neeli R, Cheesman MR, Marsollier L, Douglas P, Smith WE, Rosenkrands I, Cole ST, Leys D, Parish T, Munro AW. Characterization of active site structure in CYP121: a cytochrome P450 essential for viability of Mycobacterium tuberculosis H37Rv. J Biol Chem. 2008;283:33406–33416. doi: 10.1074/jbc.M802115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sogi KM, Holsclaw CM, Fragiadakis GK, Nomura DK, Leary JA, Bertozzi CR. Biosynthesis and regulation of sulfomenaquinone, a metabolite associated with virulence in Mycobacterium tuberculosis. ACS Infect Dis. 2016;2:800–806. doi: 10.1021/acsinfecdis.6b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLean KJ, Cheesman MR, Rivers SL, Richmond A, Leys D, Chapman SK, Reid GA, Price NC, Kelly SM, Clarkson J, Smith CE, Munro AW. (Expression, purification, and spectroscopic characterization of the cytochrome P450 CYP121 from Mycobacterium tuberculosis. J Inorg Biochem. 2002;91:527–541. doi: 10.1016/s0162-0134(02)00479-8. [DOI] [PubMed] [Google Scholar]
- 43.Gondry M, Sauguet L, Belin P, Thai R, Amouroux R, Tellier C, Tuphile K, Jacquet M, Braud S, Courçon M, Masson C, Dubois S, Lautru S, Lecoq A, Hashimoto S, Genet R, Pernodet J-L. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;6:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 44.Fonvielle M, Le Du M-H, Lequin O, Lecoq A, Jacquet M, Thai R, Dubois S, Grach G, Gondry M, Belin P. Substrate and reaction specificity of Mycobacterium tuberculosis cytochrome P450 CYP121: insights from biochemical studies and crystal structures. J Biol Chem. 2013;288:17347–17359. doi: 10.1074/jbc.M112.443853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumas VG, Defelipe LA, Petruk AA, Turjanski AG, Marti MA. QM/MM study of the C-C coupling mechanism of CYP121, an essential cytochrome P450 of Mycobacterium tuberculosis. Proteins. 2014;82:1004–1021. doi: 10.1002/prot.24474. [DOI] [PubMed] [Google Scholar]
- 46.Dornevil K, Davis I, Fielding AJ, Terrell JR, Ma L, Liu A. Cross-linking of dicyclotyrosine by the cytochrome P450 enzyme CYP121 from Mycobacterium tuberculosis proceeds through a catalytic shunt pathway. J Biol Chem. 2017;292:13645–13657. doi: 10.1074/jbc.M117.794099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad Z, Sharma S, Khuller GK, Singh P, Faujdar J, Katoch VM. Antimycobacterial activity of econazole against multidrug-resistant strains of Mycobacterium tuberculosis. Intern J Antimicrob Agents. 2006;28:543–544. doi: 10.1016/j.ijantimicag.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 48.Hudson SA, McLean KJ, Surade S, Yang Y-Q, Leys D, Ciulli A, Munro AW, Abell C. Application of fragment screening and merging to the discovery of inhibitors of the Mycobacterium tuberculosis cytochrome P450 CYP121. Angew Chem Int Ed. 2012;51:9311–9316. doi: 10.1002/anie.201202544. [DOI] [PubMed] [Google Scholar]
- 49.Hudson SA, Surade S, Coyne AG, McLean KJ, Leys D, Munro AW, Abell C. Overcoming the limitations of fragment merging: rescuing a strained merged fragment series targeting Mycobacterium tuberculosis CYP121. ChemMedChem. 2013;8:1451–1456. doi: 10.1002/cmdc.201300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudson SA, Mashlidis EH, Bender A, McLean KJ, Munro AW, Abell C. Biofragments: an approach towards predicting protein function using biologically related fragments and its application to Mycobacterium tuberculosis CYP126. ChemBioChem. 2014;15:549–554. doi: 10.1002/cbic.201300697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouellet H, Lang J, Couture M, Ortiz de Montellano PR. Reaction of Mycobacterium tuberculosis cytochrome P450 enzymes with nitric oxide. Biochemistry. 2009;48:863–872. doi: 10.1021/bi801595t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podust LM, Ouellet H, von Kries JP, Ortiz de Montellano PR. Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines. J Biol Chem. 2009;284:25211–25219. doi: 10.1074/jbc.M109.017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma A, Subbias KK, Robine O, Chaturvedi I, Nigam A, Sharma N, Chaudhary PP. Computational finding of potential inhibitor for cytochrome P450 mono-oxygenases enzyme of Mycobacterium tuberculosis. Bioinformation. 2012;8:931–937. doi: 10.6026/97320630008931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holsclaw CM, Sogi KM, Gilmore SA, Schelle MW, Leavell MD, Bertozzi CR, Leary JA. Structural characterization of a novel sulfated menaquinone produced by stf3 from Mycobacterium tuberculosis. ACS Chem Biol. 2008;3:619–624. doi: 10.1021/cb800145r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Driscoll MD, McLean KJ, Cheesman MR, Jowitt TA, Howard M, Carroll P, Parish T, Munro AW. Expression and characterization of Mycobacterium tuberculosis CYP144: common themes and lessons learned in the M. tuberculosis P450 enzyme family. Biochim Biophys Acta. 2011;1814:76–87. doi: 10.1016/j.bbapap.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Carroll P, Parish T. Deletion of cyp125 confers increased sensitivity to azoles in Mycobacterium tuberculosis. PLOS One. 2015;10:e0133129. doi: 10.1371/journal.pone.0133129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kavanagh ME, Chenge J, Zoufir A, McLean KJ, Coyne AG, Bender A, Munro AW, Abell C. A fragment profiling approach to inhibitors of the orphan M. tuberculosis P450 CYP144A1. Biochemistry. 2017;56:1559–1572. doi: 10.1021/acs.biochem.6b00954. [DOI] [PubMed] [Google Scholar]
- 58.Johnston JB, Ouellet H, Ortiz de Montellano PR. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J Biol Chem. 2010;285:36352–36360. doi: 10.1074/jbc.M110.161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnston JB, Singh AA, Clary AA, Chen C-K, Hayes PY, Chow S, De Voss JJ, Ortiz de Montellano PR. Substrate analog studies of the ω-regiospecificity of Mycobacterium tuberculosis cholesterol metabolizing cytochrome P450 enzymes CYP124A1, CYP125A1, and CYP142A1. Bioorg Med Chem. 2012;20:4064–4081. doi: 10.1016/j.bmc.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasilevskaya AV, Yantsevich AV, Sergeev GV, Lemish AP, Usanov SA, Gilep AA. Identification of Mycobacterium tuberculosis enzyme involved in vitamin D and 7-dehydrocholesterol metabolism. J Steroid Biochem Molec Biol. 2017;169:202–209. doi: 10.1016/j.jsbmb.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 61.Gatfield J, Pieters J. Essential role for cholesterol in entry of mycobacterium into macrophages. Science. 2000;288:1647–1650. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- 62.Chang JC, Harik NS, Liao RP, Sherman DR. Identification of mycobacterial genes that alter growth and pathology in macrophages and in mice. J Infect Dis. 2007;196:788–795. doi: 10.1086/520089. [DOI] [PubMed] [Google Scholar]
- 63.Chang JC, Miner MD, Pandey AK, Gill WP, Harik NS, Sassetti CM, Sherman DR. igr Genes and Mycobacterium tuberculosis cholesterol metabolism. J Bacteriol. 2009;191:5232–5239. doi: 10.1128/JB.00452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kendall SL, Withers M, Soffair CN, Moreland NJ, Gurcha S, Sidders B, Frita R, ten Bokum RA, Besra GS, Lott JS, Stoker NG. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Molec Microbiol. 2007;65:684–699. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griffin JE, Gawronski JD, DeJesus MA, Ioerger RT, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for Mycobacterial growth and cholesterol catabolism. PloS Pathogens. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosloniek KZ, Wilbrink MH, Capyk JK, Mohn WW, Ostendorf M, van der Geize R, Dijkhuisen L, Eltis LD. Cytochrome P450 125 (CYP125) catalyzes C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Molec Microbiol. 2009;74:1031–1043. doi: 10.1111/j.1365-2958.2009.06915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capyk JK, Kalscheuer R, Stewart GR, Liu J, Kwon H, Zhao R, Okamoto S, Jacobs WR, Jr, Eltis LD, Mohn WW. Mycobacterial cytochrome P450 125 (CYP125) catalyzes the terminal hydroxylation of C27-steroids. J Biol Chem. 2009;284:35534–35542. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frank DJ, Waddling CA, Ortiz de Montellano PR. Cytochrome P450 125A4, the third cholesterol C-26 hydroxylase from Mycobacterium smegmatis. Biochemistry. 2015;54:6909–6916. doi: 10.1021/acs.biochem.5b01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, Mckinney JD, Bertozzi CR, Sassetti CM. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol. 2012;19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee W, BanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sivaramakrishnan S, Ouellet H, Matsumura H, Guan S, Moënne-Loccoz P, Burlingame AL, Ortiz de Montellano PR. Proximal ligand electron donation and reactivity of the cytochrome P450 ferric-peroxo anion. J Am Chem Soc. 2012;134:6673–6684. doi: 10.1021/ja211499q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang Y, Sivaramakrishnan S, Hayashi T, Cohen S, Moënne-Loccoz P, Shaik S, Ortiz de Montellano PR. Calculated and experimental spin state of seleno cytochrome P450. Angew Chem Int Ed. 2009;48:7193–7195. doi: 10.1002/anie.200901485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Onderko EL, Silakov A, Yosca TH, Green MT. Charactrerization of a selenocysteine-ligated P450 compound I reveals direct link between electron donation and reactivity. Nat Chem. 2017;9:623–628. doi: 10.1038/nchem.2781. [DOI] [PubMed] [Google Scholar]
- 75.Frank DJ, Madrona Y, Ortiz de Montellano PR. Cholesterol ester oxidation by mycobacterial cytochromes P450. J Biol Chem. 2014;289:30417–30425. doi: 10.1074/jbc.M114.602771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kondo E, Kanai K. Accumulation of cholesterol esters in macrophages incubated with mycobacteria in vitro. Jpn J Med Sci Biol. 1976;29:123–137. doi: 10.7883/yoken1952.29.123. [DOI] [PubMed] [Google Scholar]
- 77.Frank DJ, Zhao Y, Wong SH, Basudhar D, De Voss JJ, Ortiz de Montellano PR. Cholesterol analogs with degradation-resistant alkyl side-chains are effective Mycobacterium tuberculosis growth inhibitors. J Biol Chem. 2016;291:7325–7333. doi: 10.1074/jbc.M115.708172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ouellet H, Kells PM, Ortiz de Montellano PR, Podust LM. Reverse type I inhibitor of Mycobacterium tuberculosis CYP125A1. Bioorg Med Chem Lett. 2010;21:332–337. doi: 10.1016/j.bmcl.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seward HE, Roujeinikova A, McLean KJ, Munro AW, Leys D. Crystal structure of the Mycobacterium tuberculosis P450 CYP121-fluconazole complex reveals new azole drug-P450 binding mode. J Biol Chem. 2006;281:39437–39443. doi: 10.1074/jbc.M607665200. [DOI] [PubMed] [Google Scholar]
- 80.Schmidt AW, Choi TA, Theumer G, Franzblau SG, Knölker H-J. Inhibitory effect of oxygenated cholestan-3β-ol derivatives on the growth of Mycobacterium tuberculosis. Bioorg Med Chem Lett. 2013;23:6111–6113. doi: 10.1016/j.bmcl.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 81.VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Creowe AM, Eltis LD, Perola E, Deiniger DD, Wang T, Locher CP, Russell DG. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. Plos Pathogens. 2015;11(2):e1004679. doi: 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abuhammad A. Cholesterol metabolism: a potential therapeutic target in mycobacteria. Brit J Pharmacol. 2017;174:2194–2208. doi: 10.1111/bph.13694. [DOI] [PMC free article] [PubMed] [Google Scholar]