Abstract
Humans respond to chemical exposures differently due to many factors, such as previous and concurrent stressors, age, sex, and genetic background. The vast majority of laboratory-based toxicology studies, however, have not considered the impact of population-level variability within dose-response relationships. The lack of data dealing with the influence of genetic diversity on the response to chemical exposure provides a difficult challenge for risk assessment as individuals within the population will display a wide-range of responses following toxicant challenge. Notably, the genetic background of individuals plays a major role in the variability seen in a population-level response to a drug or chemical and, thus, there is growing interest in including genetic diversity into laboratory-models. Here we outline several laboratory-based models that can be used to assay the influence of genetic variability on an individual’s response to chemicals: 1) genetically-diverse cell lines, 2) human primary cells, 3) and genetically-diverse mouse panels. We also provide a succinct review for several seminal studies to highlight the capability, feasibility, and power of each of these models. This article is intended to highlight the need to include population-level genetic diversity into toxicological study designs via laboratory-based models with the goal to provide and supplement evidence in assessing the risk posed by chemicals to the human population. As such, incorporation of genetic variability will positively impact human-based risk assessment and provide empirical data to aid and influence decision-making processes in relation to chemical exposures.
Keywords: Genetic Variability, Risk Assessment, Population-Based Models
1. Introduction
The goal of risk assessment is to characterize the potential hazardous nature of a chemical exposure within the human population. Thus, knowledge of the dose-response relationship (DRR) between any given chemical, from pharmaceuticals to environmental contaminants, and any given physiological response is valuable to accurate risk assessment. In traditional toxicology studies, DRRs are established using classical laboratory models, such as cell lines, inbred rodent, and outbred stocks that are subject to genetic bottlenecking and colony drift (Festing 2016). The results from laboratory-based toxicology studies are then extrapolated to address the potential exposure and adverse outcome risk within the human population (Collins et al. 2008). As such, DRRs are used to calculate safe-exposure limits of the respective chemicals, such as a reference dose (RfD), acceptable daily intake (ADI), or tolerable daily intake (ADI) (IPCS 2005).
In cases where interspecies and interindividual data are not available, the World Health Organization’s International Programme on Chemical Safety (IPCS) has suggested that acceptable exposure limit be adjusted by a generic total ‘uncertainty factor’ of 100. The uncertainty factor breaks down into two separate categories each consisting of a 10-fold adjustment: A) interspecies differences and B) and interindividual differences. The interspecies adjustment further breaks down into two categories: A) toxicodynamics (2.5 fold) and B) toxicokinetics (4.0 fold). Similarly, interindividual differences also break down into the same categories, but with slightly different adjustments per category: A) toxicodynamics (3.2 fold) and B) toxicokinetics (3.2 fold) (IPCS 2005). While the interspecies differences are difficult to solve without epidemiological data for the chemical exposure and timeline of interest, several aspects that drive interindividual differences, such as population-level genetic variability, are not typically assessed with common laboratory models.
The obvious reason for the exclusion of genetic diversity in classical laboratory models is to reduce experimental variability. Inclusion of genetic variability will increase noise and, from an academic standpoint, increase the risk of a poor association within a study. An excellent example of this is the use of knock-out (KO) rodent models to establish the mechanism in which a particular gene is driving a phenotype; inclusion of genetic diversity will cloud the role of the gene of interest in the response. Mechanistic studies clearly play an important role in identifying genes, proteins, pathways, and environmental factors that contribute to certain responses to chemical exposure and, thereafter, provide potential for designing therapeutics in alleviating adverse effects. However, they are a closed platform and an assessment of an individual’s exposure. Most notably, they lack the power to assess risk posed by a chemical exposure to a diverse population. This lack of data provides a unique challenge to risk-management decisions with regards to establishing safe exposure limits for a heterogeneous population (Rhomberg 2011; Zeise et al. 2013).
Recently, there has been growing interest in incorporating genetic diversity into laboratory-models used for risk assessment (Zeise et al. 2013). While many facets impact an individual’s response to chemical exposure, a profound amount of observed variation has been attributed to genetic diversity (Evans and McLeod 2003; Evans and Relling 2004; Weinshilboum 2003). The previously outlined uncertainty factors aim to adjust the safe exposure limits for interindividual variability. However, there is certainly a non-zero chance that there are susceptible individuals within the population that are not accounted for in the exposure guidelines. In addition, these uncertainty factors can result in exposure guidelines that are too conservative that may potentially cause undue harm to industrial and municipal finances. Like in the human population, individuals within laboratory-based models have differing susceptibilities to chemically-induced adverse health outcomes. For example, differing mouse strains can change the perceived risk associated with an exposure (Chapman and Schiller 1985; French et al. 2015; Poland et al. 1994; Shen et al. 1991; Weber et al. 1995). As such, incorporation of population-level genetic diversity into toxicity testing has potential to better inform risk of 1) the shape of a population-level DRR and 2) identify genetic variants within a population that may be more susceptible to a particular exposure.
A recent report published by the NRC entitled “Science and Decisions: Advancing Risk Assessment” outlines the need for incorporation of population-level genetic diversity in toxicity testing (NRC 2009). The report suggested that the incorporation of inter-individual variability present within the human population would effectively linearize the low-dose region of non-cancer DRRs which have, in the past, been considered nonlinear functions (Figure 1). Notably, the proposal by the NRC was based primarily on theoretical evidence and has not been properly tested. If put into practice for risk assessment, the low-dose linearity assumption could lead to unwanted environmental impacts. As such, there is need to incorporate genetic variability into laboratory models to ensure that risk-management decision-making is informed and grounded in empirical evidence. Furthermore, the genetic diversity within population-based models can be used to identify genetic determinants of a response to a chemical exposure. Using genome wide association, quantifiable responses that are different amongst individual’s physiological can be used to scan for regions of the genome that might be responsible for the phenotype variability. Such studies can be used to identify genomic regions that potentially drive the quantifiable differences amongst individuals. Previous studies have shown the power of genetics-based approaches in identifying genetic variants with statistically differing levels of sensitivity to exposures (Abdo et al. 2015a; Abdo et al. 2015b; French et al. 2015; Harrill et al. 2009; O’Shea et al. 2011; Venkatratnam et al. 2017). While such results provide a more-precise level of risk assessment within potential environmental exposures, they can also inform of pharmacogenetic differences that may make pharmaceuticals more effective or, on the other hand, more toxic to individuals being treated.
Figure 1. The National Research Council’s (NRC) Low-Dose Linearity Hypothesis.
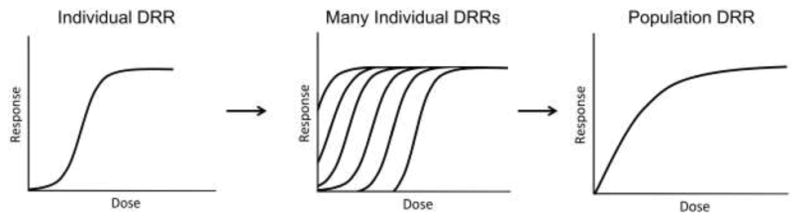
As published in a report in 2009 call “Science and Decisions: Advancing Risk Assessment,” the NRC suggest that, in considering population-level genetic variability, the low-dose region of non-cancer dose-response relationships will linearize. As such, the NRC recommends that there are no safe exposures of chemicals that induced adverse, non-cancer endpoints.
Here, we will outline several laboratory models that can provide empirical data needed to 1) establish the influence of genetic diversity within DRRs and 2) identify genetic variants and susceptible sub-populations that may have increased susceptibly to adverse effects caused by chemical exposures. Such information can be used to better assess risk associated with chemical exposures in the environment. Population-based models can be used to inform of pharmacogenetic differences as well. In understanding genetic determinants within responses, genotypes can be used to predict an adverse or enhanced response to a pharmaceutical exposure and, thus, lies at the heart of the precision medicine initiative. As such, the models outlined in this article have potential to impact the assessment and understanding of a wide-array of chemical exposures from the individual to the human population.
2. Addressing Genetic Variation with Laboratory Models
The primary goal of this article is not to advocate for the replacement of mechanistic studies, but rather to outline several laboratory-based models that can be used to account for genetic variation seen within the human population. Currently, there is much remains to be learned in regards to the variability within dose-response relationships amongst the heterogeneous human population (Rhomberg 2011). Thus, with the use of such models, laboratory-derived exposure data may better inform population-level responses to chemicals. Here, several projects are outlined showing the feasibility in assaying population-level genetic variability via three different biological models: 1) genetically-diverse cell lines, 2) human primary cells, and 3) mouse populations (Table 1).
Table 1.
Key features of the population-based models discussed in this report.
| Human Cell Lines | Human Primary Cells | Mouse Populations | |
|---|---|---|---|
| Availability | Commercial Cell Repositories | 1) Commercial Blood Banks 2) Organ Donation 3) Surgical Waste Products 4) Long-Term Cultures from Commercial Cell Repositories |
Commercial Animal Vendors (i.e. Jackson Labs) |
| Availability of Genotype Data | Available for many of the lymphoblastoid cell lines (1000 Genomes Project) | This Approach Requires Genotyping | 1) Genotypes available for many of the inbred panels such as the Collaborative Cross and Mouse Diversity Panel 2) Diversity Outbred Population requires genotyping |
| Key Advantages | 1) Cell lines provide unlimited resource to test responses over longer-periods of time 2) Easily-obtainable 3) High-throughput capability 4) Genotyping not required |
1) Primary cells likely mirror human responses better than immortalized cell lines 2) Some cells can be easily-obtained through commercial sources 3) High-throughput capable |
1) Some genetic resource panels have already been genotyped 2) Genetic polymorphisms are better randomized in mouse populations as compared to human resulting in a great statistical power to detect association 3) Minor allele frequencies are lower in mouse populations providing the need to use less animals as compared to assaying the human population |
| Key Limitations | 1) Immortalized cells respond different than primary cells 2) In vitro conditions likely do not mirror in vivo conditions 3) Need for large numbers of individuals to make an association |
1) In vitro conditions likely do not mirror in vivo conditions 2) Need to acquire primary cells for each individual experiment 3) Need to genotype each individual 4) Need for large numbers of individuals to make an association 5) Individuals have differing background exposures, disease states, ages, sexes, etc. |
1) Toxicodynamic/Toxicokinetic differences between mice and human impact responses to chemicals 2) Need to maintain a mouse colony or purchase mice for each study 3) Some populations, such as the DO stock, require genotyping for each individual |
2.1. Genetically Diverse Cell Lines
Clonal cell lines have been used in toxicological studies for decades. From an experimental standpoint, these cell lines are excellent in reducing variability in responses to chemical exposures and to gain a mechanistic understanding of chemical-induced toxicity. Furthermore, as cell lines are immortal and can be cryopreserved, they provide a resource that can be studied over long periods of time. Human lymphoblastoid cell lines (LCLs) can easily and efficiently be derived from a large number of individuals and, by assaying a large number of individual cell lines, can be used to assay population-level variability in responses to exposures (Dolan et al. 2004; Watters et al. 2004). As LCLS are immortalized cell lines, they can be readily distributed for research purposes. LCLs have been established from individuals of diverse heritages from locations throughout the world and only need to be genotyped once. The true power of these LCLs lies in the plethora of genomic and demographic data freely-available through previous studies such as the 1,000 genomes project (http://www.internationalgenome.org/) that can be used to probe links between genotype and phenotype. Specifically, LCLs can be used to link variant responses to genomic difference amongst cell lines without the need for further genotyping. These cell lines provide a powerful and high-throughput, in vitro, human platform to 1) assay population-level variability in chemical-induced toxicity while 2) performing genome-wide associations within the response of interest in vitro. Furthermore, LCLs provide a model that lies at the heart of proposed paradigm shift to incorporate high-throughput screens into toxicology to categorize and prioritize chemicals for study (Collins et al. 2008; NRC 2007)(Table 1).
Previous studies have shown that LCLs can be successfully used to probe variations in chemicals responses. For example, a past study used 87 individual LCLs from the Centre d’Etude du Polymorphisme Humain (CEPH) panel to screen responses to 14 toxicants with two different endpoints: 1) ATP production and 2) caspase-3/7 activity (O’Shea et al. 2011). Random effect modeling of the assay results suggests that cell line-to-cell line (aka interindividual) variability can account for as much as 40% of the observed variability. However, the interindividual variability was found to be dependent on the chemical exposure as well as the endpoint measured, confirming the need for chemical and assay-specific population-based data. For example, several chemicals, such as deltamethrin and rifampin, appear to elicit interindividual variability in both endpoints measured. Other chemicals, such as perfluorooctanic acid (PFOA), were found to induce interindividual variability in 1 endpoint while other chemicals, such as monoethylhexyl phthalic acid (MEHP), do not appear to induce cell line-to-cell line variability in either endpoint. The authors also leveraged the genetic information available from the LCLs to identify areas of the genome associated with changes in interindividual susceptibility to the toxicant-driven phenotype. For example, PFOA exposure was found to associate with areas on chromosome 4 and 14 and, within these areas, there are several candidate genes that may affect individual’s susceptibility to PFOA-induced injury. Such genes include FAT1 on chromosome 4 and SLC24A4, CPSF2, and RIN3 on chromosome 14. Several of these genes, such as FAT1 and SLC24A4, have been shown to be associated with PFOA exposures in previous studies (O’Shea et al. 2011).
In a recent study, 146 lymphoblastoid cell lines were exposed to a multiple concentrations of two separate mixtures of pesticides: 1) a current-use pesticide mixture (n=36 chemicals) and 2) an organic pesticide mixture (n=10 chemicals) (Abdo et al. 2015a). Curve-fitting clearly shows that, within the population of LCLs, there is a large range of susceptibility in the chemically-induced cytotoxic responses. Furthermore, the results were also used to calculate a toxicodynamic uncertainty factor (VFd) of around 3-fold for each pesticide mixture which, as noted in the report, is analogous to the level of interindividual variability for the pesticide mixtures. Within these individual differences, a polymorphism on chromosome 17 was found to be highly correlated with differences in susceptibility of the cell lines. The polymorphism was found within an open reading frame (i.e. C17orf54) and, in further detail, homozygous individuals carrying the major allele (A) were found to me more sensitive than either the heterozygous genotype (AT) or the homozygous minor allele (T). One of the most innovative aspects of this paper, however, is the use of in vitro-to-in vivo extrapolation (IVIVE) to estimate the corresponding cumulative oral equivalent dose of the chemical mixtures to reach the EC10 in the underlying cytotoxic phenotypes being assayed. Notably, a primary challenge of incorporating in vitro data within human risk assessment is determining the in vivo dose required to result in the in vitro concentrations at hand (Judson et al. 2011). Results indicated that, with data available for 31 of 36 pesticides in the current-use mixture and 4 of 10 pesticides within the organic mixture, the organic pesticide mixture required significantly less exposure to reach the EC10 of the cytotoxic phenotype. Furthermore, the authors estimate that population variability would require a 5-fold margin of safety for the organic pesticide while, in comparison, the current-use pesticide mixture would require less than a 2-fold margin of safety to account for inter-individual differences (Wetmore et al. 2014). While the IVIVE calculations required several assumptions, the methodology provides a method to estimate the chemical exposures required to reach toxicity seen in vitro and, furthermore, the influence that population variability plays in defining safe exposure limits (Abdo et al. 2015a).
In a similar study, cytotoxicity of 1,086 LCLs was assayed following exposure to 179 different chemicals found within the National Toxicology Program’s (NTP) chemical library (Abdo et al. 2015b). This study also calculated toxicodynamic variability factors (VFd) for 149 chemicals. In comparing the cumulative distribution of VFds for the current study with previously calculated VFds of 34 chemicals tested in vivo, the two studies provide very similar median values of 3.04 and 3.10, respectively. Thus, the results suggest that in vitro screens that incorporate population-level genetic variability have potential to provide valuable information for risk assessment. In assessing population variability, nearly half of the individual EC10 values have interindividual ranges that fall below the generic 3.2-fold adjustment. These results suggest that, in some cases, uncertainty factors can be too conservative. More interestingly, a subset of these chemicals within the study were found to produce EC10 value ranges that were much greater than the generic 3.2-fold adjustment indicating the need for chemical data in setting exposure limits for the human population that are accurate. These results also suggest the inherent risks associated with relying on generic uncertainty factors. Furthermore, the authors used multivariate association analysis (MAGWAS) to scan for genetic loci associated with the differences seen in the concentration-responses amongst the individual cell lines for each chemical. The results revealed several patterns and potential key players in chemical-induced cytotoxicity. For example, transmembrane proteins and solute carriers appear to play a key role in mediating chemical-induced cytotoxicity as they are consistently found in the most-significant associated loci. Similarly, a SNP (rs13120371) in 3′ UTR of SLC7A11 was found to be significantly associated with interindividual differences in 2-Amino-4-methylphenol, methyl mercuric (III) chloride, and N-methyl-p-aminophenol sulfate-induced cytotoxicity. In further detail, the results indicate which alleles appear to more sensitive to chemical-induced cytotoxicity and, thus, can be used to predict which individuals may be more susceptible to chemical-induced toxicity. For example, individuals carrying a homozygous minor allele (A) at the SLC7A11 locus were found to be more sensitive to 2-amino-4-methylphenol as compared to the hetereozygous (AT) and homozygous major allele (T) within the 3′ UTR of SLC7A11 (Abdo et al. 2015b).
2.2. Primary Human Cells
The ‘gold standard’ in analyzing human responses to chemical exposure in vitro is with primary cells taken from blood or tissue. Human primary cell cultures are more likely to mirror an in vivo response than immortalized human cell lines. On the other hand, primary cells typically can only be cultured for a short time and are more sensitive to the freeze-thaw cycles as compared to cell lines. Furthermore, individuals within a study would need to be genotyped to better understand their genetic background. The most challenging aspect of working with primary human cells to assess population variability lies with obtaining samples from a large number of individuals. While some methods are quite noninvasive, such as obtaining leukocytes from blood, other tissue samples relevant to toxicological outcomes can be quite invasive, such as the liver. However, such tissues can be obtained through many methods including biopsies, surgical waste products, organ donations, or, in some cases, long-term cultures in cell types less sensitive to cryopreservation (Table 1).
While resting primary human leukocytes are short-lived and need to processed within 24 hours of collection, previous studies have indicated that cells obtained from blood donations can be cultured within a few hours preventing the need for a freeze-thaw cycle (Phadnis-Moghe and Kaminski 2017). In a recent study, B cells were isolated from human blood sent overnight on wet ice from a commercial blood bank (Dornbos et al. 2016). Following isolation, B cells were dosed with increasing concentrations of the known immunotoxicant, 2,3,7,8-tetrachlordibenzo-p-dioxin (TCDD). Results indicated that, amongst 51 unique donors, there was some profound variability with up to 71 fold differences in the response at the high dose of 30 nM TCDD. Within the study’s population of donors, 11.5% did not respond to increasing concentrations of TCDD agreeing with previously published nonresponsive rates (Lu et al. 2010). Statistical modeling indicated that the low dose region of a TCDD-mediated DRR was best fit with a nonlinear model for two differing endpoints: 1) the number of B cells secreting and IgM and 2) the concentration of IgM secreted by B cells over a 7-day period. These results suggest that, contrary to the NRC’s assertion, the low-dose regions of all non-cancer endpoints are not linear when genetic diversity is taken into consideration (Figure 1)(NRC 2009). This study and other related reports highlight the feasibility to use human primary leukocytes for human population-level risk assessment and to identify individuals within the human population chemical-induced toxicity (Dornbos et al. 2016; Phadnis-Moghe and Kaminski 2017).
Adherent, monolayer primary tissue cells can also be used to assay interindividual variability in response to chemicals (i.e. two-dimensional) (den Braver-Sewradj et al. 2016; Martelli et al. 2003; Schuetz et al. 1995). For example, using primary hepatocyte cultures, individual’s cells were dosed with 3-methylcholanthrene and 2,3,7,8-tetrachlordibenzo-p-dioxin (TCDD) and the expression of target genes was assessed. The results showed the presence of cytochrome P450 1A1 (Cyp1A1) mRNA induction in all donors after treatment with either chemical while multidrug-resistance (mdr) mRNA induction was only seen in approximately 60% of the individuals (Schuetz et al. 1995). Similarly, a study using suspended primary human hepatocytes found a range of cell viability (83–91%) following treatment with 50 uM progesterone (Martelli et al. 2003). In a more unique study, the level of inter-donor variation in phase I and phase II metabolism was analyzed using both monolayer culture and suspension culture of hepatocytes (den Braver-Sewradj et al. 2016). Their results suggest the presence of up to 3 and 4-fold differences between donors in the cytochrome P450-mediated metabolism of diclofenac in suspension and monolayer culture, respectively. First, the report provides evidence that, even with the same cell type, the level of human interindividual variability measured can be model-specific (i.e. suspension vs. adherent cells). Secondly, this article highlights the complexity of human variability as there can be interindividual differences in the rate of metabolism of chemicals (den Braver-Sewradj et al. 2016). Thus, depending on the individual, there can be differences in the rate in which toxic chemicals are metabolized into benign compounds or, differences in the accumulation and excretion rates of toxic metabolites.
Recent advances in 3D culture models have begun to provide the opportunity to employ platforms that likely better mimic tissue microenvironments and mirror in vivo exposures (Ahmed et al. 2017; Godoy et al. 2013; Roth and Singer 2014). For example, a recent study reported on a 3D liver model in which human primary sinusoidal cells, stellate cells, and hepatocytes were co-cultured on hollow fiber (HF) membranes (Ahmed et al. 2017). Results indicated that cells retained morphology on the HF membranes and retained physiological function for up to 28 days. Most notably, the 3D cultures retained the ability to induce phase I metabolism of diazepam suggesting that this model can be used to assay individualistic differences in xenobiotic clearance can be assayed (Ahmed et al. 2017). Similarly, a different study using primary cells isolated from tumor biopsies reported that 3D, spherical cultures can be used to assay individual tumor’s susceptibility to differing cancer-therapies in a high-throughput fashion (Hagemann et al. 2017). Thus, the continuing advancement in 3D-culturing methods provides the potential to screen complex tissue micro-environments of individual’s primary cells for interindividual differences in vitro in a high-throughput fashion.
2.3 Genetically-Diverse Mouse Populations
Rodents have been used for decades in research. Rodents provide an in vivo model that is small in size providing a financially-reasonable method to control environmental factors and, yet, support complex study design. Many mouse genomes have been completely sequenced (Adams et al. 2015; Doran et al. 2016; Keane et al. 2011; Morgan et al. 2016). Thus, the use of genetically-diverse mouse panels provides the opportunity to better understand the effect of genetic variation of complex etiologies and identify genetic variants that effect susceptibility to a particular phenotype (e.g. chemical-induced toxicity) while controlling for many potentially confounding factors. The lack of genetic variation within inbred strains provide the opportunity for mice with identical genomes to be assayed over periods of time and differing experimental conditions (Bogue et al. 2015; Rusyn et al. 2010). Most importantly, there is significant diversity across strains. For example, within the Mus musculus subspecies it has been estimated that the number and distributions of polymorphisms is greater than found within human population (Ideraabdullah et al. 2004; Rusyn et al. 2010). The creation of genetic diverse reference populations, such as the Collaborative Cross (CC) and the Diversity Outbred (DO) mouse populations, provide the opportunity to assay genetic variability similar to that found in the human population and, to map the differences in responses to places within the genome with high-resolution (CCC 2012; Churchill et al. 2004; Churchill et al. 2012; Logan et al. 2013; Svenson et al. 2012; Threadgill and Churchill 2012; Threadgill et al. 2011; Welsh et al. 2012). The CC is significantly more diverse than previous, commonly used mouse panels, as it was created from 8 diverse founding strains of 3 differing Mus musculus subspecies (M. m. musculus, domesticus, castaneous) that encompass 90% of genetic variation in laboratory mice: 1) A/J, 2) C57BL/6J, 3) 129S1/SvImJ, 4) NOD/ShiltJ, 5) NZO/HILtJ, 6) CAST/EiJ, 7) PWK/PhJ, and 8) WSB/EiJ (Roberts et al. 2007; Threadgill and Churchill 2012). While the statistical power of CC is somewhat limited by the number of fully-inbred strains available, the inbred nature of the panel only requires one round of genotyping per strain (Bogue et al. 2015; Churchill et al. 2004; Threadgill and Churchill 2012; Threadgill et al. 2011). The CC panel has been used to analyze a range of complex traits (Abu-Toamih Atamni et al. 2017; Aylor et al. 2011; Kelada 2016; Kelada et al. 2012; Nashef et al. 2017; Smith et al. 2016; Xue et al. 2016). Similarly, the DO stock was created from early pre-CC strains and, while containing the same level of allelic diversity as the CC, are maintained with a high-level of heterozygosity (Bogue et al. 2015; Chesler 2014; Churchill et al. 2012). While the nature of the DO stock requires genotyping for each mouse, the heterozygosity provides the opportunity to analyze additivity while the high level of fecundity and large stock population provide an ideal model for high-resolution mapping and selective-breeding studies (Church et al. 2015; Logan et al. 2013; Svenson et al. 2012). These diverse populations of mice provide an in vivo method to analyze the impact of population-level genetic variability and to identify variants within populations that may be more or less susceptibility in chemical-induced phenotypes (Table 1).
Mouse population-based studies have already begun impacting human risk-assessment (Cichocki et al. 2017; French et al. 2015; Harrill et al. 2009; Venkatratnam et al. 2017). For example, a double-blind study in which 49 healthy humans who were exposed to maximum recommended therapeutic range of acetaminophen (4 gram/day for 7 days) found that 31% showed ≥2 fold increase of alanine aminotransferase (ALT) serum levels (Harrill et al. 2009). Thus, within the recommended therapeutic range, some human individuals appear to experience mild liver injury. To identify potential loci associated with increased risk to acetaminophen-induced toxicity, 36 inbred mouse strains were dosed with a range of acetaminophen. Results indicated interstrain differences in several endpoints including the rate of acetaminophen metabolism, ALT levels in the serum, and liver necrosis. The results show that genetic variation amongst the mouse strains profoundly changed the dose-response curves in the degree of necrosis and the level of ALT in the serum. Haplotype-association mapping suggested several genes, such as Cd44 and Capn10, were associated with inter-strain differences in ALT release. These genes were then related to human susceptibility. For example, a nonsynonymous polymorphism in the CD44 gene was found to be statistically correlated with an individual’s level of acetaminophen-induced ALT release. Similarly, a synonymous SNP in CAPN10 was found to be moderately-associated with an individual’s ALT-release. These results demonstrate the usefulness of mouse panels in identifying genes that likely contribute to human population-level variability in response to chemicals (Harrill et al. 2009).
In another study, 50 inbred strains were used to assay the influence of population variability on the toxicokinetics of cytochrome-mediated oxidation of trichloroethylene (TCE) (Venkatratnam et al. 2017). Results indicated that the levels of trichloroacetic acid (TCA), the most abundance metabolite of TCE, varied by an order of magnitude in a strain-specific manner. The results, along with another previous study of perchloroethylene, show the power of the collaborative cross in assaying toxicokinetic population variability (Cichocki et al. 2017). More interestingly, the empirical values measured in the current study were compared the predicted levels of TCA in tissues by a physiologically-based pharmacokinetic model (PBPK) based on results from 16 inbred mouse strains (Chiu et al. 2014). The CC panel suggests that the PBPK model may be under-estimating the TCA level in tissues, highlighting the need for individual-specific data for chemical exposures. More specifically, the 800 mg/kg dose groups in the CC panel were found beyond the PBPK’s predicted 95% confidence levels of TCE burden in the liver for more than half of the strains, a third of the strains in the kidney, and nearly half of the strains for the serum. As TCE and TCA are ligands of PPARα, the authors also looked at the expression of two PPARα-inducible genes, Acox1 and Cyp4a10, and found significant induction at the population level of all CC strains with notable interstrain differences as has been previously reported (Bradford et al. 2011). As the expression of Acox1 was found to be correlated with the level of TCA in the liver, the authors postulate that TCE-mediated effects may be altering TCE metabolism in a strain-specific manner. Furthermore, QTL analysis on the variability in TCA levels in the liver of the differing strains identified a list of potential genes that might explain the response differences. The gene list was narrowed down based on the function of the genes and potential links to TCEmediated effects. QRTPCR analysis indicated that expression of Acot8 and Fitm2 positively correlated with the levels of TCA in the liver and, thus, may be associated with the differences in TCE metabolism and the susceptibility to TCE-mediated toxicity (Venkatratnam et al. 2017).
In a study using DO mice, 600 mice were exposed to varying amounts of benzene via an inhalation chamber (French et al. 2015). The authors used micronucleus (MN) frequency in reticulocytes (RET) derived from the peripheral blood (PB) or bone marrow (BM) to assay the extent of chromosomal damage induced by benzene. The MN frequency was found to be significantly elevated for mice exposed to 100 ppm benzene in PB-RETs and for mice exposed to 1, 10, and 100 ppm for BM-RETs. Interestingly, a large range in the MN frequency in RETs derived from both PB and BM for the 100 ppm dose-group was reported. The most interesting aspect of this report was found in the statistical modeling of the dose-response to estimate the thresholds required to reach toxicity (Crump 1984). The results indicated that 0.205 ppm of benzene could reach the lower-bound confidence interval in the concentration-response (BMCL) found in this stock of DO mice. This BMCL was an order of magnitude lower than found in a previous report using a similar study design in the inbred B6C3F1 mouse strain (Farris et al. 1996). The difference in BMCLs between the studies suggest that incorporation of genetic variability may greatly impact safe-exposure assessments (Farris et al. 1996). In genotyping the mice and running QTL analysis, a locus on chromosome 10 was found to be significantly associated with the MN frequency derived from both the PB and BM within the 100 ppm benzene dose group. More specifically, mice that inherit a gene-duplication event in a region of chromosome 10 from the CAST/EiJ founder strain showed less benzene-induced chromosomal damage. The authors hypothesize that the gene-duplication likely leads to increased expression of several genes in this area, such as Sult3a1 and Gm4794, which are involved in sulfating toxic benzene metabolites. Notably, copy number variations is also found in human population that potentially plays a role in driving interindividual variation in the metabolism of benzene (French et al. 2015; Gaedigk et al. 2012; Yu et al. 2013).
3. Discussion
Traditionally, laboratory-based models to assess the risk associated with chemical exposures has not incorporated genetic variability. As the human population is heterogeneous, there is a need to incorporate genetic diversity into laboratory models to properly assess the risks associated with environmental exposures (Zeise et al. 2013). The lack of genetic diversity in toxicology-related laboratory models increases the potential to miss susceptible populations or, worse, misinterpret risk associated with a chemical. For example, as indicated by French et al., use of differing mouse strains can vastly impact the interpreted risk of a chemical exposure (French et al. 2015). Like human individuals, common laboratory models, such as cell lines and mouse strains, have differing sensitivities to chemicals. If the CAST/EiJ strain was used to assess the risk of benzene, our exposure assessment would differ as compared to a study using more sensitive strains such as the C57BL6/J or the WSB/EiJ (French et al. 2015). Similarly, the exposure assessment of TCDD would look quite different if the DBA/2J or AKR/J strain was used to characterize risk as compared to more sensitive strains, such as the C57BL6/J or Balb/cBy (Chapman and Schiller 1985; Poland et al. 1994; Shen et al. 1991; Weber et al. 1995).
Here, we have outlined several biological models that can be used incorporate genetic diversity into laboratory-based models: 1) genetically diverse cell lines, 2) human primary cells, and 3) mouse populations (Table 1). Each of these models have notable shortcomings. For example, the lymophoblastoid cell lines and human primary cells are, by definition, in vitro. This drastically increases the challenges with regard to extrapolating results to in vivo risk assessment. These models will most likely have differences in nutrients, temperature, O2 and CO2 concentrations, interactions with other cell types, and many other factors as compared to their counterparts found within the human body. In addition, cell lines are immortalized which likely affect responses as compared to primary cells. Primary human cells come from individuals with background exposures, diseases, ages, and many other factors that may affect the response. Mouse populations also have their limitations. While the mouse is a standardly-used in vivo model, there are obvious differences in the genetic architecture and physiology between mouse and human. However, as seen in multiple seminal projects and reports reviewed above, these laboratory-models provide the means to model human toxicokinetic and toxicodynamic population variability and identify genes and pathways involved in the responses associated with chemical toxicity. As such, these models provide a method to quantify the influence of population-level interindividual variability in a chemical- and tissue-specific manner. Beyond, these studies can be used to better predict which genetic variants may be 1) less susceptible to pharmaceutical-induced toxicity or 2) more responsive to a particular pharmaceutical as compared to another.
It should be noted that we are not arguing for the replacement of isogenic models in toxicology. As genetic variants in the population can drive differing degrees of responses or, in some cases, different physiological changes altogether, incorporation of genetic diversity into laboratory models might not always be the best method. From a purely academic standpoint, the results may be noisier when including genetic variability. Furthermore, inclusion of genetic variability may not be economically and technically feasible. Isogenic models clearly have a role to play in toxicology. For example, in mechanistic studies in which a single gene/protein/pathway is being analyzed, inclusion of genetic variability would potentially deter from the mechanism of action being studied and require an increase in sample size to reach a sufficient statistical power. However, as highlighted in the studies above, genetic variability clearly influences dose-response relationships and, from a risk-assessment point-of-view, cannot be ignored. Furthermore, many disease-states arise from complex etiologies that cannot be encapsulated by a single isogenic model.
Further incorporation of these models into toxicological screens have high potential to impact real-world risk assessment of chemical exposures from the population to the individual level. In most cases, data are not available in regards to the range of responses within the human population to environmental exposures. Risk-management decision-making relies, then, on arbitrary toxicokinetic and toxicodynamic ‘uncertainty factors’ in translating results from the laboratory into the human population. As seen in the studies outlined above, ‘uncertainty factors’ may not be stringent enough to include the susceptible individuals in the human population. In some cases, uncertainty factors may be too stringent and, therefore, may result in undue harm to municipal and industrial finances. The models outlined in this article can better inform risk-assessment so that, as opposed to arbitrary ‘uncertainty factors’, exposure limits are based on empirical data that represents the human population. As such, exposure standards will be based on evidence as opposed the NRC’s low-dose linearity assumption for non-cancer endpoints that has not been properly tested (Figure 1)(NRC 2009; Rhomberg 2009, 2011). Furthermore, these models also can identify individuals in the population that may be most at risk to chemical induce toxicity and, therefore, can help prioritize chemical-response pairs that leave certain individuals in a higher risk-category. From a pharmacogenetic standpoint, identification of variant-responders may also better prioritize which pharmaceutical are more effective or, to the contrary, more toxic to some individuals. In outlining examples of how these models have been used, we hope to not only better inform the toxicological community of the growing need to analyze population variability in chemical-responses, but to provide information on the means and feasibility to do so.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program (NIEHS SBRP P42ES4911). JJL is partially supported by AgBioResearch at Michigan State University. PD is supported by the National Institute of Environmental Health Sciences Training Grant at Michigan State University (T32 ES007255).
Footnotes
4. Conflicts of Interest.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdo N, Wetmore BA, Chappell GA, Shea D, Wright FA, Rusyn I. In vitro screening for population variability in toxicity of pesticide-containing mixtures. Environment international. 2015a;85:147–155. doi: 10.1016/j.envint.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdo N, Xia M, Brown CC, Kosyk O, Huang R, Sakamuru S, Zhou YH, Jack JR, Gallins P, Xia K, Li Y, Chiu WA, Motsinger-Reif AA, Austin CP, Tice RR, Rusyn I, Wright FA. Population-based in vitro hazard and concentration-response assessment of chemicals: the 1000 genomes high-throughput screening study. Environ Health Perspect. 2015b;123:458–466. doi: 10.1289/ehp.1408775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Toamih Atamni HJ, Ziner Y, Mott R, Wolf L, Iraqi FA. Glucose tolerance female-specific QTL mapped in collaborative cross mice. Mammalian genome: official journal of the International Mammalian Genome Society. 2017;28:20–30. doi: 10.1007/s00335-016-9667-2. [DOI] [PubMed] [Google Scholar]
- Adams DJ, Doran AG, Lilue J, Keane TM. The Mouse Genomes Project: a repository of inbred laboratory mouse strain genomes. Mammalian genome: official journal of the International Mammalian Genome Society. 2015;26:403–412. doi: 10.1007/s00335-015-9579-6. [DOI] [PubMed] [Google Scholar]
- Ahmed HMM, Salerno S, Morelli S, Giorno L, De Bartolo L. 3D liver membrane system by co-culturing human hepatocytes, sinusoidal endothelial and stellate cells. Biofabrication. 2017;9:025022. doi: 10.1088/1758-5090/aa70c7. [DOI] [PubMed] [Google Scholar]
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, de Villena FP, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome research. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue MA, Churchill GA, Chesler EJ. Collaborative Cross and Diversity Outbred data resources in the Mouse Phenome Database. Mammalian genome: official journal of the International Mammalian Genome Society. 2015;26:511–520. doi: 10.1007/s00335-015-9595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford BU, Lock EF, Kosyk O, Kim S, Uehara T, Harbourt D, DeSimone M, Threadgill DW, Tryndyak V, Pogribny IP, Bleyle L, Koop DR, Rusyn I. Interstrain differences in the liver effects of trichloroethylene in a multistrain panel of inbred mice. Toxicol Sci. 2011;120:206–217. doi: 10.1093/toxsci/kfq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCC. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DE, Schiller CM. Dose-related effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in C57BL/6J and DBA/2J mice. Toxicol Appl Pharmacol. 1985;78:147–157. doi: 10.1016/0041-008x(85)90314-x. [DOI] [PubMed] [Google Scholar]
- Chesler EJ. Out of the bottleneck: the Diversity Outcross and Collaborative Cross mouse populations in behavioral genetics research. Mammalian genome: official journal of the International Mammalian Genome Society. 2014;25:3–11. doi: 10.1007/s00335-013-9492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WA, Campbell JL, Jr, Clewell HJ, 3rd, Zhou YH, Wright FA, Guyton KZ, Rusyn I. Physiologically based pharmacokinetic (PBPK) modeling of interstrain variability in trichloroethylene metabolism in the mouse. Environ Health Perspect. 2014;122:456–463. doi: 10.1289/ehp.1307623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church RJ, Gatti DM, Urban TJ, Long N, Yang X, Shi Q, Eaddy JS, Mosedale M, Ballard S, Churchill GA, Navarro V, Watkins PB, Threadgill DW, Harrill AH. Sensitivity to hepatotoxicity due to epigallocatechin gallate is affected by genetic background in diversity outbred mice. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association. 2015;76:19–26. doi: 10.1016/j.fct.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nature genetics. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Gatti DM, Munger SC, Svenson KL. The Diversity Outbred mouse population. Mammalian genome: official journal of the International Mammalian Genome Society. 2012;23:713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki JA, Furuya S, Venkatratnam A, McDonald TJ, Knap AH, Wade T, Sweet S, Chiu WA, Threadgill DW, Rusyn I. Characterization of Variability in Toxicokinetics and Toxicodynamics of Tetrachloroethylene Using the Collaborative Cross Mouse Population. Environ Health Perspect. 2017;125:057006. doi: 10.1289/EHP788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science (New York, NY) 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump KS. A new method for determining allowable daily intakes. Fundamental and applied toxicology: official journal of the Society of Toxicology. 1984;4:854–871. doi: 10.1016/0272-0590(84)90107-6. [DOI] [PubMed] [Google Scholar]
- den Braver-Sewradj SP, den Braver MW, Vermeulen NP, Commandeur JN, Richert L, Vos JC. Inter-donor variability of phase I/phase II metabolism of three reference drugs in cryopreserved primary human hepatocytes in suspension and monolayer. Toxicology in vitro: an international journal published in association with BIBRA. 2016;33:71–79. doi: 10.1016/j.tiv.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr, Badner JA. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer research. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- Doran AG, Wong K, Flint J, Adams DJ, Hunter KW, Keane TM. Deep genome sequencing and variation analysis of 13 inbred mouse strains defines candidate phenotypic alleles, private variation and homozygous truncating mutations. Genome biology. 2016;17:167. doi: 10.1186/s13059-016-1024-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornbos P, Crawford RB, Kaminski NE, Hession SL, LaPres JJ. The Influence of Human Interindividual Variability on the Low-Dose Region of Dose-Response Curve Induced by 2,3,7,8-Tetrachlorodibenzo-p-Dioxin in Primary B Cells. Toxicol Sci. 2016;153:352–360. doi: 10.1093/toxsci/kfw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. The New England journal of medicine. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- Farris GM, Wong VA, Wong BA, Janszen DB, Shah RS. Benzene-induced micronuclei in erythrocytes: an inhalation concentration-response study in B6C3F1 mice. Mutagenesis. 1996;11:455–462. doi: 10.1093/mutage/11.5.455. [DOI] [PubMed] [Google Scholar]
- Festing MF. Genetically Defined Strains in Drug Development and Toxicity Testing. Methods in molecular biology (Clifton, NJ) 2016;1438:1–17. doi: 10.1007/978-1-4939-3661-8_1. [DOI] [PubMed] [Google Scholar]
- French JE, Gatti DM, Morgan DL, Kissling GE, Shockley KR, Knudsen GA, Shepard KG, Price HC, King D, Witt KL, Pedersen LC, Munger SC, Svenson KL, Churchill GA. Diversity Outbred Mice Identify Population-Based Exposure Thresholds and Genetic Factors that Influence Benzene-Induced Genotoxicity. Environ Health Perspect. 2015;123:237–245. doi: 10.1289/ehp.1408202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedigk A, Twist GP, Leeder JS. CYP2D6, SULT1A1 and UGT2B17 copy number variation: quantitative detection by multiplex PCR. Pharmacogenomics. 2012;13:91–111. doi: 10.2217/pgs.11.135. [DOI] [PubMed] [Google Scholar]
- Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger J, Braeuning A, Budinsky RA, Burkhardt B, Cameron NR, Camussi G, Cho CS, Choi YJ, Craig Rowlands J, Dahmen U, Damm G, Dirsch O, Donato MT, Dong J, Dooley S, Drasdo D, Eakins R, Ferreira KS, Fonsato V, Fraczek J, Gebhardt R, Gibson A, Glanemann M, Goldring CE, Gomez-Lechon MJ, Groothuis GM, Gustavsson L, Guyot C, Hallifax D, Hammad S, Hayward A, Haussinger D, Hellerbrand C, Hewitt P, Hoehme S, Holzhutter HG, Houston JB, Hrach J, Ito K, Jaeschke H, Keitel V, Kelm JM, Kevin Park B, Kordes C, Kullak-Ublick GA, LeCluyse EL, Lu P, Luebke-Wheeler J, Lutz A, Maltman DJ, Matz-Soja M, McMullen P, Merfort I, Messner S, Meyer C, Mwinyi J, Naisbitt DJ, Nussler AK, Olinga P, Pampaloni F, Pi J, Pluta L, Przyborski SA, Ramachandran A, Rogiers V, Rowe C, Schelcher C, Schmich K, Schwarz M, Singh B, Stelzer EH, Stieger B, Stober R, Sugiyama Y, Tetta C, Thasler WE, Vanhaecke T, Vinken M, Weiss TS, Widera A, Woods CG, Xu JJ, Yarborough KM, Hengstler JG. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Archives of toxicology. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann J, Jacobi C, Hahn M, Schmid V, Welz C, Schwenk-Zieger S, Stauber R, Baumeister P, Becker S. Spheroid-based 3D Cell Cultures Enable Personalized Therapy Testing and Drug Discovery in Head and Neck Cancer. Anticancer research. 2017;37:2201–2210. doi: 10.21873/anticanres.11555. [DOI] [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, Smith PC, Tennant R, Bogue M, Paigen K, Harris C, Contractor T, Wiltshire T, Rusyn I, Threadgill DW. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome research. 2009;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah FY, de la Casa-Esperon E, Bell TA, Detwiler DA, Magnuson T, Sapienza C, de Villena FP. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome research. 2004;14:1880–1887. doi: 10.1101/gr.2519704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCS. Harmonization Project Document No. 2. Geneva: World Health Organization; 2005. Chemical-Specific Adjustment Factors for Interspecies Differences in Human Variability: Guidance Document for Use of Data in Dose/Concentration–Response Assessment. [Google Scholar]
- Judson RS, Kavlock RJ, Setzer RW, Hubal EA, Martin MT, Knudsen TB, Houck KA, Thomas RS, Wetmore BA, Dix DJ. Estimating toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chemical research in toxicology. 2011;24:451–462. doi: 10.1021/tx100428e. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelada SN. Plethysmography Phenotype QTL in Mice Before and After Allergen Sensitization and Challenge. G3 (Bethesda, Md) 2016;6:2857–2865. doi: 10.1534/g3.116.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelada SN, Aylor DL, Peck BC, Ryan JF, Tavarez U, Buus RJ, Miller DR, Chesler EJ, Threadgill DW, Churchill GA, Pardo-Manuel de Villena F, Collins FS. Genetic analysis of hematological parameters in incipient lines of the collaborative cross. G3 (Bethesda, Md) 2012;2:157–165. doi: 10.1534/g3.111.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Robledo RF, Recla JM, Philip VM, Bubier JA, Jay JJ, Harwood C, Wilcox T, Gatti DM, Bult CJ, Churchill GA, Chesler EJ. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes, brain, and behavior. 2013;12:424–437. doi: 10.1111/gbb.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Crawford RB, Suarez-Martinez JE, Kaplan BL, Kaminski NE. Induction of the aryl hydrocarbon receptor-responsive genes and modulation of the immunoglobulin M response by 2,3,7,8-tetrachlorodibenzo-p-dioxin in primary human B cells. Toxicol Sci. 2010;118:86–97. doi: 10.1093/toxsci/kfq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli A, Mattioli F, Angiola M, Reimann R, Brambilla G. Species, sex and interindividual differences in DNA repair induced by nine sex steroids in primary cultures of rat and human hepatocytes. Mutation research. 2003;536:69–78. doi: 10.1016/s1383-5718(03)00036-6. [DOI] [PubMed] [Google Scholar]
- Morgan AP, Didion JP, Doran AG, Holt JM, McMillan L, Keane TM, de Villena FP. Whole Genome Sequence of Two Wild-Derived Mus musculus domesticus Inbred Strains, LEWES/EiJ and ZALENDE/EiJ, with Different Diploid Numbers. G3 (Bethesda, Md) 2016;6:4211–4216. doi: 10.1534/g3.116.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashef A, Abu-Toamih Atamni HJ, Buchnik Y, Hasturk H, Kantarci A, Stephens D, Wiess EI, Houri-Haddad Y, Iraqi FA. Collaborative Cross Mouse Population for Studying the Alveolar Bone Changes and Impaired Glucose Tolerance Comorbidity After High Fat Diet Consumption. Journal of periodontology. 2017:1–14. doi: 10.1902/jop.2017.170075. [DOI] [PubMed] [Google Scholar]
- NRC. Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press; Washington, DC: 2007. [Google Scholar]
- NRC. Science and Decisions: Advancing Risk Assessment, 2009 by the National Academy of Sciences. Washington DC: 2009. Science and Decisions: Advancing Risk Assessment. [Google Scholar]
- O’Shea SH, Schwarz J, Kosyk O, Ross PK, Ha MJ, Wright FA, Rusyn I. In vitro screening for population variability in chemical toxicity. Toxicol Sci. 2011;119:398–407. doi: 10.1093/toxsci/kfq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis-Moghe AS, Kaminski NE. Immunotoxicity testing using human primary leukocytes: An adjunct approach for the evaluation of human risk. Current Opinion in Toxicology. 2017;3:25–29. doi: 10.1016/j.cotox.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Palen D, Glover E. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Molecular pharmacology. 1994;46:915–921. [PubMed] [Google Scholar]
- Rhomberg LR. Linear low-dose extrapolation for noncancer responses is not generally appropriate. Environ Health Perspect. 2009;117:A141–142. doi: 10.1289/ehp.0800329. author reply A142–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhomberg LR. Practical Risk Assessment and Management Issues Arising were we to Adopt Low-Dose Linearity for all Endpoints. Dose Response. 2011;9:144–157. doi: 10.2203/dose-response.10-023.Rhomberg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mammalian genome: official journal of the International Mammalian Genome Society. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Singer T. The application of 3D cell models to support drug safety assessment: opportunities & challenges. Advanced drug delivery reviews. 2014;69–70:179–189. doi: 10.1016/j.addr.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Gatti DM, Wiltshire T, Kleeberger SR, Threadgill DW. Toxicogenetics: population-based testing of drug and chemical safety in mouse models. Pharmacogenomics. 2010;11:1127–1136. doi: 10.2217/pgs.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz EG, Schuetz JD, Thompson MT, Fisher RA, Madariage JR, Strom SC. Phenotypic variability in induction of P-glycoprotein mRNA by aromatic hydrocarbons in primary human hepatocytes. Molecular carcinogenesis. 1995;12:61–65. doi: 10.1002/mc.2940120202. [DOI] [PubMed] [Google Scholar]
- Shen ES, Gutman SI, Olson JR. Comparison of 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated hepatotoxicity in C57BL/6J and DBA/2J mice. Journal of toxicology and environmental health. 1991;32:367–381. doi: 10.1080/15287399109531491. [DOI] [PubMed] [Google Scholar]
- Smith CM, Proulx MK, Olive AJ, Laddy D, Mishra BB, Moss C, Gutierrez NM, Bellerose MM, Barreira-Silva P, Phuah JY, Baker RE, Behar SM, Kornfeld H, Evans TG, Beamer G, Sassetti CM. Tuberculosis Susceptibility and Vaccine Protection Are Independently Controlled by Host Genotype. mBio. 2016:7. doi: 10.1128/mBio.01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. Ten years of the collaborative cross. G3 (Bethesda, Md) 2012;2:153–156. doi: 10.1534/g3.111.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Miller DR, Churchill GA, de Villena FP. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR journal/National Research Council, Institute of Laboratory Animal Resources. 2011;52:24–31. doi: 10.1093/ilar.52.1.24. [DOI] [PubMed] [Google Scholar]
- Venkatratnam A, Furuya S, Kosyk O, Gold A, Bodnar W, Konganti K, Threadgill DW, Gillespie KM, Aylor DL, Wright FA, Chiu WA, Rusyn I. Collaborative Cross mouse population enables refinements to characterization of the variability in toxicokinetics of trichloroethylene and provides genetic evidence for the role of PPAR pathway in its oxidative metabolism. Toxicol Sci. 2017 doi: 10.1093/toxsci/kfx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber LW, Lebofsky M, Stahl BU, Smith S, Rozman KK. Correlation between toxicity and effects on intermediary metabolism in 2,3,7,8-tetrachlorodibenzo-p-dioxin-treated male C57BL/6J and DBA/2J mice. Toxicol Appl Pharmacol. 1995;131:155–162. doi: 10.1006/taap.1995.1057. [DOI] [PubMed] [Google Scholar]
- Weinshilboum R. Inheritance and drug response. The New England journal of medicine. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- Welsh CE, Miller DR, Manly KF, Wang J, McMillan L, Morahan G, Mott R, Iraqi FA, Threadgill DW, de Villena FP. Status and access to the Collaborative Cross population. Mammalian genome: official journal of the International Mammalian Genome Society. 2012;23:706–712. doi: 10.1007/s00335-012-9410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore BA, Allen B, Clewell HJ, 3rd, Parker T, Wambaugh JF, Almond LM, Sochaski MA, Thomas RS. Incorporating population variability and susceptible subpopulations into dosimetry for high-throughput toxicity testing. Toxicol Sci. 2014;142:210–224. doi: 10.1093/toxsci/kfu169. [DOI] [PubMed] [Google Scholar]
- Xue J, Schoenrock SA, Valdar W, Tarantino LM, Ideraabdullah FY. Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations. Clinical epigenetics. 2016;8:107. doi: 10.1186/s13148-016-0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Kubota T, Dhakal I, Hasegawa S, Williams S, Ozawa S, Kadlubar S. Copy number variation in sulfotransferase isoform 1A1 (SULT1A1) is significantly associated with enzymatic activity in Japanese subjects. Pharmacogenomics and personalized medicine. 2013;6:19–24. doi: 10.2147/PGPM.S36579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeise L, Bois FY, Chiu WA, Hattis D, Rusyn I, Guyton KZ. Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ Health Perspect. 2013;121:23–31. doi: 10.1289/ehp.1205687. [DOI] [PMC free article] [PubMed] [Google Scholar]


