Abstract
In an earlier study from this laboratory, Vibrio fluvialis BD146, a clinical isolate from Kolkata, India, 2002, was found to be resistant to all the fourteen antibiotics tested. It harboured a high copy number plasmid pBD146 and a low copy number plasmid. In the present study, a more detailed analysis was carried out to unravel different resistance mechanisms in this isolate. Sequencing showed that variable region of class 1 integron located on low copy number plasmid harbored arr3-cmlA-bla OXA10-aadA1 gene cassettes. Analysis for extended spectrum beta lactamases (ESBLs) revealed that BD146 was ESBL positive. Efflux pumps were involved in the drug resistance phenotype for chloramphenicol, kanamycin, streptomycin and tetracycline. Sequence analysis of pBD146 revealed the presence of genes encoding BDint an integrase with a unique sequence having little similarity to other known integrases, toxin–antitoxin (parE/parD), a replicase, trimethoprim resistance (dfrVI) and quinolone resistance (qnrVC5). Presence of cmlA, putative novel integrase and toxin–antitoxin system in V. fluvialis has been documented for the first time in this report. pBD146 showed 99% sequence similarity with pVN84 from V. cholerae O1 of Vietnam, 2004 and a plasmid from V. parahaemolyticus v110 of Hong Kong, 2010. Conjugation experiments proved the ability of pBD146 and the low copy number plasmid, to get transferred to another host imparting their antibiotic resistance traits to the transconjugants. Therefore, present study has indicated that plasmids played an important role for dissemination of drug resistance.
Electronic supplementary material
The online version of this article (10.1007/s12088-017-0687-8) contains supplementary material, which is available to authorized users.
Keywords: Plasmid, Vibrio fluvialis, Integrase, Multi drug resistance, Toxin/antitoxin system, Integron
Introduction
Vibrios have been reported to cause diarrhea in humans [1–5]. Though antibiotics are used as secondary line of treatment for diarrhea, rise of resistance to these drugs has seriously jeopardized the successful treatment. Thus, it becomes imperative to decipher the mechanisms responsible for acquisition and dissemination of drug resistance in bacteria for development of new treatment options and management of diseases.
Horizontal gene transfer (HGT) that leads to rapid dissemination of genes is a major factor responsible for evolution of multi drug resistance (MDR) [6]. HGT is mediated by mobile genetic elements such as integrons, transposons and plasmids [6]. Integrons are gene capture platforms that integrate the exogenous gene cassettes by site-specific recombination converting them into their functional forms [7]. Till date, five classes of integrons have been well defined based on ~ 40–58% identities in their integrase sequences but class 1 integrons are well characterized for their role in spread of MDR among bacteria [8]. Integrons are composed of three key elements, an integrase; a promoter and a primary recombination site attI [7]. All the exogenous gene cassettes harbour imperfect repeats called 59-base elements (attC) at their 3′ end. Thus, attI × attC recombination facilitates integration of gene cassettes while their excision is carried out by attC × attC recombination [7]. Class 1 integrons consist of two conserved segments (CS). The 5′ CS contains the integrase gene (intI1) and 3′ CS comprises of qacEΔ1 encoding resistance to quaternary ammonium compounds and sul1 encoding resistance to sulphonamides. These conserved segments are separated by a variable region that usually comprises of one or more foreign gene cassettes [7].
A previous study from this laboratory had shown the role of multiple plasmids and integrons in imparting MDR to a clinical isolate of V. fluvialis BD146, 2002, that carried a high copy number plasmid of 7.5 kb (pBD146) and a low copy number plasmid [3]. In the present study, analysis of class 1 integrons, extended spectrum beta lactamases and role of efflux pumps in drug resistance was carried out. Detailed analysis of pBD146 sequence revealed the presence of a novel integrase, Vibrio cholerae repeat (VCR) regions associated with dfrVI and qnrVC5 genes, and the toxin–antitoxin module. BLAST analysis of BD146 indicated the possibility of HGT of this plasmid between three Vibrio species from three different geographical locations in Southeast Asia.
Materials and Methods
Bacterial Isolates and DNA Isolation
Vibrio fluvialis isolates were obtained from patients with acute cholera-like diarrhea, admitted to the Infectious Diseases Hospital, Kolkata, India, between 1998 and 2006 [3, 5, 9]. Genomic DNA and plasmid DNA were isolated as described previously [3, 10]. The patients provided their written consent for participating in the study and in case of children, written consent was obtained from their parents. The consent procedure was approved by the Institutional Ethical Clearance Committee of NICED. The study was also approved by the Institutional Biosafety Committee of Institute of Advanced Research (IAR), Gandhinagar and the Review Committee on Genetic Manipulation governed by the guidelines laid down by the Department of Biotechnology, Government of India.
Polymerase Chain Reaction (PCR) and Reverse Transcription PCR (RT-PCR)
An ORF (nt 154–1062) corresponding to an integrase named BDint, was predicted in pBD146. To check its presence in other clinical isolates and transconjugants, PCR was carried out using BDint-specific primers Vcint-F/Vcint-R [3] and genomic DNA (200 ng) or plasmid DNA (10 ng) as templates. The conditions for PCR were an initial denaturation at 95 °C for 4 min, followed by 25 cycles each of denaturation at 95 °C, 0.5 min, annealing (55 °C, 1 min) and extension (72 °C, 1 min) with final polymerization at 72 °C for 7 min. Recombinant Taq DNA polymerase (Thermo-Scientific) was used. For the analysis of class 1 integron in transconjugants, PCR was carried out with the primers specific for 5′ CS, 3′ CS and variable region of class 1 integrons as described earlier [3].
RNA was isolated by RNeasy kit (Qiagen) as per manufacturer’s protocol involving lysis with lysozyme and proteinase K digestion. The RNA preparations were subsequently treated with DNaseI (Thermo-Scientific) to remove the residual genomic DNA contamination. Reverse transcription was carried out at 50 °C for 30 min using one step RT-PCR kit (Qiagen) and Vcint-F/Vcint-R primers followed by PCR to amplify cDNA. Conditions for PCR involved initial denaturation at 95 °C for 15 min, followed by 25 cycles with conditions as described above.
Antibiotic Susceptibility Test and Minimum Inhibitory Concentration (MIC) Assays
Vibrio fluvialis BD146, E. coli XL1-Blue and their transconjugants were tested for susceptibility to AMP, Ampicillin (10 μg); CHL, Chloramphenicol (30 μg); CIP, Ciprofloxacin (5 μg); COT, Co-Trimoxazole (1.25 μg trimethoprim/23.75 μg sulfamethoxazole); GEN, Gentamicin (10 μg); KAN, Kanamycin (30 μg); NAL, Nalidixic Acid (30 μg); NEO, Neomycin (30 μg); NOR, Norfloxacin (10 μg); STR, Streptomycin (10 μg); SUL, Sulfisoxazole (300 μg); TET, Tetracycline (30 μg); TRI, Trimethoprim (5 μg) and RIF, Rifampicin (5 μg) using commercial disks (HiMedia). When no interpretive criteria for V. cholerae were available based on the CLSI guidelines [11], breakpoints for enterobacteriaceae were applied. MIC for various antibiotics were determined using the Ezy MIC™ strips (HiMedia) according to manufacturer’s instruction and interpreted using the CLSI criteria. E. coli ATCC 25922 was used as control.
Efflux Pump Assay
To ascertain the role of efflux pumps in imparting resistance to drugs, synergy test was carried out as described earlier [12–14]. The test was performed using one drug for each class of antibiotics to which BD146 was resistant. The efflux pump inhibitor carbonyl cyanide-m-chlorophenyl hydrazone (CCCP) was added on MHA at 4 mg L−1 concentration. Susceptibility testing for antibiotics by MIC strip was performed as described in earlier section, both in the presence and absence of CCCP. Lowering in MIC of the isolates in the presence of CCCP indicated the role of efflux pumps in reducing the concentration of that drug inside the cell by throwing it out.
Bacterial Conjugation
BD146 was analysed for its ability to transfer resistance traits to a recipient strain by conjugation [5, 15]. BD146 donor harboured resistance to ampicillin (MIC > 256 µg mL−1) and trimethoprim (MIC > 32 µg mL−1) and intermediate resistance to tetracycline (MIC ≤ 16 µg mL−1). The recipient E. coli XL1-Blue was sensitive to ampicillin (MIC = 4 µg mL−1) and trimethoprim (MIC = 0.25 µg mL−1) and highly resistant to tetracycline (MIC > 256 µg mL−1). The recipient and the donor were mixed (2:1) on a sterile 0.45 µm nitrocellulose membrane (PALL Life Sciences) and incubated overnight for mating on LB agar at 37 °C. The transconjugants were selected on LB agar plates containing two antibiotic combinations; ampicillin (50 µg mL−1) and tetracycline (120 µg mL−1) or trimethoprim (20 µg mL−1) and tetracycline (120 µg mL−1). The transconjugants from both the selections were analysed for their plasmid profiles, presence of BDint, presence of integron and antibiotic susceptibility profiles.
Analysis of DNA Sequences
The ORF finder tool (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to predict all the possible ORFs in pBD146 sequence. These ORFs were analysed by BLAST search. I-TASSER server was used for 3D protein structure prediction [16–18]. Multiple sequence alignments were carried out using CLUSTAL Omega (1.2.1) (http://www.expasy.org/genomics/sequence_alignment).
Results
Analysis of Class 1 Integron of BD146
BD146 was found to be resistant to all the 14 antibiotics tested [3]. Integron analysis had shown that BD146 consisted of class 1 integron associated with low copy number plasmid. ORF analysis of the 4.0 kb variable region amplified from integron (KY883670) revealed the presence of genes for; rifampin ADP-ribosylating transferase, a hypothetical protein, chloramphenicol efflux pump MFS transporter, extended spectrum beta-lactamase OXA-10 and aminoglycoside 3′ adenyl transferase. BLAST analysis of 4.0 kb integron sequence showed 99% identity for 100% query coverage with segments of plasmid A from E. coli H3 (CP010168.1), segments of plasmid pNDM15-1078 from E. coli N15-01078 (CP012902.1), segments of plasmid pNDM-116-17 from V. cholerae 116-17a (LN831185.1), class 1 integron from Pseudomonas areuginosa pae G18 (EU886979.1) and part of Enterobacter aerogenes HN0711 plasmid pHN-NDM0711. This clearly established the promiscuous nature of this integron associated with multiple genera and multiple species of organisms located at different geographical locations around the world.
Extended Spectrum Beta Lactamases in BD146
As the class 1 integron carried bla OXA10 gene, ESBL and ampC beta lactamases activity was analysed using EzyMIC strip for the cefotaxime (CTX)/cefotaxime and clavulanic acid (CTX+), ceftazidime (CAZ)/ceftazidime and clavulanic acid (CAZ+) and the mixture (MIX) of ceftazidime, cefotaxime, cefepime and cloxacillin and MIX+ with clavulanic acid and tazobactam. The MIC ratio for CAZ+/CAZ and CTX+/CTX was 42.67 and 64 respectively while the MIC ratio for MIX+/MIX was 64. These results revealed that the isolate possessed high ESBL and ampC beta lactamase activity.
Role of Efflux Pumps in Imparting Drug Resistance in BD146
To ascertain the synergy between the resistance genes and the efflux pump activity, synergy test was carried out at least three times to detect the efflux pump activity as described in materials and methods. The MIC of ampicillin, chloramphenicol, ciprofloxacin, kanamycin, streptomycin, tetracycline and trimethoprim were tested in the presence and absence of efflux pump inhibitor CCCP. Fold decrease in MIC of BD146 for a particular drug in the presence of CCCP indicated the involvement of efflux pump activity in imparting resistance to that drug. Synergy tests revealed that efflux pumps were involved for the drug resistance phenotype (1.33–2 fold decrease in MIC value) for chloramphenicol, kanamycin, streptomycin and tetracycline, while they did not contribute in imparting drug resistance for ampicillin, ciprofloxacin and trimethoprim as no changes were observed in MIC values (Table 1).
Table 1.
Synergy test for efflux pump activity in V. fluvialis BD146 isolate
| Antibiotics | Antibiotics with CCCP (4 mg L−1) | BD146 (µg mL−1) | Fold change in MIC |
|---|---|---|---|
| Ampicillin | – | > 256 | 0 |
| + | > 256 | ||
| Chloramphenicol | – | 12 | 2 |
| + | 6 | ||
| Ciprofloxacin | – | 6 | 0 |
| + | 6 | ||
| Trimethoprim | – | > 32 | 0 |
| + | > 32 | ||
| Tetracycline | – | 16 | 1.33 |
| + | 12 | ||
| Kanamycin | – | 12 | 1.5 |
| + | 8 | ||
| Streptomycin | – | 64 | 1.33 |
| + | 48 |
Bold values indicate the antibiotics for which the efflux pumps were active in BD146
Sequence Analysis of pBD146 Indicated the Presence of Various Genes
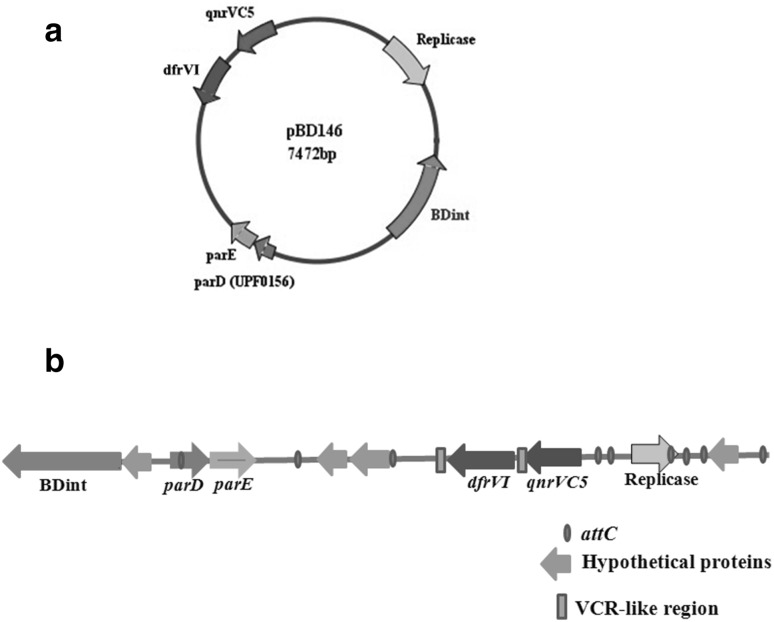
Sequence of pBD146 (EU574928) revealed that it carried genes encoding integrase (intI), replicase (repA), trimethoprim resistance (dfrVI), quinolone resistance (qnrVC5), toxin–antitoxin (parE/parD) and some hypothetical proteins (Fig. 1a). Though the presence of the first three genes was described in an earlier report [3], identification of qnrVC5 and parE/parD toxin–antitoxin genes became possible due to newer entries in GenBank leading to refinement in the BLAST search. It is possible that the hypothetical proteins would also be assigned functions in due course of time with expanding GenBank entries. Interestingly, these hypothetical proteins seemed to have their origin chiefly in the members of Vibrionaceae family barring one protein which was derived from gamma proteobacterium and has been described later in this section. For the hypothetical protein corresponding to ORF 3214–3429 nt, 87 and 84% identity was observed with the hypothetical protein VCHE16_3780 (EKG80837) from Vibrio cholerae HE-16 and another hypothetical protein from V. parahaemolyticus (WP_023622789) respectively. The hypothetical protein (ORF 7192–7428 nt) showed 86 and 85% identity with the hypothetical proteins from V. tasmaniensis (AKN38121, AKN38323, AKN39122) and V. cyclitrophicus (WP_016786352) respectively. The protein (ORF 1055–1300 nt) showed 96% identity with the hypothetical proteins from V. parahaemolyticus (KKX78394, KKX90213) and V. fluvialis (KQH89087). The ORF corresponding to 3557–3949 nt had 99 and 62% identity with the hypothetical proteins from V. cholerae HE-16 (EKG80837) and gamma proteobacterium BDW918 (EIF41853) respectively.
Fig. 1.
Schematic representation of plasmid pBD146. a Protein coding regions present on the plasmid pBD146; b Organisation of different genes such as integrase BDint (nt 154–1062), parD/parE genes (nt 2304–2546; 2539–2823), hypothetical proteins (nt 1055–1300; 3214–3429; 3557–3949;7192–7428), dfrVI (nt 4106–4579), qnrVC5 (nt 4742–5173) and a replicase (nt 6032–6922) on pBD146 with respect to 59-base elements and two VCR-like regions. The ORF finder tool was used to predict the ORFs in pBD146
The ORFs (nt 2539–2823 and nt 2304–2546) in pBD146 sequence corresponded to ParE (toxin) and ParD (antitoxin to ParE) respectively. ParE/ParD are addiction modules involved in plasmid stabilization. This is the first report of a putative toxin–antitoxin system in a plasmid from V. fluvialis.
Additionally, BLAST analysis of pBD146 revealed that some part (integrase, toxin–antitoxin system and hypothetical proteins) of this plasmid was present in V. tasmaniensis isolates (KP795520; KP795494; KP795636; KP795574 and KP795658) of 2014 from USA and V. parahaemolyticus clinical isolates from Canada (2001–2006) [LRTI01000063.1(2001); LRFP01000033.1 (2002); LRTB01000025.1(2003); LRTF01000062.1(2003); LRTF01000018.1 (2003); LRTC01000022.1(2003); LRTD01000012.1 (2003); LOHO01000004.1(2004); LOBT01000042.1 (2004); LRSU01000008(2006); LRSU01000014.1(2006)]. This fragment of pBD146 was even named as unlocalized plasmid pBD146 (the name given in this laboratory) in the V. parahaemolyticus isolates from Canada.
Analysis of pBD146 sequence revealed the presence of attC sites associated with various gene cassettes as shown in Fig. 1B. The dfrVI and qnrVC5 genes were encompassed by Vibrio cholerae repeat regions (VCRs), the attC-like regions found on the superintegron of V. cholerae, indicating the superintegron origin of these cassettes. BLAST analysis of qnrVC5 and dfrVI genes together revealed that this cassette of two genes was 99% similar to their counterparts in V. cholerae and V. parahaemolyticus mentioned below and 86% similar to a transposon from V. cholerae MCV09 (HM015627) and a part of SXT element from E. coli J53 (FJ968160). Even the VCR regions were similar suggesting that these two genes have been moving together between different organisms either as part of integrons, plasmids or SXT elements [19–22].
BLAST Analysis of pBD146 Indicated Horizontal Gene Transfer Between Three Vibrio Species
The sequence of pBD146 showed 99% identity with a plasmid pVN84 of V. cholerae O1 El Tor (2004) from Vietnam (AB200915) and another plasmid of V. parahaemolyticus v110 (2010) from Hong Kong (KC540630). While the isolates BD146 and VN84 were clinical in origin, v110 was environmental in origin [23]. This suggested horizontal transfer of a plasmid between three different vibrio species at three different locations of Southeast Asia over a period of eight years. Alternately, this plasmid could have been present in these species for a much longer time but detected recently.
pBD146 Horizontally Transferred to Escherichia coli Through Conjugation
To examine the capability of pBD146 for horizontal gene transfer, conjugation was carried out between BD146 and E. coli XL1-Blue. Transconjugants selected on ampicillin and tetracycline (amp50 + tet120) and those selected on trimethoprim and tetracycline (tri20 + tet120) were obtained with conjugation efficiency of 8 × 10−5 and 4.1 × 10−5 respectively. Both these transconjugants showed different plasmid profiles (Supplementary Fig. 1A). Transconjugants amp50 + tet120 showed resistance to ampicillin, tetracycline, rifampicin and nalidixic acid while transconjugants tri20 + tet120 harbored all the above resistance traits in addition to trimethoprim (Table 2). Only the tri20 + tet120 transconjugants showed the 657 bp amplicon of BDint (Supplementary Fig. 1B). These results revealed that pBD146 could transfer to another bacterium hitchhiking with another conjugative plasmid that harbored ampicillin and rifampicin resistance. PCR analysis of these transconjugants for 5′ CS, 3′ CS and variable regions of class 1 integrons revealed that the integron was present in both type of transconjugants and it harboured the 4.0 kb variable region having arr3-cmlA-bla OXA10-aadA1 gene cassette.
Table 2.
Antibiotic susceptibility profile of Vibrio fluvialis BD146 and its transconjugants
| Vibrio fluvialis BD146 | Transconjugants TRI20 + TET120 | Transconjugants AMP50 + TET120 | E. coli XL1-Blue | |
|---|---|---|---|---|
| Resistant | AMP, COT, CIP, SUL, NAL, TRI, RIF, CHL,GEN, STR, NOR, KAN, NEO, TET | AMP, TET, TRI, RIF, NAL | AMP, TET, RIF, NAL | TET, NAL |
Intermediate resistance and complete resistance were together considered as a resistance trait
AMP, Ampicillin (10 μg); CHL, Chloramphenicol (30 μg); CIP, Ciprofloxacin (5 μg); COT, Co-Trimoxazole (1.25 μg trimethoprim/23.75 μg sulfamethoxazole); GEN, Gentamicin (10 μg); KAN, Kanamycin (30 μg); NAL, Nalidixic Acid (30 μg); NEO, Neomycin (30 μg); NOR, Norfloxacin (10 μg); STR, Streptomycin (10 μg); SUL, Sulfisoxazole (300 μg); TET, Tetracycline (30 μg); TRI, Trimethoprim (5 μg); RIF, Rifampicin (5 μg)
TRI20, Trimethoprim (20 µg mL−1); TET120, Tetracycline (120 µg mL−1); AMP50, Ampicillin (50 µg mL−1)
Experiments were performed at least three times
BDint is a Putative Novel Integrase
BDint belonged to DNA_BRE_C superfamily consisting of DNA breaking rejoining enzymes including tyrosine recombinase. BDint showed 86, 85, 81 and 75% identity with integrases from V. alginolyticus, V. parahaemolyticus, V. cyclitrophicus and Photobacterium sp. respectively in a BLASTp analysis. The nucleotide sequence of BDint (KT182072) showed 99% identity with its counterparts in V. cholerae pVN84 and V. parahaemolyticus pv110 and 77–82% identity with the integrases from V. tasmaniensis (AKN40441; AKN39714; AKN36978). PCR showed the presence of a 657 bp BDint amplicon in twelve out of eighteen V. fluvialis isolates from 1998 to 2006 (Supplementary Fig. 1C). Three of these 100% identical sequences were submitted to GenBank (KT182073, KT182074 and KT182075). A 657 bp amplicon produced in reverse transcription confirmed the presence of mRNA for BDint in BD146. A negative control without reverse transcription step did not show any band corresponding to the expressed gene ensuring the absence of DNA contamination in RNA templates.
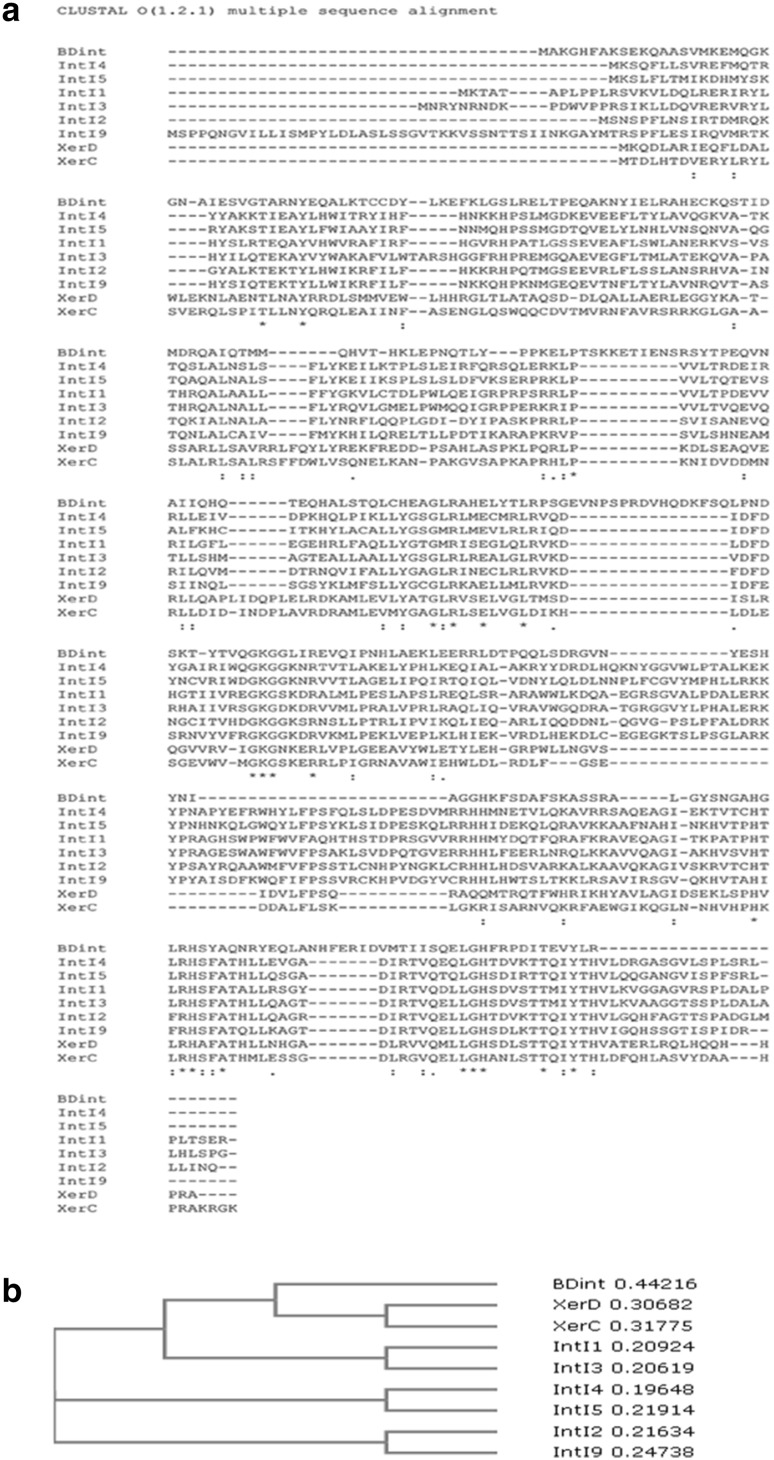
Comparison of BDint with other tyrosine recombinases such as IntI1-IntI5, IntI9, XerC and XerD using BLASTp indicated that BDint had very low similarity (19–29%) with them indicating that it was sufficiently different from all these integrases (Supplementary Table 1). This result was further confirmed by CLUSTAL analysis of BDint (Fig. 2). Alignment of all these recombinase sequences revealed that the unique additional domain (row 6 in Fig. 2a) present in the integrases related to integrons was absent in BDint establishing it to be a different entity [24]. The amino acid tail at the C-terminus of these recombinases was also absent in BDint. Additionally, BDint carried certain stretches of amino acids TSKKETIENS (row 3); EVNPSPRDVHQDKFSQ (row 4); NHFERID (row 7); that were absent in all other recombinases. Therefore, sequence analysis and phylogenetic tree (Fig. 2b) established it to be a novel integrase.
Fig. 2.
Comparison of BDint with other known integrases. Multiple sequence alignment of BDint protein sequence with other classes of integrase/recombinase sequences using Clustal O (1. 2.1) program (a); and their phylogenetic analysis (b)
The above observations were further confirmed by I-TASSER analysis (Supplementary Fig. 2). BDint showed the presence of additional helices that were distinctly missing in other integrases. In this analysis, some of the proteins structurally close to BDint included XerD recombinase from E. coli (1aOpA), XerA from archaea (4a8eA), Cre-loxP synaptic structure from Enterobacteria phage P1(3c29H), human topoisomerase I DNA complex (1ej9A) and site-specific recombinase IntI4 (2a3vB). The molecular function for BDint was predicted to be topoisomerase activity and biological process was predicted to be DNA recombination/integration (Supplementary Table 2).
Therefore, all these results mentioned above suggested that BDint and its siblings from V. cholerae and V. parahaemolyticus were novel integrases.
Discussion
An earlier study from this laboratory had described an MDR isolate of V. fluvialis BD146 (2002) from Kolkata, equipped with multiple plasmids and integrons to combat multiple antibiotics. One integron carried the putative exporter protein, while the other integron resident on a low copy number plasmid carried genes responsible for rifampicin, ampicillin, gentamicin and kanamycin resistance [3]. Apart from the low copy number plasmid, another 7.5 kb plasmid, pBD146, was also present in this bacterium carrying trimethoprim resistance gene. In other studies from the laboratory, qnrVC5, aac(6′)-Ib-cr and mutations in GyrA (S83 → I) and ParC (S85 → L) were found to be involved in quinolone resistance of BD146 [5, 25]. In the present study, the remaining drug resistance phenotypes were characterized in BD146.
The cmlA gene cassette resident on class 1 integron was for the first time reported from V. fluvilais. BLAST analysis of this integron showed 99% identity with segments of pNDM plasmids from E. coli N15-01078 (CP012902.1), V. cholerae 116-17a (LN831185.1), and Enterobacter aerogenes HN0711 (KU764665.1). ESBL and ampC beta lactamase analysis of this isolate revealed that it had high ESBL and ampC beta lacatamase activity. Presence of ESBL activity has also been reported in V. fluvialis isolates from India [26, 27]. Efflux pump activity was also involved in determining the resistance phenotype for chloramphenicol, kanamycin, streptomycin and tetracycline. There are very few studies that described the role of efflux pumps in imparting fluroquinolone resistance in V. fluvialis [9, 28] and synergy test has not been used till date in V. fluvialis.
BLAST analysis showed a high genetic relatedness between the three plasmids (pBD146, pVN84 and pV110) from clinical and environmental isolates of different species of Vibrios suggesting that these plasmids were exchanged and maintained in these Vibrio sp. from 2002 to 2010. This also indicated the possibility of this transfer via an intermediate species or descent of this plasmid from a common ancestor.
The presence of attC and VCR regions encompassing some of the genes of pBD146 were indicative of their probable origin from superintegrons. In an earlier report, presence of qnrVC2 on class 1 integron of V. cholerae plasmid pVN84 had been reported [22] but the structure of a class 1 integron was not apparent from this plasmid sequence. Similarly, pBD146 did not appear to carry an integron though the recombination sites associated with some of the gene cassettes did indicate their provenance in a superintegron.
pBD146 was able to get transferred to E. coli during conjugation with the help of another conjugative plasmid/low copy number plasmid that carried multiple drug resistance genes. The propensity of pBD146 to transfer horizontally between bacteria has also been shown in the earlier studies from this laboratory where pBD146 and similar plasmids moved from one host to another through transformation/conjugation, with concomitant expression of antibiotic resistance traits in their corresponding transformants/transconjugants [3, 5].
BLASTp, CLUSTAL and I-TASSER analysis of BDint with other integrases revealed that it was sufficiently different from other classes of integrases to be considered a novel integrase. BLAST analysis of parE and parD genes revealed that these genes constituted the first toxin–antitoxin module reported from V. fluvialis. The toxin–antitoxin modules have been attributed multiple functions such as plasmid stability, stabilization of DNA segments, protection against invading plasmids and phages and gene regulation [29]. Most interestingly, recent submissions in GenBank from mid 2015 to April 2016 have shown the presence of fragment of pBD146 harbouring the integrase, toxin–antitoxin module and a hypothetical protein in V. parahaemolyticus (2001–2006, Canada) and V. tasmaniensis (2014, USA). Therefore, there appears to be a very wide dispersal of these modules across the globe at least from 2001 to 2014. The results presented here for pBD146 show the genesis of a mosaic of genes derived from sources such as superintegrons and transposons. This plasmid/its modules have been maintained in different Vibrio spp. through the years 2001–2014, as a carrier of many traits capable of persistence in different bacteria and dissemination of drug resistance.
To summarize, in the present study multiple plasmids and integrons along with other genetic factors such as efflux pump contributed to the drug resistance phenotype of this clinical isolate (Table 3).
Table 3.
Correlation of resistance to different antibiotics with corresponding genes in clinical isolate of V. fluvialis BD146
| Antibiotic | Factor/genes responsible for resistance phenotype in V. fluvialis BD146 |
|---|---|
| Fluroquinolone | Mutation in Topoisomerase, qnrVC5, aac-6′-Ib-cr |
| Aminoglycoside | aadA1 (Aminoglycoside-3′-adenyltransferase) and efflux pump |
| Beta-lactam | blaOXA-10 (extended spactrum β-lactamase OXA) |
| Rifampicin | arr-3 (rifampicin ADP-ribosylating transferase) |
| Chloramphenicol | cmlA (MFS efflux pump) |
| Sulphonamide | sul1 gene on Class 1 integron |
| Trimethoprim | dfrVI (dihydrofolate reductase) |
| Tetracycline | Efflux pump |
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to Drs. Amit Ghosh and T. Ramamurthy, NICED, Kolkata, India, for providing the Vibrio strains and for their invaluable advice and support. The authors also thankfully acknowledge The Puri Foundation for Education in India for providing infrastructure facilities. Ms. Neha Rajpara is a recipient of senior research fellowship from Indian Council of Medical Research (ICMR), New Delhi, India (AMR/49/11-ECDI). The technical support and advice provided by Mr. Priyabrata Mohanty and Mr. K. Vinothkumar is thankfully acknowledged.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12088-017-0687-8) contains supplementary material, which is available to authorized users.
References
- 1.Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris JG., Jr Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin Infect Dis. 2003;37:272–280. doi: 10.1086/375600. [DOI] [PubMed] [Google Scholar]
- 3.Rajpara N, Patel A, Tiwari N, Bahuguna J, Antony A, Choudhury I, Ghosh A, Jain R, Bhardwaj AK. Mechanism of drug resistance in a clinical isolate of Vibrio fluvialis: involvement of multiple plasmids and integrons. Int J Antimicrob Agents. 2009;34:220–225. doi: 10.1016/j.ijantimicag.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Ramamurthy T, Sharma NC. Cholera outbreaks in India. Curr Top Microbiol Immunol. 2014;379:49–85. doi: 10.1007/82_2014_368. [DOI] [PubMed] [Google Scholar]
- 5.Singh R, Rajpara N, Tak J, Patel A, Mohanty P, Vinothkumar K, Chowdhury G, Ramamurthy T, Ghosh A, Bhardwaj AK. Clinical isolates of Vibrio fluvialis from Kolkata, India, obtained during 2006: plasmids, the qnr gene and a mutation in gyrase A as mechanisms of multidrug resistance. J Med Microbiol. 2012;61:369–374. doi: 10.1099/jmm.0.037226-0. [DOI] [PubMed] [Google Scholar]
- 6.Bhardwaj AK, Vinothkumar K. Evolution of MDRs. In: Kalia VC, editor. Quorum sensing vs quorum quenching: a battle with no end in sight. New Delhi: Springer; 2015. pp. 9–22. [Google Scholar]
- 7.Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 8.Bhardwaj AK, Vinothkumar K, Rajpara N, Mohanty P, Kutar BM (2014) Therapeutic limitations due to antibiotic drug resistance: road to alternate therapies. In: Atta-ur-Rahman, Choudhary MI (eds) Frontiers in anti-infective drug discovery, vol 3. Bentham Science, Sharjah, pp 72–141
- 9.Srinivasan VB, Virk RK, Kaundal A, Chakraborty R, Datta B, Ramamurthy T, Mukhopadhyay AK, Ghosh A. Mechanism of drug resistance in clonally related clinical isolates of Vibrio fluvialis isolated in Kolkata, India. Antimicrob Agents Chemother. 2006;50:2428–2432. doi: 10.1128/AAC.01561-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thungapathra M, Amita Sinha KK, Chaudhuri SR, Garg P, Ramamurthy T, Nair GB, Ghosh A. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob Agents Chemother. 2002;46:2948–2955. doi: 10.1128/AAC.46.9.2948-2955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical Laboratory Standard Institute (2010) Performance standards for antimicrobial susceptibility testing. In: Twentieth informational supplement, Wayne, PA: CLSI:M100-S120
- 12.Azmi IJ, Khajanchi BK, Akter F, Hasan TN, Shahnaij M, Akter M, Banik A, Sultana H, Hossain MA, Ahmed MK, Faruque SM, Talukder KA. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PLoS ONE. 2014;9:e102533. doi: 10.1371/journal.pone.0102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JY, Kim SH, Jeon SM, Park MS, Rhie HG, Lee BK. Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clin Microbiol Infect. 2008;14(8):760–765. doi: 10.1111/j.1469-0691.2008.02033.x. [DOI] [PubMed] [Google Scholar]
- 14.Taneja N, Mishra A, Kumar A, Verma G, Sharma M. Enhanced resistance to fluoroquinolones in laboratory-grown mutants & clinical isolates of Shigella due to synergism between efflux pump expression & mutations in quinolone resistance determining region. Indian J Med Res. 2015;141:81–89. doi: 10.4103/0971-5916.154508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutar BM, Rajpara N, Upadhyay H, Ramamurthy T, Bhardwaj AK. Clinical isolates of Vibrio cholerae O1 El Tor Ogawa of 2009 from Kolkata, India: preponderance of SXT element and presence of Haitian ctxB variant. PLoS ONE. 2013;8:e56477. doi: 10.1371/journal.pone.0056477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonseca EL, Vicente AC. Epidemiology of qnrVC alleles and emergence out of the Vibrionaceae family. J Med Microbiol. 2013;62:1628–1630. doi: 10.1099/jmm.0.062661-0. [DOI] [PubMed] [Google Scholar]
- 20.Kumar P, Thomas S. Presence of dfr6 gene cassette in superintegron of non-O1/non-O139 strain of Vibrio cholerae. Antimicrob Agents Chemother. 2009;53:4959–4960. doi: 10.1128/AAC.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Thomas S. Presence of qnrVC3 gene cassette in SXT and class 1 integrons of Vibrio cholerae. Int J Antimicrob Agents. 2011;37:280–281. doi: 10.1016/j.ijantimicag.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Fonseca EL, Dos Santos Freitas F, Vieira VV, Vicente AC. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg Infect Dis. 2008;14:1129–1131. doi: 10.3201/eid1407.080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu M, Chen S. Draft genome sequence of Vibrio parahaemolyticus V110, isolated from shrimp in Hong Kong. Genome Announc. 2013 doi: 10.1128/genomeA.00300-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messier N, Roy PH. Integron integrases possess a unique additional domain necessary for activity. J Bacteriol. 2001;183:6699–6706. doi: 10.1128/JB.183.22.6699-6706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinothkumar K, Kumar GN, Bhardwaj AK. Characterization of Vibrio fluvialis qnrVC5 gene in native and heterologous hosts: synergy of qnrVC5 with other determinants in conferring quinolone resistance. Front Microbiol. 2016;7:146. doi: 10.3389/fmicb.2016.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury G, Pazhani GP, Sarkar A, Rajendran K, Mukhopadhyay AK, Bhattacharya MK, Ghosh A, Ramamurthy T. Carbapenem resistance in clonally distinct clinical strains of Vibrio fluvialis isolated from diarrheal samples. Emerg Infect Dis. 2016;22:1754–1761. doi: 10.3201/eid2210.151612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chowdhury G, Pazhani GP, Nair GB, Ghosh A, Ramamurthy T. Transferable plasmid-mediated quinolone resistance in association with extended-spectrum beta-lactamases and fluoroquinolone-acetylating aminoglycoside-6′-N-acetyltransferase in clinical isolates of Vibrio fluvialis. Intl J Antimicrob Agents. 2011;38:169–173. doi: 10.1016/j.ijantimicag.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Mohanty P, Patel A, Kushwaha Bhardwaj A. Role of H- and D-MATE-type transporters from multidrug resistant clinical isolates of Vibrio fluvialis in conferring fluoroquinolone resistance. PLoS ONE. 2012;7:e35752. doi: 10.1371/journal.pone.0035752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unterholzner SJ, Poppenberger B, Rozhon W. Toxin–antitoxin systems: biology, identification, and application. Mob Genet Elem. 2013;3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.