Abstract
Previous studies have demonstrated a dynamic epigenetic regulation of genes expression in placenta trophoblasts and a dynamic imbalance of DNA methylation and hydroxymethylation. Reduced IGF-1 has been observed in preeclampsia. This study was to investigate the interactive roles between IGF-1 and the global DNA methylation/hydroxymethylation, and the status of DNA methylation/hydroxymethylation and associated enzymes such as DNMTs and TETs in peeeclamptic placentas and hypoxic trophoblasts. It was found that IGF-1 was decreased in preeclamptic placentas and hypoxic trophoblasts when compared to the control group using immunohistochemisty, western blot, qRT-PCR and ELISA. Pyrophosphate sequencing showed IGF-1 promoter was significantly hypermethylated in preeclamptic placentas, which was responsible for reduced IGF-1 expression. Preeclamptic placentas and hypoxic trophoblasts were hypermethylated and hypohydroxymethylated accompanied by remarkably higher 5mC, DNMT1 and DNMT3b, and lower DNMT3a, 5hmC, TET1, TET2 and TET3 detected by immunohistochemisty, western blot, qRT-PCR and ELISA. Pearson’s correlation confirmed a statistically significant negative correlation between IGF-1 and DNMT1. Furthermore, both treatment with 5-Aza-dc and DNMT1-siRNA significantly increased the expression of IGF-1 in HTR8 cells, indicating the potential mechanism of DNMT1-mediated DNA methylation in IGF-1 regulation. However, IGF-1 didn’t change DNA methylation or hydroxymethylation. These findings suggest that preeclampsia is associated with hypermethylation of IGF-1 promoter mediated by DNMT1 and provide new insights into the diagnosis and treatment of preeclampsia.
Keywords: Preeclampsia, IGF-1, DNA methylation/hydroxymethylation, 5mC/5hmC, DNMTs, TETs
Introduction
Preeclampsia (PE) is a serious pregnancy-induced disease characterized by hypertension, proteinuria and other systemic disorders after 20 weeks of gestation and is a leading cause of maternal and fetal morbidity and mortality affecting 2-10% pregnant women worldwide [1-4]. The main pathological characteristics of preeclampsia are poor trophoblast cell invasion and uterine spiral artery remodeling dysfunction caused by placenta ischemia and oxidative stress. Nowadays, preeclampsia is considered to be the results of the interactions of genetic and environmental factors [5,6]. However, its accurate pathogenesis remains unknown.
It was considered IGF-1 might be involved in the pathogenesis of preeclampsia. As is known, the behaviors of cells, the formation of placenta and the growth of fetus are widely regulated by insulin-like growth factor 1 (IGF-1) [7-9]. Several studies have shown that IGF-1 was significantly reduced in preeclamptic maternal placentas, umbilical cord blood and serum when compared to normal healthy pregnant women [10-12]. However, Bartha and colleagues pointed out that women with gestational hypertension had a higher level of IGF-1 than women in the control group [13]. Additionally, accumulating evidence has confirmed that IGF-1 might play a vital role in trophoblasts functions [14-16]. However, the mechanisms of IGF-1-mediated PE progression were still poorly understood.
DNA methylation is a crucial epigenetic modification of the genome that is involved in regulating many cellular processes including embryonic development, transcription, chromatin structure, X chromosome inactivation, genomic imprinting and chromosome stability [17,18]. Genomic DNA is enzymatically methylated by a family of DNA methyltransferases (DNMTs) including DNMT1, DNMT3a and DNMT3b. Given the strong evidences that gene expression is altered in preeclamptic pregnancies, it is reasonable to hypothesize that preeclampsia may also be associated with DNA methylation in key regulatory regions. Ye et al. and Tang et al. respectively found that preeclampsia was associated with decreased methylation of the GNA12 promoter and hypermethylation of the HLA-G promoter mediated by DNMT1 [19,20]. Additionally, published results showed that FABP4 might be involved in the pathogenesis of preeclampsia, and the expression of FABP4 was enhanced by miR-148a/152 mediated inhibition of DNMT1 expression [21]. Previous studies have confirmed that IGF-1 promoter region was significantly hypermethylated in many malignant tumors compared with normal tissues [22,23]. However, IGF-1 DNA methylation patterns in preeclampsia have not been investigated. A recent study published in Science reported the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in mammalian DNA by MLL partner TET1, implying that DNA hydroxymethylation mediated by ten-eleven translocaations (TETs) including TET1, TET2 and TET3 also was involved in the pathogenesis of preeclampsia [24]. Simultaneously, it was reported that hypoxia remarkably altered the epigenetic profile in cultured human placental trophoblasts [25].
In this study, we aimed to investigate the interactive roles between IGF-1 and the global DNA methylation/hydroxymethylation, and the status of DNA methylation/hydroxymethylation and associated enzymes such as DNMTs and TETs in peeeclamptic placentas and hypoxic trophoblasts. Therefore, we examined the expression of IGF-1 in preeclamptic placentas and hypoxic HTR8 and JEG3 cells, and measured the DNA methylation of IGF-1 promoter in preeclamptic placentas. In addition, we detected the levels of 5mC, DNMTs, 5hmC and TETs in preeclamptic placentas and hypoxic HTR8 and JEG3 cells. Following, we analyzed the associations of IGF-1 between 5mC, DNMTs, 5hmC and TETs using Pearson’s correlation. Furthermore, we investigated the expression of IGF-1 and its DNA methylation in promoter region in HTR8 cells treatment with 5-Aza-dc and DNMT1-siRNA. Finally, we explored the effects of IGF-1 on 5mC, DNMTs, 5hmC and TETs to study the interactive roles between IGF-1 and DNA methylation/hydroxymethylation.
Materials and methods
Ethical approval and clinical samples collection
This study was approved by the Ethics Committee of Obstetrics and Gynecology Hospital of Fudan University (Approval No. 2016-19). Written informed consents were obtained from all study subjects.
Preeclampsia was diagnosed according to the guidelines recommended by American College of Obstetricians and Gynecologists [2]. Placenta tissues associated with serious maternal complications and fetal abnormalities were excluded from this study. Ten preeclamptic patients and 10 healthy pregnant women who were in the third-trimester were enrolled into this study. Placenta tissue specimens, about 0.5 cm × 0.5 cm, were dissected from the central part of maternal side of placentas after delivery as soon as possible. Then, the specimens were washed by sterile phosphate-buffered saline (PBS) to clean the blood cells. Following, a part of the specimens were fixed in 4% paraformaldehyde for 24 h to 48 h and paraffin-embedded for immunohistochemistry (IHC). The remaining parts were snap frozen in liquid nitrogen and stored at -80°C for the extraction of protein, RNA and DNA.
Cell culture and treatment of 5-Aza-dc, DNMT1-siRNA, pEX-2-IGF-1 and IGF-1-siRNA
The human trophoblast-like cell line HTR8/SVneo (HTR8) and choriocarcinoma cell line JEG3 were cultured in DMEM/F12 medium (HyClone, USA) supplemented with 10% fetal bovine serum, 100 µg/ml penicillin, 100 µg/ml streptomycin and 100 µg/ml amphotericin B (Sangon Biotech, China) at 37°C and 5% CO2 humidified incubator. The medium was renewed every 24 h to 48 h and subcultured every 3 to 4 days by 0.25% trypsin containing EDTA. HTR8 and JEG3 cells were cultured for 24 h, 48 h, 72 h and 96 h under normal oxygen and hypoxia in a trigas cell culture incubator, respectively. The cells were harvested for isolation of protein, RNA and DNA, and the cell supernatants were collected for detection of IGF-1.
For 5-azadeoxycytidine (5-Aza-dc, Sigma Aldrich, USA) treatment, HTR8 cells were seeded at six-well culture dishes and growth in normal growth medium to achieve nearly 70% confluence. Then, the regular growth medium was replaced by the new medium containing 5-Aza-dc with different concentrations of 1 µM, 2 µM, 4 µM. After 48 h culture under normoxic conditions, the cells were scraped for extraction of protein, RNA and DNA.
Like what mentioned above, when HTR8 cells reached at a confluence of 40% to 50% and nearly 70%, siRNAs (DNMT1-siRNA and IGF-1-siRNA) and IGF-1 expression vector (pEX-2-IGF-1) were respectively added to transfect HTR8 cells. Lipofectamine 2000 transfection reagent (Invitrogen, USA) was used to transfect HTR8 cells with DNMT1-siRNA, IGF-1-siRNA and pEX-2-IGF-1 according to the manufacturer’s protocol. The regular growth medium was changed with fresh growth medium after 6 h transfection. The cells were harvested for isolation of protein, RNA and DNA after 48 h transfection. The efficiency of transfection was evaluated by quantitative real-time PCR (qRT-PCR) and western blot (WB).
Immunohistochemistry
The immunohistochemistry (IHC) was used to detect the expression of IGF-1, 5mC, DNMTs, 5hmC and TETs in placentas and carried out as described previously [26]. Briefly, the placenta tissue sections were incubated at 60°C for 1 h, deparaffinized in xylene and sequentially rehydrated in a graded series of ethanol. For antigen retrieval, the slides were immersed in 0.01 M citrate buffer (pH 6.0), incubated at above 95°C for 30 min and cooled down at room temperature. Endogenous peroxidase was eliminated with 3% H2O2 for 10 min and then blocked with 10% normal goat serum for 30 min at room temperature. Afterwards, the tissue sections were incubated overnight at 4°C with primary antibodies against IGF-1, 5mC, DNMTs, 5hmC and TETs, then placed for 45 min at room temperature. The sections were rinsed with PBS, and then secondary antibodies were added for 10 min. After washed with PBS, all sections were incubated with horseradish peroxidase (HRP) for 10 min at room temperature. Following, washed sections were incubated with 3,3-diaminobenzidine (DAB) to visualize the final product and counter-stained. Finally, the sections were dehydrated through graded alcohol and xylene in a reverse order applied in deparaffinization step, and coverslipped for observation. For negative control, the slides were incubated with PBS instead of primary antibodies. The optimal concentrations of these primary antibodies were shown in Table 1.
Table 1.
Optimal dilutions of primary antibodies for IHC
| Antibodies | Dilutions | Sources | Cat. No. |
|---|---|---|---|
| IGF-1 | 1:125 | Abcam, USA | ab9572 |
| 5mC | 1:500 | Active Motif, Japan | 39649 |
| 5hmC | 1:500 | Active Motif, Japan | 39769 |
| DNMT1 | 1:500 | Abcam, USA | ab19905 |
| DNMT3a | 1:80 | Abcam, USA | ab4897 |
| DNMT3b | 1:100 | Abcam, USA | ab119282 |
| TET1 | 1:200 | Abcam, USA | ab191698 |
| TET2 | 1:100 | Abcam, USA | ab94580 |
| TET3 | 1:50 | Novus, USA | NBP2-20602 |
Western blot analysis
The protein levels of IGF-1, DNMTs and TETs in placentas and trophoblast cells were measured by western blot. Proteins were extracted from placentas and trophoblast cells using radio immunoprecipitation assay (RIPA, Beyotime, China) lysis buffer supplemented with phenylmethanesulfonyl fluoride (PMSF, Beyotime, China) on ice. After homogenization, the supernatants were centrifuged at 12,000 rpm for 15 min at 4°C and collected for protein analysis. Protein concentrations were detected by bicinchonininc acid (BCA, Beyotime, China) protein assay kit. An equal amount of protein extracts (30 µg) were resolved in and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred onto polyvinylidene difluoride (PVDF) membranes within the transfer buffer (Beyotime, China) for 1 h to 2 h according to protein molecular weight. After blocked in 5% nonfat dry milk with phosphate-buffered saline/Tween-20 (PBST) for 1 h at room temperature, the membranes were incubated with primary antibodies overnight. Detailed information of primary antibodies was summarized in Table 2. After washing the membranes with PBST for 10 min × 3 times, the matching secondary antibodies (anti-rabbit or anti-mouse, 1:1000, Jackson, USA) were used to conjugate primary antibodies for 1 h at room temperature. The immunoreactivity was visualized by enhanced chemiluminescence (ECL) reagents (Millipore, USA). β-actin and GAPDH were used as the internal loading controls. The Quantity One software (Bio-Rad Laboratories Pty. Ltd) was used to analyze the band intensities.
Table 2.
Optimal dilutions Primary antibodies for WB
| Placentas | Trophoblasts | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Antibodies | Dilutions | Sources, Cat. No. | Antibodies | Dilutions | Sources, Cat. No. |
| IGF-1 | 1:500 | Abcam, USA, ab9572 | IGF-1 | 1:2000 | Abcam, USA, ab9572 |
| DNMT1 | 1:1000 | Abcam, USA, ab19905 | DNMT1 | 1:1000 | Abcam, USA, ab13537 |
| DNMT3a | 1:1000 | Abcam, USA, ab4897 | DNMT3a | 1:2000 | Abcam, USA, ab13888 |
| DNMT3b | 1:500 | Abcam, USA, ab119282 | DNMT3b | 1:2000 | Abcam, USA, ab13604 |
| TET1 | 1:500 | Abcam, USA, ab191698 | TET1 | 1:1000 | Abcam, USA, ab105475 |
| TET2 | 1:1000 | Abcam, USA, ab94580 | TET2 | 1:1000 | Abcam, USA, ab125084 |
| TET3 | 1:500 | Novus, USA, NBP2-20602 | TET3 | 1:1000 | Abcam, USA, ab139805 |
| β-actin | 1:1000 | Beyotime, China, AA128 | GAPDH | 1:1000 | CST, USA, #2118 |
RNA extraction, cDNA synthesis and qRT-PCR analysis
The transcript levels of IGF-1, DNMTs and TETs in the placentas and trophoblast cells were analyzed by qRT-PCR. Total RNA was extracted using TRIzol (Invitrogen, USA) reagent according to the manufacturer’s protocol. RNA (2 μl) was quantified spectrophotometrically by measuring the absorbance at 260 nm and RNA purity was determined by the 260/280 nm absorbance ratio. The complementary DNA (cDNA) was synthesized from 1 μg total RNA in 20 μl volume reactions with PrimeScript™RT Master Mix (Takara, Japan). The 20 μl cDNA product was diluted into 100 μl for later use. To quantify mRNA expression, an amount of cDNA equivalent to 20 ng of total RNA was amplified using SYBR® Premix Ex Taq II (Takara, Japan) according to manufacture’s instruction. The primers of target gene were listed in Table 3, which were designed using Primer Premier 6.0 software and were synthesized by Shanghai BioTNT Biotechnology Co., Ltd. (Shanghai, China). GAPDH served as an internal standard. Relative quantification of the target gene expression levels was conducted using the method ΔCt, ΔE = Ctexp - CtGAPDH and ΔC = Ctcon - CtGAPDH.
Table 3.
Primers sequence of targeted genes
| Gene name | Forward | Reverse |
|---|---|---|
| IGF-1 | 5’-TCCTCGCATCTCTTCTACCT-3’ | 5’-AAAAGCCCCTGTCTCCACAC-3’ |
| DNMT1 | 5’-AAGAGCCAAATCGGATGAGT-3’ | 5’-AAGCGGTCTAGCAACTCGTT-3’ |
| DNMT3a | 5’-GGTGTGGCTTTAGGAGCAGT-3’ | 5’-CTACAGGCAGGTCAGTGAGC-3’ |
| DNMT3b | 5’-GAAGGGGTGTGCTGAGTTCT-3’ | 5’-GGGTTTGAGGGGGTGTCTTA-3’ |
| TET1 | 5’-CACGCTGTGGTGAAGGACTA-3’ | 5’-AGGTAACTTTGGGGGTTGCT-3’ |
| TET2 | 5’-CGCACAGTTAGTGAACCTT-3’ | 5’-GGATTTCTTTCTTGGCTTAC-3’ |
| TET3 | 5’-CCGTGAGATGAGTCGTGAG-3’ | 5’-AGTGTGTCAAGGTCTTCGCT-3’ |
| GAPDH | 5’-GGGAAGGTGAAGGTCGGAGT-3’ | 5’-GGGGTCATTGATGGCAACA-3’ |
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay (ELISA) was used to detect the protein of IGF-1 in cell supernatants, and 5mC and 5hmC levels of placentas and trophoblast cells according to each manufacturer’s instruction. For the detection of IGF-1 (RayBio, USA), the following steps must be operated with gentle shaking: (1) Add 100 µl of each standard and sample to appropriate wells, incubate for 2.5 h at room temperature, then discard the solution and wash 4 times with 1X wash solution; (2) Add 100 µl of 1X prepared biotinylated antibody to each well, incubate for 1 h at room temperature, then discard the solution and wash 4 times; (3) Add 100 µl of prepared streptavidin solution to each well, incubate for 45 min at room temperature, then discard the solution and wash 4 times; (4) Add 100 µl of TMB one-step substrate reagent to each well, incubate for 30 min at room temperature in the dark; (5) Add 50 µl of stop solution to each well and read at 450 nm immediately. For 5mC and 5hmC quantification, methylflashTM methylated and hydroxymethylated DNA quantification kits (colorimetric) were purchased from Epigentek, USA. In brief, they were conducted as the following steps: (1) DNA binding: add 80 µl of binding solution to appropriate cells, then add 1 µl negative control, 1 µl positive control and appropriate sample DNA (100 ng for 5mC and 200 ng for 5hmC) into the designated wells, incubate at 37°C for 90 min, and remove the binding solution and wash three times; (2) Methylated and hydroxymethylated DNA capture: add 50 µl diluted capture antibody (1:1000) to each well and incubate at room temperature for 60 min, remove the solution and wash three times; then add 50 µl diluted detection antibody (1:2000 for 5mC and 1:1000 for 5hmC) to each well and incubate at room temperature for 30 min, remove the solution and wash four times; following add 50 µl diluted enhancer solution (1:5000) to each well and incubate at room temperature for 30 min, remove the solution and wash five times; (3) Signal detection: add 100 µl developer solution and incubate at room temperature for 1 to 10 min away from light, and then add 100 µl stop solution to stop enzyme reaction and read at 450 nm. (4) 5mC relative quantification: simple calculation of the percentage of 5mC in total DNA can be carried out using the following formula:
5mC% = ((Sample OD - ME3 OD)÷S)/((ME4 OD - ME3 OD) × 2*÷P) × 100%
S is the amount of input sample DNA in ng.
P is the amount of input positive control in ng.
*2 is a factor to normalize 5mC in the positive control to 100%, as the positive control contains only 50% of 5mC.
(5) 5hmC relative quantification: simple calculation of the percentage of 5hmC in total DNA can be carried out using the following formula:
5hmC% = ((Sample OD - HC4 OD)÷S)/((HC5 OD - HC4 OD) × 5*÷P) × 100%
S is the amount of input sample DNA in ng.
P is the amount of input positive control in ng.
*5 is a factor to normalize 5hmC in the positive control to 100%, as the positive control contains only 20% of 5hmC.
DNA isolation and pyrophosphate sequencing
Genomic DNA was isolated from placentas and trophoblast cells with Qiagen DNA mini kit (Qiagen, Germany). The bisulfite conversion reaction of genomic DNA and subsequent purification was performed with the use of Qiagen EpiTect Bisulfite Kit (Qiagen, Germany) following the manufacturer’s protocols. The purified DNA with an optical density value between 1.8 and 2.0 was considered to be of good quality and stored at -80°C for later experiments. To assess the methylation status of IGF-1 and quantify the percentage of methylation of each individual CpG, bisulfite-modified DNA was amplified using bisulfite PCR primers with a biotin label on the 5’ end of the foraward primer. The pyrophosphate sequencing primers were designed by PyroMark Assay Design 2.0 software, synthesized by BGI, China. The primers were as follows: IGF-1-F: 5’-AAAAATGTTTTATTTTAGTTGGG TTTTATA-3’, IGF-1-R: 5’-TCCCTTTAAAACACTATCTCATACTTTTTC-3’, IGF-1-S: 5’-ACACTATCTCATACTTTTTCT-3’. The PCR products were then processed and sequenced using PyroMark Q96 ID Pyrosequencing system (Qiagen, Germany). Pyro Q-CpG software was used to measure the percentage of methylation of each individual CpG.
Statistical analysis
All statistic analyses in this study were performed using SPSS 16.0 (SPSS, Chicago, IL, USA) and GraphPad Prism 5.0 software (GraphPad, San Diego, CA, USA). Data were presented as mean ± standard deviation (SD). Student’s t test and Mann-Whitney U test were used to compare the data in two groups. Comparisons of data among multiple groups were assessed by one-way analysis of variance (ANOVA) followed by Bonferroni correction for post hoc t-test and Kruskal-Wallis tests. The associations of IGF-1 among 5mC, DNMTs, 5hmC and TETs were determined by Pearson’s correlation. All P-values were two-sided and a P-value less than 0.05 was considered as statistical significance (P<0.05).
Results
Baseline characteristics of the study population
The clinical characteristics of the study population including 10 preeclamptic (PE) patients and 10 normal pregnant (NP) women were shown in Table 4. While no significant difference of maternal age was found between PE and NP groups, the preeclamptic mothers had shorter gestational weeks and higher systolic and diastolic blood pressure, and delivered newborns with lower birth weight. Although the expression of proteinuria with negative features (<0.3 g) was not accurately quantified in NP group, the mean value of proteinuria in PE group was 2.74 ± 0.70 and it was about ten-fold increase compared to NP group.
Table 4.
Baseline characteristics of the study population
| Items | Normal pregnancy | Preeclampsia | P-value |
|---|---|---|---|
| Maternal age (years) | 28.33 ± 0.62 | 29.37 ± 0.74 | 0.1362 |
| Gestational age (weeks) | 38.89 ± 0.13 | 37.27 ± 0.36 | 0.0009 |
| Systolic blood pressure (mmHg) | 119.0 ± 1.93 | 137.4 ± 3.69 | <0.0001 |
| Diastolic blood pressure (mmHg) | 75.75 ± 1.42 | 89.37 ± 2.81 | 0.0005 |
| Proteinuria (g/24 h) | - | 2.74 ± 0.70 | - |
| Infant birth-weight (g) | 3389 ± 71.61 | 2831 ± 126.3 | 0.0010 |
All results were analyzed by the Mann-Whitney U test and represented as mean ± SEM.
Decreased expression of IGF-1 in preeclamptic placentas and hypoxic trophoblasts
To determine the expression of IGF-1 protein, we performed IHC and WB on normal and preeclamptic placenta, and ELISA and WB on normoxic and hypoxic trophoblasts. To examine the levels of IGF-1 gene, we performed qRT-PCR on all the placenta and trophoblasts RNA samples. It was found that IGF-1 was primarily located in cytoplasm and significantly decreased in preeclamptic placentas when compared with normal placentas (Figure 1). Consistent with the changes of IGF-1 in the placentas, hypoxia remarkably inhibited the expression of IGF-1 in trophoblasts. As shown in Figure 2, the IGF-1 levels in supernatants of HTR8 cells were significantly lower in hypoxic conditions for 24 h and 48 h than those in normoxic conditions. It was also presented that the supernatants IGF-1 levels of JEG3 cells under hypoxia for 48 h and 72 h were apparently reduced compared to those under normal oxygen. The protein levels of IGF-1 in HTR8 and JEG3 cells in hypoxic conditions for 24 h were both obviously decreased compares with those in normoxic conditions, which was further confirmed by qRT-PCR analysis in HTR8 and JEG3 cells.
Figure 1.
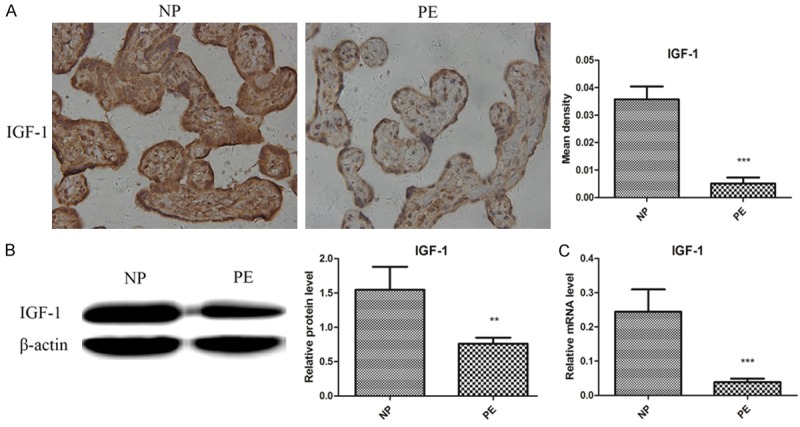
Reduced expression of IGF-1 in preeclamptic placentas. A. Immunohistochemistry revealed decreased levels of IGF-1 in PE group compared to NP group. B. Western blot showed lower expression of IGF-1 in PE group compared to NP group. C. qRT-PCR showed reduced levels of IGF-1 in PE group compared to NP group. Data were presented as Mean ± SD. **P<0.01, ***P<0.001. NP refers to normal pregnancy, PE refers to preeclampsia.
Figure 2.
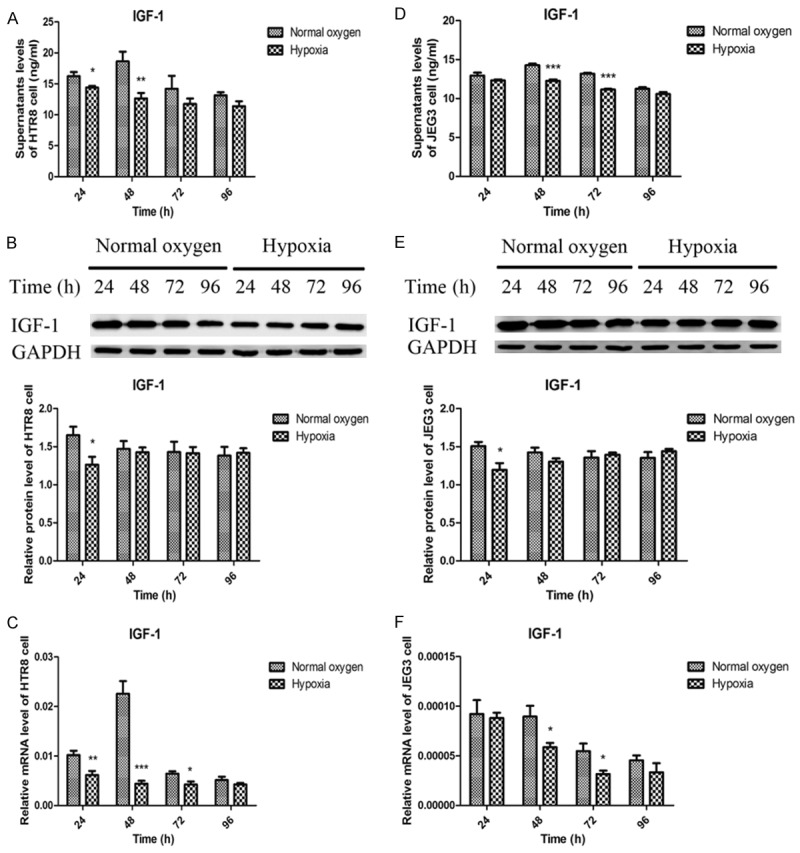
Decreased expression of IGF-1 in hypoxic HTR8 and JEG3 cells. A, D. ELISA showed decreased supernatants IGF-1 levels in hypoxic HTR8 and JEG3 cells, respectively. B, E. Western blot revealed reduced IGF-1 protein levels in hypoxic HTR8 and JEG3 cells, respectively. C, F. qRT-PCR depicted a reduction of IGF-1 mRNA levels in hypoxic HTR8 and JEG3 cells, respectively. Data were presented as Mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Hypermethylation of IGF-1 promoter leads to downregulation of IGF-1
Furthermore, we analyzed the DNA methylation status of IGF-1 promoter regions of normal and preeclamptic placentas using pyrophosphate sequencing. As shown in Figure 3, when compared to NP group, it was reported that the IGF-1 DNA methylation levels at two CpG islands had alterations with a significant upregulation at CpG_1 island and an increase trend at CpG_2 island in PE group. Analysis of IGF-1 promoter activity indicated that IGF-1 methylation plays an important role in its downregulation, and that hypermethylation may inhibit IGF-1 transcription.
Figure 3.
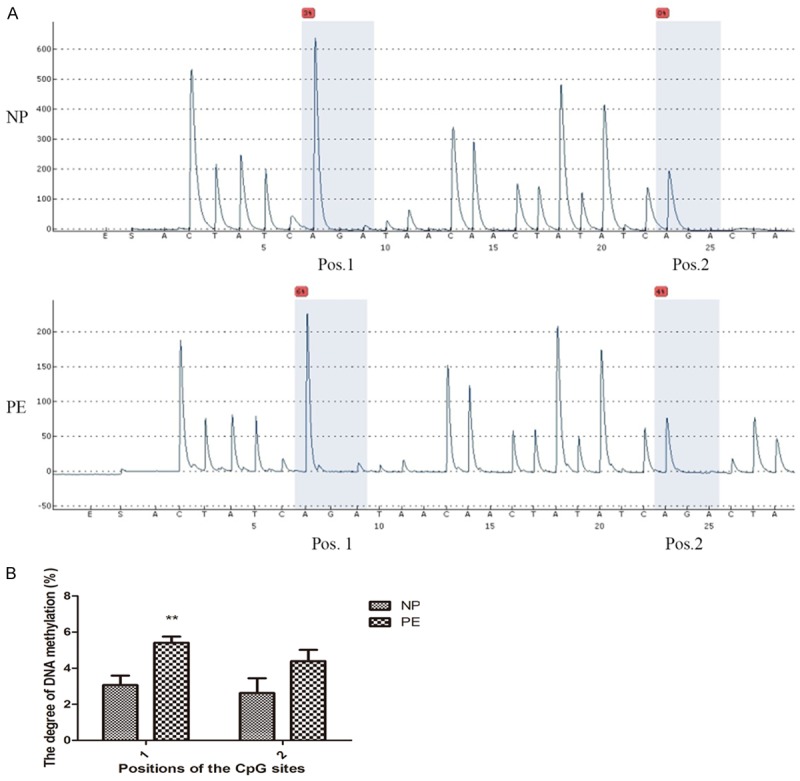
Hypermethylated IGF-1 promoter in preeclamptic placentas by pyrophosphate sequencing. A. Representative pyrosequencing images in NP and PE groups. B. The degree of DNA methylation of IGF-1 promoter at CpG_1 and CpG_2. Data were presented as Mean ± SD. **P<0.01. NP refers to normal pregnancy, PE refers to preeclampsia.
Preeclamptic placentas and hypoxic trophoblasts are hypermethylated and hypohydroxymethylated
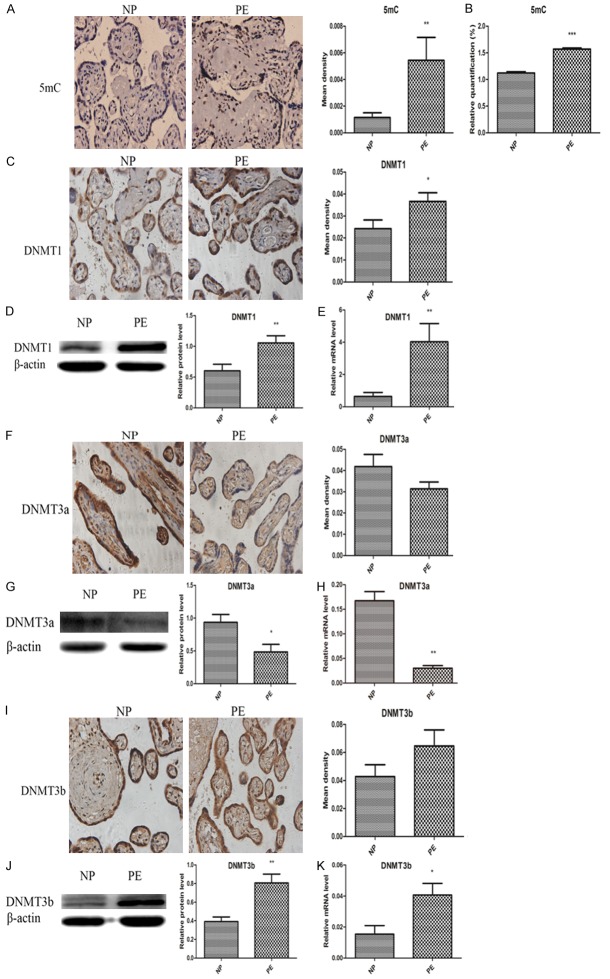
As the IGF-1 promoter region was hypermethylated in preeclamptic placentas, we further examined the overall DNA methylation and hydroxymethylation status by detecting the levels of 5mC and 5hmC, and their associated enzymes including DNMT1, DNMT3a, DNMT3b, TET1, TET2 and TET3 in the placentas and trophoblasts. As summarized in Figure 4, the results showed that the preeclamptic placentas were hypermethylated characterized by significant higher levels of 5mC, DNMT1 and DNMT3b, and lower expression of DNMT3a when compared with normal placentas, but the IHC findings of DNMT3a and DNMT3b were no different. It was also found that the preeclamptic placentas were hypohydroxymethylated with significant lower expression of 5hmC, TET1, TET2 and TET3 (Figure 5). Their alterations in HTR8 and JEG3 cells under normal oxygen and hypoxia from 24 h to 96 h were depicted in Figures 6 and 7. It was observed that the genomic DNA 5mC levels in hypoxic HTR8 and JEG3 cells were remarkably upregulated, except for the result in JEG3 cells for 24 h. The protein and mRNA levels of DNMT1 in HTR8 cells under hypoxic conditions for 48 h and 72 h, and in JEG3 cells under hypoxic conditions except for 24 h were both upregulated. Although the protein levels of DNMT3a in JEG3 cells were no significance, the protein and mRNA expression of DNMT3a in HTR8 cells and the mRNA levels of DNMT3a in JEG3 cells were almost consistent with the results in preeclamptic placentas. For DNMT3b, the protein levels in HTR8 and JEG3 cells were no difference. In contrast, the mRNA levels of DNMT3b in HTR8 and JEG3 cells under hypoxic conditions from 24 h to 72 h were apparently downregulated, however, they were both upregulated for 96 h. Likewise, the results in hypoxic HTR8 and JEG3 cells indicated significantly decreased expression of 5hmC and mRNA levels of TET1, TET2 and TET3, although varied times showed different findings. It was found that the protein levels of TET3 in hypoxic HTR8 and JEG3 cells from 48 h to 96 h were also downregulated, but no significance of the TET1 and TET2 protein levels were observed.
Figure 4.
Preeclamptic placentas were hypermethylated with higher expression of 5mC, DNMT1 and DNMT3b, and lower expression of DNMT3a compared with normal pregnant placentas. A, B. Immunohistochemistry and ELISA showed increased 5mC levels in PE group compared to NP group. C-E. Immunohistochemistry, western blot and qRT-PCR revealed higher DNMT1 in PE group than NP group. F-H. Immunohistochemistry showed no significance of DNMT3a, while western blot and qRT-PCR depicted lower DNMT3a in PE group than NP group. I-K. Immunohistochemistry showed no significance of DNMT3b, while western blot and qRT-PCR depicted higher DNMT3b in PE group than NP group. Data were shown as Mean ± SD. *P<0.05, **P<0.01, ***P<0.001. NP refers to normal pregnancy, PE refers to preeclampsia.
Figure 5.
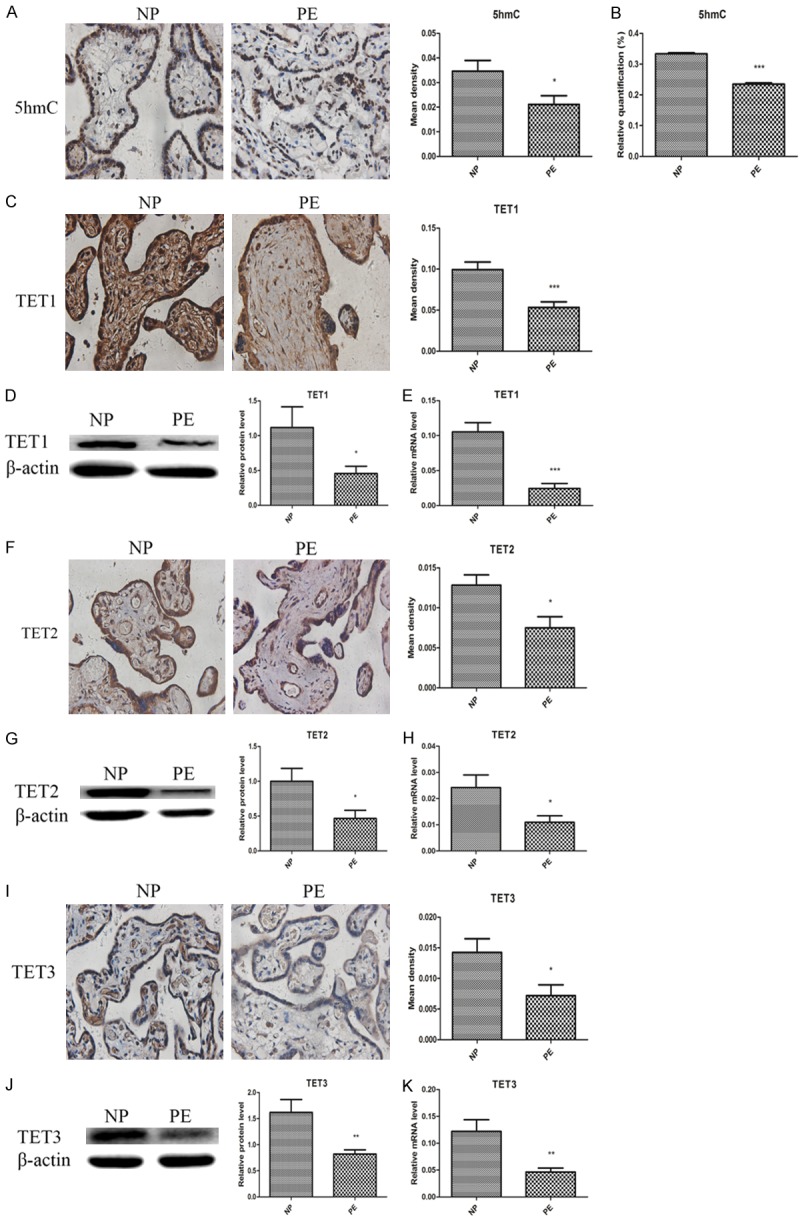
Preeclamptic placentas were hypohydroxymethylated with lower expression of 5hmC, TET1, TET2 and TET3 compared to normal pregnancy. A, B. Immunohistochemistry and ELISA showed decreased 5hmC levels in PE group compared to NP group. C-E. Immunohistochemistry, western blot and qRT-PCR revealed lower TET1 in PE group than NP group. F-H. Immunohistochemistry, western blot and qRT-PCR revealed lower TET2 in PE group than NP group. I-K. Immunohistochemistry, western blot and qRT-PCR revealed lower TET1 in PE group than NP group. Data were shown as Mean ± SD. *P<0.05, **P<0.01, ***P<0.001. NP refers to normal pregnancy, PE refers to preeclampsia.
Figure 6.
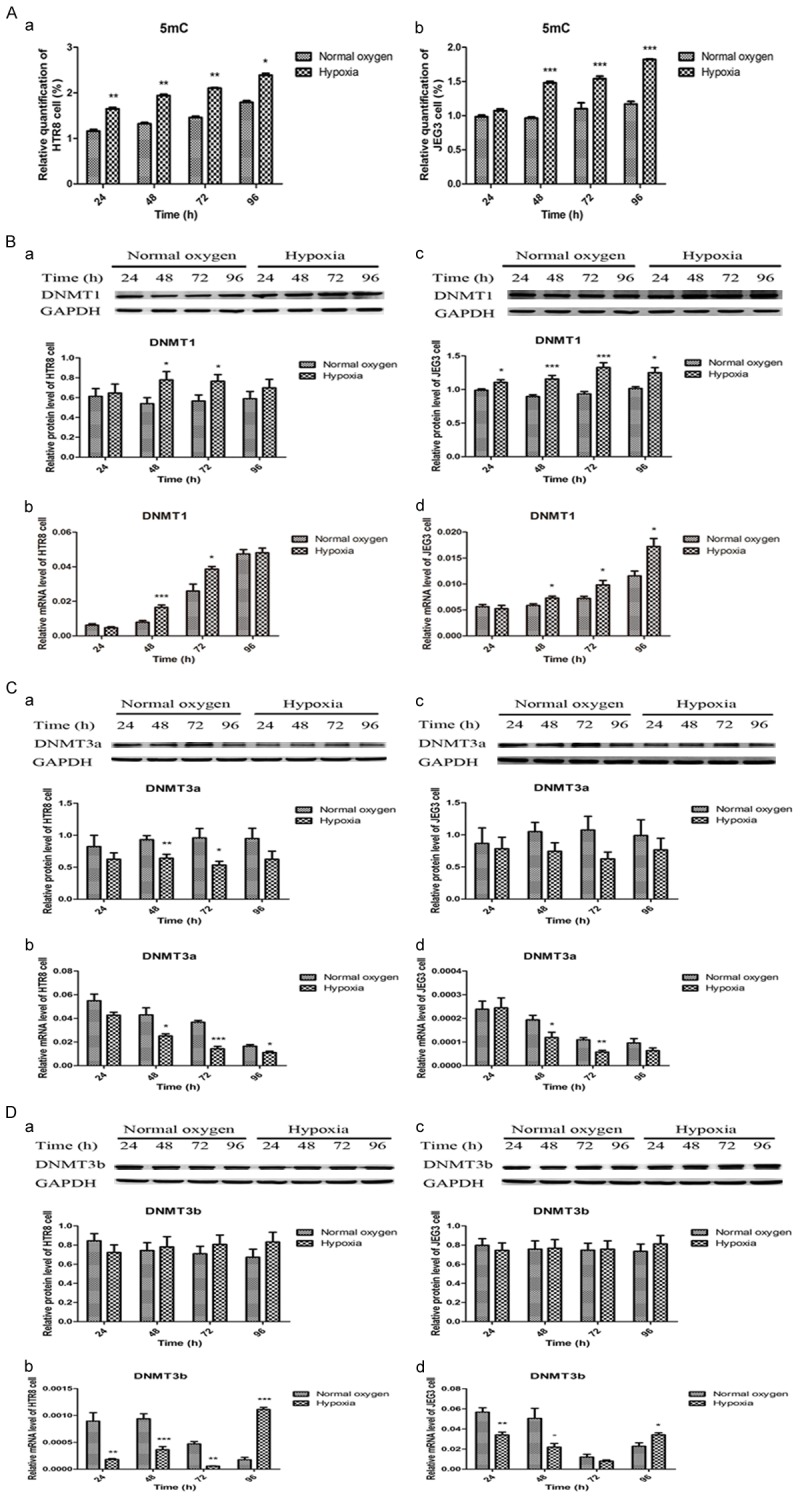
Hypoxic trophoblasts were hypermethylated with increased 5mC, DNMT1 and DNMT3b, and decreased DNMT3a compared to normoxic trophoblasts. A. Increased 5mC in hypoxic HTR8 and JEG3 cells. B. Increased DNMT1 in hypoxic HTR8 and JEG3 cells. C. Decreased DNMT3a in hypoxic HTR8 and JEG3 cells. D. Decreased DNMT3b mRNA levels from 24 h to 72 h and increased DNMT3b mRNA levels in 96 h in hypoxic HTR8 and JEG3 cells. Data were shown as Mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Figure 7.
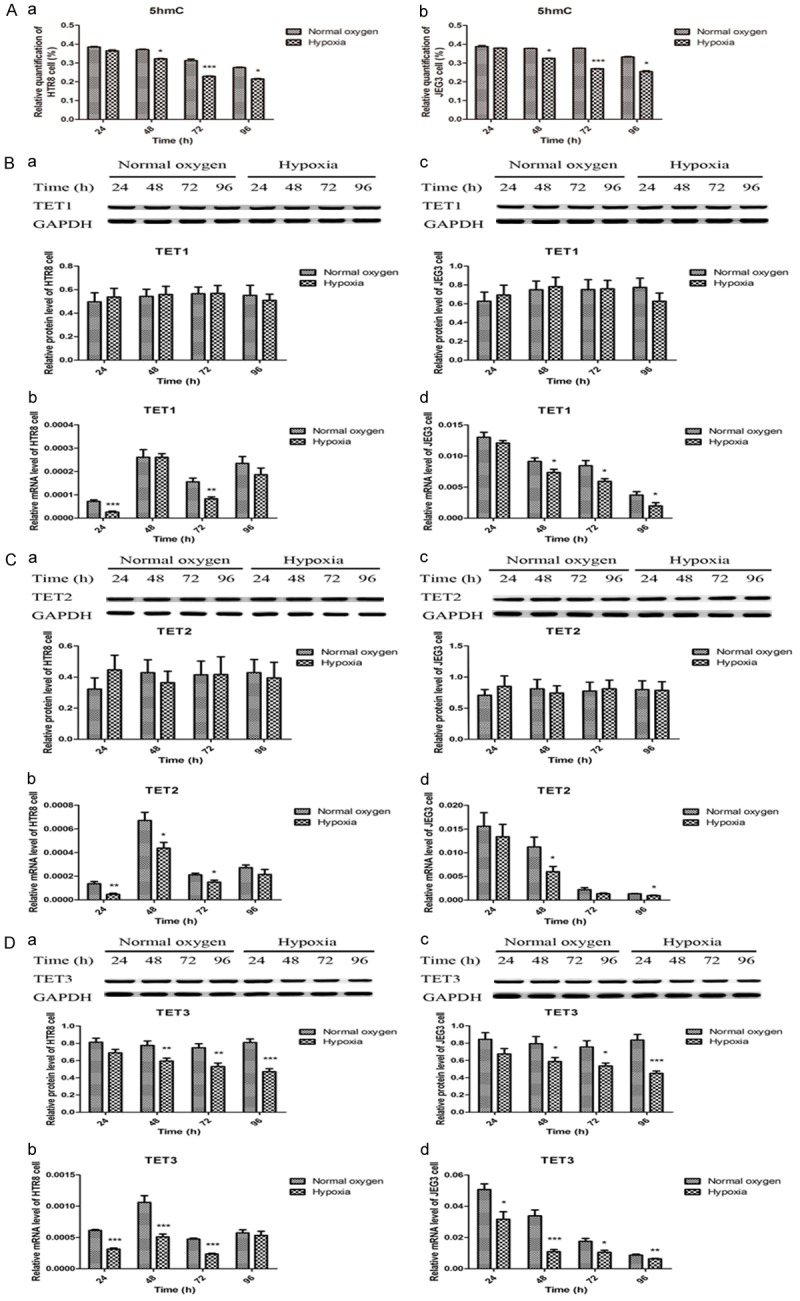
Hypoxic trophoblasts were hypohydroxyrmethylated with reduced 5mC, TET1, TET2 and TET3 compared to normoxic trophoblasts. A. Decreased 5hmC in hypoxic HTR8 and JEG3 cells. B. Decreased TET1 mRNA levels in hypoxic HTR8 and JEG3 cells. C. Decreased TET2 mRNA levels in hypoxic HTR8 and JEG3 cells. D. Decreased TET3 in hypoxic HTR8 and JEG3 cells. Data were shown as Mean ± SD. *P<0.05, **P<0.01, ***P<0.001.
Correlations between IGF-1 and 5mC, DNMTs, 5hmC, TETs
To analyze the relationships between IGF-1 and 5mC, DNMT1, DNMT3a, DNMT3b, 5hmC, TET1, TET2, TET3, we performed Pearson’s correlation analysis on the IHC data from the placenta samples. As shown in Figure 8, the results confirmed a statistically significant negative correlation between IGF-1 and 5mC, DNMT1 (R=-0.496, P=0.026; R=-0.475, P=0.034, respectively), and positive correlation between IGF-1 and 5hmC, TET1, TET2, TET3 (R=0.471, P=0.036; R=0.764, P<0.001; R=0.529, P=0.016; R=0.587, P=0.007, respectively). Therefore, we firstly focused on the potential effects of DNMT1 on IGF-1 promoter methylation.
Figure 8.
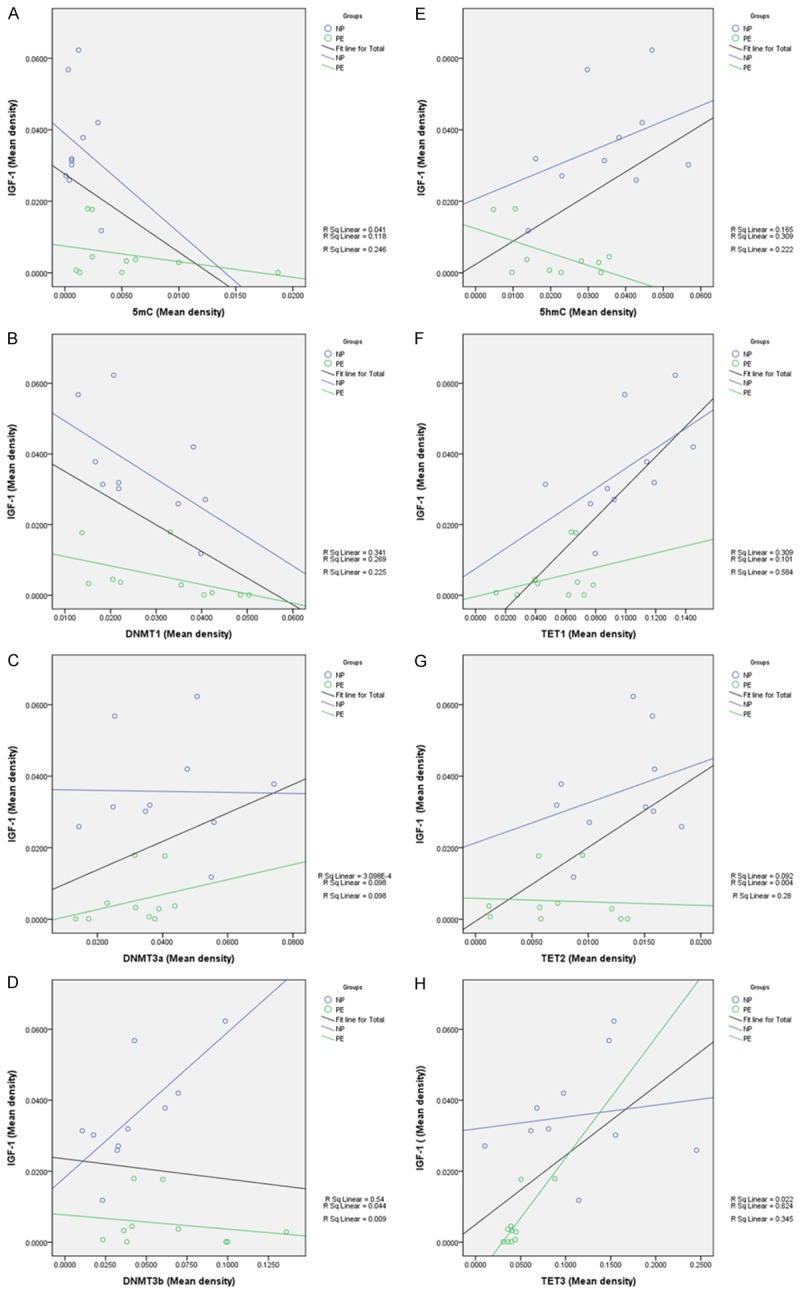
IGF-1 was negatively correlated with 5mC and DNMT1, and positively correlated with 5hmC, TET1, TET2 and TET3. A. IGF-1 and 5mC: R=-0.496, P=0.026. B. IGF-1 and DNMT1: R=-0.475, P=0.034. C. IGF-1 and DNMT3a: P=0.178. D. IGF-1 and DNMT3b: P=0.683. E. IGF-1 and 5hmC: R=0.471, P=0.036. F. IGF-1 and TET1: R=0.764, P<0.001. G. IGF-1 and TET2: R=0.529, P=0.016. H. IGF-1 and TET3: R=0.587, P=0.007.
DNMT1 downregulation results in upregulation of IGF-1 and hypomethylation of IGF-1 promoter
To investigate the potentially causal relationship between IGF-1 expression and its promoter methylation, we treated HTR8 cells with a DNMT inhibitor, 5-Aza-dc and a DNMT1 specific inhibitor, DNMT1-siRNA. As expected, 4 µM 5-Aza-dc treatment significantly increased the expression of IGF-1 and led to a significant reduction in IGF-1 promoter methylation at two CpG islands (Figure 9A, 9B and 9F) compared to the Blank control group. Moreover, an increase in IGF-1mRNA and protein, and a decrease in its promoter methylation at CpG_1 island were observed following DNMT1-siRNA treatment (Figure 9C, 9D and 9F).
Figure 9.
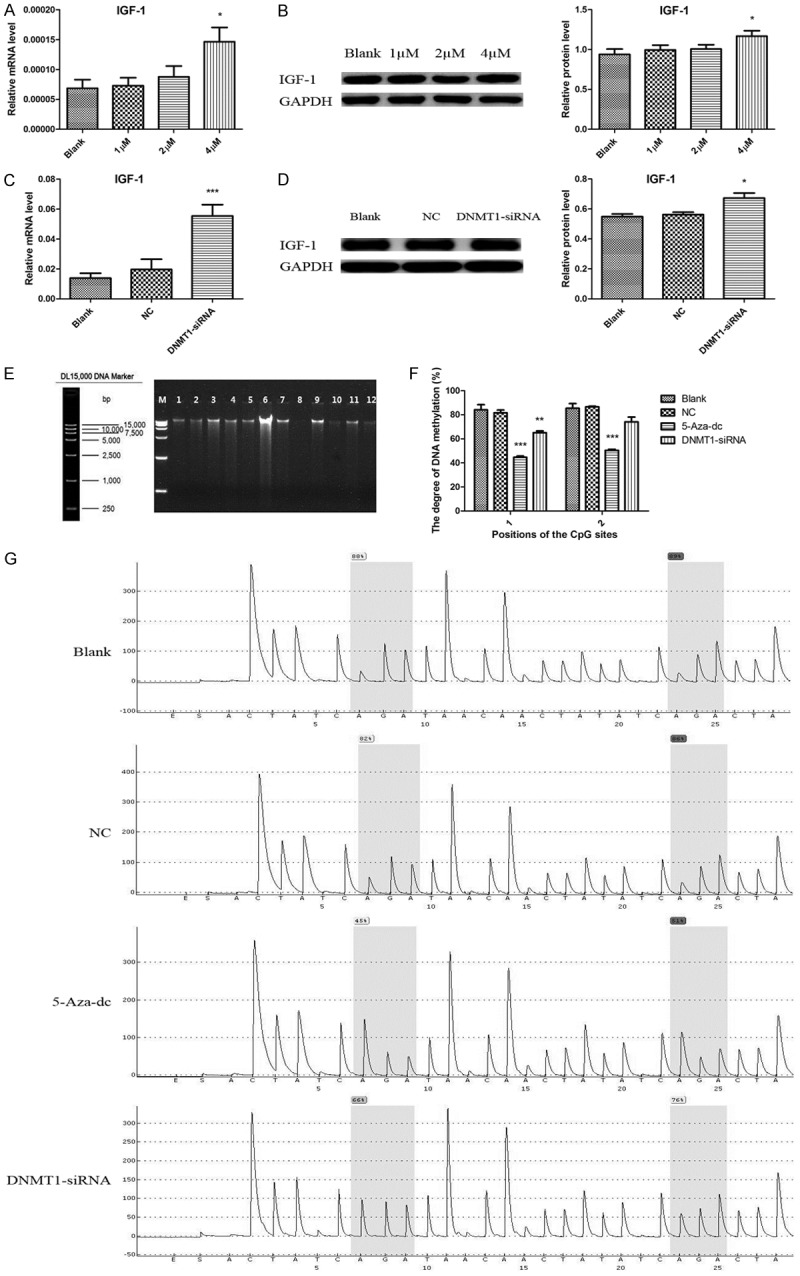
IGF-1 was upregulated caused by its hypomethylated promoter region in HTR8 cells treated with 5-Aza-dc and DNMT1-siRNA. A, B. Increased IGF-1 in HTR8 cells treated with 4µM 5-Aza-dc. C, D. Increased IGF-1 in HTR8 cells treated with DNMT1-siRNA. E. Representative image of DNA integrity verified by agarose gel electrophoresis. F. IGF-1 promoter region was hypomethylated at CpG_1 and CpG_2 in HTR8 cells treated with 5-Aza-dc and DNMT1-siRNA. G. Representative pyrosequencing images of IGF-1 in four groups. Data were shown as Mean ± SD. *P<0.05, **P<0.01, ***P<0.001. NC refers to negative control.
IGF-1 doesn’t alter genomic DNA methylation or hydroxymethylation
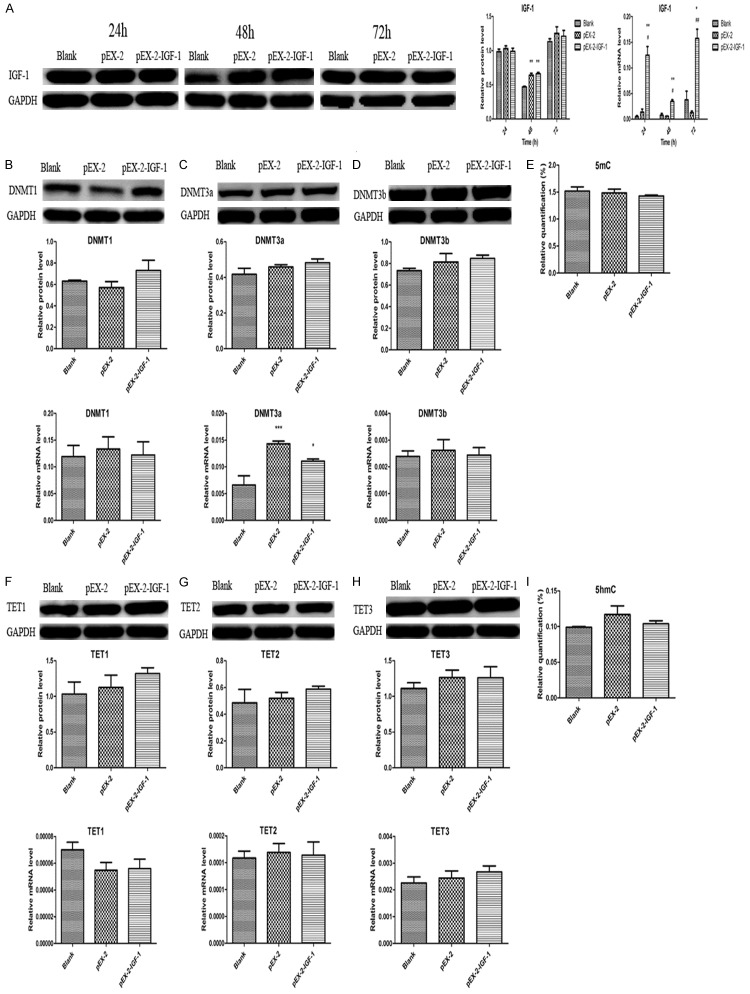
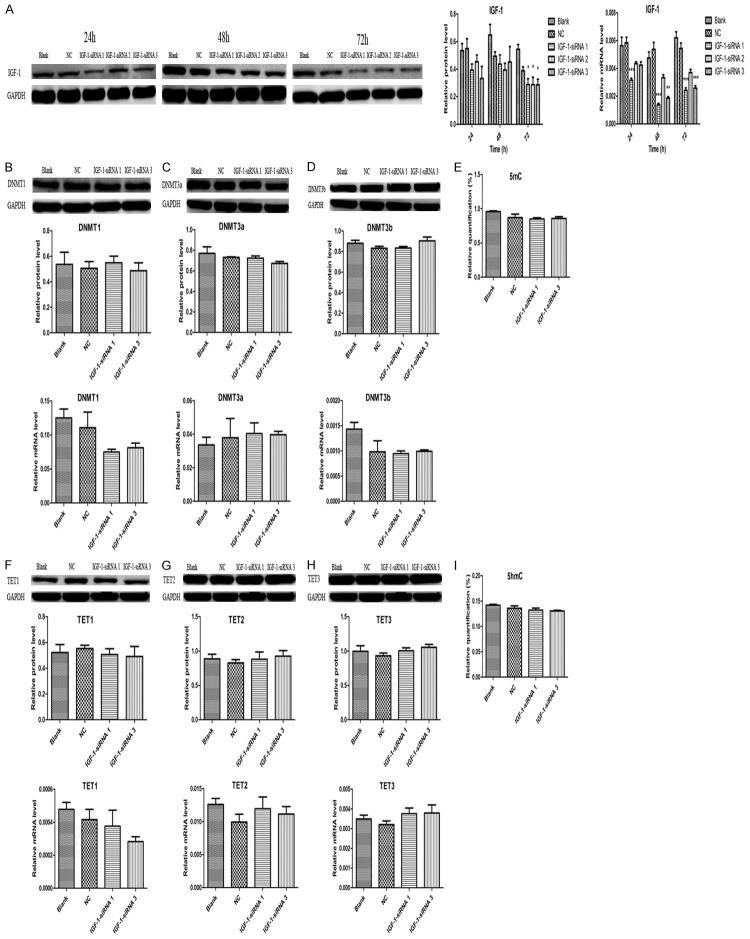
Simultaneously, we specifically upregulated and downregulated IGF-1 gene expression in HTR8 cells via a plasmid vector, pEX-2-IGF-1 and a IGF-1 inhibitor, IGF-1-siRNA to evaluate the effects of IGF-1 on genomic DNA methylation and hydroxymethylation by detecting the levels of 5mC, DNMT1, DNMT3a, DNMT3b, 5hmC, TET1, TET2 and TET3. We treated HTR8 cells with pEX-2-IGF-1 and IGF-1-siRNA for 48 h according to the results shown in Figures 10A and 11A. It was clearly found that an upregulation of IGF-1 didn’t alter DNA methylation and hydroxymethylation with no significance of 5mC, 5hmC and their related enzymes. Although the mRNA level of DNMT3a in pEX-2 and pEX-2-IGF-1 group was higher than that in Blank group, we couldn’t exclude the influence of the blank plasmid vector pEX-2 (Figure 10). The results in Figure 11 also showed that the effects of an downregulation of IGF-1 on DNA methylation and hydroxymethylation were consistent with the findings of an upregulation of IGF-1.
Figure 10.
Upregulation of IGF-1 didn’t alter genomic DNA methylation. A. IGF-1 was upregulated by pEX-2-IGF-1. B. Upregulation of IGF-1 didn’t alter DNMT1 expression. C. Upregulation of IGF-1 didn’t alter DNMT3a expression. D. Upregulation of IGF-1 didn’t alter DNMT3b expression. E. Upregulation of IGF-1 didn’t alter 5mC levels. F. Upregulation of IGF-1 didn’t change TET1 expression. G. Upregulation of IGF-1 didn’t change TET2 expression. H. Upregulation of IGF-1 didn’t change TET3 expression. I. Upregulation of IGF-1 didn’t alter 5hmC levels. Data were shown as Mean ± SD. *P<0.05, **P<0.01, #P<0.05, ##P<0.01.
Figure 11.
Downregulation of IGF-1 didn’t change genomic DNA hydroxymethylation. A. IGF-1 was downregulated by IGF-1-siRNA. B. Downregulation of IGF-1 didn’t change DNMT1 expression. C. Downregulation of IGF-1 didn’t change DNMT3a expression. D. Downregulation of IGF-1 didn’t change DNMT3b expression. E. Downregulation of IGF-1 didn’t change 5mC levels. F. Downregulation of IGF-1 didn’t alter TET1 expression. G. Downregulation of IGF-1 didn’t alter TET2 expression. H. Downregulation of IGF-1 didn’t alter TET3 expression. I. Downregulation of IGF-1 didn’t alter 5hmC levels. Data were shown as Mean ± SD. *P<0.05, **P<0.01, ***P<0.001. NC refers to negative control.
Discussion
In the present study, the findings showed a significant reduction of IGF-1 in preeclamptic placentas and hypoxic trophoblasts, and a remarkable hypermethylation of IGF-1 promoter region in preeclamptic placentas. Simultaneously, it was found that the preeclamptic placentas and hypoxic trophoblasts were hypermethylated and hypohydroxymethylated with their associated enzymes altered. Furthermore, it was observed that DNMT1 upregulation directly resulted in hypermethylation of the IGF-1 gene, eventually leading to its decreased expression. To our knowledge, this is the first study to investigate the role of IGF-1 from the perspectives of dynamic balance of DNA methylation and hydroxymethylation to explore the possible mechanisms of preeclampsia.
IGF-1 is considered to be strongly associated with the behaviors of cells, the formation of placenta and the growth of fetus [7-9]. Previous studies from domestic and abroad reported inconsistent results of IGF-1 in preeclampsia [10-13]. Most researches have concluded a significant reduction of IGF-1 in preeclamptic maternal placentas, umbilical cord blood and maternal serum compared with normal pregnant women [10-12]. This is consistent with the results of IGF-1 in preeclamptic placentas and hypoxic trophoblast cells in our current study. However, Bartha and colleagues pointed out that serum IGF-1 in preeclamptic patients and women in the control group were at the same level [13], which might be associated with the composition of the disease in the case group.
DNA methylation is a form of epigenetic regulation and commonly leads to suppressed gene expression when occurring in a regulatory region [27]. Therefore, we investigated whether IGF-1 transcription was regulated by DNA methylation in the context of CpG dinucleotides [28]. As our findings demonstrated, the hypermethylation of IGF-1 promoter region in preeclamptic placentas was responsible for the downregulation of IGF-1. This is similar to published studies in some tumor tissues [22,23], but different from some PE-related genes like GATAD1, GNA12 and HLA-G [3,19,20]. In fact, epigenetic regulation by DNA methylation is a complex issue involving concerted actions by multiple ciselements, DNA-binding factors such as transcriptional activators/repressors, methy-CpG-binding domain (MDB) factors and a range of histone modifications. It has been reported that various malignant tumors such as ovarian cancer, prostate cancer and breast cancer were hypermethylated due to overexpressed DNMTs [29-31]. Interesting, it has been studied that DNA damage might alter the specificity of DNMT1, either inhibiting the methylation of hemimethylated sites or triggering the inappropriate methylation of previously unmethylated sites and it has been confirmed that reduced DNMT1 selectively resulting from DNA damage could cause heritable changes in cytosine methylation patterns, resulting in human tumor formation [32]. Moreover, oxidative damage to DNA could therefore result in heritable, epigenetic changes in chromation organization [33]. Previous studies provided evidence that miR-148a/152 could directly target DNMT1 by binding its 3’-UTR regions, thereby suppressing DNMT1 mRNA and protein expression, and also demonstrated that overexpression of DNMT1 led to the hypermethylation of miR-148a/152 genes, and downregulation of miR-148a/152. These results established the existence of a negative feedback regulatory loop between DNMT1 and miR-148a/152 in the HTR-8 cells and the placenta of PE rats [21]. In additional, Sichen et al. [34] found that DNA methylation can affect miRNA expression. However, the mechanism responsible for DNMTs alteration is still largely unknown and needs further to be investigated in the future.
As previously known, the conversion of 5mC to 5hmC was involved in the pathogenesis of preeclampsia and many PE-related genes were hypermethylated or hypomethylated [3,19,20,24]. We proposed that the cumulative effects of these genes might alter the status of DNA methylation and hydroxymethylation determined by their associated enzymes like DNMTs and TETs. Therefore, we detected the levels of 5mC, DNMT1, DNMT3a, DNMT3b, 5hmC, TET1, TET2 and TET3 in preeclamptic placentas and hypoxic trophoblasts. It was found that the preeclamptic placentas were higher methylated and lower hydroxymethylated characterized by significant higher levels of 5mC, DNMT1 and DNMT3b, and lower expression of DNMT3a, 5hmC, TET1, TET2 and TET3 compared to normal placentas, but the IHC findings of DNMT3a and DNMT3b were no different, which were almost consistent with the results shown in hypoxic trophoblasts. In a previous study, it was not observed any place at the gene body or promoter region with opposite 5mC/5hmC levels using genome-wide mapping in the late-onset severe preeclampsia group and the control group, and the variation trend of the 5mC/5hmC levels had no significant differences between the two groups [17]. However, differential 5mC and 5hmC peaks were found showing significant difference between the two groups [17]. In agreement with our results, the findings of several studies also showed higher 5mC and lower 5hmC levels in preeclampsia [35-37]. On the contrary, Noruma et al. [38] reported that global methylation levels in the placenta were lower in patients with preeclampsia. Simultaneously, Kogg et al. also showed a reduction of 5mC in the early-onset preeclampsia [39], which might be associated with the use of antihypertensive drugs and hormones promoting fetal lung maturation. Although most studies acknowledged that DNMT1 was upregulated in preeclamptic placentas [20], the results in several researches indicated significantly decreased DNMT1 mRNA expression or no difference [21,40]. The downregulation of DNMT3a and upregulation of DNMT3b in the current study were inconsistent with previous reports with no significant difference of DNMT3a and DNMT3b [20,21]. These differences might be associated with the samples, the primary antibodies selected or the methodologies used to detected DNMTs levels. However, the levels of TET1, TET2 and TET3 were unanimously reduced in preeclampsia [35], which was reported to regulate 5hmC levels [41-43].
Furthermore, we analyzed the correlations between IGF-1 and 5mC, DNMT1, DNMT3a, DNMT3b, 5hmC, TET1, TET2 and TET3 in the placentas. The results confirmed a statistically significant negative correlation between IGF-1 and 5mC, DNMT1, and positive correlation between IGF-1 and 5hmC, TET1, TET2, TET3. Therefore, we firstly focused on the potential effects of DNMT1 on IGF-1 promoter methylation according to previous studies [20,21]. To confirm the effects of DNMT1 on IGF-1 expression and its promoter methylation, we selected a DNMT inhibitor, 5-Aza-dc and a DNMT1 specific inhibitor, DNMT1-siRNA to treat HTR8 cells. The results in the present study showed that 5-Aza-dc and DNMT1-siRNA could significantly upregulate the expression of IGF-1 by reducing the levels of its promoter methylation. In JAR cell line, 5-Aza-dc treatment lead to a significant reduction of GATAD1 methylation compared to the control group [3]. Consistently, previous researches indicated that DNMT1 participated in HLA-G gene silencing through DNA methylation and verified that DNMT1 played a vital role in FABP4 hypomethylation in HT8 cell line [20,21]. Moreover, previous studies also showed that DNMT1 overexpression contributed to promoter hypermethylation, and was associated with malignant potential and poor prognosis in human cancer [44]. Therefore, it was considered that DNMT1 was closely correlated with the methylated CpG island phenotype. Combined with other transcription factors, increased levels of DNMT1 might cause the hypermethylation of IGF-1 promoter region and thus suppress IGF-1 transcription. At the same time, we did not observe significant effects of IGF-1 on DNA methylation and hydroxymethylation with no difference of 5mC, DNMT1, DNMT3a, DNMT3b, 5hmC, TET1, TET2 and TET3 in HTR8 cell line, indicating that IGF-1 might play no role in DNA methylation and hydroxymethylation. However, further research was needed to testify this hypothesis in the future.
In the current study, we explored the new area by assessing the expression of IGF-1, its promoter methylation, and its effects on genomic DNA methylation and hydroxymethylation in clinical placenta samples as well as trophoblast cell lines. However, there are some limitations in the current study. First of all, we couldn’t conclude that IGF-1 was a cause or a consequence of preeclampsia because the study population selected were mostly in the late stages of pregnancy, while the onset of preeclampsia began during early pregnancy. Although we tend to consider IGF-1 as a cause of preeclampsia, further studies are needed to explore whether IGF-1 is a cause of preeclampsia and how to lead to the development of preeclampsia. In addition, the harvested placental samples might have an influence on the results due the purity and cell proportions [20]. Moreover, although the expression of DNMT3a and DNMT3b were controversial in different studies and the correlations between IGF-1 and DNMT3a, DNMT3b were not significant in our study, whether DNMT3a and DNMT3b could result in the reduced expression of IGF-1 or not, which was not investigated in the present study. Also, the roles of TET1, TET2 and TET3 on IGF-1 were not reported here, which should be interpreted in further studies. Last but not the least, although oxidative damage and DNA damage might be contributed to abnormal expression of DNMT1 [32,33], the possible upper stream mechanisms of abnormal expression of DNMTs and TETs were still not very clear, which still needs further studies in the future.
In summary, our findings suggest that IGF-1 were significantly reduced in preeclamptic placentas and hypoxic trophoblasts, which was caused by its hypermethylated promoter region due to increased expression of DNMT1. At the same time, it was found that preeclamptic placentas and hypoxic trophoblasts were higher rmethylated and lower hydroxymethylated accompanied by altered associated enzymes such as DNMTs and TETs. This study provided a new mechanism of reduced IGF-1 in preeclamptic placentas and hypoxic trophoblasts, implying the probable role of IGF-1 in the development of preeclampsia. These findings provided new insights into the diagnosis and treatment of preeclampsia.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No: 81300506, 81200449), National Basic Research Program of China (2015CB943300), National Key Research and Development Program of Reproductive Health & Major Birth Defects Control and Prevention (2016YFC1000400), National Science Fund of Shanghai, China (12ZR1403700), Shanghai Excellent Young Specialist Project (2016) and Shanghai Excellent Residence Project (2013). We would also like to show our great appreciation to Bank Tissue of Obstetrics and Gynecology Hospital of Fudan University for their help in collecting clinical samples.
Disclosure of conflict of interest
None.
References
- 1.Mol BW, Roberts CT, Thangaratinam S, Magee LA, de Groot CJ, Hofmeyr GJ. Pre-eclampsia. Lancet. 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 2.American College of Obstetricians and Gynecologists, Task Force on Hypertension in Pregnancy. Hypertension in pregnancy. Report of the American college of obstetricians and gynecologists’ task force on hypertension in pregnancy. Obstet Gynecol. 2013;122:1122–1131. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- 3.Ma XL, Li JP, Brost B, Cheng WJ, Jiang SW. Decreased expression and DNA methylation levels of GATAD1 in preeclamptic placentas. Cell Signal. 2014;26:959–967. doi: 10.1016/j.cellsig.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rana S, Hacker MR, Modest AM, Salahuddin S, Lim KH, Verlohren S, Perschel FH, Karumanchi SA. Circulating angiogenic factors and risk of adverse maternal perinatal outcomes in twin pregnancies with suspected preeclampsia. Hypertension. 2012;60:451–458. doi: 10.1161/HYPERTENSIONAHA.112.195065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jääskeläinen T, Heinonen S, Kajantie E, Kere J, Kivinen K, Pouta A, Laivuori H FINNPEC Study Group. Cohort profile: the finnish genetics of pre-eclampsia consortium (FINNPEC) BMJ Open. 2016;6:e013148. doi: 10.1136/bmjopen-2016-013148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang M, Du H, Han B, Xia G, Shi X, Zhang F, Fu Q, Zhang T. Hypoxia-inducible microRNA-218 inhibits trophoblast invasion by targeting LASP1: Implications for preeclampsia development. Int J Biochem Cell Biol. 2017;87:95–103. doi: 10.1016/j.biocel.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Crosley EJ, Dunk CE, Beristain AG, Christians JK. IGFBP-4 and -5 are expressed in first-trimester villi and differentially regulate the migration of HTR-8/SVneo cells. Reprod Biol Endocrinol. 2014;12:123. doi: 10.1186/1477-7827-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vatten LJ, Nilsen TI, Juul A, Jeansson S, Jenum PA, Eskild A. Changes in circulating level of IGF-1 and IGF-binding protein-1 from the first to second trimester as predictors of preeclampsia. Eur J Endocrinol. 2008;158:101–105. doi: 10.1530/EJE-07-0386. [DOI] [PubMed] [Google Scholar]
- 9.Lauszus FF, Fuglsang J. IGF-1 is associated with fetal growth and preterm delivery in type 1 diabetic pregnancy. Gynecol Endocrinol. 2016;32:488–491. doi: 10.3109/09513590.2015.1134477. [DOI] [PubMed] [Google Scholar]
- 10.Dubova EA, Pavlov KA, Lyapin VM, Kulikova GV, Shchyogolev AI, Sukhikh GT. Expression of insulin-like growth factors in the placenta in preeclampsia. Bull Exp Biol Med. 2014;157:103–107. doi: 10.1007/s10517-014-2502-4. [DOI] [PubMed] [Google Scholar]
- 11.Kharb S, Nanda S. Patterns of biomarkers in cord blood during pregnancy and preeclampsia. Curr Hypertens Rev. 2017;13:57–64. doi: 10.2174/1573402113666170126101914. [DOI] [PubMed] [Google Scholar]
- 12.Sifakis S, Akolekar R, Kappou D, Mantas N, Nicolaides KH. Maternal serum insulin-like growth factor-I at 11-13 weeks in preeclampsia[J] . Prenatal Diagnosis. 2010;30:1026–1031. doi: 10.1002/pd.2555. [DOI] [PubMed] [Google Scholar]
- 13.Bartha JL, Romero-Carmona R, Torrejon-Cardoso R, Comino-Delgado R. Insulin, insulin-like growth factor-1, and insulin resistance in women with pregnancy-induced hypertension. Am J Obstet Gynecol. 2002;187:735–740. doi: 10.1067/mob.2002.126283. [DOI] [PubMed] [Google Scholar]
- 14.Forbes K, Shah VK, Siddals K, Gibson JM, Aplin JD, Westwood M. Statins inhibit insulin-like growth factor action in first trimester placenta by altering insulin-like growth factor 1 receptor glycosylation. Mol Hum Reprod. 2015;21:105–114. doi: 10.1093/molehr/gau093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayama R, Izawa T, Sakai K, Suciu N, Iwashita M. Improvement of insulin sensitivity promotes extravillous trophoblast cell migration stimulated by insulin-like growth factor-I. Endocr J. 2013;60:359–368. doi: 10.1507/endocrj.ej12-0241. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Tan R, MacCalman CD, Eastabrook G, Park SH, Dutz JP, von Dadelszen P. IFN-gamma-mediated extravillous trophoblast outgrowth inhibition in first trimester explant culture: a role for insulin-like growth factors. Mol Hum Reprod. 2008;14:281–289. doi: 10.1093/molehr/gan018. [DOI] [PubMed] [Google Scholar]
- 17.Zhu L, Lv R, Kong L, Cheng H, Lan F, Li X. Genome-wide mapping of 5mC and 5hmC identified differentially modified genomic regions in late-onset severe preeclampsia: a pilot study. PLoS One. 2015;10:e0134119. doi: 10.1371/journal.pone.0134119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 19.Ye W, Shen L, Xiong Y, Zhou Y, Gu H, Yang Z. Preeclampsia is associated with decreased methylation of the GNA12 promoter. Ann Hum Genet. 2016;80:7–10. doi: 10.1111/ahg.12136. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Liu HY, Li H, Peng T, Gu WR, Li XT. Hypermethylation of the HLA-G promoter is associated with preeclampsia. Mol Hum Reprod. 2015;21:736–744. doi: 10.1093/molehr/gav037. [DOI] [PubMed] [Google Scholar]
- 21.Yang A, Zhang H, Sun Y, Wang Y, Yang X, Yang X, Zhang H, Guo W, Zhu G, Tian J, Jia Y, Jiang Y. Modulation of FABP4 hypomethylation by DNMT1 and its inverse interaction with miR-148a/152 in the placenta of preeclamptic rats and HTR-8 cells. Placenta. 2016;46:49–62. doi: 10.1016/j.placenta.2016.08.086. [DOI] [PubMed] [Google Scholar]
- 22.Christopoulos PF, Msaouel P, Koutsilieris M. The role of the insulin-like growth factor-1 system in breast cancer. Mol Cancer. 2015;14:43–56. doi: 10.1186/s12943-015-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brahmkhatri VP, Prasanna C, Atreya HS. Insulin-like growth factors system in cancer: novel targeted therapies. Biomed Res Int. 2015;2015:538019. doi: 10.1155/2015/538019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuen RK, Chen B, Blair JD, Robinson WP, Nelson DM. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics. 2013;8:192–202. doi: 10.4161/epi.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma M, Chen XY, Li B, Li XT. Melatonin protects premature ovarian insufficiency induced by tripterygium glycosides: role of SIRT1. Am J Transl Res. 2017;9:1580–1602. [PMC free article] [PubMed] [Google Scholar]
- 27.Mao LY, Zhou QJ, Zhou SF, Wlibur RR, Li XT. Roles of apolipoprotein E (ApoE) and inducible nitric oxide synthase (iNOS) in inflammation and apoptosis in preeclampsia pathogenesis and progression. PLoS One. 2013;8:e58168. doi: 10.1371/journal.pone.0058168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–628. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardenas H, Vieth E, Lee J, Segar M, Liu Y, Nephew KP, Matei D. TGF-beta induces global changes in DNA methylation during the epithelial-tomesenchymal transition in ovarian cancer cells. Epigenetics. 2014;9:1461–1472. doi: 10.4161/15592294.2014.971608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue G, Ren ZX, Chen YX, Zhu JY, Du YR, Pan D, Li XM, Hu BR. A feedback regulation between miR-145 and DNA methyltransferase 3b in prostate cancer cell and their responses to irradiation. Cancer Lett. 2015;361:121–127. doi: 10.1016/j.canlet.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 31.Kala R, Shah HN, Martin SL, Tollefsbol TO. Epigenetic-based combinatorial resveratrol and pterostilbene alters DNA damage response by affecting SIRT1 and DNMT enzyme expression, including SIRT1-dependent gamma-H2AX and telomerase regulation in triple-negative breast cancer. BMC Cancer. 2015;15:672. doi: 10.1186/s12885-015-1693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 33.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Chowdhury R, Liu F, Chou AP, Li T, Mody RR, Lou JJ, Chen W, Reiss J, Soto H, Prins R, Liau LM, Mischel PS, Nghiemphu PL, Yong WH, Cloughesy TF, Lai A. Tumor-suppressive miR148 is silenced by CpG island hypermethylation in IDH1-mutant gliomas. Clin Cancer Res. 2014;20:5808–5822. doi: 10.1158/1078-0432.CCR-14-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Tang Y, Liu X, Zhou Q, Xiao X, Lan F, Li X, Hu R, Xiong Y, Peng T. 14-3-3 tau (YWHAQ) gene promoter hypermethylation in human placenta of preeclampsia. Placenta. 2014;35:981–988. doi: 10.1016/j.placenta.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Kulkarni A, Mehendale S, Pisal H, Kilari A, Dangat K, Salunkhe S, Taralekar V, Joshi S. Association of omega-3 fatty acids and homocysteine concentrations in preeclampsia. Clin Nutr. 2011;30:60–64. doi: 10.1016/j.clnu.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Gao WL, Li D, Xiao ZX, Liao QP, Yang HX, Li Y, Ji L, Wang YL. Dection of global DNA methylation and paternally imprinted H19 gene methylation in preeclamptic placntas. Hypertens Res. 2011;34:655–661. doi: 10.1038/hr.2011.9. [DOI] [PubMed] [Google Scholar]
- 38.Nomura Y, Lambertini L, Rialdi A, Lee M, Mystal EY, Grabie M, Manaster I, Huynh N, Finik J, Davey M, Davey K, Ly J, Stone J, Loudon H, Eglinton G, Hurd Y, Newcorn JH, Chen J. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci. 2014;21:131–137. doi: 10.1177/1933719113492206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogg K, Blair JD, von Dadelszen P, Robinson WP. Hypomethylation of the LEP gene in placenta and elevated maternal leptin concentration in early onset preeclampsia. Mol Cell Endocrinol. 2013;367:64–73. doi: 10.1016/j.mce.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta. 2007;28:487–497. doi: 10.1016/j.placenta.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JJ, Zhu Y, Zhu Y, Wu JL, Liang WB, Zhu R, Xu ZK, Du Q, Miao Y. Association of increased DNA methyltransferase expression with carcinogenesis and poor prognosis in pancreatic ductal adenocarcinoma. Clin Transl Oncol. 2012;14:116–124. doi: 10.1007/s12094-012-0770-x. [DOI] [PubMed] [Google Scholar]