Abstract
Phylogenetic analyses of a combined DNA data matrix containing ITS, LSU, rpb2 and tub2 sequences of representative Xylariales revealed that the genus Barrmaelia is a well-defined monophylum, as based on four of its described species (B. macrospora, B. moravica, B. oxyacanthae, B. rhamnicola) and the new species B. rappazii. The generic type of Entosordaria, E. perfidiosa, is revealed as the closest relative of Barrmaelia, being phylogenetically distant from the generic type of Clypeosphaeria, C. mamillana, which belongs to Xylariaceae sensu stricto. Entosordaria and Barrmaelia are highly supported and form a distinct lineage, which is recognised as the new family Barrmaeliaceae. The new species E. quercina is described. Barrmaelia macrospora, B. moravica and B. rhamnicola are epitypified and E. perfidiosa is lecto- and epitypified. Published sequences of Anthostomella and several Anthostomella-like species from the genera Alloanthostomella, Anthostomelloides, Neoanthostomella, Pseudoanthostomella and Pyriformiascoma are evaluated, demonstrating the necessity of critical inspection of published sequence data before inclusion in phylogenies. Verified isolates of several species from these genera should be re-sequenced to affirm their phylogenetic affinities. In addition, the generic type of Anthostomella should be sequenced before additional generic re-arrangements are proposed.
Electronic supplementary material
The online version of this article (doi:10.1007/s11557-017-1329-6) contains supplementary material, which is available to authorized users.
Keywords: Anthostoma, Ascomycota, Clypeosphaeria, Phylogenetic analysis, Pyrenomycetes, Sordariomycetes, Stereosphaeria, Xylariaceae
Introduction
Xylariaceae have long been treated in a conservative, morphology-based concept, and only informal subgroupings like Hypoxyloideae and Xylarioideae were accepted despite polyphyly of several genera. Recently, Wendt et al. (2017) subdivided Xylariaceae into three families based on multigene phylogeny of an ITS–LSU–rpb2–tub2 matrix. They resurrected and emended the family Hypoxylaceae, widened the Graphostromataceae to include the genera Biscogniauxia, Camillea, Obolarina and Vivantia, and restricted Xylariaceae mostly to genera with geniculosporium-like asexual morphs. This facilitates phylogenetic placement of other genera affiliated with Xylariaceae sensu lato. One example is the genus Anthostomella, which houses a number of species, whose morphological traits vary considerably and may, thus, be phylogenetically uninformative. Ascomata are usually immersed in the host tissue, covered by a clypeus or not, have amyloid or sometimes non-amyloid ascus apices and brown amerosporous ascospores with or without a hyaline appendage cell, with or without a gelatinous sheath. One major challenge to study them on hosts other than palms is the difficulty to spot them, as they cannot be collected regularly, and, often, they produce very limited material. Francis (1975) performed a study on the systematics of Anthostomella species on the stems and leaves of herbaceous plants and gymnosperms based on morphology alone. A similar but more voluminous study was carried out by Lu and Hyde (2000). Using a few newly collected specimens, Daranagama et al. (2015, 2016) determined that Anthostomella is polyphyletic within Xylariaceae and described several new genera.
There is some confusion in the literature about the generic type of Anthostomella. Eriksson (1966) pointed out that lectotypification of Anthostomella with A. phaeosticta by Clements and Shear (1931) was in error and that A. limitata is the true generic type of Anthostomella. According to the ICN, this lectotypification is valid and has to be followed unless conservation with a different type is formally approved, and A. limitata is correctly listed as the generic type in Index Fungorum. In arguing that A. limitata does not exhibit several morphological characters then considered typical for the genus, Francis (1975) proposed A. tomicoides as the generic type, but this change has never been formally proposed and approved to become in effect. However, in the subsequent publications cited above, Francis (1975) was followed and A. tomicoides was accepted as the generic type. Neither A. limitata nor A. tomicoides have yet been sequenced.
Several genera have been segregated from Anthostomella or newly described, or subgenera were elevated to the generic rank. One of the latter is Lopadostoma (Jaklitsch et al. 2014) and another Entosordaria. The generic type of Entosordaria, E. perfidiosa, is characterised by non-amyloid asci and ascospores, which have a unique apical germ apparatus consisting of radiating slits (Eriksson 1966; Eriksson and Hawksworth 1986). Nonetheless, the genus was subsumed by Barr (1989) under Clypeosphaeria (see also Jaklitsch et al. 2016).
A transition to and now a member of the Diatrypaceae is the genus Anthostoma, which currently encompasses the single lignicolous species A. decipiens (Rappaz 1992; Jaklitsch et al. 2014). In a study designed to assess Anthostomella on hardwoods, Rappaz (1995) described the genus Barrmaelia, whose species, in part, also resemble Diatrypaceae, particularly in ascospore features, but, in contrast, have short-pedicellate asci and non-amyloid ascus apices. Furthermore, Barrmaelia species are typically characterised by ascomata that are immersed in the wood or bark and stromata that tend to blacken the host surface, in combination with light to dark brown, one-celled, smooth, ellipsoid to allantoid ascospores without sheath or appendages and with or without a germ slit. Rappaz (1995) combined six species in Barrmaelia (B. macrospora, B. moravica, B. oxyacanthae, B. picacea, B. pseudobombarda and B. sustenta) and described one new species, which he also selected as the generic type, B. rhamnicola. No new taxa have been added to this genus since then.
Although Rappaz (1995) only had morphology at hand, his concept withstands molecular phylogenetic analyses, as we show below. We, therefore, describe the new species B. rappazii to honour him, present the molecular systematics of five species of Barrmaelia and two of Entosordaria, including the new species E. quercina. The genera Barrmaelia and Entosordaria form a distinct lineage, which we name as the new family Barrmaeliaceae.
Materials and methods
Isolates and specimens
All newly prepared isolates used in this study originated from ascospores of fresh specimens. The numbers of strains including NCBI GenBank accession numbers of gene sequences used to compute the phylogenetic trees are listed in Table 1. Isolates have been deposited at the Westerdijk Fungal Biodiversity Institute (CBS-KNAW), Utrecht, the Netherlands. Details of the specimens used for morphological investigations are listed in the Taxonomy section under the respective descriptions. Herbarium acronyms are according to Thiers (2017). Specimens have been deposited in the Fungarium of the Institute of Botany, University of Vienna (WU).
Table 1.
Isolates and accession numbers used in the phylogenetic analyses. Isolates/sequences in bold were isolated/sequenced in the present study. For details about sequence accessions retrieved from GenBank, see Jaklitsch and Voglmayr (2012), Jaklitsch et al. (2014, 2016), Daranagama et al. (2016), Hernández-Restrepo et al. (2016) and Wendt et al. (2017)
| Species | Specimen or strain numbera | Origin | Statusb | GenBank accession numbersc | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | rpb2 | tub2 | tef1 | ||||
| Amphirosellinia fushanensis | HAST 91111209 | Taiwan | HT | GU339496 | N/A | GQ848339 | GQ495950 | |
| Amphirosellinia nigrospora | HAST 91092308 | Taiwan | HT | GU322457 | N/A | GQ848340 | GQ495951 | |
| Annulohypoxylon annulatum | CBS 140775 | Texas | ET | KY610418 | KY610418 | KY624263 | KX376353 | |
| Annulohypoxylon atroroseum | ATCC 76081 | Thailand | AJ390397 | KY610422 | KY624233 | DQ840083 | ||
| Annulohypoxylon michelianum | CBS 119993 | Spain | KX376320 | KY610423 | KY624234 | KX271239 | ||
| Annulohypoxylon moriforme | CBS 123579 | Martinique | KX376321 | KY610425 | KY624289 | KX271261 | ||
| Annulohypoxylon nitens | MFLUCC 12-0823 | Thailand | KJ934991 | KJ934992 | KJ934994 | KJ934993 | ||
| Annulohypoxylon stygium | MUCL 54601 | French Guiana | KY610409 | KY610475 | KY624292 | KX271263 | ||
| Annulohypoxylon truncatum | CBS 140778 | Texas | ET | KY610419 | KY610419 | KY624277 | KX376352 | |
| Anthostomella formosa | MFLUCC 14-0170 | Italy | KP297403 | KP340544 | KP340531 | N/Ad | ||
| Anthostomella helicofissa | MFLUCC 14-0173 | Italy | HT | KP297406 | KP297406e | KP340534 | KP406617 | |
| Anthostomella obesa | MFLUCC 14-0171 | Italy | HT | KP297405 | KP340546 | KP340533 | N/Ad | |
| Anthostomella rubicola | MFLUCC 16-0479 | Italy | KX533455 | KX533456 | KX789493 | KX789494 | ||
| Anthostomelloides forlicesenica | MFLUCC 14-0007 | Italy | HT | KP297396 | KP297396e,f | N/Af | KP406607 | |
| Anthostomelloides krabiensis | MFLUCC 15-0678 | Thailand | HT | KX305927 | KX305928 | KX305929 | N/A | |
| Astrocystis concavispora | MFLUCC 14-0174 | Italy | KP297404 | KP340545 | KP340532 | KP406615 | ||
| Barrmaelia macrospora | BM = CBS 142768 | Austria | ET | KC774566 | KC774566 | MF488995 | MF489014 | MF489005 |
| Barrmaelia moravica | Cr1 = CBS 142769 | Austria | ET | MF488987 | MF488987 | MF488996 | MF489015 | MF489006 |
| Barrmaelia oxyacanthae | BO = CBS 142770 | Austria | MF488988 | MF488988 | MF488997 | MF489016 | MF489007 | |
| Barrmaelia rappazii | Cr2 = CBS 142771 | Norway | HT | MF488989 | MF488989 | MF488998 | MF489017 | MF489008 |
| Barrmaelia rhamnicola | BR = CBS 142772 | France | ET | MF488990 | MF488990 | MF488999 | MF489018 | MF489009 |
| Barrmaelia rhamnicola | BR1 | France | MF488991 | MF488991 | MF489000 | MF489019 | MF489010 | |
| Biscogniauxia arima | WSP 122 | Mexico | IT | EF026150 | N/A | GQ304736 | AY951672 | |
| Biscogniauxia atropunctata | Y.M.J. 128 | USA | JX507799 | N/A | JX507778 | AY951673 | ||
| Biscogniauxia marginata | MFLUCC 12-0740 | France | KJ958407 | KJ958408 | KJ958409 | KJ958406 | ||
| Biscogniauxia nummularia | MUCL 51395 | France | ET | KY610382 | KY610427 | KY624236 | KX271241 | |
| Biscogniauxia repanda | ATCC 62606 | USA | KY610383 | KY610428 | KY624237 | KX271242 | ||
| Brunneiperidium gracilentum | MFLUCC 14-0011 | Italy | HT | KP297400 | KP340542 | KP340528 | KP406611 | |
| Calceomyces lacunosus | CBS 633.88 | Japan | HT | KY610397 | KY610476 | KY624293 | KX271265 | |
| Camillea obularia | ATCC 28093 | Puerto Rico | KY610384 | KY610429 | KY624238 | KX271243 | ||
| Camillea tinctor | Y.M.J. 363 | Martinique | JX507806 | N/A | JX507790 | JX507795 | ||
| Clypeosphaeria mamillana | CLM = CBS 140735 | France | ET | KT949897 | KT949897 | MF489001 | N/A | |
| Collodiscula bambusae | GZUH 0102 | China | KP054279 | KP054280 | KP276675 | KP276674 | ||
| Collodiscula fangjingshanensis | GZUH 0109 | China | HT | KR002590 | KR002591 | KR002592 | KR002589 | |
| Collodiscula japonica | CBS 124266 | China | JF440974 | JF440974 | KY624273 | KY624316 | ||
| Creosphaeria sassafras | ST.MA. 14087 | Argentina | KY610411 | KY610468 | KY624265 | KX271258 | ||
| Daldinia andina | CBS 114736 | Ecuador | HT | AM749918 | KY610430 | KY624239 | KC977259 | |
| Daldinia bambusicola | CBS 122872 | Thailand | HT | KY610385 | KY610431 | KY624241 | AY951688 | |
| Daldinia caldariorum | MUCL 49211 | France | AM749934 | KY610433 | KY624242 | KC977282 | ||
| Daldinia concentrica | CBS 113277 | Germany | AY616683 | KY610434 | KY624243 | KC977274 | ||
| Daldinia dennisii | CBS 114741 | Australia | HT | JX658477 | KY610435 | KY624244 | KC977262 | |
| Daldinia eschscholtzii | MUCL 45435 | Benin | JX658484 | KY610437 | KY624246 | KC977266 | ||
| Daldinia loculatoides | CBS 113279 | UK | ET | AF176982 | KY610438 | KY624247 | KX271246 | |
| Daldinia macaronesica | CBS 113040 | Spain | PT | KY610398 | KY610477 | KY624294 | KX271266 | |
| Daldinia petriniae | MUCL 49214 | Austria | ET | AM749937 | KY610439 | KY624248 | KC977261 | |
| Daldinia placentiformis | MUCL 47603 | Mexico | AM749921 | KY610440 | KY624249 | KC977278 | ||
| Daldinia pyrenaica | MUCL 53969 | France | KY610413 | KY610413 | KY624274 | KY624312 | ||
| Daldinia steglichii | MUCL 43512 | Papua New Guinea | PT | KY610399 | KY610479 | KY624250 | KX271269 | |
| Daldinia theissenii | CBS 113044 | Argentina | PT | KY610388 | KY610441 | KY624251 | KX271247 | |
| Daldinia vernicosa | CBS 119316 | Germany | ET | KY610395 | KY610442 | KY624252 | KC977260 | |
| Diatrype disciformis | CBS 197.49 | Netherlands | N/A | DQ470964 | DQ470915 | N/A | ||
| Entoleuca mammata | J.D.R. 100 | France | GU300072 | N/A | GQ844782 | GQ470230 | ||
| Entonaema liquescens | ATCC 46302 | USA | KY610389 | KY610443 | KY624253 | KX271248 | ||
| Entosordaria perfidiosa | BW3 | Germany | MF488992 | MF488992 | MF489002 | MF489020 | MF489011 | |
| Entosordaria perfidiosa | EPE = CBS 142773 | Austria | ET | MF488993 | MF488993 | MF489003 | MF489021 | MF489012 |
| Entosordaria quercina | RQ = CBS 142774 | Greece | HT | MF488994 | MF488994 | MF489004 | MF489022 | MF489013 |
| Euepixylon sphaeriostomum | J.D.R. 261 | USA | GU292821 | N/A | GQ844774 | GQ470224 | ||
| Eutypa lata | UCR-EL1 | USA | JGI | JGI | JGI | JGI | ||
| Graphostroma platystomum | CBS 270.87 | France | HT | JX658535 | DQ836906 | KY624296 | HG934108 | |
| Hypocreodendron sanguineum | J.D.R. 169 | Mexico | GU322433 | N/A | GQ844819 | GQ487710 | ||
| Hypoxylon carneum | MUCL 54177 | France | KY610400 | KY610480 | KY624297 | KX271270 | ||
| Hypoxylon cercidicola | CBS 119009 | France | KC968908 | KY610444 | KY624254 | KC977263 | ||
| Hypoxylon crocopeplum | CBS 119004 | France | KC968907 | KY610445 | KY624255 | KC977268 | ||
| Hypoxylon fendleri | MUCL 54792 | French Guiana | KF234421 | KY610481 | KY624298 | KF300547 | ||
| Hypoxylon fragiforme | MUCL 51264 | Germany | ET | KC477229 | KM186295 | KM186296 | KX271282 | |
| Hypoxylon fuscum | CBS 113049 | France | ET | KY610401 | KY610482 | KY624299 | KX271271 | |
| Hypoxylon griseobrunneum | CBS 331.73 | India | HT | KY610402 | KY610483 | KY624300 | KC977303 | |
| Hypoxylon haematostroma | MUCL 53301 | Martinique | ET | KC968911 | KY610484 | KY624301 | KC977291 | |
| Hypoxylon howeanum | MUCL 47599 | Germany | AM749928 | KY610448 | KY624258 | KC977277 | ||
| Hypoxylon hypomiltum | MUCL 51845 | Guadeloupe | KY610403 | KY610449 | KY624302 | KX271249 | ||
| Hypoxylon investiens | CBS 118183 | Malaysia | KC968925 | KY610450 | KY624259 | KC977270 | ||
| Hypoxylon lateripigmentum | MUCL 53304 | Martinique | HT | KC968933 | KY610486 | KY624304 | KC977290 | |
| Hypoxylon lenormandii | CBS 119003 | Ecuador | KC968943 | KY610452 | KY624261 | KC977273 | ||
| Hypoxylon monticulosum | MUCL 54604 | French Guiana | ET | KY610404 | KY610487 | KY624305 | KX271273 | |
| Hypoxylon musceum | MUCL 53765 | Guadeloupe | KC968926 | KY610488 | KY624306 | KC977280 | ||
| Hypoxylon ochraceum | MUCL 54625 | Martinique | ET | KC968937 | N/A | KY624271 | KC977300 | |
| Hypoxylon papillatum | ATCC 58729 | USA | HT | KC968919 | KY610454 | KY624223 | KC977258 | |
| Hypoxylon perforatum | CBS 115281 | France | KY610391 | KY610455 | KY624224 | KX271250 | ||
| Hypoxylon petriniae | CBS 114746 | France | HT | KY610405 | KY610491 | KY624279 | KX271274 | |
| Hypoxylon pilgerianum | ST.MA. 13455 | Martinique | KY610412 | KY610412 | KY624308 | KY624315 | ||
| Hypoxylon porphyreum | CBS 119022 | France | KC968921 | KY610456 | KY624225 | KC977264 | ||
| Hypoxylon pulicicidum | CBS 122622 | Martinique | HT | JX183075 | KY610492 | KY624280 | JX183072 | |
| Hypoxylon rickii | MUCL 53309 | Martinique | ET | KC968932 | KY610416 | KY624281 | KC977288 | |
| Hypoxylon rubiginosum | MUCL 52887 | Germany | ET | KC477232 | KY610469 | KY624266 | KY624311 | |
| Hypoxylon samuelsii | MUCL 51843 | Guadeloupe | ET | KC968916 | KY610466 | KY624269 | KC977286 | |
| Hypoxylon submonticulosum | CBS 115280 | France | KC968923 | KY610457 | KY624226 | KC977267 | ||
| Hypoxylon ticinense | CBS 115271 | France | JQ009317 | KY610471 | KY624272 | AY951757 | ||
| Hypoxylon trugodes | MUCL 54794 | Sri Lanka | ET | KF234422 | KY610493 | KY624282 | KF300548 | |
| Hypoxylon vogesiacum | CBS 115273 | France | KC968920 | KY610417 | KY624283 | KX271275 | ||
| Jackrogersella cohaerens | CBS 119126 | Germany | KY610396 | KY610497 | KY624270 | KY624314 | ||
| Jackrogersella minutella | CBS 119015 | Portugal | KY610381 | KY610424 | KY624235 | KX271240 | ||
| Jackrogersella multiformis | CBS 119016 | Germany | ET | KC477234 | KY610473 | KY624290 | KX271262 | |
| Kretzschmaria deusta | CBS 163.93 | Germany | KC477237 | KY610458 | KY624227 | KX271251 | ||
| Lopadostoma dryophilum | CBS 133213 | Austria | ET | KC774570 | KC774570 | KC774526 | MF489023 | |
| Lopadostoma turgidum | CBS 133207 | Austria | ET | KC774618 | KC774618 | KC774563 | MF489024 | |
| Microdochium lycopodinum | CBS 122885 | Austria | HT | JF440979 | JF440979 | KP859125 | KP859080 | |
| Microdochium phragmitis | CBS 285.71 | Poland | ET | KP859013 | KP858949 | KP859122 | KP859077 | |
| Microdochium seminicola | CBS 139951 | Switzerland | HT | KP859038 | KP858974 | KP859147 | KP859101 | |
| Nemania abortiva | BISH 467 | USA | HT | GU292816 | N/A | GQ844768 | GQ470219 | |
| Nemania beaumontii | HAST 405 | Martinique | GU292819 | N/A | GQ844772 | GQ470222 | ||
| Nemania bipapillata | HAST 90080610 | Taiwan | GU292818 | N/A | GQ844771 | GQ470221 | ||
| Nemania maritima | HAST 89120401 | Taiwan | ET | N/A | N/A | GQ844775 | GQ470225 | |
| Nemania maritima | ST.MA. 04019 = J.F. 03075 | France | KY610414 | KY610414 | N/A | N/A | ||
| Nemania primolutea | HAST 91102001 | Taiwan | HT | EF026121 | N/A | GQ844767 | EF025607 | |
| Neoanthostomella viticola | MFLUCC 16-0243 | Italy | HT | KX505957 | KX505958 | KX789496 | KX789495 | |
| Obolarina dryophila | MUCL 49882 | France | GQ428316 | GQ428316 | KY624284 | GQ428322 | ||
| Podosordaria mexicana | WSP 176 | Mexico | GU324762 | N/A | GQ853039 | GQ844840 | ||
| Podosordaria muli | WSP 167 | Mexico | HT | GU324761 | N/A | GQ853038 | GQ844839 | |
| Poronia pileiformis | WSP 88113001 | Taiwan | ET | GU324760 | N/A | GQ853037 | GQ502720 | |
| Poronia punctata | CBS 656.78 | Australia | HT | KT281904 | KY610496 | KY624278 | KX271281 | |
| Pseudoanthostomella delitescens | MFLUCC 16-0477 | Italy | KX533451 | KX533452 | KX789491 | KX789490 | ||
| Pseudoanthostomella pini-nigrae | MFLUCC 16-0478 | Italy | HT | KX533453 | KX533454 | KX789492 | N/A | |
| Pseudoanthostomella senecionicola | MFLUCC 15-0013 | Italy | HT | KX505960 | KX505959 | KX789489 | N/A | |
| Pyrenopolyporus hunteri | MUCL 52673 | Ivory Coast | ET | KY610421 | KY610472 | KY624309 | KU159530 | |
| Pyrenopolyporus laminosus | MUCL 53305 | Martinique | HT | KC968934 | KY610485 | KY624303 | KC977292 | |
| Pyrenopolyporus nicaraguensis | CBS 117739 | Burkina Faso | AM749922 | KY610489 | KY624307 | KC977272 | ||
| Pyriformiascoma trilobatum | MFLUCC 14-0012 | Italy | HT | KP297402 | KP340543 | KP340530 | KP406613 | |
| Rhopalostroma angolense | CBS 126414 | Ivory Coast | KY610420 | KY610459 | KY624228 | KX271277 | ||
| Rosellinia aquila | MUCL 51703 | France | KY610392 | KY610460 | KY624285 | KX271253 | ||
| Rosellinia buxi | J.D.R. 99 | France | GU300070 | N/A | GQ844780 | GQ470228 | ||
| Rosellinia corticium | MUCL 51693 | France | KY610393 | KY610461 | KY624229 | KX271254 | ||
| Rosellinia necatrix | CBS 349.36 | Argentina | AY909001 | KF719204 | KY624275 | KY624310 | ||
| Rostrohypoxylon terebratum | CBS 119137 | Thailand | HT | DQ631943 | DQ840069 | DQ631954 | DQ840097 | |
| Ruwenzoria pseudoannulata | MUCL 51394 | D. R. Congo | HT | KY610406 | KY610494 | KY624286 | KX271278 | |
| Sarcoxylon compunctum | CBS 359.61 | South Africa | KT281903 | KY610462 | KY624230 | KX271255 | ||
| Stilbohypoxylon elaeicola | Y.M.J. 173 | French Guiana | EF026148 | N/A | GQ844826 | EF025616 | ||
| Stilbohypoxylon quisquiliarum | Y.M.J. 172 | French Guiana | EF026119 | N/A | GQ853020 | EF025605 | ||
| Thamnomyces dendroidea | CBS 123578 | French Guiana | HT | FN428831 | KY610467 | KY624232 | KY624313 | |
| Xylaria acuminatilongissima | HAST 95060506 | Taiwan | HT | EU178738 | N/A | GQ853028 | GQ502711 | |
| Xylaria adscendens | J.D.R. 865 | Thailand | GU322432 | N/A | GQ844818 | GQ487709 | ||
| Xylaria arbuscula | CBS 126415 | Germany | KY610394 | KY610463 | KY624287 | KX271257 | ||
| Xylaria bambusicola | WSP 205 | Taiwan | HT | EF026123 | N/A | GQ844802 | AY951762 | |
| Xylaria brunneovinosa | HAST 720 | Martinique | HT | EU179862 | N/A | GQ853023 | GQ502706 | |
| Xylaria curta | HAST 494 | Martinique | GU322444 | N/A | GQ844831 | GQ495937 | ||
| Xylaria discolour | HAST 131023 | USA | ET | JQ087405 | N/A | JQ087411 | JQ087414 | |
| Xylaria hypoxylon | CBS 122620 | Sweden | ET | KY610407 | KY610495 | KY624231 | KX271279 | |
| Xylaria multiplex | HAST 580 | Martinique | GU300098 | N/A | GQ844814 | GQ487705 | ||
| Xylaria polymorpha | MUCL 49884 | France | KY610408 | KY610464 | KY624288 | KX271280 | ||
aATCC, American Type Culture Collection, Manassas, USA; BISH, Bishop Museum, Honolulu, USA; CBS, Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; GZUH, Guizhou University, Guiyang, China; HAST, Academia Sinica, Taipei, Taiwan; J.D.R., Jack D. Rogers, Washington State University, Pullman, USA; J.F., Jacques Fournier, Rimont, France; MFLUCC, Mae Fah Luang University, Chiang Rai, Thailand; MUCL, Université Catholique de Louvain, Louvain-la-Neuve, Belgium; ST.MA., Marc Stadler, Helmholtz-Zentrum für Infektionsforschung, Braunschweig, Germany; UCR, University of California, Riverside, USA; Y.M.J., Yu-Ming Ju, Academia Sinica, Taipei, Taiwan; WSP, Washington State University, Pullman, USA
bET, epitype; HT, holotype; IT, isotype; PT, paratype
cN/A, not available; JGI, sequences retrieved from JGI-DOE (http://genome.jgi.doe.gov/)
d tub2 sequences of Daranagama et al. (2015) not included, as they are erroneous, actually representing rpb2 sequences of an unidentified fungus
ePartial LSU of the deposited ITS sequences used for analyses, as the LSU sequences of Daranagama et al. (2015) are not from the same fungus (highly distinct from the LSU part of the ITS)
fGenBank sequences of LSU and rpb2 of Daranagama et al. (2015) not included, as they are erroneous, representing sequences of an unidentified pleosporalean fungus
Culture preparation, growth rate determination and phenotype analysis
Cultures were prepared and maintained as described previously (Jaklitsch 2009). Microscopic observations were made in tap water, except where noted. Morphological analyses of microscopic characters were carried out as described earlier (Jaklitsch 2009). Methods of microscopy included stereomicroscopy using Nikon SMZ1500, Olympus SZX10 and Euromex Novex RZ 65.560, light microscopy using Euromex XHR MIC 625, Olympus BX51 and Nomarski differential interference contrast (DIC) using the compound microscopes Nikon Eclipse E600 and Zeiss Axio Imager.A1. Images and data were gathered with Nikon Coolpix 4500, Nikon DS-U2, Nikon D90, Olympus DP72 and Zeiss Axiocam 506 colour digital cameras and measured directly with the microscope, or with Olympus cellSens Dimension, NIS-Elements D v.3.0 and Zeiss ZEN Blue Edition softwares. Amyloidity of asci was assessed using Lugol or Melzer reagent. Measurements are reported as maximum and minimum in parentheses and the range representing the mean plus and minus the standard deviation of a number of measurements given in parentheses.
DNA extraction and sequencing methods
The extraction of genomic DNA was performed as reported previously (Voglmayr and Jaklitsch 2011; Jaklitsch et al. 2012) using the DNeasy Plant Mini Kit (QIAgen GmbH, Hilden, Germany). The following loci were amplified and sequenced: the complete internally transcribed spacer region (ITS1–5.8S–ITS2) and a ca. 1.3-kb fragment of the large subunit nuclear ribosomal DNA (nLSU rDNA), amplified and sequenced as a single fragment with primers V9G (de Hoog and Gerrits van den Ende 1998) and LR5 (Vilgalys and Hester 1990); a ca. 1.2-kb fragment of the RNA polymerase II subunit 2 (rpb2) gene with primers fRPB2-5f and fRPB2-7cr (Liu et al. 1999) or dRPB2-5f and dRPB2-7r (Voglmayr et al. 2016a); a ca. 1.3–1.5-kb fragment of the translation elongation factor 1-alpha (tef1) gene with primers EF1-728F (Carbone and Kohn 1999) and TEF1LLErev (Jaklitsch et al. 2005) or EF1-2218R (Rehner and Buckley 2005); and a ca. 1.6-kb fragment of the beta-tubulin (tub2) gene with primers T1 and T22 (O’Donnell and Cigelnik 1997). Polymerase chain reaction (PCR) products were purified using an enzymatic PCR cleanup (Werle et al. 1994) as described in Voglmayr and Jaklitsch (2008). DNA was cycle-sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit v.3.1 (Applied Biosystems, Warrington, UK) with the same primers as in PCR; in addition, primers ITS4 (White et al. 1990), LR2R-A (Voglmayr et al. 2012) and LR3 (Vilgalys and Hester 1990) were used for the ITS–LSU region, TEF1_INTF (Jaklitsch 2009) and TEFD_iR (5′ GTCTGGCCATCCTTGGAGAT 3′) for tef1 and BtHV2r (Voglmayr et al. 2016b, 2017) for tub2. Sequencing was performed on an automated DNA sequencer (3730xl Genetic Analyser, Applied Biosystems).
Analysis of sequence data
Following the phylogenetic placement of Barrmaelia macrospora within the Xylariaceae sensu lato clade in earlier analyses (Jaklitsch et al. 2014, 2016), sequences of Barrmaelia and Entosordaria were analysed within the combined ITS, LSU rDNA, rpb2 and tub2 matrix of Wendt et al. (2017). As only a few tef1 sequences are available for Xylariales, this marker was not included in the matrix but the sequences were deposited at GenBank as a secondary barcode marker. To obtain a more representative taxon sampling, selected sequences were added to this matrix from Hernández-Restrepo et al. (2016) and from Daranagama et al. (2015, 2016). From the latter two publications dealing with Anthostomella-like representatives, only accessions for which at least three of the four loci are available were included; before addition, it proved necessary to check these sequences with NCBI nucleotide BLAST searches for their correct gene and lineage identity, and obviously erroneous sequences as well as regions of poor sequence quality were excluded. For Eutypa lata, sequences were retrieved from the genome of strain UCR-EL1 deposited at JGI-DOE (http://genome.jgi.doe.gov/). Following the analyses of Jaklitsch et al. (2016), sequences of Microdochium (Microdochiaceae) were selected as the outgroup to root the trees. Familial classification of Xylariaceae sensu lato follows Wendt et al. (2017). All alignments were produced with the server version of MAFFT (http://www.ebi.ac.uk/Tools/msa/mafft), checked and refined using BioEdit version 7.0.9.0 (Hall 1999). After exclusion of ambiguously aligned regions and long gaps, the final matrix contained 4668 nucleotide characters, i.e. 600 from the ITS, 1359 from the LSU, 1162 from rpb2 and 1547 from tub2.
Maximum parsimony (MP) analysis of the combined matrix was performed using a parsimony ratchet approach. For this, a nexus file was prepared using PRAP v.2.0b3 (Müller 2004), implementing 1000 ratchet replicates with 25% of randomly chosen positions upweighted to 2, which was then run with PAUP v.4.0a151 (Swofford 2002). The resulting best trees were then loaded in PAUP and subjected to heuristic search with TBR branch swapping (MULTREES option in effect, steepest descent option not in effect). Bootstrap analysis with 1000 replicates was performed using five rounds of replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping (MULTREES option in effect, steepest descent option not in effect) during each bootstrap replicate. In all MP analyses, molecular characters were unordered and given equal weight; analyses were performed with gaps treated as missing data; the COLLAPSE command was set to minbrlen.
Maximum likelihood (ML) analyses were performed with RAxML (Stamatakis 2006) as implemented in raxmlGUI 1.3 (Silvestro and Michalak 2012), using the ML + rapid bootstrap setting and the GTRGAMMA substitution model with 1000 bootstrap replicates. The matrix was partitioned for the individual gene regions, and substitution model parameters were calculated separately for them.
Results
Assessment of published sequences
NCBI Nucleotide BLAST searches revealed serious problems for some sequences of Daranagama et al. (2015), which were, therefore, excluded from the analyses (Table 1). LSU sequences KP340547 (Anthostomella helicofissa) and KP340538 (Anthostomelloides forlicesenica) were not added to the matrix, as they did not correspond to the LSU part (ca. 540 bp) included in the ITS sequences KP297406 and KP297396 of the same accessions. Whereas LSU sequence KP340547 was revealed as xylarialean by BLAST searches but differed in 60 positions (3 gaps and 57 substitutions) from the LSU part of KP297406, a BLAST search of KP340538 revealed various Pleosporales (Kalmusia, Coniothyrium, Dendrothyrium) as the closest match (84% sequence similarity). Therefore, for these two accessions, only the LSU part of the ITS sequences was included in the LSU matrix. rpb2 sequence KP340524 (Anthostomelloides forlicesenica) was excluded as well, as a BLAST search also revealed pleosporalean affinities (80% similarity to sequence LK936413 of Leptosphaerulina chartarum, 77% similarity to sequences DQ677970 of Phaeodothis winteri and DQ677956 of Coniothyrium palmarum). tub2 sequences KP406614 (Anthostomella formosa) and KP406616 (Anthostomella obesa) were also excluded, as BLAST searches actually revealed them as rpb2 sequences. This was also confirmed in an alignment containing the rpb2 sequences included in the present study, where they were highly similar to rpb2 sequences of various Anthostomella species (not shown); however, both were different from the rpb2 sequences KP340531 and KP340533 published for the same isolates in the same publication.
Molecular phylogeny
Of the 4668 nucleotide characters of the combined matrix, 2210 are parsimony informative (338 of ITS, 422 of LSU, 638 of rpb2 and 812 of tub2). Figure 1 shows a simplified phylogram of the best ML tree (lnL = −136212.706) obtained by RAxML. Maximum parsimony analyses revealed four MP trees 32,311 steps long, which were identical except for a polytomy within the three terminal taxa of Anthostomella and an unresolved position of Hypoxylon ochraceum and H. pilgerianum relative to each other; the strict consensus tree of the four MP trees is provided in the Supplementary Information. The backbone of the MP trees was similar to the ML tree, except for a sister group relationship of Lopadostomataceae and Diatrypaceae and a slightly different position of the Calceomyces–Neoanthostomella clade; in addition, there were a few minor topological differences within the Xylariaceae and Graphostromataceae.
Fig. 1.
Simplified phylogram of the best ML trees (lnL = −136212.706) revealed by RAxML from an analysis of the combined ITS–LSU–rpb2–tub2 matrix of selected Xylariales. Strains in bold were sequenced in the current study. The Hypoxylaceae clade, which is not treated in detail, is collapsed to provide sufficient space for the other clades of interest. ML and MP bootstrap support above 50% are given at the first and second positions, respectively, above or below the branches. The arrows denote topological conflict with previous phylogenies (Anthostomelloides forlicesenica) or major incongruence with the morphology of the clade in which it is placed (Pyriformiascoma trilobatum)
All families received high to maximum support in all analyses, as did the Xylariaceae sensu lato (Fig. 1). The genera Barrmaelia and Entosordaria were revealed as the closest relatives with maximum support but formed a separate lineage within Xylariaceae sensu lato, and are classified here within the new family Barrmaeliaceae. Within the Xylariaceae sensu lato, the basal position of Hypoxylaceae was highly supported, but the phylogenetic relationships between the other three families (Barrmaeliaceae, Graphostromataceae and Xylariaceae sensu stricto) remain uncertain due to the lack of significant backbone support. Clypeosphaeria mamillana is revealed as the closest relative of Anthostomelloides krabiensis with high (99% MP BS) to maximum (ML) support, and both are sister clade to the rest of the Xylariaceae sensu stricto with high support as well (Fig. 1). The second species of Anthostomelloides, A. forlicesenica, is not closely related to A. krabiensis but sister species of Brunneiperidium gracilentum within Xylariaceae sensu stricto with high (98% MP BS) to maximum (ML) support. The genera Anthostomella and Pseudoanthostomella are placed outside Xylariaceae sensu lato and form a highly supported lineage; sister group relationship to the highly supported Calceomyces–Neoanthostomella clade is revealed with medium support only in the ML analyses. Pyriformiascoma trilobatum is placed within Microdochium with maximum support in both analyses.
Taxonomy
Barrmaeliaceae Voglmayr & Jaklitsch, fam. nov.
MycoBank MB 822042
Type genus: Barrmaelia Rappaz.
Other genus in the family: Entosordaria Höhn.
Saprobic on wood or bark. Stroma if present mostly in wood and blackening the surface in wide areas or in elongate bands, sometimes darker around the ostioles; entostroma prosenchymatous, poorly developed, without KOH-extractable pigments. Ascomata (perithecia) globose, sometimes raising the substrate, singly, in small groups or gregarious. Peridium melanised, pseudoparenchymatous to prosenchymatous. Hamathecium of numerous persistent, hyaline, septate paraphyses. Asci eight-spored, cylindrical, persistent, with inamyloid or infrequently amyloid apical ascus apparatus. Ascospores yellow to dark brown; unicellular with or without germ slit (Barrmaelia), or two-celled with septum near one end, the small cell hyaline, the large cell dark brown and with an apical germ apparatus consisting of radial slits (Entosordaria); allantoid or ellipsoid, inequilateral, slightly inequilateral or nearly equilateral, with narrowly or broadly rounded ends. Asexual morph libertella-like where known (Barrmaelia; Rappaz 1995).
Barrmaelia Rappaz, Mycol. Helv. 7(1): 130 (1995).
Type species: Barrmaelia rhamnicola Rappaz, Mycol. Helv. 7(1): 131 (1995).
Stromata mostly in wood, usually discolouring the wood surface grey to black, entostroma poorly developed (Rappaz 1995). Ascomata perithecial, immersed in wood or bark, rarely erumpent, often blackening the host surface, globose, ellipsoid or pyriform; ostiolar pore rounded. Peridium melanised, pseudoparenchymatous to prosenchymatous. Hamathecium of apically free, hyaline paraphyses. Asci unitunicate, cylindrical, with a short stipe, generally eight-spored; with an inamyloid apical apparatus. Ascospores light to dark brown, one-celled, smooth, asymmetrically ellipsoid to allantoid, without sheath or appendages, with or without a germ slit. Asexual morph (fide Rappaz 1995) libertella-like, only known from pure culture, conidiomata globose, more or less melanised, up to 1 mm in diam., exuding the conidia in white to pinkish droplets. Conidiophores erect, branched. Conidiogenous cells hyaline, conical or cylindrical, arranged in palisades, apex sometimes with faint annellations. Conidiogenesis holoblastic, proliferation percurrent or sympodial. Conidia hyaline, falcate, one end truncate, the other rounded or slightly acute.
Notes: As we did not observe an asexual morph in pure culture, its description is adapted from Rappaz (1995).
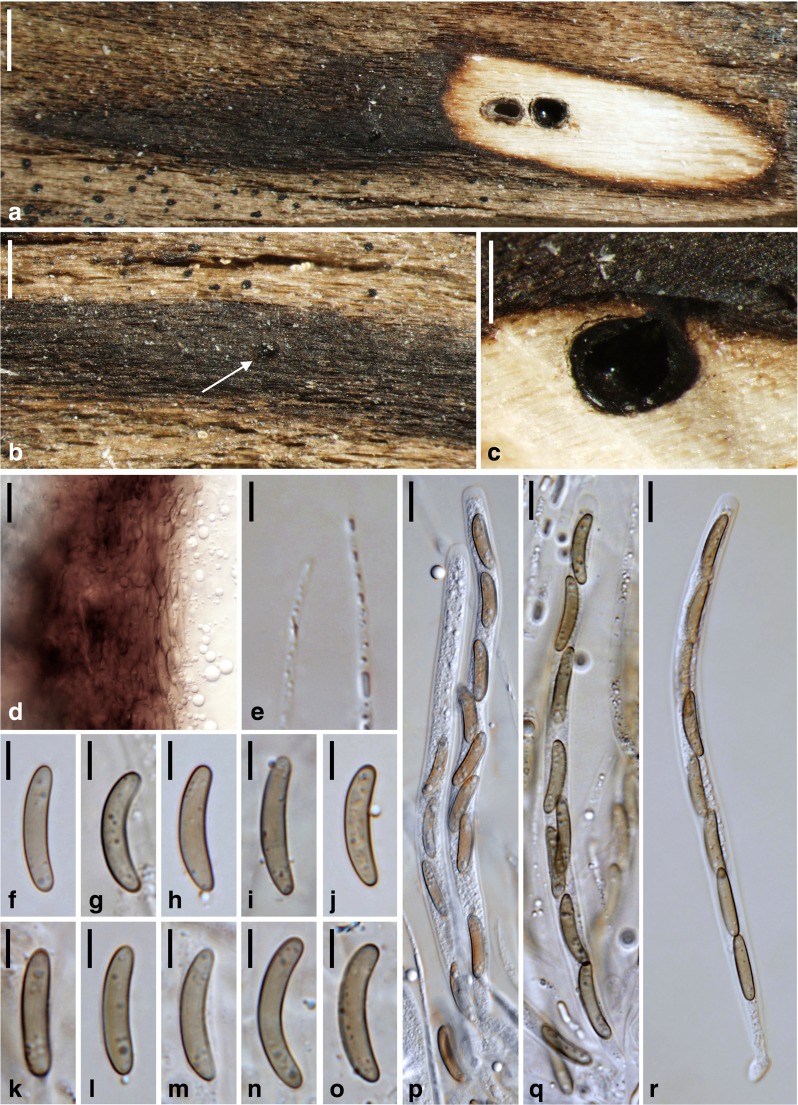
Barrmaelia macrospora (Nitschke) Rappaz, Mycol. Helv. 7(1): 135 (1995). Fig. 2.
Fig. 2.
Barrmaelia macrospora (a, b, d, e, h–q: WU 36920, epitype; c, f, g, r–t: B 70 0009349). a Stroma with blackened wood surface. b, c Perithecia in vertical section. d, e Perithecia in transverse section. f, g Ostioles protruding through the blackened wood surface (g with perithecia in transverse section). h Vertical section of perithecial wall. i Paraphyses apices. j, k Asci. l–t Ascospores; arrow denoting germ slit (t). All in water. Scale bars: a, c, f, g = 500 μm; b, d, e = 250 μm; h, i = 5 μm; j–t = 10 μm
Basionym. Valsa macrospora Nitschke, Pyrenomyc. Germ. 1: 145 (1867).
For synonyms, see Rappaz (1995).
Stromata blackening the wood surface in areas of up to 5 × 1.5 cm. Wood usually unchanged among ascomata, sometimes slightly pale brown. Ascomata perithecial, 400–600 μm diam., 300–500 μm high (n = 10), usually gregarious but separate, rarely two in contact, immersed, depressed globose to ellipsoid. Ostiolar apices inconspicuous, sometimes slightly raised, circular. Peridium 20–35 μm thick (n = 10), pseudoparenchymatous at the outer side and consisting of moderately thick-walled cells encrusted with brown material, tending to be prosenchymatous, lighter coloured and thinner-walled at the inner side, partly filled with oil drops. Paraphyses numerous, filled with oil drops, 2–4 μm wide, slightly tapering towards the apex, obtuse. Asci 108–143 × 9–11 μm, spore part 91–123 μm long, stipe 5–21 μm long (n = 20), cylindrical, containing eight biseriate or obliquely uniseriate ascospores, with short stipe and an inamyloid apical apparatus. Ascospores (18.2–)20.5–24.0(−26.0) × (4.0–)4.8–5.9(−6.5) μm, l/w = (3.1–)3.7–4.7(−5.4) μm (n = 60), one-celled, narrowly ellipsoid to fusoid, asymmetric, ends sometimes slightly pointed, brown, germ slit hard to observe, with a lighter coloured band at the concave side, apically also sometimes lighter coloured, filled with minute oil drops, smooth.
Colonies on CMD and MEA white; aerial hyphae abundant. No asexual morph observed.
Habitat: In wood of (partly) decorticated twigs and branches of Populus spp., also on Ligustrum (fide Rappaz 1995).
Distribution: Europe (Czech Republic, France, Germany, Netherlands, Norway, Sweden, Switzerland, United Kingdom), possibly also the USA (fide Rappaz 1995).
Typification. Germany, Nordrhein-Westfalen, Münsterland, [Münster-] Handorf, on Sarothamnus scoparius, without date, Th. Nitschke, (B 70 0009297; sub Valsa macrospora, holotype, labelled as “Lectotype”). Epitype of Valsa macrospora, here designated: France, Côte-d’Or (21), Marcilly-sur-Tille, les Creux, on branch of Populus aff. nigra, 2 Sep. 2012, A. Gardiennet A.G. 12107 (WU 36920; ex-epitype culture CBS 142768 = BM; MBT377828).
Other material examined: Germany, Nordrhein-Westfalen, Münsterland, [Münster-] Nienberge; on wood of Populus sp. (originally given as Quercus). Dec. 1865, Th. Nitschke (B 70 0009349).
Notes: For synonyms, see Rappaz (1995). Concerning typification, Nitschke (1867) only cited material from Handorf on Sarothamnus in his protologue. In their list of type specimens of Nitschke deposited in B, Gerhardt and Hein (1979) mention two envelopes mounted on a sheet without a place or date on the envelopes. However, the holotype B 70 0009297 now only contains a single envelope with an asexual morph with hyaline conidia, i.e. no sexual morph is present. Therefore, epitypification became necessary. Rappaz (1995) selected B 70 0009349 as the lectotype, but that material was not cited in the protologue. It is, however, authentic material of Valsa macrospora (collected by Nitschke before publication), as both Nitschke and Rappaz considered it to be the fungus described in the protologue.
Barrmaelia macrospora is usually easy to identify due to its large and relatively narrow ascospores with one lighter coloured side. The inconspicuous germ slit was best visible in B 70 0009349 (Fig. 2t). Cannon (2015) provides a description of a slightly deviating British collection with larger, occasionally one-septate ascospores measuring (23.5–)26–29 × 7–8.5 μm, which may represent a distinct species.
Barrmaelia moravica (Petr.) Rappaz, Mycol. Helv. 7(1): 134 (1995). Fig. 3.
Fig. 3.
Barrmaelia moravica (a–c, f–o, v–y: WU 36924; d, e, p–u: W 1970-0024077, lectotype). a, d Ostioles protruding through the periderm. b Stroma beneath the periderm. c, e Perithecia in transverse section. f, g Perithecium in vertical section. h, i Vertical section of perithecial wall. j–u Ascospores. v, w Paraphyses apices. x, y Asci. All in water. Scale bars: a, b, d = 500 μm; c = 250 μm; e = 300 μm; f = 200 μm; g = 100 μm; h, i, x, y = 10 μm; j–w = 5 μm
Basionym. Eutypa moravica Petr., Ann. Mycol. 25(3/4): 224 (1927).
Stromata immersed in bark, covered by the periderm except for the ostiolar openings; in areas lacking periderm visible as black spots of up to 6 mm diam., not discolouring the periderm but sometimes blackening the bast around the perithecia. Ascomata perithecial, 300–700 μm (n = 15) diam., 200–500 mm high (n = 10), usually crowded to gregarious, rarely solitary, globose, ellipsoid to pyriform, contents whitish when immature, brown when mature. Ostioles conspicuous, papillate, often elongate, ostiolar pore rounded. Peridium 15–25 μm thick (n = 10), pseudoparenchymatous at the outer side and consisting of thick-walled dark brown cells, tending to be prosenchymatous, lighter coloured and thinner-walled at the inner side, partly filled with oil drops. Paraphyses numerous, narrowly thread-like, ca. 1–2 μm wide in the middle, filled with oil drops, tapering towards the apex. Asci 90–109 × 6–7 μm, spore part 63–100 μm, stipe 7–41 μm long (n = 20), cylindrical, containing eight obliquely uniseriate ascospores, with an inamyloid apical apparatus. Ascospores (12.3–)13.0–15.0(−16.3) × (2.3–)2.5–3.0(−3.3) μm, l/w = (4.2–)4.5–5.4(−6.0) (n = 60), one-celled, allantoid, without germ slit, light brown, filled with oil drops in the poles, smooth.
Colonies on CMD and MEA white; aerial hyphae abundant. No asexual morph observed.
Habitat: In bark of thin dead branches of Salix caprea attached to the tree.
Distribution: Europe (Austria, Czech Republic, Spain).
Typification. Lectotype of Eutypa moravica designated by Rappaz (1995): Czech Republic, Hranice (“Mährisch Weisskirchen“), Usti, on Salix caprea, Dec. 1925, F. Petrak (W 1970-0024077). Isotype: W 1978-0010895. Epitype of Eutypa moravica, here designated: Austria, Kärnten, Millstatt, Hinterdellach, on dead attached branch of Salix caprea, soc. Platystomum compressum, Cyphellopsis sp., Capronia sp., 3 Nov. 2015, W. Jaklitsch & H. Voglmayr (WU 36924; ex-epitype culture CBS 142769 = Cr1; MBT377829).
Notes: This species is well characterised by its light brown, allantoid and relatively small ascospores. Barrmaelia rappazii is superficially similar but differs morphologically mainly by larger and darker brown ascospores, and in having effused, black stromata with sparsely distributed perithecia. Barrmaelia rhamnicola also has allantoid ascospores but they are larger, filled with bigger oil drops and have a slightly darker colour, and it occurs on a different host.
Barrmaelia oxyacanthae (Mont.) Rappaz, Mycol. Helv. 7(1): 137 (1995). Fig. 4.
Fig. 4.
Barrmaelia oxyacanthae (a, d, e, g–n, p–u: WU 36925; b, c, f, o: PC 0706585, holotype). a–c Ostioles protruding through the blackened wood surface. d Perithecium in vertical section. e, f Perithecia in transverse section. g Vertical section of perithecial wall. h–o Ascospores (o in KOH + Melzer) with germ slits of spore-length (j–n). p–r Paraphyses apices. s–u Asci. All in water, except where noted. Scale bars: a–c = 500 μm; d, f = 250 μm; e = 350 μm; g, o, s–u = 10 μm; h–n, p–r = 5 μm
Basionym. Sphaeria oxyacanthae Mont., in Castagne, Suppl. Cat. Pl. Mars.: 48 (1851).
For synonyms, see Rappaz (1995).
Stromata discolouring the wood surface grey to black; wood usually showing no or only slight discolouration around the ascomata. Ascomata perithecial, 300–700 μm wide (n = 15), 300–500 μm high (n = 15), often closely spaced and arranged in lines, subglobose, ellipsoid to pyriform. Ostiolar necks with circular outline, ostioles shiny and slightly raised. Peridium 15–20 μm thick (n = 15), pseudoparenchymatous at the outer side and consisting of moderately thick-walled, dark brown cells, prosenchymatous, lighter coloured and thinner-walled at the inner side, partly filled with oil drops. Paraphyses numerous, 2–3.5 μm wide in the middle, filled with oil drops, slightly tapering towards the apex, obtuse. Asci 98–130 × 8–9 μm, spore part 73–100 μm long, stipe 15–34 μm long (n = 20), cylindrical, containing eight obliquely uniseriate ascospores, with an inamyloid apical apparatus. Ascospores (11.5–)12.3–14.2(−16.2) × (4.6–)5.3–6.3(−7.5) μm, l/w = (1.9–)2.1–2.5(−3.2) (n = 151), one-celled, ellipsoid, slightly inequilaterally, with a straight germ slit of spore-length (sometimes slightly shorter), brown to dark brown, filled with several small oil drops in the poles, smooth.
Colonies on CMD and MEA white; aerial hyphae abundant. No asexual morph observed.
Habitat: In wood of twigs and branches of various hardwoods.
Distribution: Widespread (Africa, Asia, Europe and North America); for details, see Cannon and Minter (2013).
Holotype: France, place and date unknown, in branches of Crataegus oxyacantha, soc. Sphaeria lata var. corticalis, L. Castagne (PC 0706585 ex herb. C. Montagne).
Other material examined: Austria, Steiermark, Deutschlandsberg, Koralpe, near the parking place of the walking path to the Grünanger- and Bärentalhütte; 15°00′52″E 46°49′37″N, on dead attached branches of Salix cf. caprea, 6 May 2016, G. Friebes (WU 36925; culture CBS 142770 = BO); Schadminger Tauern: Kleinsölk-Obertal, Schwarzensee, 1163 m, on Salix sp., 18 Sep. 1991, Ch. Scheuer 2897 (GZU 000317705). Germany, Sachsen, Königstein, on dead branches of Salix purpurea, Oct. 1880 and Apr. 1881, W. Krieger (GZU 000317701; as Anthostoma schmidtii); Schkeuditz, on dead branches of Fraxinus excelsior, spring 1874, G. Winter (GZU 000317700; as Anthostoma schmidtii). Italy, Venetia, Treviso, Selva, on decorticated dead branches of Castanea vesca, autumn 1873, P.A. Saccardo (GZU 000317702; as Anthostoma schmidtii). USA, South Dakota, Mellette, in a glacial valley, on branches of Fraxinus sp., Aug. 1950, F. Petrak (GZU 000317704; as Anthostoma melanotes); same data, 9 Aug. 1950, F. Petrak (GZU 000317703; as Anthostoma melanotes).
Notes: For synonyms, see Rappaz (1995). He found a libertella-like asexual morph in pure culture. Rappaz (1995) recognised three groups based on ascospore size within his broad concept of B. oxyacanthae. The first group with the smallest ascospores (“mean length between 12.5 and 13”) contains the type of B. oxyacanthae and agrees very well with GZU 000317702, whose mean length of 12.6 μm (n = 30) corresponds exactly with our measurements of the type collection. The sequenced collection WU 36925 has a mean length of 13.3 μm (n = 31) and, thus, appears to be an intermediate between the first and second group, the latter of which has a “mean length between 13.5–14”. The group with the longest ascospores (“between 14.5–15”) is said to mostly contain material on Salix. Of the three collections studied on this substrate, WU 36925 belongs to either the first or second group (see above), whereas GZU 000317705 falls in the second group (mean length 13.6 μm, n = 30) and GZU 000317701 best fits in the third group (mean length 14.3 μm, n = 30). GZU 000317700 does not contain mature ascomata. GZU 000317704 and GZU 000317703 from South Dakota (USA) have mean lengths of 16.2 and 19.6 (n = 30), respectively; thus, they likely represent different, probably undescribed, species.
Barrmaelia oxyacanthae differs from other Barrmaelia species in its relatively dark brown, ellipsoid ascospores with a well-visible germ slit. It is most similar to B. pseudobombarda, which has narrower ascospores (Rappaz 1995; Mathiassen et al. 2015). Cannon and Minter (2013) give a morphological description and illustrations of B. oxyacanthae and details on its ecology and distribution.
Barrmaelia rappazii Jaklitsch, Friebes & Voglmayr, sp. nov. Fig. 5.
Fig. 5.
Barrmaelia rappazii, holotype (WU 36926). a Stroma with perithecia in transverse section. b Stroma (arrow denoting ostiole). c Perithecium in vertical section. d Vertical section of perithecial wall. e Paraphyses apices. f–o Ascospores. p–r Asci. All in water. Scale bars: a = 1 mm; b, c = 500 μm; d, p–r = 10 μm; e–o = 5 μm
MycoBank no.: MB 822043
Etymology: In honour of F. Rappaz, who established the genus Barrmaelia.
Stromata discolouring the wood surface grey to black in areas extending up to 6 × 0.6 cm; wood internally either nearly white between ascomata or darkened in patches. Ascomata perithecial, (450–)560–795(−900) μm (n = 14) diam., (420–)480–635(−660) μm (n = 9) high, sparsely distributed within the stromata and distantly spaced, immersed, depressed globose to ellipsoid. Ostioles forming minute, shiny black, rounded papillae above the wood surface. Peridium 20–45 μm thick (n = 7), pseudo- to prosenchymatous, cells moderately thick-walled and encrusted with brown material. Paraphyses up to 3.2 μm wide in the lower part, tapering, ca. 1 μm wide at the apex, filled with numerous oil drops when vital. Asci 117–158 × 5.8–8.5 μm, spore part 95–136 μm long, stipe 11.5–29.5 μm long (n = 11), cylindrical, containing eight uniseriate ascospores, with an inamyloid apical apparatus. Ascospores (12.8–)15.5–18.0(−19.5) × (2.8–)3.0–3.5(−3.8) μm, l/w = (3.8–)4.5–5.7(−6.5) (n = 39), one-celled, allantoid, brown, without germ slit, filled with few small oil drops, smooth.
Colonies on CMD and MEA white; aerial hyphae abundant. No asexual morph observed.
Habitat: In wood of twigs and branches of Populus tremula.
Distribution: Europe, only known from the type location in Norway.
Holotype: Norway, Stange, Hedmark, Rotlia Naturreservat, 7.5 km S Stange Kirke, on decorticated wood of Populus tremula, soc. Platystomum compressum, 30 Nov. 2015, P. Vetlesen PV-R221 (WU 36926; ex-holotype culture CBS 142771 = Cr2).
Other material examined: USA, North Dakota, Nylands Grove, on Populus deltoides, 29 Mar. 1914, J.F. Brenckle (W 1978-0018347, as Anthostoma flavoviride).
Notes: Barrmaelia rappazii can be recognised by its allantoid, brown and relatively narrow ascospores without a germ slit, and its black stromata with sparsely distributed ascomata. Morphological differences to the most similar species, B. moravica, are given there. Barrmaelia rhamnicola is another species with allantoid ascospores without a germ slit but they contain larger oil drops and are somewhat longer and wider.
Rappaz (1995) mentions a collection (W 1978-0018347) similar to B. moravica but growing on Populus and having larger and darker ascospores, thus apparently resembling B. rappazii. However, the examination of this collection revealed it to be a different species with shorter and wider ascospores measuring (12.0–)14.0–16.0(−16.8) × (3.3–)3.7–4.5(−5.0) μm, l/w = (2.5–)3.2–4.3(−4.9) (n = 30), as was already indicated by the congruent measurements given in Rappaz (1995). In the absence of sequence data, we currently refrain from describing it as a new species.
Barrmaelia rhamnicola Rappaz, Mycol. Helv. 7(1): 130 (1995). Fig. 6.
Fig. 6.
Barrmaelia rhamnicola (a, c, e, g–l, s–u: WU 36927; d, m–o: WU 36928, epitype; b, f, p–r: F. Rappaz no 890611-2, LAU, holotype). a, b Ostioles protruding through the blackened wood surface. c Perithecia in transverse section. d–f Perithecia in vertical section. g Vertical section of perithecial wall. h Paraphyses apices. i–r Ascospores. s–u Asci (with paraphyses in u). All in water. Scale bars: a = 500 μm; b = 1 mm; c = 250 μm; d = 150 μm; e = 200 μm; f = 300 μm; g, h, s–u = 10 μm; i–r = 5 μm
Stromata discolouring the wood surface grey to black; wood showing no discolouration among ascomata. Ascomata perithecial, 200–500 μm wide (n = 15), 200–400 μm high (n = 10), usually gregarious but separate, rarely two in contact, immersed, outline in vertical section elliptic to broadly pyriform, in horizontal section broadly elliptic to round, contents whitish when immature, brown when mature. Ostiolar area often somewhat elongate but ostiolar necks with circular outline, ostioles rarely shiny and slightly raised. Peridium 20–25 μm thick (n = 15), pseudoparenchymatous at the outer side and consisting of thick-walled and dark brown cells, prosenchymatous, lighter coloured and thinner-walled at the inner side, partly filled with oil drops. Paraphyses numerous, 2–3 μm wide in the middle, filled with oil drops, slightly tapering towards the apex, obtuse. Asci 112–147 × 7–8 μm, spore part 81–110 μm long, stipe 14–32 μm long (n = 20), cylindrical, containing eight uni- or biseriate ascospores, with an inamyloid apical apparatus. Ascospores (14.8–)16.3–19.3(−21.3) × (3.3–)3.8–4.5(−5.0) μm, l/w = (3.4–)4.0–4.8(−5.3) (n = 90), smooth, one-celled, no sheath or appendages observed, without germ slit, usually slightly allantoid, light brown, filled with numerous oil drops (or two oil drops when dead).
Colonies on CMD and MEA white; aerial hyphae abundant. No asexual morph observed.
Habitat: In wood of decorticated dead twigs and branches of Rhamnus alpina.
Distribution: Europe (France, Spain, Switzerland).
Typification. Holotype: Switzerland, Vaud, les Rochers-de-Naye, Sautaudon, on Rhamnus alpina, June 1989, F. Rappaz no 890611-2 (LAU). Epitype of Barrmaelia rhamnicola, here designated: France, Côte-d’Or (21), Etaules, le Plain d’Avaux, on dead branch of Rhamnus alpina, 21 Jan. 2016, A. Gardiennet A.G. 16009 (WU 36927; ex-epitype culture CBS 142772 = BR; MBT377830).
Other specimens examined: France, Côte-d’Or (21), Chenôve, le Plateau, on dead branch of Rhamnus alpina, 26 Jan. 2016, A. Gardiennet A.G. 16011 (WU 36928; culture BR1). Spain, Asturias, Somiedo, way up to Altu de la Farrapona, Carboneo, ca. 1400 m s.m., on decorticated branches of Rhamnus alpina, 9 Jun. 2017, J. Fournier J.F. 17014 (WU 35984).
Notes: Barrmaelia rhamnicola is distinguished from other species of the genus by the often slightly curved, relatively large ascospores, which are filled with conspicuous oil drops and lack a germ slit. For comparison with the other allantoid-spored species without germ slit, see notes under B. moravica and B. rappazii. Rappaz (1995) observed a libertella-like asexual morph in pure cultures.
Entosordaria (Sacc.) Höhn., Sber. Akad. Wiss. Wien, Math.-naturw. Kl., Abt. 1129: 167 (1920), emend.
Synonym. Stereosphaeria Kirschst., Ann. Mycol. 37(1/2): 96 (1939).
Type species: Entosordaria perfidiosa (De Not.) Höhn.
Ascomata perithecial, scattered, immersed to erumpent, depressed globose to ellipsoid, circular in transverse section. Peridium brown. Hamathecium of apically free, thin, sparsely branched paraphyses. Asci unitunicate, cylindrical, with uniseriate ascospores; apex inamyloid without distinct ring or amyloid with a discoid ring. Ascospores two-celled with septum near one end, the small cell hyaline, the large cell dark brown and with an apical germ apparatus consisting of radial slits. Asexual morph unknown.
Notes: Entosordaria was first described as a subgenus of Anthostomella (Saccardo 1882) and raised to the generic rank by Höhnel (1920), with E. perfidiosa as the generic type. Eriksson (1966) outlined the fundamental morphological differences from Anthostomella, i.e. inamyloid asci and dorsiventrally flattened ascospores with an apical germ apparatus consisting of radiating slits. He confined Entosordaria to the generic type and removed the genus from the Xylariaceae. Later, he (in Eriksson and Hawksworth 1986) argued that Stereosphaeria is the valid generic name to be used, considering Entosordaria (Sacc.) Höhn. to be a younger heterotypic homonym of Entosordaria Speg. However, Entosordaria Speg. has not been validly described according to ICN Art. 38.1, as Spegazzini (1910) neither provided a diagnosis nor a reference to a previous valid description. Therefore, Entosordaria (Sacc.) Höhn. remains available and, based on priority, is the generic name to be used.
Barr (1989) classified E. perfidiosa in Clypeosphaeria, based on similarities of their ascospores, apical ascus apparatus, ascomata, clypei and peridium structure. However, molecular phylogenies do not support a close relationship, as the generic type, Clypeosphaeria mamillana, is placed in Xylariaceae s. str. with high support (Fig. 1).
With the addition of the closely related E. quercina, the genus Entosordaria also includes a species with an amyloid ascus ring, which shows that this character is not suitable for generic classification within Xylariales.
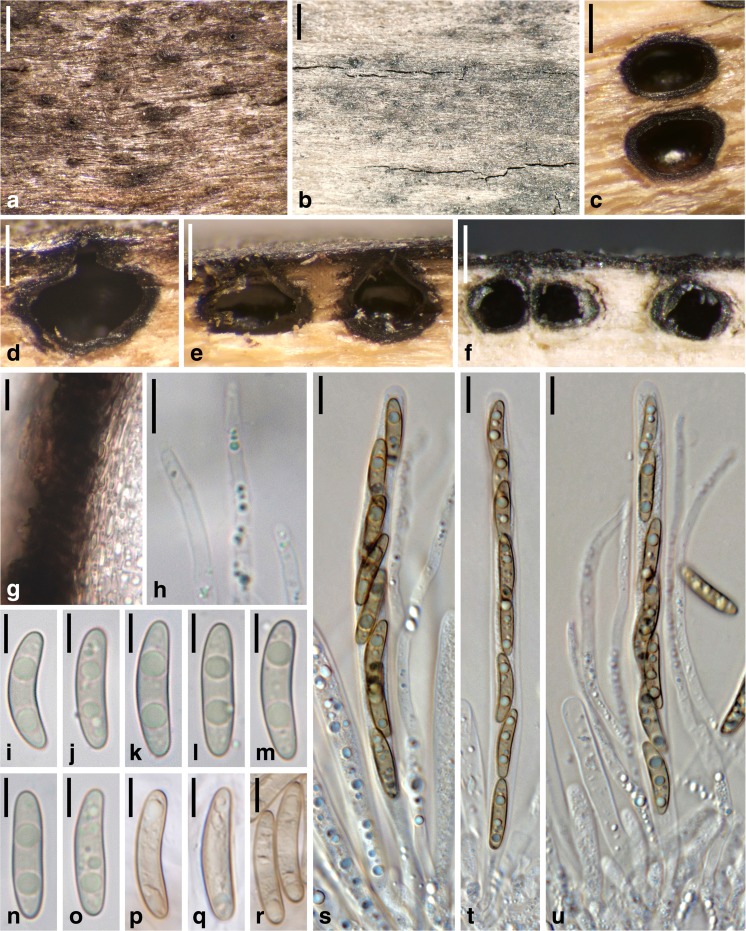
Entosordaria perfidiosa (De Not.) Höhn., Sber. Akad. Wiss. Wien, Math.-naturw. Kl., Abt. 1129: 166 (1920). Fig. 7.
Fig. 7.
Entosordaria perfidiosa (a–n, p–v: WU 35981, epitype; o, w–a1: WU 35982). a–c Erumpent ascomata with apical papilla in face view (two fused ascomata in c). d Two ascomata in transverse section. e Ascoma with apical papilla in side view. f Ascoma in vertical section. g Transverse section of perithecial wall (in 3% KOH). h–j Asci (in 3% KOH; with paraphyses in h). k, l Ascus apices (in 3% KOH + Lugol). m–a1 Ascospores (m–p immature; v in 3% KOH); the arrows denote radial slits of the apical germ apparatus (v, a1). All in water, except where noted. Scale bars: a = 1 mm; b–f = 200 μm; g, k–a1 = 10 μm; h–j = 20 μm
Basionym. Sordaria perfidiosa De Not., Comm. Soc. crittog. Ital. 2(fasc. 3): 481 (1867).
For synonyms, see Barr (1989).
Ascomata perithecial, scattered, solitary or in groups, partly immersed to erumpent, depressed globose to ellipsoid, circular in transverse section, 400–800 μm diam., with a distinct central apical papilla 120–200 μm wide. Peridium 20–40 μm wide, brown, pseudoparenchymatous, of dark brown isodiametric to elongate cells 2–12 μm diam. Paraphyses numerous, sparsely branched, 1–2 μm wide. Asci in 3% KOH (185–)200–220(−225) × (11–)12–14(−15) μm (n = 27), unitunicate, cylindrical, with a short stipe, with eight (partly obliquely) uniseriate ascospores, with an inamyloid apical apparatus, no apical ring seen. Ascospores (20.5–)21.8–25.8(−29.5) × (8.7–)10.0–11.2(−12.0) μm, l/w = (1.8–)2.1–2.5(−2.7) (n = 71), ellipsoid, with subacute to rounded ends, two-celled with septum near one end, the small cell hyaline, the large cell dark brown at maturity and with an apical germ apparatus consisting of radial slits, multiguttulate when young, at maturity often with a large and several small guttules.
Colonies on CMD and MEA white; aerial hyphae abundant. No asexual morph observed.
Habitat: In bark of old trunks of living Acer pseudoplatanus.
Distribution: Europe.
Typification. Lectotype of Sordaria perfidiosa, here designated: Italy, Riva, Corteccia dell’Acer pseudoplatanus, 30. Oct. 1863, Ab. Carestia, no. 413 (RO; MBT377831). Syntype: Riva, Sulla corteccia dell’Acer pseudoplatanus, 22 Apr. 1858, Ab. Carestia, no. 222 (RO). Epitype of Sordaria perfidiosa, here designated: Austria, Kärnten, St. Margareten im Rosental, at Brici (“Writze”), on bark of Acer pseudoplatanus, 10 Apr. 2016, H. Voglmayr & W. Jaklitsch (WU 35981; ex-epitype culture CBS 142773 = EPE; MBT377832).
Other material examined: Germany, Baden-Württemberg, Hornberg, on bark of Acer pseudoplatanus, Dec. 2015, B. Wergen (WU 35982; culture BW3). France, Hautes-Alpes (05), Vallouise-Pelvoux, Ailefroide, on bark of living trunk of Acer pseudoplatanus, soc. Decaisnella mesascium, 28 Jul. 2017, A. Gardiennet A.G. 17056.
Notes: Entosordaria perfidiosa is well characterised by the ascospores with an apical germ apparatus consisting of radiating slits in combination with inamyloid asci and its growth on bark of old Acer pseudoplatanus trees. It has been classified in Clypeosphaeria by Barr (1989); however, it is only distantly related with C. mamillana, the generic type (see Fig. 1). For comparison with E. quercina, see below.
Two syntypes of Sphaeria perfidiosa are present at RO, which were studied in detail by O. Eriksson (see Eriksson and Hawksworth 1986), but he did not select a lectotype. Type specimens at RO are no longer sent out for study, but detailed photographic documentation of the two syntypes was generously provided by Mrs. A. Millozza (pers. comm.). Based on the abundance of ascomata, we select no. 413 as the lectotype. For nomenclatural stability, a recent Austrian collection for which a culture and DNA sequences are available is designated as the epitype.
Entosordaria quercina Voglmayr & Jaklitsch, sp. nov. Fig. 8.
Fig. 8.
Entosordaria quercina, holotype (WU 35983). a, b Ascomata immersed in bark in face view. c Ascoma in transverse section. d Ascoma in vertical section. e–h Asci (with paraphyses in e, f). i–l Ascus apices (j in Lugol; k, l in 3% KOH + Lugol; note the gelatinous sheath surrounding the ascospores, the arrows in k and l denote the radial slits of the apical germ apparatus of the ascospore). m Transverse section of perithecial wall. n–y Ascospores; the arrows denote the short radial slits of the apical germ apparatus. All in 3% KOH, except where noted. Scale bars: a = 500 μm; b–d = 200 μm; e–h = 20 μm; i–y = 10 μm
MycoBank no.: MB 822044
Etymology: Referring to the host genus Quercus.
Ascomata perithecial, scattered, solitary, immersed below bark and raising it, depressed globose to ellipsoid, circular in transverse section, 400–800 μm diam., without an apical papilla, ostiole not to slightly protruding above cortex. Peridium 20–30 μm wide, brown, pseudoparenchymatous, of dark brown isodiametric to elongate cells 3–11 μm diam. Paraphyses numerous, sparsely branched, 1.5–2.5 μm wide. Asci in 3% KOH (258–)270–293(−310) × (17–)18.5–21.5(−22) μm (n = 17), unitunicate, cylindrical, with a short stipe, with eight (partly obliquely) uniseriate ascospores, ascus apex containing a discoid amyloid apical ring 5.3–6.8 × 1.3–1.8 μm (n = 15; in 3% KOH + Lugol). Ascospores (31–)34–38(−40) × 12–13.5(−16) μm, l/w = (2.4–)2.7–3.0(−3.2) (n = 50), ellipsoid to allantoid, two-celled with septum near one end, the small cell hyaline, the large cell dark brown at maturity and with an apical germ apparatus consisting of radial slits, multiguttulate when young, at maturity often with a large and several small guttules, surrounded by a gelatinous sheath quickly dissolving in water.
Colonies on CMD white, on MEA a reddish and yellowish pigment developing; aerial hyphae abundant. No asexual morph observed.
Habitat: In bark of dead twigs of Quercus coccifera.
Distribution: Only known from the type locality in Crete (Greece).
Holotype: Greece, Crete, Chania, Omalos, 920 m s.m., 35.37° N, 23.897° E, in bark of Quercus coccifera, 5 June 2015, H. Voglmayr & W. Jaklitsch (WU 35983; ex-holotype culture CBS 142774 = RQ).
Notes: Ascospore morphology of Entosordaria quercina fits E. perfidiosa, from which it differs in an amyloid ascus ring, much larger ascospores and asci, immersed ascomata without a distinct apical papilla and the host. In addition, the ascospores of E. quercina are commonly allantoid. The radiating slits of the apical germ apparatus are shorter than in E. perfidiosa; thus, they are less distinct.
Discussion
Phylogenetic relationships and familial classification within Xylariaceae sensu lato
Our phylogenetic analyses are fully concordant with Wendt et al. (2017) in revealing Hypoxylaceae, Graphostromataceae and Xylariaceae sensu stricto as highly supported distinct lineages within the former Xylariaceae sensu lato, with Hypoxylaceae being placed basal to the rest (Fig. 1). The highly supported Barrmaelia–Entosordaria clade is also contained within the highly supported Xylariaceae sensu lato, but not affiliated with any of these families; a sister group relationship to Xylariaceae sensu stricto receives only moderate support (71%) in ML analyses and is unsupported in the MP analyses. Therefore, to be consistent with the new familial classification of Wendt et al. (2017), the Barrmaelia–Entosordaria clade is classified here within the new family Barrmaeliaceae.
Our data also demonstrate that the genus Entosordaria is phylogenetically distinct from Clypeosphaeria, disproving the generic concept of Barr (1989). In the phylogenetic analyses of Jaklitsch et al. (2016) based on ITS–LSU sequence data, the generic type Clypeosphaeria mamillana was contained within Xylariaceae sensu lato, but its closest relatives remained unclear due to the lack of internal backbone support. In our multigene analyses, the phylogenetic position of Clypeosphaeria mamillana is now resolved to belong to Xylariaceae sensu stricto, where it forms a highly supported basal clade together with Anthostomelloides krabiensis, the generic type of Anthostomelloides (Fig. 1). Although both species differ in their ascospore characters, they share a similar wedge-shaped amyloid apical apparatus (Jaklitsch et al. 2016; Tibpromma et al. 2017). The second species of Anthostomelloides included in our analyses, A. forlicesenica, is not revealed as being closely related to the generic type, but as a sister species to Brunneiperidium gracilentum with high to maximum support (Fig. 1), with which it shares a discoid amyloid apical apparatus. This discrepancy in phylogenetic placement compared to Daranagama et al. (2016) may be caused by their obviously erroneous LSU and rpb2 sequences (see above), which were excluded from our analyses. This has been confirmed by an MP analysis of the matrix including the erroneous (pleosporalean) rpb2 and LSU sequences of A. forlicesenica, which result in an unsupported phylogenetic position of the latter as sister to the A. krabiensis–Clypeosphaeria clade (not shown).
As, apart from the commonly sequenced ITS–LSU rDNA, few sequence data are available for most lineages of Xylariales, the phylogenetic position of many taxa of putative xylariaceous affinities remains unresolved (Wendt et al. 2017). Whereas the ITS–LSU sequences are useful for barcoding purposes, molecular phylogenies solely based on these markers commonly do not provide sufficient phylogenetic resolution, and backbone support of many deeper nodes is often low (e.g. Jaklitsch and Voglmayr 2012; Jaklitsch et al. 2014, 2016). Considering the substantial increase of phylogenetic resolution observed in the multigene analyses of Wendt et al. (2017) and the current study (Fig. 1), rpb2 and tub2 should be included as standard markers in future phylogenetic studies of Xylariales, in addition to the usually sequenced ITS–LSU rDNA.
Anthostomella and Anthostomella-like genera
Recently, several investigations were published on Anthostomella (Daranagama et al. 2015, 2016; Tibpromma et al. 2017). In these publications, the genus Anthostomella was recognised to be polyphyletic, and several new genera and species were established.
Due to the lack of sequence data for rpb2 and tub2, only a subset of these taxa could be incorporated in our analyses. However, for most new Anthostomella-like genera, at least the generic type could be included, and we believe that some of the results are conclusive and should encourage more detailed studies and a critical evaluation of the published data. There are some topological differences between our analyses and those of Daranagama et al. (2015, 2016), which may be caused by the inclusion of some obviously erroneous sequences in the latter (see the Results section above). In our analyses, Pseudoanthostomella and Anthostomella in the sense of Daranagama et al. (2016) were united in a highly supported clade clearly placed outside Xylariaceae sensu lato (Fig. 1), whereas in Daranagama et al. (2016), Pseudoanthostomella and Anthostomella formed separate clades (clades A and C in their fig. 2). However, they only included members of Xylariaceae sensu lato in their analyses, with a single distantly related sordariomycete, Sordaria fimicola, as the outgroup, and internal support of the tree backbone relevant for the topology of Anthostomella-like fungi was low or absent. In our ML analysis, the Pseudoanthostomella–Anthostomella clade is the sister group of a highly supported clade containing Neoanthostomella viticola and Calceomyces lacunosus, the latter representing a genus of uncertain affinities within Xylariales (Wendt et al. 2017). The monotypic genus Alloanthostomella, introduced by Daranagama et al. (2016) for Anthostomella rubicola, is not supported in our analyses, as it is placed within the highly supported Anthostomella clade (Fig. 1), a position which was also revealed in Daranagama et al. (2015).
Our phylogenies suggest that the Pseudoanthostomella–Anthostomella clade may represent a distinct family (Fig. 1). However, we refrain from formally establishing a new family because the generic type, Anthostomella limitata, has not been sequenced, and it is, as yet, unclear whether Anthostomella in the sense of Daranagama et al. (2015, 2016) phylogenetically includes the generic type. Therefore, the correct application and circumscription of Anthostomella remains uncertain until sequences of the generic type become available.
In our molecular phylogenetic analyses, the Anthostomella-like Pyriformiascoma trilobatum is placed within Microdochium with maximum support (Fig. 1). The sexual morphs of Microdochium have thin-walled, hyaline to pale brown, fusoid ascospores with commonly variable but more or less regular septation and asci with a distinct funnel-shaped amyloid apical ascus apparatus (Parkinson et al. 1981; Jaklitsch and Voglmayr 2012; Hernández-Restrepo et al. 2016). Pyriformiascoma trilobatum differs substantially from all known sexual morphs of Microdochium by two-celled, inequilateral, oblong-ellipsoid ascospores consisting of a large olivaceous-brown cell and a hyaline dwarf cell and by an indistinct inamyloid apical ascus apparatus (Daranagama et al. 2015). Considering that Microdochium is morphologically homogeneous, it is unlikely that Pyriformiascoma belongs there, and the sequences of the latter may, rather, originate from a Microdochium contaminant. The “conidia” illustrated for P. trilobatum in Daranagama et al. (2015) recall unicellular terminal chlamydospores which are known from Microdochium species (Hernández-Restrepo et al. 2016). Pyriformiascoma trilobatum should be re-sequenced from verified cultures to ascertain its phylogenetic position.
An evaluation of published sequences reveals that sequence data quality should be critically checked by BLAST searches and detailed inspection of alignments before inclusion into phylogenetic analyses. An indicator for problems in the sequence data used for phylogenetic analyses are exceptionally long branch lengths in phylograms like, for example, those seen for some clades in Daranagama et al. (2016). Marked topological differences between our analyses and those of previous publications (Daranagama et al. 2015, 2016), but also between the latter, may, at least partly, be due to the inclusion of obviously inaccurate, dubious or erroneous sequences which have been identified and removed from our matrix. These errors may cast general doubts on the accuracy of the sequences published for these species, and all markers should be re-sequenced from verified material to corroborate their phylogenetic affinities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 11 kb)
Acknowledgements
Open access funding provided by Austrian Science Fund (FWF). We thank Rosella Marcucci at PAD for access to the fungarium and support, Anna Millozza at RO for providing detailed documentation and photographs of the syntypes of Sphaeria perfidiosa, the herbarium curators of B, LAU, PC and W for sending specimens and Christian Scheuer at GZU for managing collections. Per Vetlesen, Jacques Fournier and Björn Wergen are gratefully acknowledged for providing fresh specimens. Financial support by the Austrian Science Fund (FWF; project P27645-B16) to HV is gratefully acknowledged.
Footnotes
This article is part of the “Special Issue on ascomycete systematics in honour of Richard P. Korf who died in August 2016”.
Electronic supplementary material
The online version of this article (doi:10.1007/s11557-017-1329-6) contains supplementary material, which is available to authorized users.
References
- Barr ME. Clypeosphaeria and the Clypeosphaeriaceae. Syst Ascomycet. 1989;8:1–8. [Google Scholar]
- Cannon PF (2015) Barrmaelia macrospora. Fungi of Great Britain and Ireland. Available online at: http://fungi.myspecies.info/taxonomy/term/6795/descriptions
- Cannon PF, Minter DW (2013) Barrmaelia oxyacanthae. IMI Descriptions of Fungi and Bacteria. Set 195 No. 1941. 4 pp. CABI, Egham, Surrey, UK
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- Clements FE, Shear CL. The genera of fungi. New York: H.W. Wilson; 1931. [Google Scholar]
- Daranagama DA, Camporesi E, Tian Q, Liu XZ, Chamyuang S, Stadler M, Hyde KD. Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Divers. 2015;73:203–238. doi: 10.1007/s13225-015-0329-6. [DOI] [Google Scholar]
- Daranagama DA, Camporesi E, Jeewon R, Liu XZ, Stadler M, Lumyong S, Hyde KD. Taxonomic rearrangement of Anthostomella (Xylariaceae) based on a multigene phylogeny and morphology. Cryptogam Mycol. 2016;37:509–538. doi: 10.7872/crym/v37.iss4.2016.509. [DOI] [Google Scholar]
- de Hoog GS, Gerrits van den Ende AHG. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Eriksson O. On Anthostomella Sacc., Entosordaria (Sacc.) Höhn. and some related genera (Pyrenomycetes) Svensk Bot Tidskr. 1966;60:315–324. [Google Scholar]
- Eriksson O, Hawksworth DL. Notes on ascomycete systematics. Nos. 1–224. Syst Ascomycet. 1986;5:113–174. [Google Scholar]
- Francis SM. Anthostomella Sacc. (part I) Mycol Pap. 1975;139:1–97. [Google Scholar]
- Gerhardt E, Hein B. Die nomenklatorischen Typen der von Th. Nitschke beschriebenen Arten im Pilzherbar des Botanischen Museums Berlin-Dahlem [De herbario berolinensi notulae 12] Willdenowia. 1979;9:313–330. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Crous PW. Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia. 2016;36:57–82. doi: 10.3767/003158516X688676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhnel F. Fragmente zur Mykologie XXIV. Sitzungsber Akad Wiss Wien. Math-naturw Kl, Abt 1. 1920;129:137–184. [Google Scholar]
- Jaklitsch WM. European species of Hypocrea Part I. The green-spored species. Stud Mycol. 2009;63:1–91. doi: 10.3114/sim.2009.63.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales) Fungal Divers. 2012;52:75–98. doi: 10.1007/s13225-011-0104-2. [DOI] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 2005;97:1365–1378. doi: 10.1080/15572536.2006.11832743. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Stadler M, Voglmayr H. Blue pigment in Hypocrea caerulescens sp. nov. and two additional new species in sect. Trichoderma. Mycologia. 2012;104:925–941. doi: 10.3852/11-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Fournier J, Rogers JD, Voglmayr H. Phylogenetic and taxonomic revision of Lopadostoma. Persoonia. 2014;32:52–82. doi: 10.3767/003158514X679272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Gardiennet A, Voglmayr H. Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia. 2016;37:82–105. doi: 10.3767/003158516X690475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Lu BS, Hyde KD (2000) A world monograph of Anthostomella. Fungal Diversity Series 4:1–376. Fungal Diversity Press, Hong Kong
- Mathiassen G, Granmo A, Rämä T. Barrmaelia pseudobombarda (Ascomycota), a rare European species with a peculiar disjunct distribution. Österr Z Pilzk. 2015;24:9–13. [Google Scholar]
- Müller K. PRAP—computation of Bremer support for large data sets. Mol Phyl Evol. 2004;31:780–782. doi: 10.1016/j.ympev.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Nitschke TRJ (1867) Pyrenomycetes Germanici. Die Kernpilze Deutschlands bearbeitet von Dr. Th. Nitschke 1: i–ii, 1–160. Eduard Trewendt, Breslau
- O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phyl Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- Parkinson VO, Sivanesan A, Booth C. The perfect state of the rice leaf scald fungus and the taxonomy of both the perfect and imperfect states. Trans Brit Mycol Soc. 1981;76:59–69. doi: 10.1016/S0007-1536(81)80009-5. [DOI] [Google Scholar]
- Rappaz F. Anthostoma decipiens et sa position systématique. Mycol Helv. 1992;5:21–32. [Google Scholar]
- Rappaz F. Anthostomella and related xylariaceous fungi on hard wood from Europe and North America. Mycol Helv. 1995;7:99–168. [Google Scholar]
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- Saccardo PA. Sylloge fungorum 1:1–766. 1882. [Google Scholar]
- Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- Spegazzini CL (1910) Fungi Chilenses. Contribución al Estudio de los Hongos Chilenos [Chilean fungi]. Rev Fac Agron Vet Univ Nac La Plata ep. 2 6(1):1–205
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP* 4.0b10: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Massachusetts
- Thiers B. Index Herbariorum: a global directory of public herbaria and associated staff. 2017. [Google Scholar]
- Tibpromma S, Daranagama DA, Boonmee S, Promputtha I, Nontachaiyapoom S, Hyde KD. Anthostomelloides krabiensis gen. et sp. nov. (Xylariaceae) from Pandanus odorifer (Pandanaceae) Turkish J Bot. 2017;41:107–116. doi: 10.3906/bot-1606-45. [DOI] [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM. Prosthecium species with Stegonsporium anamorphs on Acer. Mycol Res. 2008;112:885–905. doi: 10.1016/j.mycres.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM. Molecular data reveal high host specificity in the phylogenetically isolated genus Massaria (Ascomycota, Massariaceae) Fungal Divers. 2011;46:133–170. doi: 10.1007/s13225-010-0078-5. [DOI] [Google Scholar]
- Voglmayr H, Rossman AY, Castlebury LA, Jaklitsch WM. Multigene phylogeny and taxonomy of the genus Melanconiella (Diaporthales) Fungal Divers. 2012;57:1–44. doi: 10.1007/s13225-012-0175-8. [DOI] [Google Scholar]
- Voglmayr H, Akulov OY, Jaklitsch WM. Reassessment of Allantonectria, phylogenetic position of Thyronectroidea, and Thyronectria caraganae sp. nov. Mycol Prog. 2016;15:921–937. doi: 10.1007/s11557-016-1218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Gardiennet A, Jaklitsch WM. Asterodiscus and Stigmatodiscus, two new apothecial dothideomycete genera and the new order Stigmatodiscales. Fungal Divers. 2016;80:271–284. doi: 10.1007/s13225-016-0356-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Castlebury LA, Jaklitsch WM. Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales) Persoonia. 2017;38:136–155. doi: 10.3767/003158517X694768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt L, Sir EB, Kuhnert E, Heitkämper S, Lambert C, Hladki AI, Romero AI, Luangsa-ard JJ, Srikitikulchai P, Peršoh D, Stadler M (2017) Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol Prog 1–40. doi:10.1007/s11557-017-1311-3
- Werle E, Schneider C, Renner M, Völker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 11 kb)