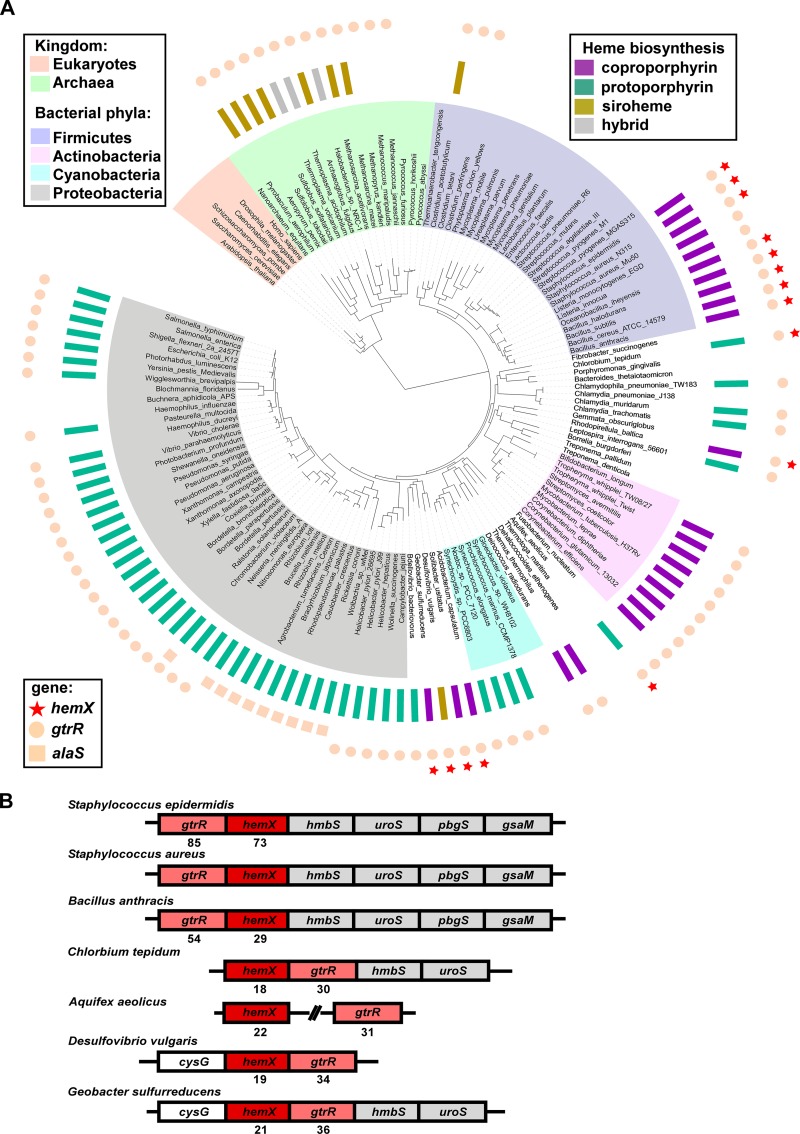
FIG 7 .
hemX is conserved across bacterial phyla and invariably co-occurs (A) and colocalizes (B) with gtrR. (A) The occurrence of hemX (stars), gtrR (circles), and alaS (squares) homologues (outermost rings) was mapped onto the tree of life (53). The pathway by which protoheme is synthesized in each of the analyzed organisms is presented in the middle ring as follows (adapted from reference 14): the classic protoporphyrin-dependent pathway (teal), coproporphyrin-dependent path (purple), or siroheme-dependent path (gold). Gray rectangles mark the organisms that contain unusual combinations of genes normally involved in different pathways for protoheme synthesis (hybrid paths [14]). The absence of a rectangle in the middle ring indicates the absence of any known route for protoheme synthesis in an organism. Likewise, the absence of a circle (gtrR) or square (alaS) in the outermost ring shows the inability of an organism to produce tetrapyrroles of any kind. Note that hemX does not occur in such organisms. (B) The immediate genomic neighborhood of the hemX gene in seven representative genomes, with ClustalW alignment scores for HemX and GtrR for each organism relative to S. aureus.