Abstract
Background:
There is increasing evidence that environmental, rather than genetic, factors are the major causes of most chronic diseases. By measuring entire classes of chemicals in archived biospecimens, exposome-wide association studies (EWAS) are being conducted to investigate associations between a myriad of exposures received during life and chronic diseases.
Objectives:
Because the intraindividual variability in biomarker levels, arising from changes in environmental exposures from conception onwards, leads to attenuation of exposure–disease associations, we posit that saliva can be collected repeatedly in longitudinal studies to reduce exposure–measurement errors in EWAS.
Methods:
From the literature and an open-source saliva–metabolome database, we obtained concentrations of 1,233 chemicals that had been detected in saliva. We connected salivary metabolites with human metabolic pathways and PubMed Medical Subject Heading (MeSH) terms, and performed pathway enrichment and pathway topology analyses.
Results:
One hundred ninety-six salivary metabolites were mapped into 49 metabolic pathways and connected with human metabolic diseases, central nervous system diseases, and neoplasms. We found that the saliva exposome represents at least 14 metabolic pathways, including amino acid metabolism, TCA cycle, gluconeogenesis, glutathione metabolism, pantothenate and CoA biosynthesis, and butanoate metabolism.
Conclusions:
Saliva contains molecular information worthy of interrogation via EWAS. The simplicity of specimen collection suggests that saliva offers a practical alternative to blood for measurements that can be used to characterize individual exposomes. https://doi.org/10.1289/EHP1011
Introduction
Because genetic factors typically account for only about 18% of chronic disease risks, it is reasonable to infer that nongenetic factors (i.e., exposures) are major causes of chronic diseases (Rappaport 2016). Given the myriad exposures from both exogenous and endogenous sources that an individual experiences during life [the “exposome” (Wild 2005)], investigators are performing exposome-wide association studies (EWAS) that interrogate levels of chemicals in biospecimens to discover causes of chronic diseases (Patel et al. 2010; Rappaport 2012; Wild et al. 2013). By measuring entire classes of chemicals (e.g., small molecules, protein modifications, antigens) in archived biospecimens from incident disease cases and matched controls, EWAS can pinpoint discriminating features that then generate hypotheses for targeted follow-up studies (Rappaport 2011; Rappaport et al. 2014). For example, Hazen and coworkers employed this avenue to implicate joint microbial/human metabolism of the nutrient choline as a potentially major cause of coronary heart disease (Wang et al. 2011; Tang et al. 2013; Koeth et al. 2013).
An important challenge to designing EWAS is the intraindividual variability in levels of circulating molecules arising from changes in diet, lifestyle factors, and sources of pollutants during decades of life that precede disease onset. This within-person variability in biomarker levels leads to exposure measurement errors that attenuate causal signals and obscure disease associations (Lin et al. 2005; Sampson et al. 2013). One way to circumvent such measurement errors is to perform longitudinal studies with repeated biospecimens, collected from subjects during critical stages of life (Rappaport 2011; Robinson and Vrijeheid 2015). The most logical approach for doing this relies on prospective cohorts that archived blood or other biospecimens repeatedly from the same subjects. However, such cohorts are rare and repeated collection of blood, the main archival specimen, is difficult to perform (Hansen et al. 2007; Randell et al. 2016).
Saliva (also referred as oral fluid) is a natural filtrate of blood that contains omic features (small molecules, metals, proteins, and DNA) worthy of interrogation via EWAS. Because collection is “stress-free,” repeated specimens of saliva are routinely obtained for determination of steroid hormones in psychobiological studies (Hjortskov et al. 2004; Kajantie and Phillips 2006; Hunter et al. 2011). Sampling of saliva is straightforward and protocols are available that allow subjects to collect their own samples and ship them to a laboratory or repository.
Metabolomics is recognized as a powerful top-down approach for detecting small molecules in biological matrices (Nicholson and Wilson 2003; German et al. 2005). These small molecules can be either substrates or end products of cellular metabolism and can originate from exogenous sources via inhalation, ingestion and dermal absorption, or from endogenous processes including human and microbial metabolism. Adductomics is another top-down technique that employs modifications of blood proteins like hemoglobin or human serum albumin (HSA) to characterize exposures to reactive electrophiles that are inherently toxic but cannot be measured directly in biospecimens (Rubino et al. 2009; Li et al. 2011; Carlsson et al. 2014; Grigoryan et al. 2016; Rappaport 2012). Because blood is in equilibrium with the tissues and saliva is in equilibrium with blood, both blood and saliva represent dynamic reservoirs of small molecules that are present in the body at a given time. Given the potential utility of saliva as a biospecimen for EWAS, we will evaluate the linkages between salivary metabolites and human metabolic pathways, as well as those between these pathways and chronic diseases. We will also consider methods for collection and analysis of saliva via untargeted metabolomics and adductomics.
Methods
Saliva Metabolome
Salivary metabolites () were obtained from the saliva metabolome database (http://www.salivametabolome.ca/) that was recently integrated into the Human Metabolome Database (HMDB) (Wishart et al. 2013; Dame et al. 2015). This database compiles important physical, chemical and biological information of metabolites derived from peer-reviewed articles, including chemical structure, chemical class, origin, biological function, and cellular location. Normal and abnormal concentrations are also reported for metabolites previously quantified in humans as well as standard deviations of the measurements that can be used in some cases to estimate the intra- and interindividual variability. It is important to note that this database was built from a recent study by Dame et al. who combined contemporary measurements of salivary metabolites in healthy subjects with literature data derived from the healthy and/or diseased subjects (Dame et al. 2015). Those investigators measured 308 salivary metabolites in 16 healthy adult subjects sampled once and two subjects sampled three times in a day (morning, midday, and afternoon/evening). They also reported levels of an additional 708 metabolites from peer-reviewed articles. All metabolites included in the saliva metabolome database were used for analyses.
Visualization of Human Metabolic Pathways
Salivary metabolites were connected to their human metabolic pathways using the Metscape 3.1 App (Karnovsky et al. 2012) for Cytoscape 3.2.1. (Shannon et al. 2003). The network of metabolites and reactions was built using the internal Metscape database that integrates metabolomics and gene-expression data derived from all available biospecimens, and compiled in the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Edinburgh Human Metabolic Network (EHMN) (Karnovsky et al. 2012). Nodes (i.e., metabolites) were colored according to their source (e.g., host, microbial). Edges connecting nodes represent KEGG and EHMN biochemical reactions. Only salivary metabolites with a KEGG IDs (196 metabolites) were retained with “human” as the model organism. The network type “compound” was used to map metabolic connections between small molecules.
Pathway Enrichment and Pathway Topology Analysis
Pathway enrichment and topology analyses of the saliva metabolome were performed using MetaboAnalyst 3.0 (Xia et al. 2015) with “Homo sapiens” as the model organism. Pathway enrichment analysis was conducted using the hypergeometric test to determine whether saliva metabolites were represented in a particular KEGG metabolic pathway more than expected by chance. indicate statistically significant representations of metabolic pathways. Pathway topology analysis was performed using the “relative-betweenness” centrality measure to estimate the importance of saliva metabolites relative to the biological pathway structure. Metabolite centrality is reported on a scale from 0 (isolated metabolite) to 1 (key metabolite).
Connections between the Saliva Metabolome and Human Chronic Diseases
Salivary metabolites were connected with human chronic diseases using the MetDisease App for Cytoscape 3.2.1 (Duren et al. 2014). MetDisease is a text-mining App that queried metabolites previously reported in any biospecimen as associated with PubMed Medical Subject Heading (MeSH) terms in peer-reviewed human and/or animal studies. For each disease, the total number of salivary metabolites, associated with a particular MeSH term, was determined. Salivary metabolites were queried using their KEGG IDs as identifiers and their shared names as select attributes. MeSH terms related to both chronic and nonchronic diseases (e.g., bacterial infections) were included in the network. The network of MeSH terms was built with Cytoscape 3.2.1 (Shannon et al. 2003) using the organic layer. The number of salivary metabolites associated with MeSH terms was used as the node attributes. Edges connecting nodes represented interconnections between MeSH terms.
Results and Discussion
The Saliva Metabolome
Saliva is a mixture of fluids originating mainly from the parotid, submandibular, sublingual, and minor salivary glands, and to a lesser extent from oral and nasal mucosa (de Almeida et al. 2008). Saliva consists of approximately 99% water with the remaining 1% comprised of electrolytes, mucus, cellular debris, proteins, and small molecules (Humphrey and Williamson 2001; de Almeida et al. 2008). Transfer of small molecules from blood to saliva relies on passive diffusion of lipophilic compounds and ultrafiltration of hydrophilic compounds of low molecular weight () (Gallardo and Queiroz 2008; de Almeida et al. 2008; Thieme 2012; Higashi 2012). The of a molecule plays an important role in transfer from blood to saliva. For example, basic molecules accumulate in saliva due to ion-trapping phenomena associated with their transfer from neutral blood (pH 7.4) to acidic saliva (pH 6) (Thieme 2012). Also, several factors including disease states, diet, drug consumption, and physical activity can significantly affect saliva excretion and saliva pH can be influenced by variation in bicarbonate concentrations (Aps and Martens, 2005).
Several studies have evaluated the correlation between saliva and blood concentrations for compounds with diverse physical and chemical properties, as summarized in Table 1. The median value of the Pearson correlation coefficients was 0.92 with a range of ). High correlations were observed between blood and saliva concentrations for neutral molecules that are not affected by changes in saliva pH (Hill et al. 2001; Sakaguchi and Hasegawa 2005; Juniarto et al. 2011; Gunnala et al. 2015). Lower correlations for acidic and basic compounds have been attributed to intraindividual variation in saliva pH as well as buccal contamination (for nicotine) (Fisher et al. 2013). However, these studies have been conducted only on a small subset of compounds (mainly drugs and pollutants). Because the blood–saliva transfer of small molecules relies on physical and chemical processes, more in-depth studies are needed to determine effects of , polarity, physical activity, diet, and disease state.
Table 1.
Blood:saliva concentration ratios and correlations for various classes of small molecules.
| Compound | Log Pa | Blood: saliva ratio (mean) | Correlation coefficient () | Reference | |
|---|---|---|---|---|---|
| Therapeutic drugs | |||||
| Clozapine | 3.7 | 15.9 | 3.6b | 0.728 | Fisher et al. 2013 |
| Norclozapine | 3.2 | 15.9 | 3.6b | 0.806 | Fisher et al. 2013 |
| Quetiapine | 2.9 | 15.1 | 3.0b | 0.843 | Fisher et al. 2013 |
| Risperidone | 3.4 | 8.8 | 2.6b | 0.954 | Fisher et al. 2013 |
| 9-hydroxyrisperidone | 2.3 | 13.7 | 2.5b | 0.640 | Fisher et al. 2013 |
| Alprazolam | 2.2 | 18.3 | 2.3 | ND | Gjerde et al. 2014 |
| Clonazepam | 2.8 | 11.9 | 7.1 | ND | Gjerde et al. 2014 |
| Diazepam | 2.6 | 2.9 | 27.0 | ND | Gjerde et al. 2014 |
| Nordiazepam | 2.8 | 12.3 | 22.0 | ND | Gjerde et al. 2014 |
| Busulfan | −0.9 | NA | 0.92b | 0.980 | Rauh et al. 2006 |
| Methylphenidate | 1.5 | 8.9 | 0.5b | ND | Seçilir et al. 2013 |
| Mycophenolic acid | 2.4 | 3.6 | NRb | 0.922 | Shen et al. 2009 |
| Voriconazole | 1.6 | 12.7 | 2.04b | 0.943 | Vanstraelen et al. 2015 |
| Illicit drugs | |||||
| Cocaine | 1.9 | 8.8 | 0.4b | ND | Moolchan et al. 2000 |
| Benzoylecgonine | 1.7 | 3.1 | 5b | ND | Moolchan et al. 2000 |
| Ecgonine methyl ester | 0.1 | 14.6 | 1b | ND | Moolchan et al. 2000 |
| Pollutants | |||||
| Nicotine | 0.9 | 8.9 | NRb | 0.300 | Shin et al. 2002 |
| Cotinine | 0.4 | 4.8 | 2.3b | 0.980 | Shin et al. 2002 |
| Perchlorate | −0.9 | −6.9 | 0.07c | 0.927 | Oldi et al. 2009 |
| Endogenous | |||||
| Cortisol | 1.8 | 12.6 | 0.87 | 0.933 | Gunnala et al. 2015 |
| 7-HydroxyDHEA | 1.8 | 18.2 | 4.5c | 0.975 | Hill et al. 2001 |
| Androstenedione | 2.9 | 19.0 | NRb | 0.781 | Juniarto et al. 2011 |
| 17-Hydroxyprogesterone | 2.9 | 12.7 | NRb | 0.964 | Juniarto et al. 2011 |
| Testosterone | 2.9 | 19.1 | NRc | 0.843 | Sakaguchi and Hasegawa 2005 |
Note: ND, not determined.
From in silico prediction using ALOGPS 2.1(http://www.vcclab.org/lab/alogps/).
Plasma:saliva ratio.
Serum:saliva ratio.
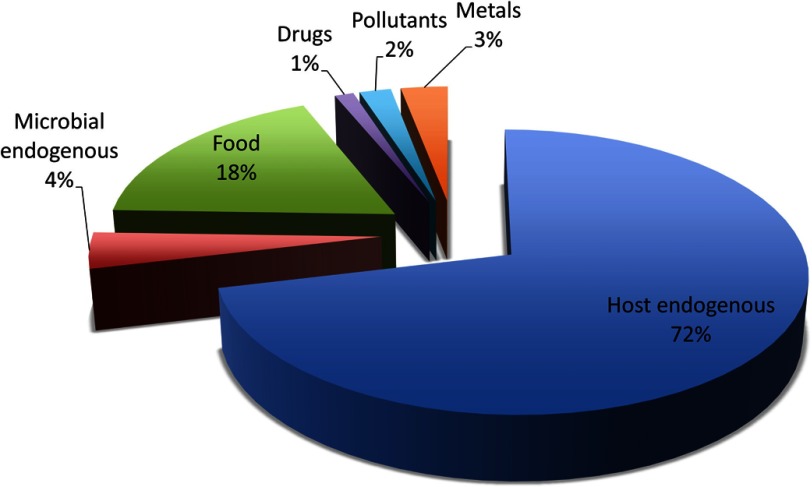
Dame et al. (2015) recently combined results from metabolomic analyses of human saliva with a literature review of salivary metabolites. This characterization of the saliva metabolome included 1,233 small molecules, metals and ions, which is about one-fourth of the 4,549 metabolites that have been reported in human blood (Wishart et al. 2013). The difference in coverage of the two metabolomes reflects the lower metabolite concentrations observed in saliva (nM to low ) compared with blood ( to mM). We classified the sources of these salivary molecules and metals as follows: a) host endogenous (879), b) microbial endogenous (52), c) food (225), d) drugs (15), e) pollutants (25), and f) metals (37) (Figure 1).
Figure 1.
Source category of salivary metabolites compiled from the saliva metabolome database (Dame et al. 2015).
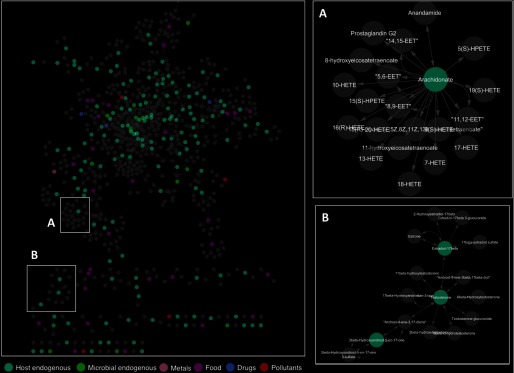
Using Metscape, we mapped the 1,233 salivary metabolites into biological pathways (Figure 2). Metscape uses an internal database that integrates metabolomic and gene-expression data from KEGG and EHMN (Karnovsky et al. 2012). Because only 196 of the 1,233 (16%) metabolites detected in saliva were included in these databases, an additional 529 metabolites were added to the network through linkages (i.e., direct neighbor in metabolic pathways) with the 196 salivary metabolites through 49 recognized pathways. Most of these additional 529 metabolites were either phase-II conjugates or compounds tightly bound to proteins. In saliva, metabolites are mainly present in the free form (i.e., unbound fraction) due to blood-saliva transfer processes. Phase-II conjugation increases the molecular weight, acidity, and hydrophilicity of each parent compound (Figure 2A), whereas protein binding increases the size of the compound. Both of these processes limit the efficiency of blood–saliva transfer. For compounds tightly bound to proteins, such as eicosanoids (markers of inflammation) (Figure 2A), the free form represents only about 1% of the total concentration in the systemic circulation (Brodersen et al. 1990; Fujiwara and Amisaki 2013; Bessonneau et al. 2015b). Salivary levels of these compounds are very low (nM) and, therefore, difficult to detect with untargeted analyses.
Figure 2.
Network of metabolic pathways reconstructed from metabolites detected in human saliva. Gray nodes represent metabolites that had not previously been detected in saliva but have direct neighbors in metabolic pathways. Edges represent biochemical connections between metabolites.
Next, we performed pathway enrichment and pathway topology analyses to identify the most significant pathways in the saliva exposome. Table 2 summarizes the 14 most significant metabolic pathways (), which prominently includes amino acid metabolism, TCA cycle, gluconeogenesis, glutathione metabolism, pantothenate and CoA biosynthesis, and butanoate metabolism.
Table 2.
Metabolic pathways represented by the saliva metabolome (metaboanalyst – homosapiens) for pathways having from pathway enrichment analysis. Impact values are derived from pathway topology analysis.
| Pathway | Total metabolitesa | Hitsb | p-valuec | Impactd |
|---|---|---|---|---|
| Alanine, aspartate, and glutamate metabolism | 24 | 13 | 0.88 | |
| beta-Alanine metabolism | 28 | 13 | 0.38 | |
| Phenylalanine metabolism | 45 | 18 | 0.45 | |
| Arginine and proline metabolism | 77 | 26 | 0.73 | |
| Nitrogen metabolism | 39 | 15 | 0.22 | |
| Citrate cycle (TCA cycle) | 20 | 9 | 0.43 | |
| Glycolysis or gluconeogenesis | 31 | 12 | 0.32 | |
| D-Glutamine and D-glutamate metabolism | 11 | 6 | 0.64 | |
| Glycine, serine, and threonine metabolism | 48 | 16 | 0.56 | |
| Glutathione metabolism | 38 | 13 | 0.35 | |
| Pantothenate and CoA biosynthesis | 27 | 10 | 0.35 | |
| Valine, leucine, and isoleucine biosynthesis | 27 | 10 | 0.36 | |
| Butanoate metabolism | 40 | 13 | 0.21 | |
| D-Arginine and D-ornithine metabolism | 8 | 4 | 0.50 |
Total number of metabolites involved in the pathway.
Number of salivary metabolites involved in the pathway.
Fisher’s exact test was used to measure the association between input metabolites (i.e., salivary metabolites) and pathways. indicate that associations are less likely due to random chance.
Impact represents, on a scale from 0 to 1, the centrality of salivary metabolites in their respective pathways.
Connections between the Saliva Metabolome and Human Diseases
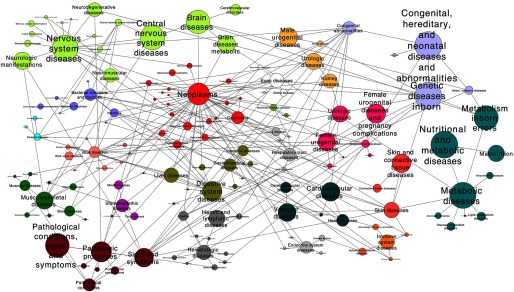
In order to provide additional biological context for the saliva metabolome, we evaluated associations between salivary metabolites and MeSH terms using MetDisease. The resulting network (Figures 3 and 4) indicates that salivary metabolites were associated with most human diseases (Table 3). For example, substantial evidence from breast cancer epidemiology shows the influence of hormones at different stages of women’s development. A nested case–control study found that premenopausal concentrations of testosterone were associated with breast cancer risk (Zeleniuch-Jacquotte et al. 2012). Another case–control study reported lower salivary testosterone levels in women with breast cancer, suggesting a protective effect of testosterone (Dimitrakakis et al. 2010). The carboxylic acid ester phenylacetate and its analogs have also been linked to breast neoplasms (Sawatsri et al. 2001) due to their antiproliferative activities on human breast cancer cells. Recently, targeted metabolomics of salivary levels of polyamines such as spermidine has discriminated breast cancer patients from healthy subjects (Takayama et al. 2016). Polyamines are essential molecules for normal cell growth and regulate gene expression by modulating ligand–receptor interactions, including estradiol binding to estrogens receptors (Cervelli et al. 2014). Previous studies have attributed the proliferation of highly invasive breast cancer tumor cells to the upregulation of polyamine metabolism (Cervelli et al. 2014).
Figure 3.
Network of PubMed Medical Subject Headings (MeSH) terms reported associated with salivary metabolites. The size of a node and text reflects the number of metabolites associated with MeSH terms. Edges represent links between MeSH terms.
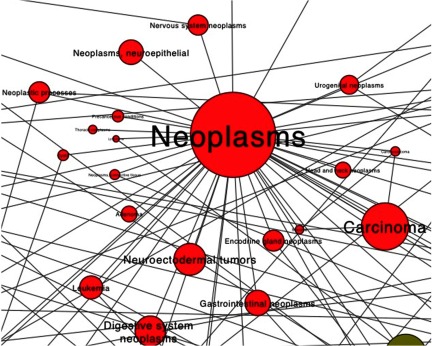
Figure 4.
Subnetwork of neoplasm-related PubMed Medical Subject Headings (MeSH) terms reported associated with salivary metabolites. The size of a node and text reflects the number of metabolites associated with MeSH terms. Edges represent links between MeSH terms.
Table 3.
Summary of the most important human chronic diseases associated with salivary metabolites.
| Human chronic disease | Number of salivary metabolites |
|---|---|
| Congenital, hereditary, and neonatal diseases and abnormalities | 145 |
| Nutritional and metabolic diseases | 144 |
| Central nervous system diseases | 138 |
| Neoplasms | 118 |
| Digestive system diseases | 105 |
| Urogenital diseases | 103 |
| Cardiovascular diseases | 101 |
| Immune system diseases | 76 |
| Respiratory tract diseases | 69 |
| Endocrine system diseases | 64 |
There is also growing evidence that the human gut microbiome and its complex interactions with exogenous exposures play roles in disease processes (Jiménez et al. 2008; Orešič et al. 2008; Nicholson et al. 2012). Of the 14 metabolites of known microbial origin, 12 were associated with congenital, hereditary, and neonatal diseases, 11 with nervous system diseases, 10 with nutritional and metabolic diseases, eight with digestive system diseases, and seven each with neoplasms and urogenital and pregnancy complications.
The above analyses suggest that the saliva metabolome captures a biologically meaningful fraction of exposures that are associated with human diseases. Thus, any discriminating salivary molecules detected by EWAS should be well worth following up in targeted studies to examine sources, causality, disease mechanisms, and interventions (Rappaport 2012). We envision that creating exposure terms, similar to MeSH terms indexed to the peer-reviewed literature, would allow annotation of metabolites based on their origins and facilitate investigation of exposure sources and temporality of exposure–disease associations. For example, Scalbert et al. (2014) performed a comprehensive review of metabolites associated with specific food groups (e.g., red meat, fish, vegetables) from human dietary interventions and cross-sectional studies. They reported that many metabolites were significantly correlated (Pearson correlation , ) with food consumption; e.g., apple consumption was associated with kaempferol, isorhamnetin, m-coumaric acid and phloretin, carrot consumption with α-carotene, and soy consumption with daidzein, genistein, isoflavones, and O-desmethylangolensin. Likewise, Rappaport et al. (2014) compiled literature values for 94 environmental chemicals and nutrients that have been measured in the general population.
Saliva as a Convenient Biospecimen for Longitudinal Studies
Saliva offers several advantages over traditional biospecimens archived in population-based studies. As with blood, saliva provides a snapshot of the internal exposome at the time of collection. However, whereas blood sampling usually requires a trained phlebotomist, saliva can be collected by the subjects themselves, using commercial kits (Shirtcliff et al. 2000; Abraham et al. 2012). This advantage should increase participation rates in cohort studies, particularly when repeated sampling is desired. In a recent study of inflammatory bowel disease, Randell et al. (2016) reported that the participation rate was significantly higher across 591 participants when they were asked to contribute a saliva sample (38% participation), compared with blood sample (23% participation). Similarly, Hansen et al. (2007) found a participation rate of 72% when individuals were asked to deliver saliva samples for DNA analysis, compared with 31% for venous blood samples collected at a medical facility. Another investigation revealed that the participation rates for collection of saliva samples and dried blood spots (DBS) were equal at 71.0% for 4,600 women (Sakhi et al. 2015). Although DBS specimens are also minimally invasive and amenable to self-collection by participants, the volume of blood is small (approximately per drop) and the analytical matrix consists of both serum and hemolysate, thereby complicating untargeted analysis (Vuckovic 2012). Also, it is unclear whether subjects would be amenable to repeated collection of DBS, which does involve some discomfort. Thus, saliva specimens offer an attractive alternative to blood sampling for investigating individual exposomes with the high temporal resolution (i.e., repeated measurements) needed to explore health trajectories. Several studies have found low day-to-day variability in salivary levels of tightly regulated metabolites such as steroid hormones (Gann et al. 2001; Viardot et al. 2005). For example, Gann et al. (2001) observed that the intraclass correlation for peak progesterone between two consecutive menstrual cycles in healthy premenopausal women ranged from 0.72 to 0.76, indicating that cycle-to-cycle variability within women was smaller than variability across women.
Analytical Considerations
Saliva can easily be collected with a variety of commercial devices such as Salivette® or Drugwipe®. The most common approach is to collect saliva with an absorbent pad or swab, from which saliva is then recovered in the laboratory by centrifugation (Drummer 2008). Prior to sampling, saliva production can be stimulated with citric acid and by chewing of gum or sterile pads (Gröschl et al. 2008; Lund et al. 2011). This approach is often used to increase the volume of saliva collected and to control the pH so as to standardize the transfer of acidic and basic molecules. Unstimulated saliva can be collected passively with a collection tube or an oral swab (Drummer 2008). Collection devices should be selected carefully because some media, including cotton and polypropylene pads, can nonspecifically bind small molecules (Gallardo and Queiroz 2008; de Almeida et al. 2008; Gröschl et al. 2008; Higashi 2012). Given that the buccal cavity can be contaminated by components originating from previous oral ingestion (Shin et al. 2002), it is useful for subjects to refrain from eating and drinking for a few hours prior to saliva collection. Once collected, saliva samples can be easily transported by mail to the laboratory for analysis. Because a large number of salivary metabolites such as amino acids, steroids, or fatty acids are under circadian control (i.e., high diurnal variation) (Dallmann et al. 2012; Dame et al. 2015), it is important to collect repeated samples at similar times (e.g., all samples collected during early morning). Saliva is also an attractive alternative matrix for pediatric populations, due to the noninvasiveness of the sample collection. Although several collection devices have been specifically developed for children 6 mo–6 y old, more studies are required to address the safety of saliva sampling devices and procedures in young children.
Several studies have demonstrated that various classes of compounds, including illicit drugs (Lund et al. 2011), therapeutic drugs (Mendonza et al. 2006; Moore et al. 2007; Ogawa et al. 2014), pollutants (Bentley et al. 1999; Wang and Lu 2009; Gherardi et al. 2010), and endogenous steroids (Higashi et al. 2011; Alvi et al. 2013) are stable in saliva for a few days at room temperature and for at least 1 year when stored at low temperature ().
Sample Preparation
The strategy for preparing samples plays an important role in the quality of metabolomics data. Issues related to the quenching of metabolism prior to analysis, ion suppression during mass spectrometry and metabolite instability can adversely impact the interpretation of data from untargeted analysis. The most common techniques for preparing saliva samples for mass spectrometry are liquid–liquid extraction (LLE), protein precipitation (PP), and solid-phase extraction (SPE) (Higashi 2012). Although LLE and PP provide exhaustive extraction of small molecules, such procedures are labor intensive, difficult to automate, require multistep sample handling, and are prone to suppression or enhancement of ionization in mass spectrometric analysis. Most of metabolites in the saliva metabolome database have been obtained through LLE or PP of saliva samples. These analytical techniques often used an internal standard in order to correct for possible loss and/or degradation during sample preparation and for matrix effects during analysis.
In contrast to LLE and PP, solid-phase microextraction (SPME) offers a simple, fast, and sensitive technique for preparing saliva and other biological fluids for metabolomics analysis (Bojko et al. 2014; Bessonneau et al. 2013, 2015a). In fact, when coupled to the Concept-96 system (employing 96-well plates), thin-film SPME (TF-SPME) motivates automation of all preconditioning, extraction, rinsing, and desorption steps (Jiang and Pawliszyn 2012). For example, preparation for saliva via TF-SPME can be achieved in less than 2 min per sample and prevents matrix effects and adsorption of macromolecules compared with LLE (Bessonneau et al. 2015a).
The small size and biocompatibility of SPME materials also permits in vivo sampling of saliva (Bessonneau et al. 2015a). By placing the SPME probe in the mouth for a short period of time, this technique can be used for rapid extraction and stabilization of metabolites for analysis (Bessonneau et al. 2015b). For example, SPME coatings were able to rapidly stabilize highly reactive metabolites (i.e., eicosanoids) formed in vivo and prevented their autooxidation during storage (Bessonneau et al. 2015b). Although promising, in vivo sampling of saliva with SPME is a new technique that will require substantial validation to address variability arising from salivary flow rate, pH, sample volume, and agitation.
The Saliva Adductome
Biotransformation of dietary chemicals and pollutants leads to the formation of reactive electrophiles, including reactive oxygen and nitrogen species, aldehydes, oxiranes, and quinones that can modify DNA, proteins, and lipids and can lead to dysregulation of homeostasis (Farmer and Davoine 2007; Fritz and Petersen 2013). Because reactive species are unstable, it is difficult to measure them in their free-circulating form. However, one can detect adducts formed from reactions between reactive electrophiles and nucleophilic residues on DNA and proteins. Several targeted studies have reported correlations between adducts of reactive electrophiles from endogenous and exogenous sources with DNA (Sturla 2007,) and prominent blood proteins, that is, hemoglobin (Hb) and human serum albumin (HSA) [reviewed by Törnqvist et al. 2002; Rubino et al. 2009].
Both DNA and proteins are also present in saliva, albeit at lower concentrations than in blood. For example, HSA is found in saliva at concentrations of about compared with in serum (Wang et al. 2012; Shaila et al. 2013; Metgud and Patel 2014; Nam et al. 2015); whereas Hb is present at a 50,000-fold lower concentration in saliva () compared with blood () (Nomura et al. 2012). For DNA, mean salivary levels () are 2-fold lower than those in blood () (Abraham et al. 2012). Although the analysis of salivary adducts is still in its infancy, targeted studies have reported adducts of salivary DNA with electrophiles from smoking and dietary sources (Bessette et al. 2010; Chen and Lee 2014; Chen and Lin 2014), and of HSA adducts in nasal lavage fluid from workers exposed to reactive electrophiles (Kristiansson et al. 2004; Jeppsson et al. 2009).
Untargeted analysis of adducts of DNA, HAS, and Hb has motivated adductomics for untargeted characterization of exposures to reactive electrophiles (Rappaport et al. 2012). That is, an adductome represents the totality of adducts generated from reactions between reactive electrophiles and a particular nucleophilic locus on one of these molecules. By characterizing a complete adductome, it is possible to map systemic exposures that occurred over the in vivo residence time of the nucleophile, which can range from hours to months. Particular attention has been paid to mass spectrometric characterization of DNA adductomes in human tissue samples (reviewed by Balbo et al. 2014) and the blood proteins, HSA (Li et al. 2011; Grigoryan et al. 2016) and Hb (Carlsson et al. 2014).
Here, we posit that salivary HSA and Hb residence times would be similar to those in blood (i.e., 30 days for HSA and 60 days for Hb), which are much longer than those for DNA (a few days). Consequently, by performing untargeted mass spectrometry of HSA and Hb adducts in saliva samples, it should be possible to characterize systemic exposures to reactive electrophiles during 1–2 mo prior to specimen collection. With repeated saliva sampling every few years, one can construct an individual exposure history of relevance to a person’s health trajectory.
Conclusions
Saliva contains a rich set of molecular information for circulating chemicals that can be interrogated via EWAS, with metabolomics and adductomics, to discover exposure–risk factors for chronic diseases. Although the number of metabolites detected in saliva is smaller than that in blood, we can anticipate that analytical improvements will uncover many additional salivary metabolites, present at low concentrations.
Given the simplicity and noninvasiveness of specimen collection, saliva offers a practical alternative to blood for longitudinal measurements of individual’s exposomes. Several studies have shown that participants are more willing to donate saliva specimens than venous blood, and saliva can be collected by the subjects themselves. This would eventually improve participation rates in cohort studies and, therefore, generate larger sets of biospecimens.
Data-driven studies that utilize repeated omic measurements from individuals at different stages of life should reduce exposure measurement errors and thereby increase power to discover unknown causes of chronic diseases. Given the ease of collection, saliva could well be the specimen of choice for obtaining repeated samples to profile small molecules, DNA, and proteins.
Acknowledgments
Support for this work was provided by the Natural Sciences and Engineering Research Council of Canada Industrial Research program and the Canada Research Chairs program (V.B. and J.P.) and by the U.S. National Institutes of Health through grants P42ES04705 and R33CA191159 (S.M.R.).
References
- Abraham JE, Maranian MJ, Spiteri I, Russell R, Ingle S, Luccarini C, et al. 2012. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics 5:19, 10.1186/1755-8794-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi SN, Al Dgither S, Hammami MM. 2013. Development and validation of LC-MS/MS method for determination of testosterone level in human saliva using lovastatin as internal standard. J Bioequiv Availab 5:228–232. [Google Scholar]
- Aps JK, Martens LC. 2005. Review: the physiology of saliva and transfer of drugs into saliva. Forensic Sci Int 150:119–131, PMID: 15944052, 10.1016/j.forsciint.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Balbo S, Turesky RJ, Villalta PW. 2014. DNA adductomics. Chem Res Toxicol 27:356–366, PMID: 24437709, 10.1021/tx4004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley MC, Abrar M, Kelk M, Cook J, Phillip K. 1999. Validation of an assay for the determination of cotinine and 3-hydroxycotinine in human saliva using automated solid-phase extraction and liquid-chromatography with tandem mass spectrometric detection. J Chromatogr B Biomed Sci Appl 723:185–194, 10.1016/S0378-4347(98)00494-0. [DOI] [PubMed] [Google Scholar]
- Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, et al. 2010. Identification of carcinogen DNA adducts in human saliva by linear quadrupole ion trap/multistage tandem mass spectrometry. Chem Res Toxicol 23:1234–1244, PMID: 20443584, 10.1021/tx100098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonneau V, Bojko B, Pawliszyn J. 2013. Analysis of human saliva metabolome by direct immersion solid-phase microextraction LC and benchtop orbitrap MS. Bioanalysis 5:783–792, PMID: 23534423, 10.4155/bio.13.35. [DOI] [PubMed] [Google Scholar]
- Bessonneau V, Boyaci E, Maciazek-Jurczyk M, Pawliszyn J. 2015a. In vivo solid-phase microextraction sampling of human saliva for non-invasive and on-site monitoring. Anal Chim Acta 856:35–45. [DOI] [PubMed] [Google Scholar]
- Bessonneau V, Zhan Y, Lannoy IAM, Saldivia V, Pawliszyn J. 2015b. In vivo solid-phase microextraction liquid-chromatography-tandem mass spectrometry for monitoring blood eicosanoids time profile after lipopolysaccharide-induced inflammation in Sprague-Dawley rats. J Chromatogr A 1424:134–138. [DOI] [PubMed] [Google Scholar]
- Bojko B, Reyes-Garcés N, Bessonneau V, Goryński K, Mousavi F, Souza Silva EA, et al. 2014. Solid-phase microextraction in metabolomics. Trends Anal Chem 61:168–180, 10.1016/j.trac.2014.07.005. [DOI] [Google Scholar]
- Brodersen R, Andersen S, Vorum H, Nielsen SU, Pedersen O. 1990. Multiple fatty acid binding to albumin in human blood plasma. Eur J Biochem 189:343–349, PMID: 2338079. [DOI] [PubMed] [Google Scholar]
- Carlsson H, von Stedingk H, Nilsson U, Törnqvist M. 2014. LC-MS/MS screening strategy for unknown adducts to N-terminal valine in hemoglobin applied to smokers and nonsmokers. Chem Res Toxicol 27:2062–2070, PMID: 25350717, 10.1021/tx5002749. [DOI] [PubMed] [Google Scholar]
- Cervelli M, Pietropaoli S, Signore F, Amendola R, Mariottini P. 2014. Polyamines metabolism and breast cancer: state of the art and perspectives. Breast Cancer Res Treat 148:233–248, PMID: 25292420, 10.1007/s10549-014-3156-7. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Lee CR. 2014a. Detection and simultaneous quantification of three smoking-related ethylthymidine adducts in human salivary DNA by liquid chromatography tandem mass spectrometry. Toxicol Lett 224:101–107. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Lin CR. 2014b. Noninvasive measurement of smoking-associated N(3)-ethyladenine and N(7)-ethylguanine in human salivary DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Toxicol Lett 225:27–33. [DOI] [PubMed] [Google Scholar]
- Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. 2012. The human circadian metabolome. Proc Natl Acad Sci U S A 109:2625–2629, PMID: 22308371, 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame ZT, Aziat F, Mandal R, Krishnamurthy R, Bouatra S, Borzouie S, et al. 2015. The human saliva metabolome. Metabolomics 11:1864–1883, 10.1007/s11306-015-0840-5. [DOI] [Google Scholar]
- de Almeida Pdel V, Grégio AM, Machado MA, de Lima AA, Azevedo LR. 2008. Saliva composition and functions: a comprehensive review. J Contemp Dent Pract 9:72–80, PMID: 18335122. [PubMed] [Google Scholar]
- Dimitrakakis C, Zava D, Marinopoulos S, Tsigginou A, Antsaklis A, Glaser R. 2010. Low salivary testosterone in patients with breast cancer. BMC Cancer 10:547–554, PMID: 20937135, 10.1186/1471-2407-10-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummer OH. 2008. Introduction and review of collection techniques and applications of drug testing of oral fluid. Ther Drug Monit 30:203–206, PMID: 18367981, 10.1097/FTD.0b013e3181679015. [DOI] [PubMed] [Google Scholar]
- Duren W, Weymouth T, Hull T, Omenn GS, Athey B, Burant C, et al. 2014. MetDisease—connecting metabolites to diseases via literature. Bioinformatics 30:2239–2241, PMID: 24713438, 10.1093/bioinformatics/btu179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Davoine C. 2007. Reactive electrophile species. Curr Opin Plant Biol 10:380–386, PMID: 17646124, 10.1016/j.pbi.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Fisher DS, van Schalkwyk GI, Seedat S, Curran SR, Flanagan RJ. 2013. Plasma, oral fluid, and whole-blood distribution of antipsychotics and metabolites in clinical samples. Ther Drug Monit 35:345–351, PMID: 23666566, 10.1097/FTD.0b013e318283eaf2. [DOI] [PubMed] [Google Scholar]
- Fritz KS, Petersen DR. 2013. An overview of the chemistry and biology of reactive aldehydes. Free Radic Biol Med 59:85–91, PMID: 22750507, 10.1016/j.freeradbiomed.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Amisaki T. 2013. Fatty acid binding to serum albumin: molecular simulation approaches. Biochim Biophys Acta Gen Subj 1830:5427–5434, 10.1016/j.bbagen.2013.03.032. [DOI] [PubMed] [Google Scholar]
- Gann PH, Giovanazzi S, Van Horn L, Branning A, Chatterton RT Jr. 2001. Saliva as a medium for investigating intra- and interindividual differences in sex hormone levels in premenopausal women. Cancer Epidemiol Biomarkers Prev 10:59–64, PMID: 11205490. [PubMed] [Google Scholar]
- Gallardo E, Queiroz JA. 2008. The role of alternative specimens in toxicological analysis. Biomed Chromatogr 22:795–821, PMID: 18506679, 10.1002/bmc.1009. [DOI] [PubMed] [Google Scholar]
- German JB, Hammock BD, Watkins SM. 2005. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics 1:3–9, PMID: 16680201, 10.1007/s11306-005-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardi M, Gordiani A, Gatto M. 2010. Development and validation of method for analysis of some ototoxic solvents in saliva matrix by headspace gas chromatography/mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 878:2391–2396, PMID: 20727840, 10.1016/j.jchromb.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Gjerde H, Langel K, Favretto D, Verstraete AG. 2014. Detection of 4 benzodiazepines in oral fluid as biomarker for presence in blood. Ther Drug Monit 36:252–256, PMID: 24061449, 10.1097/FTD.0b013e3182a3ab42. [DOI] [PubMed] [Google Scholar]
- Gröschl M, Köhler H, Topf HG, Rupprecht T, Rauh M. 2008. Evaluation of saliva collection devices for the analysis of steroids, peptides and therapeutic drugs. J Pharm Biomed Anal 47:478–486, PMID: 18325706, 10.1016/j.jpba.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Grigoryan H, Edmands W, Lu SS, Yano Y, Regazzoni L, Iavarone AT, et al. 2016. An adductomics pipeline for untargeted analysis of modifications to Cys34 of human serum albumin. Anal Chem , 10.1021/acs.analchem.6b02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnala V, Guo R, Minutti C, Durazo-Arvizu R, Laporte C, Mathews H, et al. 2015. Measurement of salivary cortisol level for the diagnosis of critical illness-related corticosteroid insufficiency in children. Pediatr Crit Care Med 6(4):e101–e106, 10.1097/PCC.0000000000000361. [DOI] [PubMed] [Google Scholar]
- Hansen TO, Simonsen MK, Nielsen FC, Hundrup YA. 2007. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol Biomarkers Prev 16:2072–2076, 10.1158/1055-9965.EPI-07-0611. [DOI] [PubMed] [Google Scholar]
- Higashi T. 2012. Salivary hormone measurement using LC/MS/MS: specific and patient-friendly for assessment of endocrine function. Bio Pharm Bull 35:1401–1408, 10.1248/bpb.b212009. [DOI] [PubMed] [Google Scholar]
- Higashi T, Ito K, Narushima M, Sugiura T, Inagaki S, Min JZ, et al. 2011. Development and validation of stable-isotope dilution liquid chromatography–tandem mass spectrometric method for determination of salivary progesterone. Biomed Chromatogr 25:1175–1180, PMID: 21294140, 10.1002/bmc.1586. [DOI] [PubMed] [Google Scholar]
- Hill M, Lapcík O, Havlíková H, Morfin R, Hampl R. 2001. 7-Hydroxydehydroepiandrosterone epimers in human serum and saliva. Comparison of gas chromatography-mass spectrometry and radioimmunoassay. J Chromatogr A 935:297–307, PMID: 11762781. [DOI] [PubMed] [Google Scholar]
- Hjortskov N, Garde AH, Ørbæk P, Hansen AM. 2004. Evaluation of salivary cortisol as a biomarker of self-reported mental stress in field studies. Stress Health 20:91–98, 10.1002/smi.1000. [DOI] [Google Scholar]
- Humphrey SP, Williamson RT. 2001. A review of saliva: normal composition, flow and function. J Prosthet Dent 85:162–169, PMID: 11208206, 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- Hunter AL, Minnis H, Wilson P. 2011. Altered stress responses in children exposed to early adversity: a systematic review of salivary cortisol studies. Stress 14:614–626, PMID: 21675865, 10.3109/10253890.2011.577848. [DOI] [PubMed] [Google Scholar]
- Jeppsson MC, Lindh CH, Kristiansson MH, Nielsen J, Jönsson BA. 2009. Methylhexahydrophthalic anhydride adducted albumin tryptic peptides in nasal lavage fluid. Inhal Toxicol 21:1013–1020, PMID: 19772480, 10.1080/08958370802715997. [DOI] [PubMed] [Google Scholar]
- Jiang R, Pawliszyn J. 2012. Thin-film microextraction offers another geometry for solid-phase microextraction. Trends Anal Chem 39:245–253, 10.1016/j.trac.2012.07.005. [DOI] [Google Scholar]
- Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, et al. 2008. Is meconium from healthy newborns actually sterile? Res Microbiol 159:187–193, PMID: 18281199, 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Juniarto AZ, Goosens K, Setyawati BA, Drop SL, de Jong FH, Faradz SM. 2011. Correlation between androstenedione and 17-hydroxyprogesterone levels in saliva and plasma of patients with congenital adrenal hyperplasia. Singapore Med J 52:810–813. [PubMed] [Google Scholar]
- Kajantie E, Phillips DIW. 2006. The effects of sex and hormonal status on the physiological response to acute psychosocial stress. Psychoneuroendocrinology 31:151–178, 10.1016/j.psyneuen.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, et al. 2012. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 28:373–380, PMID: 22135418, 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. 2013. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19:576–585, 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansson MH, Lindh CH, Jönsson BA. 2004. Correlations between air levels of hexahydrophthalic anhydride (HHPA) and HHPA-adducted albumin tryptic peptides in nasal lavage fluid from experimentally exposed volunteers. Rapid Commun Mass Spectrom 18:1592–1598, PMID: 15282784, 10.1002/rcm.1527. [DOI] [PubMed] [Google Scholar]
- Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, et al. 2011. Profiling Cys34 adducts of human serum albumin by fixed-step selected reaction monitoring. Mol Cell Proteomics 10:M110.004606, 10.1074/mcp.M110.004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Kupper LL, Rappaport SM. 2005. Air samples versus biomarkers for epidemiology. Occup Environ Med 62:750–760, PMID: 16234400, 10.1136/oem.2004.013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund HME, Øiestad EL, Gjerde H, Christophersen AS. 2011. Drugs of abuse in oral fluid collected by two different sample kits – stability testing and validation using ultra performance tandem mass spectrometry analysis. J Chromatogr B 879:3367–3377, 10.1016/j.jchromb.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Mendonza AE, Gohh RY, Akhlaghi F. 2006. Analysis of mycophenolic acid in saliva using liquid chromatography tandem mass spectrometry. Ther Drug Monit 28:402–406, PMID: 16778726, 10.1097/01.ftd.0000211826.65607.05. [DOI] [PubMed] [Google Scholar]
- Metgud R, Patel S. 2014. Serum and salivary levels of albumin as diagnostic tools for oral pre-malignancy and oral malignancy. Biotech Histochem 89:8–13, PMID: 23738795, 10.3109/10520295.2013.793394. [DOI] [PubMed] [Google Scholar]
- Moolchan ET, Cone EJ, Wstadik A, Huestis MA, Preston KL, et al. 2000. Cocaine and metabolite elimination patterns in chronic cocaine users during cessation: plasma and saliva analysis. J Anal Toxicol 24:458–466, PMID: 11043647. [DOI] [PubMed] [Google Scholar]
- Moore C, Rana S, Coulter C. 2007. Determination of meperidine, tramadol and oxycodone in human oral fluid using solid phase extraction and gas chromatography-mass spectrometry. J Chromatogr B Anlyt Technol Biomed Life Sci 850:370–375, 10.1016/j.jchromb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Nam SH, Jung HI, Kang SM, Inaba D, Kwon HK, Kim BI. 2015. Validity of screening methods for periodontitis using salivary hemoglobin level and self-report questionnaires in people with disabilities. J Periodontol 86:536–545, PMID: 25569125, 10.1902/jop.2015.140457. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267, PMID: 22674330, 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Wilson ID. 2003. Understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov 2:668–676, PMID: 12904817, 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- Nomura Y, Tamaki Y, Eto A, Kakuta E, Ogino D, Nakamura Y, et al. 2012. Screening for periodontal diseases using salivary lactate dehydrogenase, hemoglobin level, and statistical modeling. J Dent Sci 7:379–383, 10.1016/j.jds.2012.09.005. [DOI] [Google Scholar]
- Ogawa S, Tadokoro H, Sato M, Higashi T. 2014. Enantioselective determination of ibuprofen in saliva by liquid chromatography/tandem mass spectrometry with chiral electrospray ionization-enhancing and stable isotope-coded derivatization. J Pharm Biomed Anal 98:387–392, PMID: 24999866, 10.1016/j.jpba.2014.06.024. [DOI] [PubMed] [Google Scholar]
- Oldi JF, Kannan K. 2009. Perchlorate in human blood serum and plasma: Relationship to concentrations in saliva. Chemosphere 77(1):43–47, PMID: 19564037, 10.1016/j.chemosphere.2009.05.047. [DOI] [PubMed] [Google Scholar]
- Orešič M, Simell S, Sysi-Aho M, Näntö-Salonen K, Seppänen-Laakso T, Parikka V, et al. 2008. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med 205:2975–2984, PMID: 19075291, 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, Bhattacharya J, Butte AJ. 2010. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS One 5(5):e10746, PMID: 20505766, 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell RL, Gulati AS, Cook SF, Martin CF, Chen W, Jaeger EL, et al. 2016. Collecting biospecimens from an internet-based prospective cohort study of inflammatory bowel disease (CCFA Partners): a feasibility study. JMIR Res Protoc 5(1):e3, 10.2196/resprot.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM. 2011. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol 21:5–9, PMID: 21081972, 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Rappaport SM. 2012. Biomarkers intersect with the exposome. Biomarkers 17:483–489, PMID: 22672124, 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM. 2016. Genetic factors are not the major causes of chronic diseases. PLoS One 11(4):e0154387, PMID: 27105432, 10.1371/journal.pone.0154387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A. 2014. The blood exposome and its role in discovering causes of disease. Environ Health Perspect 122:769–774, PMID: 24659601, 10.1289/ehp.1308015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Li H, Grigoryan H, Funk W, Williams ER. 2012. Adductomics: characterizing exposures to reactive electrophiles. Toxicol Lett 213:83–90, PMID: 21501670, 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh M, Stachel D, Kuhlen M, Groschl M, Holter W, Rascher W. 2006. Quantification of busulfan in saliva and plasma in haematopoietic stem cell transplantation in children: validation of liquid chromatography tandem mass spectrometry method. Clin Pharmacokinet 45(3):305–316, PMID: 16509762, 10.2165/00003088-200645030-00006. [DOI] [PubMed] [Google Scholar]
- Robinson O, Vrijeheid M. 2015. The pregnancy exposome. Curr Environ Health Rep 2:204–213, PMID: 26231368, 10.1007/s40572-015-0043-2. [DOI] [PubMed] [Google Scholar]
- Rubino FM, Pitton M, Di Fabio D, Colombi A. 2009. Toward an “omic” physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom Rev 28:725–784, PMID: 19127566, 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Hasegawa T. 2005. Analysis of salivary testosterone by liquid chromatography-tandem mass spectrometry: correlation with serum bioavailable testosterone and aging. Rinsho Byori 53:388–394. [PubMed] [Google Scholar]
- Sakhi AK, Bastani NE, Ellingjord-Dale M, Gundersen TE, Blomhoff R, Ursin G. 2015. Feasibility of self-sampled dried blood spot and saliva samples sent by mail in population-based study. BMC Cancer 15:265–273, PMID: 25886002, 10.1186/s12885-015-1275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JN, Boca SM, Shu XO, Stolzenberg-Solomon RZ, Matthews CE, Hsing AW, et al. 2013. Metabolomics in epidemiology: sources of variability in metabolite measurements and implications. Cancer Epidemiol Biomarkers Prev 22:631–640, PMID: 23396963, 10.1158/1055-9965.EPI-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawatsri S, Samid D, Malkapuram S, Sidell N. 2001. Inhibition of estrogen-dependent breast cell responses with phenylacetate. Int J Cancer 93:687–692, PMID: 11477579. [DOI] [PubMed] [Google Scholar]
- Scalbert A, Brennan L, Manach C, Andres-Lacueva C, Dragsted LO, Rappaport SM, et al. 2014. The food metabolome: a window over dietary exposure. Am J Clin Nutr 99:1286–1308, PMID: 24760973, 10.3945/ajcn.113.076133. [DOI] [PubMed] [Google Scholar]
- Seçilir A, Schrier L, Bijleveld YA, Toersche JH, Jorjani S, Burggraaf J, et al. 2013. Determination of methylphenidate in plasma and saliva by liquid chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 923–924:22–28, PMID: 23454305, 10.1016/j.jchromb.2013.01.027. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504, PMID: 14597658, 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaila M, Pai GP, Shetty P. 2013. Salivary protein concentration, flow rate, buffer capacity and pH estimation: a comparative study among young and elderly subjects, both normal and with gingivitis and periodontitis. J Indian Soc Periodontol 17:42–46, 10.4103/0972-124X.107473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Li S, Zhang Y, Yuan X, Fan Y, Liu Z, et al. 2009. Determination of total, free and saliva mycophenolic acid with a LC-MS/MS method: application to pharmacokinetic study in healthy volunteers and renal transplant patients. J Pharm Biomed Anal 50(3):515–521, PMID: 19574013, 10.1016/j.jpba.2009.05.030. [DOI] [PubMed] [Google Scholar]
- Shin HS, Kim JG, Shin YJ, Jee SH. 2002. Sensitive and simple method for the determination of nicotine and cotinine in human urine, plasma and saliva by gas chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 769:177–183, PMID: 11936690. [DOI] [PubMed] [Google Scholar]
- Shirtcliff EA, Granger DA, Schwartz EB, Curran MJ, Booth A, Overman WH. 2000. Assessing estradiol in biobehavioral studies using saliva and blood spots: simple radioimmunoassay protocols, reliability, and comparative validity. Horm Behav 38:137–147, PMID: 10964528, 10.1006/hbeh.2000.1614. [DOI] [PubMed] [Google Scholar]
- Sturla SJ. 2007. DNA adduct profiles: chemical approaches to addressing the biological impact of DNA damage from small molecules. Curr Opin Chem Biol 11:293–299, PMID: 17574899, 10.1016/j.cbpa.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368:1575–1584, PMID: 23614584, 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama T, Tsutsui H, Shimizu I, Toyama T, Yoshimoto N, Endo Y, et al. 2016. Diagnostic approach to breast cancer patients based on target metabolomics in saliva by liquid chromatography with tandem mass spectrometry. Clin Chim Acta 452:18–26, PMID: 26523874, 10.1016/j.cca.2015.10.032. [DOI] [PubMed] [Google Scholar]
- Thieme D. 2012. Potential and limitations of alternative specimens in doping control. Bioanalysis 4:1613–1622, PMID: 22831477, 10.4155/bio.12.150. [DOI] [PubMed] [Google Scholar]
- Törnqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, Rydberg P. 2002. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J Chromatogr B Analyt Technol Biomed Life Sci 778:279–308, PMID: 12376136. [DOI] [PubMed] [Google Scholar]
- Vanstraelen K, Maertens J, Augustijns P, Lagrou K, de Loor H, Mols R, et al. 2015. Investigation of saliva as an alternative to plasma monitoring of voriconazole. Clin Pharmacokinet 54(11):1151–1160, PMID: 25910879, 10.1007/s40262-015-0269-z. [DOI] [PubMed] [Google Scholar]
- Viardot A, Huber P, Puder JJ, Zulewski H, Keller U, Müller B. 2005. Reproducibility of nighttime salivary cortisol and its use in the diagnosis of hypercortisolism compared with urinary free cortisol and overnight dexamethasone suppression test. J Clin Endocrinol Metab 90:5730–5736, 10.1210/jc.2004-2264. [DOI] [PubMed] [Google Scholar]
- Vuckovic D. 2012. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal Bioanal Chem 403:1523–1548, PMID: 22576654, 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- Wang RE, Tian L, Chang YH. 2012. A homogeneous fluorescent sensor for human serum albumin. J Pharm Biomed Anal 63:165–169, PMID: 22326845, 10.1016/j.jpba.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VS, Lu MY. 2009. Application of solid-phase microextraction and gas chromatography-mass spectrometry for measuring chemical in saliva of synthetic leather workers. J Chromatogr B Analyt Technol Biomed Life Sci 877:24–32, PMID: 19036646, 10.1016/j.jchromb.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63, PMID: 21475195, 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP. 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev 14:1847–1850, PMID: 16103423, 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Wild CP, Scalbert A, Herceg Z. 2013. Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ Mol Mutagen 54:480–499, PMID: 23681765, 10.1002/em.21777. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. 2013. HMDB 3.0—the Human Metabolome Database in 2013. Nucleic Acids Res 41:D801–D807, PMID: 23161693, 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov I, Han B, Wishart DS. 2015. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucl Acids Res 43(W1):W251–W257, , 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeleniuch-Jacquotte A, Afanasyeva Y, Kaaks R, Rinaldi S, Scarmo S, Liu M, et al. 2012. Premenopausal serum androgens and breast cancer risk: a nested case-control study. Breast Cancer Res 14:R32, 10.1186/bcr3117. [DOI] [PMC free article] [PubMed] [Google Scholar]