Abstract
Background
Outbreaks of hypersensitivity pneumonitis (HP) are not uncommon in workplaces where metal working fluid (MWF) is used to facilitate metal turning. Inhalation of microbe-contaminated MWF has been assumed to be the cause, but previous investigations have failed to establish a spatial relationship between a contaminated source and an outbreak.
Objectives
After an outbreak of five cases of HP in a UK factory, we carried out blinded, molecular-based microbiological investigation of MWF samples in order to identify potential links between specific microbial taxa and machines in the outbreak zone.
Methods
Custom-quantitative PCR assays, microscopy and phylogenetic analyses were performed on blinded MWF samples to quantify microbial burden and identify potential aetiological agents of HP in metal workers.
Measurements and main results
MWF from machines fed by a central sump, but not those with an isolated supply, was contaminated by mycobacteria. The factory sump and a single linked machine at the centre of the outbreak zone, known to be the workstation of the index cases, had very high levels of detectable organisms. Phylogenetic placement of mycobacterial taxonomic marker genes generated from these samples indicated that the contaminating organisms were closely related to Mycobacterium avium.
Conclusions
We describe, for the first time, a close spatial relationship between the abundance of a mycobacterium-like organism, most probably M. avium, and a localised outbreak of MWF-associated HP. The further development of sequence-based analytic techniques should assist in the prevention of this important occupational disease.
Keywords: hypersensitivity pneumonitis, occupational lung disease, allergic alveolitis
Key messages.
What is the key question?
Inhalation of microbially contaminated aerosols is considered to be a primary cause of hypersensitivity pneumonitis (HP) in metal workers, but there is considerable uncertainty over which organisms are responsible.
What is the bottom line?
Through novel molecular detection methods, in the setting of a small outbreak of HP in a UK factory, we identified a close spatial relationship with probable exposure to Mycobacterium avium.
Why read on?
Our findings suggest a way forward in the prevention of what is now the most common cause of occupational HP in the UK.
Introduction
During the cutting, milling and grinding processes used in the mechanical manufacture of metal components, metal working fluids (MWFs) provide lubrication, cooling and the removal of debris. Typically, a single sump feeds several machines with MWF, which is re-circulated; microbial contamination is contained through the use of biocides and regular monitoring through, for example, dipslide analysis. During machining processes, there is aerosolisation of MWF and inhalation of contaminated fluid mists is believed to be a primary cause of hypersensitivity pneumonitis (HP) in metal workers.1
It is over 20 years since the publication of the first reports of HP attributed to MWF.2 In the interim, several hundred further cases have been identified, most frequently in North America and Europe. Cases appear to develop in temporal clusters or outbreaks, suggesting that they occur in response to a change in exposure. Prompted by analogy with other occupational causes of HP, investigators have sought evidence for microbial contamination of MWF in workplaces where cases have occurred. The findings have been mixed with bacterial,3 fungal4 and mycobacterial5 species, and endotoxin6 each being implicated on the basis of traditional microbial analysis of MWF samples and, in some reports, associated immunological responses in affected workers. The latter—in most cases the detection of precipitating IgG antibodies to contaminated MWF samples—have however failed, as with other causes of occupational HP, to distinguish reliably between cases and healthy, but exposed, coworkers.7
Particular attention has been paid to the role of contaminating non-tuberculous mycobacteria (NTM). Notably, molecular techniques identified a single strain of Mycobacterium immunogenum in all but 2 of 107 MWF samples collected from 10 US workplaces with cases of HP,8 a finding that led some to claim that this species has a particular capacity for colonisation and infection in this setting; a subsequent outbreak in France provided some support for this position.5 Other reports, however, suggest that a greater diversity of NTM species may be relevant.9
Here, we describe a small outbreak of HP in a workforce of UK metal turners. We hypothesised that a mycobacterium-like organism was the causative organism of HP in this instance and aimed to provide evidence to support a causal, spatial relationship using molecular methods in combination with microscopy, phylogenetic analysis and immunology.
Methods
Case identification
In 2012, two men were diagnosed with HP acquired through their work as metal turners in an aircraft parts factory. Each described cough and breathlessness that improved away from work, with an onset around 2010. Further information on their clinical presentations and on those of a third colleague identified subsequently and with similar symptoms (see Findings section) is available elsewhere.10 On high-resolution CT scan, two had changes indicative of fibrotic HP; the third had a mosaic pattern suggestive of small airways disease. None had a lung biopsy. As a result, we screened, in 2013, all current employees of the factory who had potential exposure to aerosolised MWF mist. The survey comprised a symptoms questionnaire, a clinical examination of the chest and spirometry; workers with any of the following were referred for further investigation at Royal Brompton Hospital, London:
asthma symptoms of onset since employment at the factory
other respiratory symptoms that improved away from work, were worse at work or were recurrent
any established interstitial lung disease
repeated ‘chest infections’ or ‘flu’
unexplained exertional breathlessness
unexplained weight loss (even in the absence of respiratory symptoms)
crackles on auscultation of the chest
abnormal spirometry
Hospital investigations included chest radiography, spirometry and, where considered appropriate, full lung function testing and high-resolution CT of the chest. Following a multidisciplinary team discussion, we identified HP in employees who had otherwise unexplained, work-related respiratory symptoms, reductions in FVC and/or carbon monoxide transfer factor and abnormal findings (ground glass shadowing, nodularity, mosaicism or a pattern of usual interstitial pneumonitis) on high-resolution CT scan.
Environmental sampling and microbiological analysis
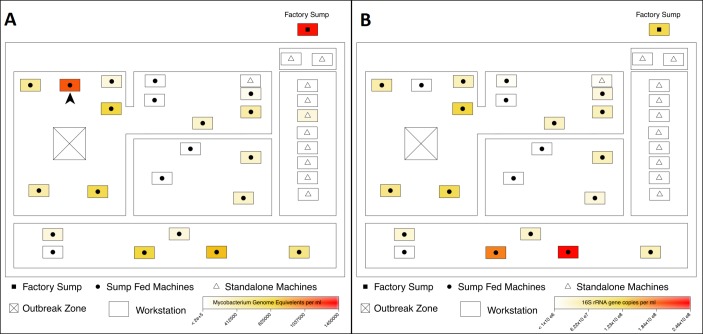
A scheme of the factory production areas is provided in figure 1A; there were no physical barriers between the areas and they shared a common system of roof-based exhaust ventilation. At the time of case identification (and for at least 5 years previously), the factory used the same MWF in all its workstations, a water-based semisynthetic product that was regularly treated with both a fungicide and a biocide. At the start of the health survey, samples of MWF were collected from 33 machine sumps and stored at 4°C. The samples were labelled in such a way that their source could not be identified before microbiological analysis.
Figure 1.
(A) Indication of mycobacterial burden per millilitre of metal working fluid based on a HSP65 gene-targeted quantitative PCR (QPCR). HSP65 gene copy numbers per millilitre of fluid were greatest in the factory sump and stations associated with the outbreak. All cases of hypersensitivity pneumonitis worked at or near a single station, indicated with an arrow, believed to be the focal point of exposure. (B) Indication of bacterial burden per millilitre of metal working fluid based on 16S rRNA gene targeted QPCR. Zones with the highest 16S rRNA gene copies per millilitre were not associated with the outbreak zone.
DNA extraction
One millilitre of emulsified MWF from each sample was spun at 14 000xg for 30 min to pellet debris and biological material. The supernatant was removed and the pellet resuspended in 978 µL of sodium phosphate buffer from the Fast DNA spin kit for soil extraction kit (MP Biomedicals, Ohio, USA). The resuspended pellet was transferred to a lysing matrix E tube and extraction carried out following the manufacturer’s instructions.
Quantitative PCR
We developed a Sybr green quantitative PCR (QPCR) assay to enumerate mycobacterial genome equivalents in the MWF samples. In addition, a second assay, enumerating the 16S rRNA gene copy number, was used to estimate total bacterial burden in the same MWF samples. The details are provided in online supplementary file 1.
thoraxjnl-2017-210226supp001.pdf (491KB, pdf)
Microscopy
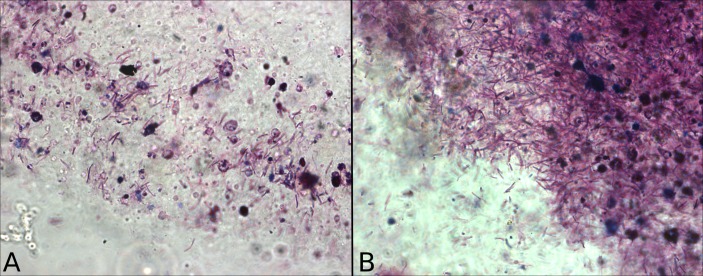
To assess visually the presence of mycobacteria-like organisms, we used a cold acid-fast Kinyoun stain (RAL Diagnostics, Martillac, France) to detect mycolic acid-rich cell walls. One millilitre of MWF per sample was spun at 14 000 g for 30 min and a smear of the pelted debris was fixed on a glass slide using 100% methanol. The stain and counterstain were applied according to the manufacturer’s instructions and examined under a 100× oil immersion lens.
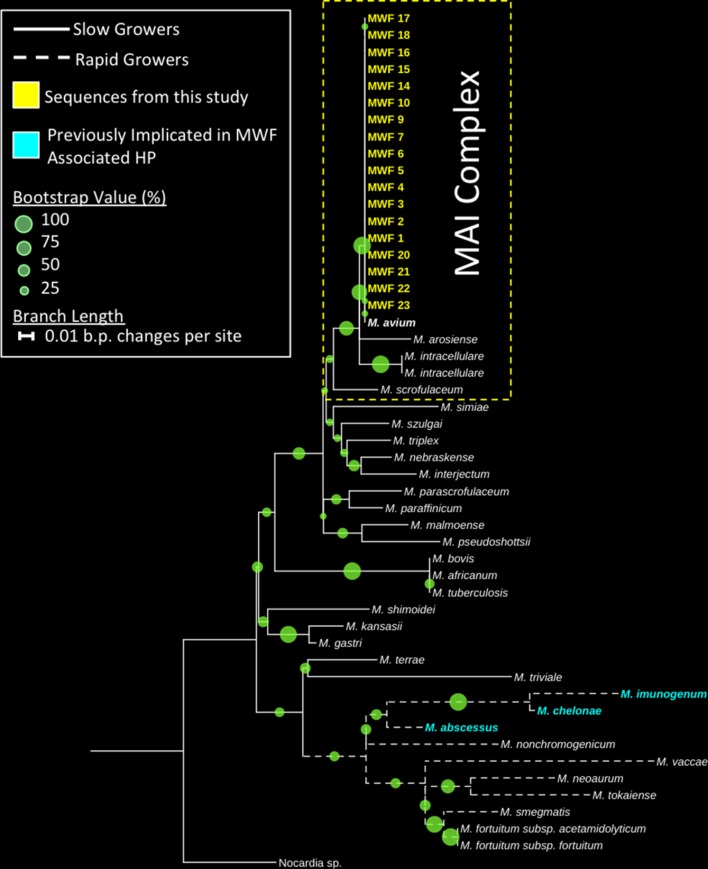
Phylogenetic analysis
Bootstrapped maximum-likelihood trees were constructed from HSP65 gene sequences obtained from MWF samples; details are provided in online supplementary file 1.
Spatial analysis
To examine the distribution of mycobacterial and total bacterial burden across the factory, we assigned each sample a spatial coordinate for their machine of origin and calculated the mycobacterial genome and 16S rRNA gene equivalents per millilitre of MWF. We used the software package R11 with the libraries maptools12 and ggplot213 to create maps of the factory displaying the data. Colour ramp scales were truncated to the lowest standard of each assay denoting the limits of detection for each assay after normalisation for sample volume. We used QuantumGIS to create custom shapefiles of the factory layout from schematics drawn at the site. We used the Kruskal-Wallis test in comparisons of mycobacterial and 16S rRNA counts between sump-fed and stand-alone machines, and between machines in the outbreak zone and elsewhere.
Precipitating antibodies to MWF
Using the Ouchterlony double-diffusion method in agar gel, we searched for precipitating antibodies to used and unused MWF in serum from cases (n=5) and a random sample of exposed but healthy workers (n=5).
Findings
Two hundred and forty-six employees (99% of those with potential exposure to MWF) were surveyed at work and 69 referred for hospital investigation as a consequence; at this stage, their precise work locations were unknown to the clinician. In this way, a further three cases of HP were identified; each noted that their symptoms began in around 2010. One had received the diagnosis at another hospital but a cause had not been identified. The remaining two had histories of characteristic respiratory symptoms that developed in 2009/2010, improved away from work and could not be explained otherwise; their radiology and lung function at assessment were normal and their diagnoses were considered ‘probable’ rather than definite. None of the five identified cases reported weight loss. All worked in close proximity (‘outbreak zone’ in figure 1A); we identified no cases of HP in employees working in other areas.
Samples of MWF from the factory sump and sump-fed workstations were contaminated by organisms containing an amplifiable HSP65 gene fragment. The highest level of contamination (>1.4 million genome equivalents per millilitre of MWF) was found in the sample from the main sump, but one sump-fed workstation in the outbreak zone showed a very high level of contamination, in excess of 1.2 million genome equivalents per millilitre of fluid (figure 1A). In contrast, workstations isolated from the main sump had the lowest levels of contamination, with little or no detectable PCR targets.
Microscopic inspection of MWF samples taken from the factory sump and the heavily contaminated workstation within the outbreak zone revealed rod-shaped organisms of approximately 2 µL in length, which accepted a stain specific for mycolic acids (figure 2). This strongly suggests the presence of mycobacterium-like organisms, at high abundance, within these samples.
Figure 2.
Mycobacterium-like organisms, visualised with a cold acid-fast stain, from samples originating from (A) the outbreak zone and (B) the factory sump.
Phylogenetic analysis of high-quality HSP65 gene Sanger sequences from contaminated MWF samples were shown to group tightly with a representative sequence from the M. avium type strain (figure 3). High bootstrap support and phylogenetic positioning of these sequences suggest that these organisms were distinct from closely related members of the M. avium–intracellulare complex and other immunogenic mycobacteria previously implicated in MWF-associated HP such as the ‘rapid-growing’ M. imunogenum, M. abcessus and M. chelonae. Furthermore, the mycobacterial sequences obtained from the contaminated MWF in this study were positioned wholly within clades representing slow-growing mycobacteria.
Figure 3.
Bootstrapped (n=500) maximum-likelihood tree showing the phylogenetic positioning of HSP65 genes originating from contaminated metal working fluid (MWF) clustering with a Mycobacterium avium type strain. Bootstrap values are denoted by circle size at the branch nodes and organisms previously identified as causative organisms of hypersensitivity pneumonitis (HP) in metal workers are highlighted in red. Sequences from this study are highlighted in blue. The tree was rooted using a closely related member of the Nocardia genus. MAI, M. avium–intracellulare.
Mapping of 16S rRNA gene copies, used as a proxy for total bacterial burden, showed no spatial association with the outbreak zone (figure 1B). In addition, there was a significant difference between sump-fed workstations and stand-alone workstations in relation to both mycobacterial (p<0.001) and total bacterial burden (p<0.001) (online supplementary figure S1A), suggesting that microbial contamination of workstations originated from the sump via circulating MWF. Furthermore, there were significant differences between mycobacterial genome equivalents (p=0.01) and 16S rRNA gene copy numbers (p=0.02) in workstations associated with the outbreak zone compared with all other zones combined (online supplementary figure S1B).
thoraxjnl-2017-210226supp002.jpg (141.7KB, jpg)
We observed a weak positive precipitin to the used (but not the clean) MWF in serum from two of the cases, but none of the exposed controls. No precipitins, in either cases or controls, were detected in assays with the unused MWF sample.
Discussion
We present a hypothesis-driven, detailed spatial analysis of an outbreak of HP caused by exposure to MWF. The recirculating fluid originating from a single factory sump was linked to high levels of mycobacterial contamination in associated—but not stand-alone—workstations. All identified cases could be located to work in an area with demonstrable contamination by a biologically plausible causative agent; conversely, no cases occurred in the absence of contamination. While we did not have access to airborne samples, we suggest that the single workstation with the highest level of contamination (figure 1A) was the focal point of aerosol-based exposure to an HP-causing agent. The temporal clustering of cases with the onset of work-related symptoms in around 2010 followed a significant increase in production.
We used the 16S rRNA gene targeted QPCR measurements as a proxy for total bacterial burden in each of the MWF samples. The results showed a statistical link between 16S rRNA gene copy numbers and sump-fed machines across the factory, suggesting that recirculating MWF through a centralised sump may predispose to growth and dissemination of biological agents. In contrast to the mycobacterial data, there was no significant difference in general 16S rRNA gene copy numbers between the outbreak zone and other machines, suggesting that a specific aspect of the microbiota, most likely here mycobacterial, is responsible. Studies of HP in the metal working environment and elsewhere have suggested fungal contaminants may play a significant role in the development of the disease. We cannot rule this out as we did not attempt to quantify fungal biomass, but our initial microscopic screenings of MWF resulted in no obvious fungal-like structures.
An important limitation of our findings arises from our analysis of MWF samples that were collected after the onset of the outbreak; no contemporary samples were available. Our interpretation thus rests on the assumption that the contamination and its spatial distribution that we observed had been present for a few years. While we cannot rule out a role for a co-located organism, this appears to be the first implication of M. avium in this setting and is the first time a mycobacterium has been linked to a UK outbreak of HP in metal workers. The organism is a well-known cause of ‘hot tub lung’, which clinically may mimic both acute and chronic HP in a manner that is similar to the varied presentations of our cases.14 Other NTM have been identified in other MWF-associated outbreaks, notably M. immunogenum and M. chelonae, and they are often cited as a potential cause of HP in other settings where aerosol exposure has occurred.1 15 16
We deliberately chose a strategy of high sensitivity in workplace screening but a subsequent case designation that was specific to HP on the grounds that this would give us the best chance of identifying its cause. All the employees referred for hospital investigation were assessed with no more knowledge of their workplace than that it was on the production floor; indeed, two of the original cases were originally diagnosed (elsewhere) without an occupational attribution being made.10 During the investigation, we identified a small number of workers with alternative respiratory diagnoses including asthma and allergic bronchopulmonary aspergillosis, but we did not include these in our outbreak analysis. Finally, we were careful to arrange analysis of the MWF samples without knowledge of their source. While HP resulting from exposure to a workstation fed by a single sump contaminated by M. chelonae has been previously documented,3 there has been no detailed description of the methods used or of the spatial relationship between exposure and cases.
Unlike mycobacterial taxa previously identified as agents of HP in MWF, M. avium has slow growth properties.17 Due to the general difficulty in culturing slowly growing organisms,18 it is possible that culture-based studies of mycobacteria in MWF may preferentially identify mycobacteria with rapid growth strategies. Specific short-term culture methods require harsh decontamination of environmental samples to prevent overgrowth by fast-growing organisms, reducing their sensitivity.19 Slow-growing mycobacteria may be missed through standard dipslide analysis typically lasting less than 48 hours. DNA-based analysis of the 16S rRNA gene can provide ambiguous results when delineating the NTM genus into species,20 and our use of the HSP65 gene as a taxonomic marker provides greater phylogenetic resolution than 16S rRNA gene sequences.21 Chemical lysis procedures often used in DNA extraction often fail to break apart the tough waxy cell walls of mycobacteria22; in contrast, physical lysis procedures, such as the bead beating method we used, efficiently lyse mycobacterial cells.23
The method by which NTM elicit an immune response is unclear. Whole-cell challenges of mouse macrophages in cell culture by a range of mycobacterial species, including M. avium, have been shown to induce a dose-dependent production of inflammatory markers such as interleukin 6, tumour necrosis factor α, nitric oxide and reactive oxygen species showing that both pathogenic and potentially non-pathogenic mycobacteria are capable of producing an inflammatory response in vitro.24 Furthermore, cellular components such as the heat shock protein (product of the gene measured here) have been shown to induce Th1-type responses, airway hyper-responsiveness and airway inflammation.25 In addition, cellular components unique to mycobacterial species produce immune responses in animal models.26 However, we identified precipitating antibodies to contaminated MWF in only two cases (but no exposed controls), suggesting that this is not a sensitive approach to diagnosis, an experience that is common to other outbreaks.5 7
Biocides are commonly added to MWF to control contamination but may not deter the emergence of resistant mycobacteria.9 Our evidence suggests the presence of mycobacterial taxa and a range of other bacteria residing in the factory sump and sump-fed machines despite treatment with biocides. Indeed, ‘island biogeography’ theory27 predicts such defined ecological niches of biocide-tolerant communities in MWF systems and a cocktail or rotation of biocides with different modes of action may be required to prevent resistant microbial selection and further outbreaks of HP. However, since it is unclear whether mycobacterial cells, their cellular components, metabolites or a combination are responsible for eliciting the immune responses associated with HP outbreaks, control mechanisms may need to be focused on the prevention of mycobacterial contamination and/or exposure rather than remediation of contaminated fluids. Cellular fractions and metabolites, which persist after cell death, could remain immunogenic and, due to their size, evade clean-up procedures such as filtering. Following the outbreak in this factory, a programme of de-contamination, improved containment and replacement of the MWF with an alternative appears to have prevented the onset of new cases.
It is likely that numerous micro-organisms, including a number of mycobacterial species, have immunogenic properties that can result in HP in a subset of people. A number of environmental NTMs can survive and grow in MWF and specific mycobacteria taxa, including M. avium, may elicit an immune response in a proportion of workers exposed to contaminated MWF aerosols during standard factory processes. The causative organisms may vary from outbreak to outbreak, but we suggest that molecular-based screening of MWF and the monitoring of spatially clustered distributions of contamination could be useful in the early detection and prevention of what is now the most common cause of occupational HP in the UK.28
Acknowledgments
The authors are grateful to the factory management and employees for their cooperation during this investigation.
Footnotes
MFM and PC contributed equally.
Contributors: The outbreak investigation was devised by PC, CMB, LC, HH and JC. JS assisted in the clinical assessment of referred cases. MJ performed the immunological investigations. The microbial analysis was undertaken by PLJ with guidance from MFM, SC and WOCC.
Funding: Wellcome Trust, The Asmarley Trust and the BRU Royal Brompton and Harefield NHS Foundation Trust.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Murat JB, Grenouillet F, Reboux G, et al. . Factors influencing the microbial composition of metalworking fluids and potential implications for machine operator’s lung. Appl Environ Microbiol 2012;78:34–41. 10.1128/AEM.06230-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muilenberg ML, Burge HA, Sweet T. Hypersensitivity pneumonitis and exposure to acid-fast Bacilli in coolant aerosols. J Allergy Clin Immun 1993;91:311. [Google Scholar]
- 3.Kreiss K, Cox-Ganser J. Metalworking fluid-associated hypersensitivity pneumonitis: a workshop summary. Am J Ind Med 1997;32:423–32. [DOI] [PubMed] [Google Scholar]
- 4.Fox J, Anderson H, Moen T, et al. . Metal working fluid-associated hypersensitivity pneumonitis: an outbreak investigation and case–control study. Am J Ind Med 1999;35:58–67. [DOI] [PubMed] [Google Scholar]
- 5.Tillie-Leblond I, Grenouillet F, Reboux G, et al. . Hypersensitivity pneumonitis and metalworking fluids contaminated by mycobacteria. Eur Respir J 2011;37:640–7. 10.1183/09031936.00195009 [DOI] [PubMed] [Google Scholar]
- 6.Abrams L, Seixas N, Robins T, et al. . Characterization of metalworking fluid exposure indices for a study of acute respiratory effects. Appl Occup Environ Hyg 2000;15:492–502. 10.1080/104732200301287 [DOI] [PubMed] [Google Scholar]
- 7.Trout D, Weissman DN, Lewis D, et al. . Evaluation of hypersensitivity pneumonitis among workers exposed to metal removal fluids. Appl Occup Environ Hyg 2003;18:953–60. 10.1080/10473220390237683 [DOI] [PubMed] [Google Scholar]
- 8.Wallace RJ Jr, Zhang Y, Wilson RW, et al. . Presence of a single genotype of the newly described species Mycobacterium immunogenum in industrial metalworking fluids associated with hypersensitivity pneumonitis. Appl Environ Microbiol 2002;68:5580–4. 10.1128/AEM.68.11.5580-5584.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan IU, Selvaraju SB, Yadav JS. Occurrence and characterization of multiple novel genotypes of Mycobacterium immunogenum and Mycobacterium chelonae in metalworking fluids. FEMS Microbiol Ecol 2005;54:329–38. 10.1016/j.femsec.2005.04.009 [DOI] [PubMed] [Google Scholar]
- 10.Cullinan P, D’Souza E, Tennant R, et al. . Lesson of the month: extrinsic allergic (bronchiolo)alveolitis and metal working fluids. Thorax 2014;69:1059–60. 10.1136/thoraxjnl-2014-205251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ihaka R. Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat 1996;5:299–314. [Google Scholar]
- 12.Bivand R, Lewin-Koh N. Maptools: tools for reading and handling spatial objects. 2013. http://cran.r-project.org/package=maptools
- 13.Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat 2011;3:180–5. 10.1002/wics.147 [DOI] [Google Scholar]
- 14.Verma G, Jamieson F, Chedore P, et al. . Hot tub lung mimicking classic acute and chronic hypersensitivity pneumonitis: two case reports. Can Respir J 2007;14:354–6. 10.1155/2007/138270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkinham JO. Mycobacterial aerosols and respiratory disease. Emerg Infect Dis 2003;9:763–7. 10.3201/eid0907.020415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacasse Y, Girard M, Cormier Y. Recent advances in hypersensitivity pneumonitis. Chest 2012;142:208–17. 10.1378/chest.11-2479 [DOI] [PubMed] [Google Scholar]
- 17.Philley JV, Griffith DE. Treatment of slowly growing mycobacteria. Clin Chest Med 2015;36:79–90. 10.1016/j.ccm.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 18.Burdz TV, Wolfe J, Kabani A. Evaluation of sputum decontamination methods for Mycobacterium tuberculosis using viable colony counts and flow cytometry. Diagn Microbiol Infect Dis 2003;47:503–9. 10.1016/S0732-8893(03)00138-X [DOI] [PubMed] [Google Scholar]
- 19.Radomski N, Cambau E, Moulin L, et al. . Comparison of culture methods for isolation of nontuberculous mycobacteria from surface waters. Appl Environ Microbiol 2010;76:3514–20. 10.1128/AEM.02659-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth A, Fischer M, Hamid ME, et al. . Differentiation of phylogenetically related slowly growing mycobacteria based on 16S–23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol 1998;36:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNabb A, Eisler D, Adie K, et al. . Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J Clin Microbiol 2004;42:3000–11. 10.1128/JCM.42.7.3000-3011.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branch DW, Smith GT, Vreeland EC, Blakemore R, Alland D, Nucleic acid extraction using a rapid, chemical free, ultrasonic technique for point-of-care diagnostics. IEEE Int Ultrason Symp. (IUS), 2014. [Google Scholar]
- 23.Travis ER, Gaze WH, Pontiroli A, et al. . An inter-laboratory validation of a real time PCR assay to measure host excretion of bacterial pathogens, particularly of Mycobacterium bovis. PLoS One 2011;6:e27369 10.1371/journal.pone.0027369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huttunen K, Ruotsalainen M, Iivanainen E, et al. . Inflammatory responses in RAW264.7 macrophages caused by mycobacteria isolated from moldy houses. Environ Toxicol Pharmacol 2000;8:237–44. 10.1016/S1382-6689(00)00047-8 [DOI] [PubMed] [Google Scholar]
- 25.Rha YH, Taube C, Haczku A, et al. . Effect of microbial heat shock proteins on airway inflammation and hyperresponsiveness. J Immunol 2002;169:5300–7. 10.4049/jimmunol.169.9.5300 [DOI] [PubMed] [Google Scholar]
- 26.Clark-Curtiss JE. Identification of virulence determinants in pathogenic mycobacteria. Curr Top Microbiol Immunol 1998;225:57–79. [DOI] [PubMed] [Google Scholar]
- 27.van der Gast CJ, Lilley AK, Ager D, et al. . Island size and bacterial diversity in an archipelago of engineering machines. Environ Microbiol 2005;7:1220–6. 10.1111/j.1462-2920.2005.00802.x [DOI] [PubMed] [Google Scholar]
- 28.Barber CM, Wiggans RE, Carder M, et al. . Epidemiology of occupational hypersensitivity pneumonitis; reports from the SWORD scheme in the UK from 1996 to 2015. Occup Environ Med 2017;74:528–30. 10.1136/oemed-2016-103838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
thoraxjnl-2017-210226supp001.pdf (491KB, pdf)
thoraxjnl-2017-210226supp002.jpg (141.7KB, jpg)