Abstract
Background
The impact of folate deficiency on global DNA methylation is uncertain. It also is unclear whether global DNA methylation is associated with outcome in HCC. LINE-1 methylation levels, as a surrogate marker of global methylation, may be influenced by folate deficiency. However, the interaction between LINE-1 methylation and folate level on overall survival (OS) in hepatocellular carcinoma (HCC) patients is unknown. We evaluated whether LINE-1 hypomethylation and folate deficiency are associated with HCC prognosis.
Methods
We prospectively recruited 172 HCC patients between 2008 and 2012. LINE-1 methylation levels in plasma and white blood cells (WBC) were measured by pyrosequencing, and plasma folate levels by a radioprotein-binding assay.
Results
Patients with plasma LINE-1 methylation <70.0% (hypomethylation) had significantly worse OS compared with those with ≥70.0% methylation (hypermethylation) [hazard ratio (HR) = 1.77; 95% confidence interval (CI) 1.12–2.79; P = 0.015]. HCC patients with lower plasma folate levels also had worse survival (<27.7 vs. ≥27.7 nmol/L; HR = 1.96; 95% CI, 1.24–3.09; P = 0.004). Furthermore, survival was poor in patients in whom both plasma LINE-1 methylation and folate levels were low compared with those patients in whom both levels were high (HR = 3.36; 95%CI, 1.77–6.40; P <0.001). This interaction neared statistical significance (P = 0.057). No significant association was found between WBC LINE-1 methylation levels and survival.
Conclusions
These findings suggest that both lower plasma levels of LINE-1 methylation and folate are associated with worse survival in HCC patients.
Keywords: LINE-1 methylation, Folate, Hepatocellular carcinoma, Overall survival
Genetic and epigenetic alterations accumulate gradually and act together to promote the development of hepatocellular carcinoma (HCC).1,2 The most common epigenetic change associated with human cancers is in DNA methylation, which includes site-specific CpG island promoter hypermethylation and global DNA hypomethylation.3 Interestingly, global hypomethylation has recently been postulated to be an important contributor to hepatocarcinogenesis. Hypomethylation in repetitive elements, which enhances their activity as retrotransposons, has been suggested to have deleterious effects, through insertion, deletions and genomic rearrangements introducing genome instability.4,5 In particular, long interspersed nucleotide elements-1 (LINE-1) is the most common repetitive DNA element and constitutes about 17% of the human genome.6 Due to its high genomic content, LINE-1 methylation is used as a surrogate marker for estimating the level of global DNA methylation.6,7 Although controversial, evidence suggest that LINE-1 hypomethylation may be associated with shorter disease-free survival and poor prognosis.8 Studies have demonstrated that tumor-derived nucleic acids can be detected in plasma or serum samples in cancer patients and appear to serve as surrogate biomarkers for epigenetic alterations present within the tumor microenvironment.9 Limited data also suggest that changes in global DNA methylation levels within blood may serve as prognostic biomarkers in HCC.10,11
Methyl groups for DNA methylation reactions are primarily supplied by S-adenosylmethionine through folate and vitamin B12-depedent one-carbon metabolism. Folate is a methyl donor in a number of molecular pathways (DNA methylation, synthesis and repair) that are necessary for cell survival and replication. Folate deficiency can lead to global hypomethylation, inducing carcinogenesis.12,13 However, studies examining the association between plasma folate and survival in HCC are limited.
In this study, we quantified LINE-1 methylation and plasma folate levels in a prospective cohort of 172 HCC patients. We evaluated whether LINE-1 hypomethylation and folate deficiency independently or multiplicatively associated with HCC prognosis.
MATERIALS AND METHODS
Patients
From October 2008 to July 2014, patients (≥18 years old) with newly diagnosed or with recurrent HCC at Columbia University Medical Center were recruited for this prospective study. We excluded patients who had any other previous malignancy within the past 5 years, those who had undergone previous liver transplantation, and those with Child-Pugh (CP) class C disease. The demographic and clinicopathological characteristics of a subset of the patients were described previously.14 During the study period, 187 patients were invited to participate and 12 patients refused. In addition, one patient without evidence of HCC on pathological examination and two patients who were lost of follow-up also were excluded. Finally, 172 HCC patients were included in the study and followed until March 2016. The study protocol was approved by the Institutional Review Board at Columbia University, and all patients provided written, informed consent.
Clinical and Questionnaire Data
Subjects completed an epidemiologic questionnaire, underwent a physical examination, and provided a fasting morning blood sample at the time of enrollment. Information on biochemical blood analysis was obtained from the medical records. Simultaneously, an additional 30 mL of blood was collected and processed immediately by the Irving Institute’s Biomarker Core at Columbia University. Plasma was separated from EDTA anticoagulated whole blood, which was centrifuged for 10 min at room temperature at 3000 rpm. The plasma containing upper phase was removed and then recentrifgued under the same conditions. The upper plasma phase was isolated, making sure not to disturb the pellet to minimize contamination with buffy coat. All blood samples were divided into aliquots and stored at −80 °C until analysis. Laboratory staff was blinded to the study hypotheses and outcomes.
Measurement of LINE-1 and Folate
Plasma DNA was isolated using QIAmp UltraSense Virus Kits (Qiagen, Valencia, CA). Genomic DNA was extracted from the white blood cells (WBC) fraction by a standard salting out procedure. Aliquots of DNA (500 ng) were bisulfite-treated with the EZ DNA methylation kit (Zymo Research, Orange, CA) following the manufacturer’s protocol. Pyrosequencing for LINE-1 methylation levels in plasma DNA and WBC was performed using PCR with sequencing primers targeting three CpG sites (positions 318–331, accession no. X58075) (Supplementary Table S1). The biotinylated PCR products were purified and made single-stranded to act as a template in the pyrosequencing reaction using the PyroMark Q96 Workstation (Qiagen). Then, the pyrosequencing was run on a PyroMark Q96 MD instrument (Qiagen), with subsequent quantitation of methylation levels determined with the PyroMark CpG 1.0.11 software. The degree of methylation was determined by averaging across all three interrogated CpG sites in the analysis. Non-CpG cytosine residues were used as internal controls to verify efficient sodium bisulfite DNA conversion; all samples contained no detectable nonconverted C. The assay for LINE-1 failed on two plasma DNA samples and thirteen WBC DNA samples. The interassay coefficient of variation (CV) was 0.5% for plasma DNA and 1.0% for WBC DNA.
Plasma concentrations of folate were measured using a radioprotein-binding assay (SimulTRAC-S; MP Biomedicals). To determine folate concentration, we used folic acid as pteroylglutamic acid for calibration, and its 125I-labeled analog was used as the tracer. Twelve samples failed in the assay. The intra-assay and inter-assay CVs were 6% and 14%, respectively. All measurements were conducted in duplicate.
Statistical Analysis
Continuous variables were expressed as categorical variables using median values as cutoffs. The following variables were used in the data analysis: age (≥62.7 or <62.7 years), gender, race/ethnicity, viral hepatitis B and C infection (yes/no), alcohol use (yes/no), cigarette smoking (yes/no), diabetes (yes/no), BMI (≥25.0 or <25.0 kg/m2), waist to hip ratio (WHR) (≥0.952 or < 0.952), Child-Pugh class (A/B), metastasis (yes/no), BCLC stage (0, A/B, C), Milan criteria (within/outside), and serum alpha-fetoprotein (AFP) (≥47.85 or <47.85 ng/mL) and carbohydrate antigen 19–9 (CA 19–9) levels (≥38.0 or <38.0 U/mL). Experimental results for plasma LINE-1, WBC LINE-1, and plasma folate were expressed as median with interquartile range (IQR). The Wilcoxon rank-sum test or Kruskal–Wallis test was used to compare values between demographic and tumor characteristics. We also categorized these biomarkers using quartiles of baseline levels. Kaplan–Meier plots and univariate Cox proportional hazards models were used to determine the association between variables of interest and time to death.
Variables that were clinically relevant and showed statistical significance in univariate analyses were included in the multivariate Cox proportional hazards models to estimate the adjusted hazard ratios (HRs) and 95% confidence intervals (CIs). According to our previous study, covariates, such as age, CP class, Milan criteria, AFP, and liver transplantation, were included in the multivariate Cox proportional hazards models.14 Overall survival (OS) was measured from the date of enrollment to the date of death/date of censoring. Patients who were alive at the last follow-up evaluation were censored at that time. A likelihood ratio test comparing models with and without an interaction term was used to evaluate whether LINE-1 methylation level interacted with folate level to modify the risk of survival. All analyses were performed using Statistical Analysis System 9.4 (SAS Institute, Cary, NC), and two-sided P values < 0.05 were considered statistically significant.
RESULTS
Patients’ Demographic and Tumor Characteristics associated with LINE-1 Methylation and Folate Levels
The median age of the study participants was 62.7 years. Among the cohort, 78% were men, 48% were non-Hispanic white, 17% had HBV, 58% had HCV, and 44% were diabetic. A majority of the patients were CP class A (62%), 38% were within the Milan criteria, and 64% were within BCLC stage B and C. Median follow-up was 1.12 years, and median survival time was 1.50 years. During the follow-up period, 12.4% underwent a liver resection and 21.2% a liver transplantation.
Median (IQR) baseline levels of plasma LINE-1 methylation, WBC LINE-1 methylation, and plasma folate were 72.3% (4.0), 76.5% (5.3), and 27.7 nmol/L (18.6), respectively. The levels of these three biomarkers according to the subjects’ demographic and tumor characteristics are summarized in Table 1. Higher WBC LINE-1 levels were associated with subjects who were younger than 62.7 years old (P = 0.02) and with a waist to hip ratio ≥0.952 (P = 0.023). Higher plasma LINE-1 levels were observed among subjects who were older than 62.7 years (P = 0.037). However, plasma folate level was not associated with any demographic or tumor characteristics.
TABLE 1.
Median levels and interquartile ranges of plasma LINE-1 methylation, WBC LINE-1 methylation, and plasma folate according to demographic and tumor characteristics
| Variablea | Nb | Plasma LINE-1 methylation level (%) |
Pc | Nb | WBC LINE-1 methylation level (%) |
Pc | Nb | Plasma folate level (nmol/L) |
Pc |
|---|---|---|---|---|---|---|---|---|---|
| All cases | 170 | 72.3 (4.0) | 159 | 76.5 (5.3) | 160 | 27.7 (18.6) | |||
| Age (year) | 0.037 | 0.020 | 0.416 | ||||||
| <62.7 | 85 | 71.6 (3.9) | 78 | 77.8 (5.6) | 80 | 26.7 (17.0) | |||
| ≥62.7 | 85 | 73.2 (3.7) | 81 | 75.5 (4.7) | 80 | 29.4 (20.0) | |||
| Gender | 0.964 | 0.283 | 0.463 | ||||||
| Female | 37 | 71.8 (3.4) | 37 | 76.3 (5.1) | 33 | 25.4 (16.5) | |||
| Male | 133 | 72.3 (4.2) | 122 | 76.8 (5.4) | 127 | 28.5 (19.9) | |||
| Race | 0.575 | 0.729 | 0.353 | ||||||
| Non-Hispanic white | 81 | 72.0 (4.4) | 73 | 76.9 (5.6) | 75 | 29.8 (21.6) | |||
| Non-Hispanic black | 18 | 73.0 (4.2) | 18 | 75.8 (5.5) | 18 | 32.2 (22.0) | |||
| Hispanic | 43 | 71.7 (4.3) | 42 | 76.5 (5.3) | 43 | 24.4 (15.3) | |||
| Asia and other | 28 | 72.7 (3.0) | 26 | 75.2 (4.2) | 24 | 24.9 (19.8) | |||
| Hepatitis B virus | 0.988 | 0.558 | 0.247 | ||||||
| Negative | 141 | 72.3 (4.3) | 131 | 76.8 (5.3) | 134 | 28.9 (17.8) | |||
| Positive | 29 | 71.9 (2.9) | 28 | 74.5 (5.3) | 26 | 24.0 (20.9) | |||
| Hepatitis C virus | 0.758 | 0.749 | 0.979 | ||||||
| Negative | 72 | 72.2 (3.6) | 65 | 76.8 (5.4) | 65 | 27.9 (20.9) | |||
| Positive | 98 | 72.3 (4.3) | 94 | 76.4 (5.2) | 95 | 27.4 (17.3) | |||
| BMI at enrollment (kg/m2) | 0.077 | 0.081 | 0.659 | ||||||
| <25.0 | 52 | 71.3 (5.0) | 52 | 74.9 (4.8) | 46 | 30.0 (15.1) | |||
| ≥25.0 | 103 | 72.6 (4.2) | 92 | 76.9 (5.6) | 99 | 27.2 (19.4) | |||
| Waist-to-hip ratio | 0.075 | 0.023 | 0.748 | ||||||
| <0.952 | 73 | 71.8 (4.6) | 70 | 76.3 (5.2) | 67 | 28.1 (16.2) | |||
| ≥0.952 | 78 | 72.6 (4.0) | 70 | 77.4 (5.4) | 74 | 27.1 (21.1) | |||
| Diabetes | 0.933 | 0.514 | 0.054 | ||||||
| Negative | 95 | 72.1 (4.2) | 87 | 76.4 (5.4) | 88 | 26.1 (18.0) | |||
| Positive | 75 | 72.3 (4.0) | 72 | 76.8 (5.6) | 72 | 33.5 (21.8) | |||
| Alcohol history | 0.978 | 0.130 | 0.150 | ||||||
| Negative | 124 | 72.1 (3.9) | 116 | 76.5 (4.7) | 115 | 26.8 (18.0) | |||
| Positive | 46 | 72.3 (4.3) | 43 | 78.1 (6.3) | 45 | 32.4 (15.1) | |||
| Cigarette smoking | 0.182 | 0.039 | 0.203 | ||||||
| Never | 63 | 72.6 (3.7) | 59 | 74.7 (5.5) | 59 | 27.5 (17.3) | |||
| Ever | 100 | 72.0 (4.2) | 93 | 76.8 (5.1) | 94 | 29.6 (19.7) | |||
| CP class | 0.748 | 0.969 | 0.066 | ||||||
| A | 104 | 72.3 (4.0) | 97 | 76.3 (5.2) | 97 | 30.7 (20.7) | |||
| B | 65 | 71.9 (3.9) | 61 | 76.8 (5.6) | 62 | 25.0 (17.0) | |||
| Metastatic disease | 0.247 | 0.971 | 0.110 | ||||||
| Negative | 141 | 72.3 (3.9) | 131 | 76.5 (5.3) | 132 | 27.2 (16.7) | |||
| Positive | 19 | 71.8 (6.7) | 19 | 76.8 (5.4) | 19 | 39.2 (28.4) | |||
| Milan criteria | 0.337 | 0.314 | 0.556 | ||||||
| Within | 64 | 72.4 (3.4) | 60 | 76.9 (5.2) | 61 | 26.8 (20.3) | |||
| Outside | 106 | 72.1 (4.2) | 99 | 75.7 (5.6) | 99 | 28.1 (17.2) | |||
| BCLC stage | 0.723 | 0.383 | 0.378 | ||||||
| 0, A | 61 | 72.4 (3.2) | 57 | 77.2 (5.4) | 59 | 25.8 (19.8) | |||
| B, C | 109 | 72.1 (4.4) | 102 | 76.3 (4.7) | 101 | 29.5 (17.8) | |||
| AFP (ng/mL) | 0.157 | 0.501 | 0.809 | ||||||
| <47.85 | 85 | 72.6 (3.4) | 79 | 76.3 (5.6) | 80 | 27.4 (18.6) | |||
| ≥47.85 | 85 | 71.5 (5.0) | 80 | 76.8 (5.3) | 80 | 28.0 (17.7) | |||
| CA 19–9 (U/mL) | 0.307 | 0.782 | 0.431 | ||||||
| <38.0 | 81 | 72.3 (3.5) | 75 | 76.5 (5.1) | 76 | 28.7 (21.4) | |||
| ≥38.0 | 87 | 72.1 (4.4) | 82 | 77.0 (5.9) | 82 | 27.3 (18.0) |
Median (interquartile range)
Numbers not being equal to the total number was due to missing data
Wilcoxon rank-sum test or Kruskal–Wallis test
Association between Demographic and Tumor Characteristics and Overall Survival
The univariate analysis for the association between demographic and tumor characteristics and overall survival in HCC patients is shown in Supplementary Table S2. A worse survival was significantly associated with CP class B (HR = 1.57), metastatic disease (HR = 3.73), outside Milan criteria (HR = 3.56), BCLC stage B or C (HR = 3.01), and AFP ≥ 47.85 ng/mL (HR = 2.71). In contrast, liver transplantation conferred an improved OS (HR = 0.12). These variables were selected as potential covariates in the multivariate analyses.
Association Between LINE-1 Methylation and Folate Levels and Overall Survival
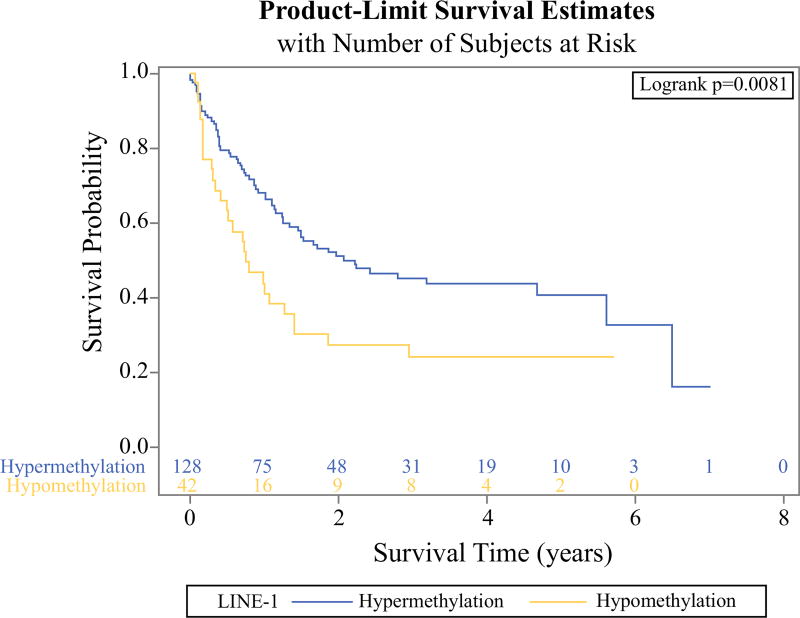
Using quartile levels of plasma LINE-1, WBC LINE-1, and plasma folate, we evaluated their associations with HCC survival (Supplementary Table S2). For plasma LINE-1, compared to Q4 (≥74.0%), the HRs were lower in Q3 (72.26–74.0%) and Q2 (70.0–72.26%) but higher in Q1 (<70.0%). Thus, we defined Q1 as “hypomethylation” and combined Q2, Q3, and Q4 into “hypermethylation.” The patients with plasma LINE-1 hypomethylation experienced a shorter survival than those with plasma LINE-1 hypermethylation (Kaplan–Meier analysis; log-rank test, P = 0.0081; Fig. 1). The corresponding HR was 1.81 (95% CI 1.16–2.83; P = 0.009).
FIG. 1.
Kaplan–Meier curve for overall survival (OS) of hepatocellular carcinoma (HCC) patients according to plasma LINE-1 levels. HCC patients with LINE-1 hypomethylation (<70%) had a significantly higher risk of death compared to those with LINE-1 hypermethylation (≥70%) (log-rank test, P = 0.0081)
For WBC LINE-1 methylation, patients in Q1 (<73.7%), Q2 (73.7–76.5%), and Q3 (76.5–79.0%) had a 30% increased risk of death compared to those in Q4 (≥79.0%). However, the higher risk was not statistically significant when we combined the Q1, Q2, and Q3 quartiles.
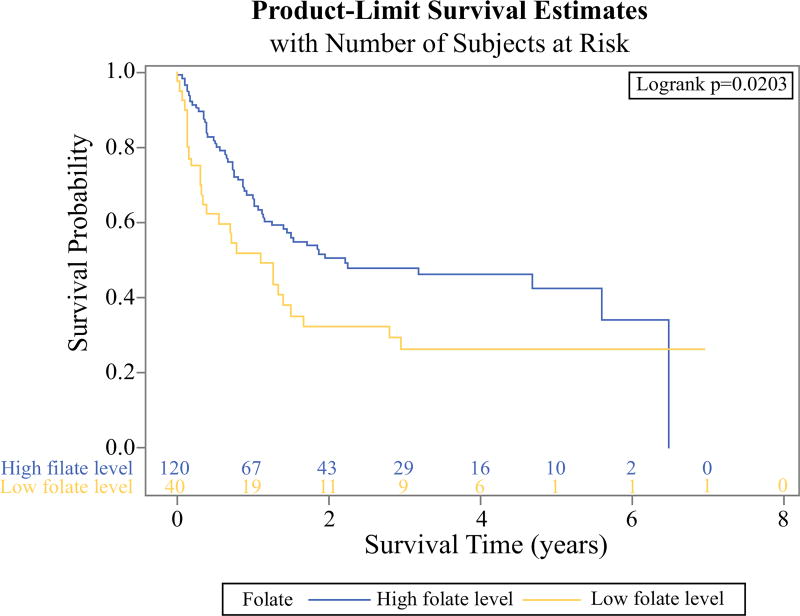
For plasma folate, compared with Q4 (≥39.2 nmol/L), the death rate was decreased in patients in Q3 (27.7–39.2 nmol/L) but increased in Q1 (<20.6 nmol/L) and Q2 (20.6–27.7 nmol/L). Therefore, we divided the plasma folate levels into low (Q1 and Q2) and high plasma folate (Q3 and Q4). The low plasma folate group had a significantly higher risk of death compared with the high plasma folate group (log-rank test, P = 0.0203; Fig. 2). The corresponding HR was 1.74 (95% CI 1.13–2.68; P = 0.013).
FIG. 2.
Kaplan–Meier curve for overall survival (OS) of hepatocellular carcinoma (HCC) patients according to plasma folate levels. HCC patients with low plasma folate level (<27.7 nmol/L) had a significantly higher risk of death compared to those with high plasma folate level (≥27.7 nmol/L) (log-rank test, P = 0.0203)
Multivariate Analyses for the association between LINE-1 Methylation and Folate Levels and Overall Survival
In the multivariate Cox proportional hazards Model 1, patients with low plasma LINE-1 methylation had shorter OS compared with patients with high LINE-1 methylation (HR = 1.77; 95% CI 1.12–2.79; P = 0.015; Table 2). Similarly, Model 2 shows that patients with low folate level had a poorer OS than those with high folate level (HR = 1.96; 95% CI 1.24–3.09; P = 0.004). We also analyzed the combination of plasma LINE-1 methylation and plasma folate on survival in Model 3. There was significantly worse OS for patients with low levels of both plasma LINE-1 methylation and folate compared with those with both high levels of LINE-1 methylation and folate (HR = 3.36; 95% CI 1.77–6.40; P <0.001). However, this interaction was of borderline significance (P = 0.057).
TABLE 2.
Multivariate Cox proportional hazards model for overall survival in HCC patients
| Variable | Model 1 | Model 2 | Model 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| HR | (95% CI) | P | HR | (95% CI) | P | HR | (95% CI) | P | |
| Age (year) | |||||||||
| ≥62.7 vs. <62.7 | 1.06 | (0.88–1.27) | 0.527 | 1.04 | (0.86–1.25) | 0.725 | 1.09 | (0.90–1.33) | 0.396 |
| CP class | |||||||||
| B vs. A | 1.92 | (1.26–2.94) | 0.003 | 1.43 | (0.92–2.23) | 0.109 | 1.61 | (1.03–2.52) | 0.038 |
| Milan criteria | |||||||||
| Outside vs. within | 2.09 | (1.24–3.53) | 0.006 | 2.42 | (1.39–4.22) | 0.002 | 2.32 | (1.33–4.02) | 0.003 |
| AFP (ng/mL) | |||||||||
| ≥47.85 vs. <47.85 | 1.97 | (1.27–3.06) | 0.003 | 2.37 | (1.48–3.78) | < 0.001 | 2.38 | (1.49–3.80) | <0.001 |
| Transplant | |||||||||
| Yes vs. no | 0.13 | (0.06–0.31) | < 0.001 | 0.15 | (0.07–0.36) | < 0.001 | 0.17 | (0.31–1.88) | <0.001 |
| Plasma LINE-1 methylation (%) | |||||||||
| Q1 (<70.0) vs. Q2–4 (≥70.0) | 1.77 | (1.12–2.79) | 0.015 | ||||||
| Plasma folate (nmol/L) | |||||||||
| Q1–2 (< 27.7) vs. Q3–4 (≥ 27.7) | 1.96 | (1.24–3.09) | 0.004 | ||||||
| Plasma LINE-1 (%)/Plasma folate (nmol/L) | |||||||||
| Q2–4 (≥70.0)/Q3–4 (≥27.7) | 1.00 | ||||||||
| Q1 (<70.0)/Q3–4 (≥27.7) | 0.76 | (0.31–1.88) | 0.556 | ||||||
| Q2–4 (≥70.0)/Q1–2 (<27.7) | 1.52 | (0.90–2.56) | 0.116 | ||||||
| Q1 (<70.0)/Q1–2 (<27.7) | 3.36 | (1.77–6.40) | <0.001 | ||||||
DISCUSSION
We examined the hypothesis that global DNA hypomethylation and folate deficiency are associated with worse overall survival in a prospective cohort study of 172 HCC patients. We found that both lower levels of plasma LINE-1 methylation and plasma folate could serve as prognostic markers in HCC in our dataset. In addition, we found an interaction between LINE-1 and folate, although of borderline significance.
Several studies have investigated the association between global DNA methylation and prognostic outcome in HCC patients. Using liver tissue specimens, most studies have demonstrated that LINE-1 hypomethylation is an independent risk factor for cancer recurrence and worse outcome among HCC patients.15–19 However, Lee et al. failed to find a significant association, which may be due to inadequate statistical power (n = 20).20 In addition, two studies also indicated that serum LINE-1 hypomethylation was significantly associated with poor survival.10,11 We first demonstrated that LINE-1 hypomethylation in plasma but not in WBC DNA was associated with worse survival in HCC patients. Detecting methylation profiles of circulating cell-free DNA (cfDNA) has emerged as a promising noninvasive approach for the diagnosis, prognosis, and monitoring of cancers.9 The presence of hypermethylated tumor suppressor genes in cfDNA has been described in plasma samples from HCC patients as well.21–23 These studies also have shown a high concordance of DNA methylation patterns in plasma and tumor DNA and indicate that plasma DNA may be used as a reliable source for testing methylation profiles in liver cancer patients.22,23 Therefore, our findings support a potential role of plasma LINE-1 hypomethylation as a useful surrogate marker to evaluate clinical outcomes in HCC patients.
Folate, as a methyl donor in the synthesis of S-adenosylmethionine, has been shown to mediate hepatocarcinogenesis by participating in DNA methylation and also in nucleotide synthesis andDNA repair.24,25 Studies have shown an inverse association between blood folate levels and HCC risk.26–30 Therefore, folate status also may be associated with HCC prognosis. More advanced stage HCC patients have lower folate levels than those with earlier stage disease, but no study examined the role of circulating folate in survival of HCC patients.31,32 Our results, similar to Rossi et al. and Yang et al., show patients with lower levels of circulating folate were at increased risk of all-cause mortality after cancer diagnosis.12,13
The combined analysis of LINE-1 and folate showed a borderline significant interaction associated with OS in HCC patients. Given the importance of folate in DNA methylation and synthesis, it is plausible that chronic folate deficiency may be associated with global DNA hypomethylation. Measuring global DNA methylation indirectly through the incorporation of 3H-methyl S-adenosylmethionine, a number of epidemiological studies have suggested a positive correlation between folate and global DNA methylation in healthy subjects.33 In addition, a recent study also reported that folate deficiency is associated with LINE-1 hypomethylation.34 Although no prior study focused on the interaction between folate status and global DNA methylation on cancer survival, our current findings of a strong association in the combination of low folate and LINE-1 hypomethylation are supported by a prior report that folate supplementation could limit the aggressiveness of glioma through the remethylation of DNA repeat elements via the Sp1/Sp3-mediated transcriptional up-regulation of genes coding for Dnmt3a and Dnmt3b proteins.35 Those data suggest that the combined analysis of LINE-1 hypomethylation and folate deficiency can improve their sensitivity as prognostic biomarkers.
The current study had several notable strengths. This prospective cohort study collected information on a wide variety of factors, such as demographic, lifestyle, and clinicopathological factors, and long-term follow-up. In addition, all survival events in the study were confirmed by a medical oncologist. Despite these considerable strengths, the study has several limitations. The foremost limitation was that LINE-1 levels and folate status were determined only at baseline so changes over time could not be assessed. We also had a limited sample size to detect interactions.
In conclusion, our findings suggest that lower plasma levels of global DNA methylation and folate are associated with worse OS in patients with HCC. The prognostic significance of LINE-1 methylation and folate status requires further validation in a large-scale, prospective study.
Supplementary Material
Acknowledgments
This work is supported by NIH Grants R01 ES005116 (RMS), P30 ES009089 (RMS) and by the Ministry of Science and Technology, Taiwan MOST 104-2918-I-038-001 (CCY).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1245/s10434-017-5913-4) contains supplementary material, which is available to authorized users.
CONFLICT OF INTEREST There is no interest confliction involved for this submission.
References
- 1.Lee SM, Kim-Ha J, Choi WY, et al. Interplay of genetic and epigenetic alterations in hepatocellular carcinoma. Epigenomics. 2016;8(7):993–1005. doi: 10.2217/epi-2016-0027. [DOI] [PubMed] [Google Scholar]
- 2.Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011;12(2):130–7. doi: 10.2174/138920211795564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303(5664):1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 5.Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419(1–2):75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miousse IR, Koturbash I. The Fine LINE: Methylation Drawing the Cancer Landscape. Biomed Res Int. 2015;2015:131547. doi: 10.1155/2015/131547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 10.Ramzy II, Omran DA, Hamad O, Shaker O, Abboud A. Evaluation of serum LINE-1 hypomethylation as a prognostic marker for hepatocellular carcinoma. Arab J Gastroenterol. 2011;12(3):139–42. doi: 10.1016/j.ajg.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Tangkijvanich P, Hourpai N, Rattanatanyong P, Wisedopas N, Mahachai V, Mutirangura A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta. 2007;379(1–2):127–33. doi: 10.1016/j.cca.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 12.Rossi E, Hung J, Beilby JP, Knuiman MW, Divitini ML, Bartholomew H. Folate levels and cancer morbidity and mortality: prospective cohort study from Busselton, Western Australia. Ann Epidemiol. 2006;16(3):206–12. doi: 10.1016/j.annepidem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Bostick RM, Friedman JM, Flanders WD. Serum folate and cancer mortality among U.. adults: findings from the Third National Health and Nutritional Examination Survey linked mortality file. Cancer Epidemiol Biomark Prev. 2009;18(5):1439–47. doi: 10.1158/1055-9965.EPI-08-0908. [DOI] [PubMed] [Google Scholar]
- 14.Siegel AB, Goyal A, Salomao M, et al. Serum adiponectin is associated with worsened overall survival in a prospective cohort of hepatocellular carcinoma patients. Oncology. 2015;88(1):57–68. doi: 10.1159/000367971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada K, Baba Y, Ishimoto T, et al. LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann Surg Oncol. 2015;22(4):1280–7. doi: 10.1245/s10434-014-4134-3. [DOI] [PubMed] [Google Scholar]
- 16.Anwar SL, Krech T, Hasemeier B, et al. Loss of DNA methylation at imprinted loci is a frequent event in hepatocellular carcinoma and identifies patients with shortened survival. Clin Epigenetics. 2015;7:110. doi: 10.1186/s13148-015-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C, Utsunomiya T, Ikemoto T, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann Surg Oncol. 2014;21(Suppl 4):S729–35. doi: 10.1245/s10434-014-3874-4. [DOI] [PubMed] [Google Scholar]
- 18.Gao XD, Qu JH, Chang XJ, et al. Hypomethylation of long interspersed nuclear element-1 promoter is associated with poor outcomes for curative resected hepatocellular carcinoma. Liver Int. 2014;34(1):136–46. doi: 10.1111/liv.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Xu Y, Zhao J, et al. Elevated expression of the stem cell marker CD133 associated with Line-1 demethylation in hepatocellular carcinoma. Ann Surg Oncol. 2011;18(8):2373–80. doi: 10.1245/s10434-011-1599-1. [DOI] [PubMed] [Google Scholar]
- 20.Lee HS, Kim BH, Cho NY, et al. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res. 2009;15(3):812–20. doi: 10.1158/1078-0432.CCR-08-0266. [DOI] [PubMed] [Google Scholar]
- 21.Zhang YJ, Wu HC, Shen J, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13(8):2378–84. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]
- 22.Iyer P, Zekri AR, Hung CW, et al. Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp Mol Pathol. 2010;88(1):107–111. doi: 10.1016/j.yexmp.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong IH, Lo YM, Zhang J, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59(1):71–3. [PubMed] [Google Scholar]
- 24.Pogribny IP, James SJ, Beland FA. Molecular alterations in hepatocarcinogenesis induced by dietary methyl deficiency. Mol Nutr Food Res. 2012;56(1):116–25. doi: 10.1002/mnfr.201100524. [DOI] [PubMed] [Google Scholar]
- 25.Butler LM, Arning E, Wang R, et al. Prediagnostic levels of serum one-carbon metabolites and risk of hepatocellular carcinoma. Cancer Epidemiol Biomark Prev. 2013;22(10):1884–93. doi: 10.1158/1055-9965.EPI-13-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welzel TM, Katki HA, Sakoda LC, et al. Blood folate levels and risk of liver damage and hepatocellular carcinoma in a prospective high-risk cohort. Cancer Epidemiol Biomark Prev. 2007;16(6):1279–82. doi: 10.1158/1055-9965.EPI-06-0853. [DOI] [PubMed] [Google Scholar]
- 27.Wu MY, Kuo CS, Lin CY, Lu CL, Syu Huang RF. Lymphocytic mitochondrial DNA deletions, biochemical folate status and hepatocellular carcinoma susceptibility in a case-control study. Br J Nutr. 2009;102(5):715–21. doi: 10.1017/S0007114509243054. [DOI] [PubMed] [Google Scholar]
- 28.Chang SC, Goldstein BY, Mu L, et al. Plasma folate, vitamin B12, and homocysteine and cancers of the esophagus, stomach, and liver in a Chinese population. Nutr Cancer. 2015;67(2):212–23. doi: 10.1080/01635581.2015.989375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui LH, Quan ZY, Piao JM, et al. Plasma Folate and Vitamin B12 Levels in Patients with Hepatocellular Carcinoma. Int J Mol Sci. 2016;17(7):1032. doi: 10.3390/ijms17071032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng SB, Liu HT, Lin PT, Lai CY, Huang YC. Folate and vitamin B-6 status are not associated with homocysteine, oxidative stress and antioxidant capacities in patients with hepatocellular carcinoma. Eur J Clin Nutr. 2016;70(7):855–8. doi: 10.1038/ejcn.2015.222. [DOI] [PubMed] [Google Scholar]
- 31.Kuo CS, Lin CY, Wu MY, Lu CL, Huang RF. Relationship between folate status and tumour progression in patients with hepatocellular carcinoma. Br J Nutr. 2008;100(3):596–602. doi: 10.1017/S0007114508911557. [DOI] [PubMed] [Google Scholar]
- 32.Lin CC, Yin MC. B vitamins deficiency and decreased anti-oxidative state in patients with liver cancer. Eur J Nutr. 2007;46(5):293–9. doi: 10.1007/s00394-007-0665-8. [DOI] [PubMed] [Google Scholar]
- 33.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23(8):853–9. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agodi A, Barchitta M, Quattrocchi A, et al. Low fruit consumption and folate deficiency are associated with LINE-1 hypomethylation in women of a cancer-free population. Genes Nutr. 2015;10(5):480. doi: 10.1007/s12263-015-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hervouet E, Debien E, Campion L, et al. Folate supplementation limits the aggressiveness of glioma via the remethylation of DNA repeats element and genes governing apoptosis and proliferation. Clin Cancer Res. 2009;15(10):3519–29. doi: 10.1158/1078-0432.CCR-08-2062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.