Abstract
Studies show that the continuous consumption of fructose can lead to nonalcoholic fatty liver disease (NAFLD) and steatohepatitis. We aimed to investigate the role of Metformin in an animal model of liver injury caused by fructose intake, focusing on the molecular markers of lipogenesis, beta-oxidation, and antioxidant defenses. Male three months old C57BL/6 mice were divided into control group (C) and fructose group (F, 47% fructose), maintained for ten weeks. After, the groups received Metformin or vehicle for a further eight weeks: control (C), control + Metformin (CM), fructose (F), and fructose + Metformin (FM). Fructose resulted in hepatic steatosis, insulin resistance and lower insulin sensitivity in association with higher mRNA levels of proteins linked with de novo lipogenesis and increased lipid peroxidation. Fructose diminished mRNA expression of antioxidant enzymes, and of proteins responsible for mitochondrial biogenesis. Metformin reduced de novo lipogenesis and increased the expression of proteins related to mitochondrial biogenesis, thereby increasing beta-oxidation and decreasing lipid peroxidation. Also, Metformin upregulated the expression and activity of antioxidant enzymes, providing a defense against increased reactive oxygen species generation. Therefore, a significant reduction in triglyceride accumulation in the liver, steatosis and lipid peroxidation was observed in the FM group. In conclusion, fructose increases de novo lipogenesis, reduces the antioxidant defenses, and diminishes mitochondrial biogenesis. After an extended period of fructose intake, Metformin treatment, even in continuing the fructose intake, can reverse, at least partially, the liver injury and prevents NAFLD progression to more severe states.
Keywords: Steatosis, Lipogenesis, beta-oxidation, Oxidative stress, Stereology
Highlights
-
•
Fructose increases lipogenesis and lipid peroxidation, reduces the antioxidant defenses, and mitochondrial biogenesis.
-
•
Metformin mechanism of action remains partially understood and controversial.
-
•
Metformin can reverse the liver injury preventing the progression to more severe states.
1. Introduction
The fructose consumption has increased dramatically in recent years incorporated in industrial products and sugary drinks [1]. Fructose is metabolized to triose phosphates by hepatocytes, enterocytes, and kidney tubular cells. In contrast to glucose, fructose metabolism is not tightly regulated by cellular energy status and fructose consumption leads to an overflow of triose phosphates into hepatocytes and a subsequent disposal of these compounds, leading to increased lactic acid production, gluconeogenesis and de novo lipogenesis [2]. Studies have demonstrated that the continuous consumption of fructose can lead to nonalcoholic fatty liver disease (NAFLD) in both humans [3] and rodents [4].
NAFLD is a highly prevalent condition, as population studies indicate that 10–50% of the worldwide population possess a reversible form of hepatic steatosis [5]. However, in a small percentage of individuals, it can progress to hepatocellular death and inflammation, a condition known as nonalcoholic steatohepatitis (NASH), and in more severe cases, cirrhosis and hepatocellular carcinoma [6]. An increased oxidative stress (OStress) is one of the major factors that trigger liver inflammation and NAFLD progression to NASH [7]. A fructose-rich diet may lead to reactive oxygen species (ROS) generation [8], at the same time that reduced antioxidant potential [9].
The mechanisms involved in the continuum of hepatic insult are still widely investigated, but it has traditionally been thought to result from two distinct events. The first event is an increased rate of lipid influx and reduced lipid clearance, leading to fat accumulation in the liver [10]. The second event is an inflammatory process caused by increased liver ROS and cytokine activation [11], probably resulted from the exposure of hepatocytes to a greater concentration of lipids and/or carbohydrates. Metformin improves hyperglycemia mainly through the suppression of hepatic gluconeogenesis along with the improvement of insulin signaling. However, its mechanism of action remains partially understood and controversial [12].
In this study, we have focused on three main actions involved in liver injury of NAFLD: lipogenesis and biogenesis of mitochondria, beta-oxidation (BOxid), and OStress. Although much has been investigated about these measures in the liver, many questions remain unanswered. Additionally, we use an experimental model that is relevant due to increased fructose intake in beverages and soft drinks, with the consequent increase of NAFLD in the population.
2. Materials and methods
2.1. Animals and diet
The Ethics Committee for the Care and Use of Experimental Animals of the State University of Rio de Janeiro approved the experimental protocol (protocol number CEUA/022/2015). The experiment was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH Publication number 85-23, revised in 1996). The animals have been maintained in ventilated cages under controlled conditions (Nexgen system, Allentown Inc., PA, USA, 20 ± 2° C and 12 h/12 h dark/light cycle), with free access to food and water.
Initially, 40 three-months old male C57BL/6 mice were randomly divided into two groups (n = 20/group) and fed control diet (C) or fructose diet (F, 47% of fructose), during ten weeks. Both diets had the same amount of total carbohydrates, but in the F diet, part of the starch was replaced by pure fructose (PragSolucoes, Jau, SP, Brazil, following the recommendations for rodents of the American Institute of Nutrition, Table 1) [13].
Table 1.
Composition and energy content of the diets (control, C, and fructose, F). Mineral and vitamin mixtures are in accordance with the formulation of the American Institute of Nutrition (AIN93M) [13].
| Content (g) | Diets |
|
|---|---|---|
| C | F | |
| Casein | 140 | 140 |
| Corn starch | 620.7 | 146.4 |
| Sucrose | 100 | 100 |
| Fructose | – | 474.3 |
| Soybean oil | 40 | 40 |
| Fibers | 50 | 50 |
| Vitamin mix | 10 | 10 |
| Minerals mix | 35 | 35 |
| Cystine | 1.8 | 1.8 |
| Choline | 2.5 | 2.5 |
| Antioxidant | 0.008 | 0.008 |
| Total | 1000 | 1000 |
| Energy (kcal) | 3804 | 3804 |
| Carbohydrates (% energy) | 76 | 76 |
| Proteins (% energy) | 14 | 14 |
| Lipids (% energy) | 10 | 10 |
After the early ten weeks, the animals were randomly separated into two additional groups (n = 10/group) to include Metformin hydrochloride treatment (250 mg/kg/day, Pharmanostra, GO, Brazil) for a further eight weeks:
-
a)
C group: control diet for ten weeks, followed by control diet and vehicle (NaCl, orogastric gavage) for eight weeks;
-
b)
CM group: control diet for ten weeks, followed by control diet and Metformin (orogastric gavage) for eight weeks;
-
c)
F group: Fructose diet for ten weeks, followed by fructose diet and vehicle (NaCl, orogastric gavage) for eight weeks;
-
d)
FM group: Fructose diet for ten weeks, followed by fructose diet and Metformin (orogastric gavage) for eight weeks.
2.2. Body mass, food and energy intake
Body mass (BM) was measured weekly. Food intake was monitored daily, determined as the difference between the food supplied and the amount of food left in the cage. The diets were renewed daily, and the remaining chow was discarded.
2.3. Oral glucose tolerance test (OGTT)
We performed OGTT one day before the administration of Metformin, and two days before euthanasia, in 6 h fasted animals that received glucose (25% in sterile 0.9% NaCl) at a dose of 1 g/kg by orogastric gavage. The glycemia was measured at fasting (time 0) and 15, 30, 60 and 120 min after glucose administration (Glucometer Accu-Chek, Roche, SP, Brazil). We assessed glucose tolerance based on the area under the curve (AUC) (GraphPad Prism version 7.0 for Windows; La Jolla, CA, USA).
2.4. Euthanasia
The animals were food-deprived from 1 AM to 7 AM, then deeply anesthetized (sodium pentobarbital, 150 mg/kg intraperitoneal). Blood was collected, plasma was obtained (120 g/15 min at room temperature), and stored at −20° C. The liver was dissected, weighed and fragments from all lobes were collected and fixed for 48 h (formaldehyde 4% w/v, 0.1 M phosphate buffer, pH 7.2). Alternatively, fragments were frozen at −80° C.
2.5. Plasma analysis, insulin resistance, and insulin sensitivity
Total cholesterol (TC) and triglycerides (TG) were measured by an automatic spectrophotometer using its recommended commercial kit (Bioclin System II, Quibasa, Belo Horizonte, MG, Brazil). Plasma concentrations of insulin were measured using the Single Plex kit (EZRMI-13K Rat/Mouse Insulin ELISA, Millipore Merck, Darmstadt, Germany). The homeostasis model assessment of insulin resistance (HOMA-IR) was estimated: HOMA-IR = fasting blood glucose (mmol/L) × fasting serum insulin (IU/mL)/22.5 [14], as well as the quantitative insulin sensitivity check index (QUICKI): [1/log (fasting insulin μU/mL) + log (fasting glucose mg/dL)] [15].
2.6. Liver
Liver fragments were embedded in Paraplast Plus (Sigma-Aldrich, St. Louis, MO, USA), sectioned (5-μm-thick), and stained with hematoxylin and eosin. Digital images of the sections were analyzed (Leica DMRBE microscope, Wetzlar, Germany; Lumenera Infinity 1-5c camera, Ottawa, Canada). Five fields per animal, 36-test-points per field, were sufficient to estimate the volume density of hepatic steatosis by point-counting with a standard error of 5% [16]: Vv [steatosis, liver] = Pp [steatosis, liver]/PT (Pp is the number of points that hit the fat drops, PT is the total test-points) [17]. In frozen fragments, we measured hepatic TG (Bioclin System II, Quibasa, BH, Brazil).
2.7. RT-qPCR
Total RNA was extracted from approximately 50 mg of liver tissue using Trizol reagent (Invitrogen, CA, USA). RNA amount was determined using Nanovue spectroscopy (GE Life Sciences), and 1 mg of RNA was treated with DNAse I (Invitrogen, CA, USA). Synthesis of the first strand cDNA was performed using Oligo (dT) primers for mRNA and Superscript III reverse-transcriptase (both Invitrogen). Quantitative real-time PCR (RT-qPCR) used a BioRad CFX96 cycler and the SYBR Green mix (Invitrogen, CA, USA). Endogenous control beta-actin normalized the selected gene expressions. Efficiencies of RT-qPCR for the target gene and the endogenous control were approximately equal, calculated through dilution series of cDNA. After a pre-denaturation and polymerase-activation program (4 min at 95° C), 44 cycles (each one consisting of 95° C for 10 s and 60° C for 15 s) were followed by a melting curve program (60–95° C with a heating rate of 0.1° C/s). Negative controls consisted of wells in which cDNA was substituted for deionized water. The relative expression ratio of mRNA was calculated by the equation 2−ΔΔCT, in which −ΔCT expresses the difference between the number of cycles (CT) of the target genes and the endogenous control. The sequences of the sense and antisense primers used for amplification are detailed in Table 2. We analyze the following gene expressions: CAT, catalase; CHREBP, carbohydrate response element-binding protein; FAT/CD36, fatty acid translocase; GPx, glutathione peroxidase; GR, glutathione reductase; PGC1alpha, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; Plin2, lipid droplet protein Perilipin 2; PPARgamma, peroxisome proliferator activator receptor gamma; SOD2, superoxide dismutase 2; SREBP-1c, sterol regulatory element-binding protein-1c. The sequences of the sense and antisense primers used for amplification are detailed in Table 2.
Table 2.
Primers used in RTq-PCR and their sequence for the evaluation of gene expression.
| Gene | Sequence forward 5′→3′ | Sequence reverse 5′→3′ |
|---|---|---|
| beta-actin | TGTTACCAACTGGGACGACA | GGGGTGTTGAAGGTCTCAAA |
| CAT | TTGACAGAGAGCGGATTCCT | TCTGGTGATATCGTGGGTGA |
| CD36 | CCCTCCAGAATCCAGACAAC | TGCATTTGCCAATGTCTAGC |
| ChREBP | CACTCAGGGAATACACGCCTAC | ATCTTGGTCTTAGGGTCTTCAGG |
| GPx | CCCGTGCGCAGGTACAG | CAGCAGGGTTTCTATGTCAGGTT |
| GR | GGGATTGGCTGTGATGAGAT | GGTGACCAGCTCCTCTGAAG |
| PGC1alpha | AACCACACCCACAGGATCAGA | TCTTCGCTTTATTGCTCCATGA |
| PLIN2 | AATATGCACAGTGCCAACCA | CGATGCTTCTCTTCCACTCC |
| PPARalpha | CAAGGCCTCAGGGTACCACTAC | GCCGAATAGTTCGCCGAAA |
| PPARgamma | CACAATGCCATCAGGTTTGG | GCTGGTCGATATCACTGGAGATC |
| SOD | CAGGACCCATTGCAAGGAA | GTGCTCCCACACGTCAATCC |
| SREBP1c | AGCAGCCCCTAGAACAAACA | TCTGCCTTGATGAAGTGTGG |
Abbreviations: CAT, Catalase; CD36, Cluster of differentiation 36; ChREBP, Carbohydrate responsive element binding protein; GPx, Glutathione peroxidase; GR, Glutathione reductase; PGC, Peroxisome proliferator-activated receptor coactivator-1alpha; PLIN, lipid droplet protein Perilipin; PPAR, Peroxisome proliferator activator receptor; SOD, Superoxide dismutase; SREBP, Sterol regulatory element binding protein.
2.8. Antioxidant enzyme activity assays
SOD, Catalase and GPx activity were determined in liver homogenate by spectrophotometry (Genesys 10S UV–Vis Spectrophotometer, Thermo Scientific, CA, USA). SOD activity was assayed based on its ability to inhibit pyrogallol autoxidation [18]. Catalase activity was measured by the rate of decrease in hydrogen peroxide concentration [19]. GPx activity was measured by monitoring the oxidation of NADPH at 340 nm in the presence of hydrogen peroxide [20]. The total protein content of each sample was determined by BCA protein assay kit (Thermo Scientific, Rockford, IL, USA).
2.9. Malondialdehyde assay
As an index of lipid peroxidation, we used the thiobarbituric acid reactive substances (TBARS) method for analyzing malondialdehyde (MDA). MDA levels were assessed based on its reaction with thiobarbituric acid, which forms a colored complex that can be quantified spectrophotometrically at 532 nm (Genesys 10S UV–Vis Spectrophotometer, Thermo Scientific, CA, USA). 1,1,3,3-tetramethoxypropane was used as a standard, and the results are expressed as MDA equivalents (nmol/mg protein) [21].
2.10. Western blot
Liver fragments (100 mg) were added to a lysis buffer containing protease inhibitors, homogenized and centrifuged (4500 rpm during 20 min at 4° C), and supernatants were collected. Protein concentration was then determined using the BCA protein assay kit (Thermo Scientific, Rockford, IL, USA). After denaturation, proteins were separated by electrophoresis on a polyacrylamide gel (SDS-PAGE) and transferred to a PVDF membrane. Membranes were then blotted with primary antibodies for AMPK (adenosine monophosphate-activated protein kinase) and phospho-AMPK followed by incubation with secondary antibodies. Bands were detected by chemiluminescence (ECL Prime, Amersham, UK) using the ChemiDoc system (Bio-Rad, Hercules, CA, USA). The intensity of the bands was quantified using ImageJ software (NIH, imagej.nih.gov/ij, USA). The expression of the structural protein β-actin was used to correct the blot data. Both primary and secondary antibodies were purchased from Santa Cruz Biotechnology, CA, USA.
2.11. Data analysis
Data were tested for normality and homoscedasticity of the variances and then expressed as the mean and standard deviation (SD). We tested the differences between the groups in the pre-treatment period with t-test (C and F groups). We tested the contribution of diet and Metformin in the post-treatment period with a two-way ANOVA (posthoc test of Holm-Sidak) (GraphPad Prism version 7.02 for Windows, La Jolla, CA, USA). We accepted P-values < 0.05 as significant.
3. Results
3.1. Fructose diet and Metformin on body mass, food intake, and glucose
The groups C and F began the treatment with no difference in their BM (P = 0.345). During the treatment, we did not observe a difference in food intake (P = 0.87, Table 3). After treatment, BM and food intake remain without difference among the groups (P = 0.99, Table 3).
Table 3.
Data from the experimental groups. Data are expressed as the mean and SD (n = 10/group). P < 0.05 when compared to the † C group; § F group; ‡ CM group (t-test in the pre-treatment period, and two-way ANOVA and the posthoc test of Holm–Sidak in post-treatment period). AUC, the area under the curve; C, control group; F, fructose group; OGTT, oral glucose tolerance test.
| Pre-treatment | C | F | ||
|---|---|---|---|---|
| Initial body mass (g) | 25.8 ± 1.34 | 25.7 ± 1.32 | ||
| Final body mass (g) | 29.7 ± 1.12 | 29.6 ± 1.30 | ||
| Fasting glucose (mmol/L) | 6.4 ± 0.26 | 8.6 ± 0.39† | ||
| OGTT (AUC, mmol/L/min) |
710.8 ± 11.41 |
1012.0 ± 47.68† |
||
| Post-treatment |
C |
CM |
F |
FM |
| Final body mass (g) | 31.9 ± 0.44 | 31.3 ± 1.34 | 31.4 ± 0.88 | 31.1 ± 1.77 |
| Food intake (g/day/mouse) | 2.8 ± 0.33 | 2.9 ± 0.66 | 2.6 ± 0.28 | 2.8 ± 0.37 |
| Liver mass/Tibia length (g/cm) | 0.5 ± 0.01 | 0.5 ± 0.01 | 0.6 ± 0.02† | 0.6 ± 0.01§‡ |
| Fasting glucose (mmol/L) | 7.5 ± 0.79 | 7.3 ± 0.61 | 9.5 ± 0.13† | 8.2 ± 0.20§‡ |
| Insulin (pmol/L) | 89.2 ± 9.25 | 89.6 ± 4.86 | 187.9 ± 9.41† | 95.2 ± 4.54§ |
| HOMA-IR | 4.37 ± 0.65 | 4.25 ± 0.29 | 11.59 ± 0.51† | 5.02 ± 0.19§‡ |
| QUICKI | 0.195 ± 0.003 | 0.194 ± 0.001 | 0.179 ± 0.001† | 0.192 ± 0.001§‡ |
| Total cholesterol (mmol/L) | 2.9 ± 0.09 | 2.8 ± 0.12† | 3.5 ± 0.07† | 3.1 ± 0.16§‡ |
| Plasma triglyceride (mmol/L) | 4.65 ± 0.11 | 4.59 ± 0.20 | 5.05 ± 0.31† | 4.50 ± 0.05§ |
| Hepatic triglyceride (mmol/L) | 0.72 ± 0.10 | 0.75 ± 0.12 | 1.91 ± 0.13† | 1.36 ± 0.07§‡ |
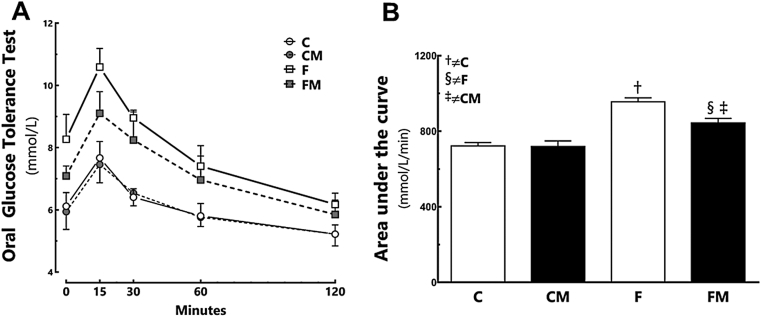
In the pre-treatment period, the F group was hyperglycemic (+33% compared to the C group; P = 0.002), and showed greater OGTT AUC (+42% compared to the C group; P = 0.0003, Table 3). In the post-treatment period, the F group remained hyperglycemic (+27% than the C group; P = 0.005, Table 3), with greater OGTT AUC (+32% compared to the C group; P < 0.0001, Fig. 1). Metformin decreased glycemia and OGTT AUC in the FM group (−12% than the F group; P < 0.0001, Table 3 and Fig. 1). However, glycemia was still greater in the FM group (+13% compared to the CM group; P < 0.0001, Fig. 1). Diet and Metformin had interaction affecting the glucose levels (two-way ANOVA; P = 0.029).
Fig. 1.
Oral glucose tolerance test (A) and area under the curve (B) in the post-treatment period. Data are expressed as the mean and SD (n = 10 each group). P < 0.05 when: † compared to the C group; ‡ compared to the CM group (two-way ANOVA and the posthoc test of Holm–Sidak). Groups: C, control diet; F, fructose diet; M, Metformin.
Fructose increased HOMA-IR and reduced QUICKI in the F group (HOMA-IR +165%, P < 0.0001; QUICK −10% compared to the C group, P < 0.0001). Metformin diminished HOMA-IR and increased QUICKI in the FM group (HOMA-IR, −57%, P < 0.0001; QUICKI, +10% than the F group, P < 0.0001). HOMA-IR was still high in the FM group (+18% compared to the CM group, P = 0.0446).
3.2. Fructose diet and Metformin on plasma determinations
Fructose elevated TC levels in the F group (+19% compared to the C group; P < 0.0001, Table 3). Metformin reduced TC in the FM group (−12% compared to the F group; P < 0.0001). The FM group had the TC level still higher than the CM group (+12%; P = 0.0006, Table 3). Diet and Metformin had interaction affecting TC levels (two-way ANOVA; P = 0.022).
Fructose augmented TG concentration in the F group (+8% compared to the C group; P = 0.001, Table 3). Metformin diminished TG level in the FM group (−10% than the F group; P < 0.0001, Table 3). Diet and Metformin had interaction affecting TG levels (two-way ANOVA; P = 0.038).
Fructose elevated plasma insulin in the F group (+47% compared to the C group; P < 0.0001). Metformin diminished plasma insulin in the FM group (−11% compared to the F group; P < 0.0001). Diet and Metformin had interaction in plasma insulin (two-way ANOVA; P < 0.0001, Table 3).
3.3. Metformin decreases fructose-induced steatosis and improves hepatic biochemistry
Fructose augmented the liver mass in the F group (+18% compared to the C group, P < 0.0001). Metformin reduced liver mass in the FM group (−10% compared to the F group, P = 0.0002), but the liver mass was still greater in the FM group (+8% compared to the CM group; P = 0.0024, Table 3). Diet and Metformin had an interaction affecting liver mass (two-way ANOVA; P = 0.0006).
Fructose elevated hepatic TG in the F group (+165% compared to the C group; P < 0.0001). Metformin reduced hepatic TG in the FM group (−29% compared to the F group; P < 0.0001, Table 3), but it remained higher in the FM group (+80% compared to the CM group; P < 0.0001). Diet and Metformin showed an interaction affecting hepatic TG (two-way ANOVA; P < 0.0001).
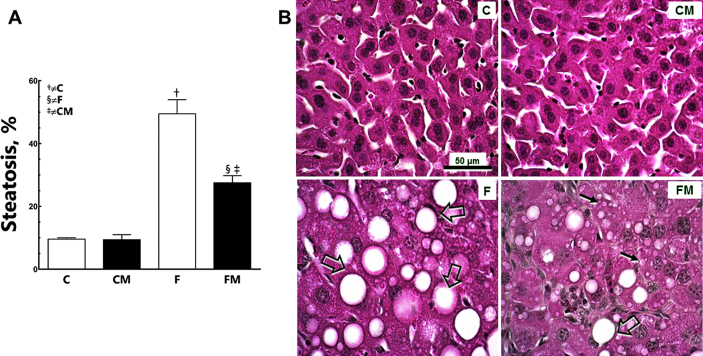
Fructose worsened steatosis in the F group (+532% compared to the C group; P < 0.0001, Fig. 2). Metformin reduced steatosis in the FM group (−55% compared to the F group; P < 0.0001). However, steatosis was still greater in the FM group (+187% than the CM group; P < 0.0001). Diet and Metformin showed interaction, but also affected hepatic steatosis independently (two-way ANOVA; P < 0.0001).
Fig. 2.
Hepatic steatosis (A), and photomicrographs of the liver tissue (B) in post-treatment period (HE staining). Data are expressed as the mean and SD (n = 10 each group). P < 0.05 when: † compared to the C group; ‡ compared to the CM group; § compared to the F group (two-way ANOVA and the posthoc test of Holm–Sidak). Groups: C, control diet; F, fructose diet; M, Metformin. Mice that received F diet present numerous hepatocytes with fat droplets characterizing macro- (open arrows) and micro-steatosis (arrows). The treatment with Metformin reduced fat droplets in the liver even with the F diet.
3.4. Metformin improves beta-oxidation and decreases liver lipogenesis
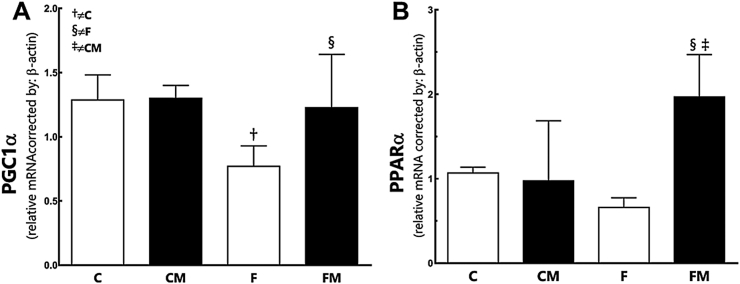
Fructose diminished PGC1alpha in the F group (−39.8% compared to the C group; P = 0.02, Fig. 3). Metformin had increased PGC1alpha in the FM group (+37% compared to the F group; P = 0.03) and restored PGC1alpha at the level of the CM group (P = 0.95 comparing the FM group with the CM group). Diet and Metformin did not interact, but affected the findings independently (two-way ANOVA; P = 0.01).
Fig. 3.
Hepatic gene expression of PGC1α (A) and PPARα (B) from the experimental groups in the post-treatment period. Endogenous control beta-actin was used to normalize the expression of the selected genes. Data are expressed as the mean and SD (n = 10 each group). P < 0.05 when: † compared to the C group; ‡ compared to the CM group; § compared to the F group (two-way ANOVA and the posthoc test of Holm–Sidak). Groups: C, control diet; F, fructose diet; M, Metformin. Abbreviations: peroxisome proliferator activator receptor alpha (PPARα); Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α).
Fructose did not alter PPARalpha in the F group, although it was 38% lower compared to the C group (P = 0.4, Fig. 3). Metformin enhanced PPARalpha in the FM group (+201% compared to the F group; P = 0.001). PPARalpha was higher in the FM group than in the CM group (+102%; P = 0.01). Diet and Metformin affected the findings independently and showed interaction (two-way ANOVA; P = 0.002).
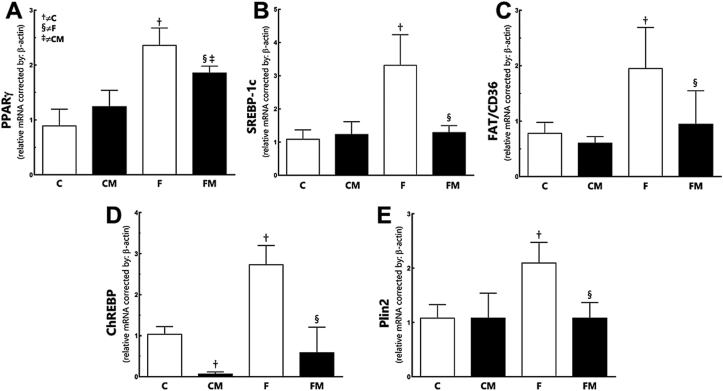
Fructose increased PPARgamma in the F group (+164% compared to the C group; P < 0.0001, Fig. 4). Metformin diminished PPARgamma in the FM group (−21% compared to the F group, P = 0.001). However, PPARgamma was still high in the FM group (+49% compared to the CM group; P = 0.008). Metformin affected the result (two-way ANOVA; P = 0.0005).
Fig. 4.
Hepatic gene expression of PPARγ (A), SREBP-1c (B), FAT/CD36 (C), ChREBP (D) and Plin2 (E) from the experimental groups in the post-treatment period. Endogenous control beta-actin was used to normalize the expression of the selected genes. Data are expressed as the mean and SD (n = 10 each group). P < 0.05 when: † compared to the C group; ‡ compared to the CM group; § compared to the F group (two-way ANOVA and the posthoc test of Holm–Sidak). Groups: C, control diet; F, fructose diet; M, Metformin. Abbreviations: peroxisome proliferator activator receptor gamma (PPARγ); sterol regulatory element-binding protein-1c (SREBP-1c); carbohydrate response element-binding protein (CHREBP); Fatty acid translocase (FAT/CD36) and perilipin-2 (Plin-2).
Fructose enhanced SREBP-1c in the F group (+206% compared to the C group, P < 0.0001, Fig. 4). Metformin reduced SREBP-1c in the FM group (−157% in relation to the F group, P < 0.0001). Metformin restored the SREBP-1c with no difference to the CM group (P = 0.90). Diet and Metformin showed interaction in the findings (two-way ANOVA; P = 0.0003).
Fructose enhanced ChREBP in the F group (+162% compared to the C group, P < 0.0001, Fig. 4). Metformin diminished ChREBP in the FM group (−440% in relation to the F group, P < 0.0001), and no difference remains between the groups CM and FM (P = 0.10). Metformin affected the result (two-way ANOVA; P < 0.0001).
Fructose elevated FAT/CD36 expression in the F group (+150% compared to the C group; P = 0.009). Metformin reduced FAT/CD36 expression in the FM group (−52% compared to the F group; P = 0.02, Fig. 4), the FM group and the CM group were equivalent (P = 0.64). There was not a significant interaction between diet and treatment (two-way ANOVA; P = 0.07). Diet and Metformin affected independently FAT/CD36 expression; diet (P = 0.003), Metformin (two-way ANOVA; P = 0.01).
Perilipin-2 (PLIN2) was upregulated after the fructose intake in the F group (+94% compared to the C group; P = 0.002, Fig. 4). Metformin reduced PLIN2 expression in the FM group (−48%, compared to the F group, P = 0.002). No difference in PLIN2 was observed between the groups FM and CM (P = 0.9). Diet and Metformin showed an interaction in the results (two-way ANOVA; P = 0.005).
3.5. Metformin increases hepatic antioxidant defense and reduces lipid peroxidation
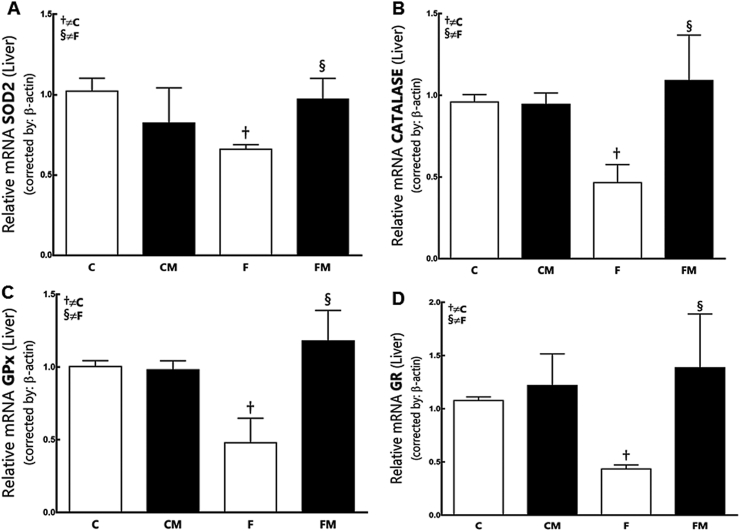
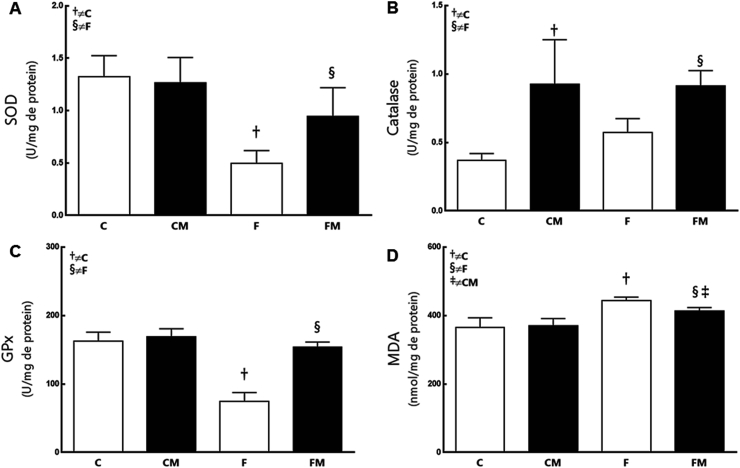
Fructose diminished SOD2 expression (−35%) and its enzymatic activity in the F group (−53%) compared to the C group (P = 0.0159, Fig. 5, Fig. 6). Metformin had increased SOD2 expression (+47%) and its enzymatic activity in the FM group (+76%) compared to the F group (P = 0.0349). SOD2 expression and its enzymatic activity were not different between the groups FM and CM (P = 0.31). Diet and Metformin showed interaction in the results (two-way ANOVA; P = 0.002).
Fig. 5.
Hepatic gene expression of SOD2 (A), Catalase (B), GPx (C) and GR (D) from the experimental groups in the post-treatment period. Endogenous control beta-actin was used to normalize the expression of the selected genes. Data are expressed as the mean and SD (n = 10 each group). P < 0.05 when: † compared to the C group; ‡ compared to the CM group; § compared to the F group (two-way ANOVA and the posthoc test of Holm–Sidak). Groups: C, control diet; F, fructose diet; M, Metformin. Abbreviations: superoxide dismutase 2 (SOD2); glutathione peroxidase (GPx); glutathione reductase (GR).
Fig. 6.
The hepatic enzymatic activity of SOD (A), Catalase (B), GPx (C) and MDA (D) from the experimental groups in the post-treatment period. Data are expressed as the mean and SD (n = 10 mice each group). P < 0.05 when: † compared to the group C; § compared to the F group (two-way ANOVA and the posthoc test of Holm–Sidak). Groups: C, control diet; F, fructose diet; M, Metformin. Abbreviations: superoxide dismutase (SOD); glutathione peroxidase (GPx); malondialdehyde (MDA).
Fructose only reduced CAT expression in the F group (−52%) compared to the C group (P = 0.003, Fig. 5). Metformin had increased CAT expression (+134%) and its enzymatic activity in the FM group (+59%) compared to the F group (P = 0.02). CAT enzymatic expression was also increased in the CM group (+151% compared the group C; P = 0.001).
CAT expression and its enzymatic activity were not different in the groups FM and CM (P = 0.5060).
Fructose reduced GPx expression and its enzymatic activity in the F group (−54%) compared to the C group (P = 0.0009, Fig. 5, Fig. 6). Metformin had increased GPx expression (+146%) and its enzymatic activity in the FM group (+106%) compared to the F group (P < 0.0001). The groups FM and CM did not show differences in GPx. Diet and Metformin interacted with the results (two-way, P < 0.0001).
Fructose diminished GR expression in the F group (−60% compared to the C group; P = 0.03). Metformin had increased GR expression in the FM group (+187% compared to the F group, P = 0.03, Fig. 5). GR expression was not different between the groups FM and CM (P = 0.68). Metformin affected the result (two-way ANOVA; P = 0.002).
Regarding lipid peroxidation in the liver, Fructose increased MDA levels in the F group (+21% compared to C group; P < 0.0001, Fig. 6). Metformin changed MDA concentrations in the FM group (−7% compared to the F group, P = 0.03; +11% in relation to the CM group, P = 0.007).
3.6. Metformin improves AMPK activation
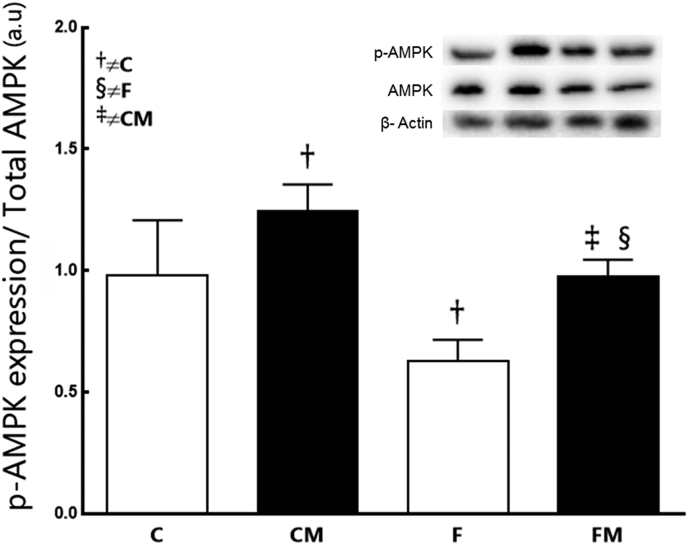
Fructose decreased the AMPK phosphorylation in the F group (−36% compared to C group; P = 0.0044, Fig. 7). Metformin elevated the AMPK phosphorylation in the FM group (+56% compared to the F group, P = 0.004). However, the AMPK phosphorylation has not been completely restored in the FM group (−22% than the CM group; P = 0.02). Metformin had an action increasing the AMPK phosphorylation in the CM group (+28% compared to the C group, P = 0.0217). Thus, Metformin affected the result (two-way ANOVA; P < 0.001).
Fig. 7.
Hepatic protein expression of AMPK from the experimental groups in the post-treatment period. Endogenous control beta-actin was used to normalize the expression of the selected genes. Data are expressed as the mean and SD (n = 10 mice each group). P < 0.05 when: † compared to the C group; ‡ compared to the CM group; § compared to the F group (two-way ANOVA and the posthoc test of Holm–Sidak). Groups: C, control diet; F, fructose diet; M, Metformin. Abbreviations: AMP-activated protein kinase (AMPK).
4. Discussion
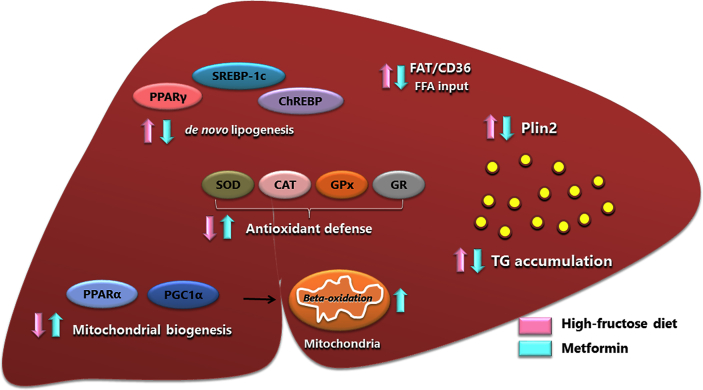
We illustrated our conception of the Metformin action in the fructose model in Fig. 8. The study identified three major events related to the fructose diet: a) liver injury (NAFLD, with steatosis, an increase in lipogenesis and lipid peroxidation and reduced BOxid), b) impaired carbohydrate metabolism (IR, reduced IS, augmented TG), c) increased OStress (impairment of enzymes). Metformin improved IR, despite a continuously elevated intake of fructose. Metformin decreased lipogenesis, steatosis and lipid peroxidation, made more efficient BOxid and increased the antioxidant defenses.
Fig. 8.
F diet stimulates SREBP-1c and CHREBP, increasing de novo lipogenesis. Also, PPARγ and its transcript FAT/CD36 are activated, increasing fatty acid influx from adipose tissue. There are reduced antioxidant defenses and inhibition in PGC1α, impairing mitochondrial biogenesis. These factors result in triglyceride accumulation within the hepatocyte (justified by elevated Plin2), featuring hepatic steatosis. The treatment with Metformin reverses all these processes. There is a decrease in de novo lipogenesis; reduced PPARγ and its transcript FAT/CD36; increase in PGC1α and PPARα expression, which promote mitochondrial biogenesis with a consequent increased β-oxidation. Metformin also accentuates antioxidant enzymes, factors that can prevent the progression of liver disease. Metformin consequently decreases triglyceride accumulation in the liver. Abbreviations: sterol regulatory element-binding protein-1c (SREBP-1c); carbohydrate response element-binding protein (CHREBP); peroxisome proliferator activator receptor gamma (PPARγ); fatty acid translocase (FAT/CD36); peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α); superoxide dismutase (SOD); catalase (CAT); glutathione peroxidase (GPx); glutathione reductase (GR); triglycerides (TG); lipid droplet protein Perilipin 2 (Plin2).
It is imperative to start commenting on Metformin's actions on health in general and then move on to the analysis of our findings related to NAFLD. Metformin has a beneficial action, not only for the promotion of healthy aging, but also on the prevention of sedentariness damages [22]. Epidemiological studies have identified an association between Metformin use and a beneficial effect on cancer prevention and treatment [23]. Likely, Metformin enhances the Forkhead box O3 (FOXO3), transcription factors involved in protection from OStress by upregulating antioxidants such as catalase and SOD, and associated with longevity in humans [24]. Also, Metformin attenuates hepatic OStress in fructose-fed rats [25], which might be associated with enhancement of the catalase [26] and/or mitochondrial activity [27].
At a molecular level, besides Metformin increases AMP-activated protein kinase (AMPK) activity and increases antioxidant protection, resulting in reductions in both oxidative damage accumulation and chronic inflammation is beneficial on Healthspan and Lifespan [28].
However, Metformin, besides being the most widely used oral anti-diabetic drug worldwide, has its action only partially understood and controversial, perhaps because the studies were different about the pharmacological concentrations (doses) of Metformin, sometimes much higher than maximally achievable therapeutic doses [29]. In the current study, we are concerned with the problem of a high dose of Metformin, and so we used an average of the prescriptions found in the literature for rodents, 250 mg/kg of body mass per day, which was lower than we used previously [30].
Our panel two is easy-to-read. The photomicrographs of liver tissue in animals clearly demonstrate the effects of fructose intake causing hepatic steatosis, and metformin decreasing steatosis. Everything was analyzed and confirmed with the use of techniques of quantitative morphology (stereology).
The balance between insulin resistance/sensitivity to insulin is central in NAFLD. In the fructose diet model, Metformin improved insulin sensitivity, which agrees with previous reports [31], although multiple and not well-known signaling pathways are defined [32]. For instance, Metformin upregulates the insulin receptor (IR) beta expression and downstream IRS2/phosphatidylinositol-4,5-bisphosphate 3-kinase/protein kinase B (PI3K/Akt) signaling transduction, enhancing hepatic glycogen storage and improving insulin resistance [33].
Also, besides the direct effects on hepatic insulin signaling increasing FOXO3 phosphorylation [34], Metformin protects the liver from the onset of fructose-induced NAFLD through altering intestinal permeability and subsequently the endotoxin-dependent activation of hepatic Kupffer cells [35].
In panel three we indicated how the gene expression of two transcription factors related to the biogenesis of mitochondria and lipid regulation behave in the fructose animals and the Metformin animals. We can clearly see that the fructose diet was responsible for depressing the expression of PGC1alpha and that Metformin was in charge of enhancing both factors, even in animals continuing to receive the fructose diet. PGC-1alpha is a transcription factor that increases the number and function of mitochondria in steatotic livers [36], a known pivotal regulator of mitochondrial BOxid and liver lipid metabolism. A selective modulation of hepatic PGC-1alpha functions might be a novel mechanism involved in the therapeutic action of Metformin [37]. Also, PPARalpha is a transcription factor and major regulator of lipid metabolism in the liver. PPARalpha is associated with BOxid that is activated under conditions of energy deprivation, being necessary for the process of ketogenesis, a key adaptive response to prolonged fasting [38]. Our findings are in line with these reports. Recently, a dual activation of PPARgamma and PPARalpha was seen with effect in ameliorating NASH by modulation of some hepatic and adipose tissue gene expressions [39].
In panel 4 we can see that all the data are significantly increased in the fructose animals, and Metformin reduced all the data, sometimes to the control levels. PPARgamma is related to lipogenesis [40]. SREBP-1c regulates the genes required for glucose metabolism and fatty acid synthesis, and this is a relation between OStress and NAFLD via SREBP-1c [41]. In this study, an increase in AMPK phosphorylation promoted by Metformin suppresses SREBP-1c expression, justifying the action of Metformin in regulating lipid and glucose metabolism [42]. The fatty acid translocase (FAT/CD36) belongs to the class B scavenger receptor family, which is used by the hepatocytes to take up free fatty acids via transport proteins [43]. Hepatic FAT/CD36 upregulation is significantly associated with insulin resistance, hyperinsulinemia and increased steatosis in patients with NASH and hepatitis C virus with fatty liver. Translocation of FAT/CD36 to the plasma membrane of hepatocytes may contribute to liver fat accumulation in patients with NAFLD [44]. ChREBP is another important transcription factor in the hepatic response to excess dietary carbohydrate and targeting ChREBP may prevent fructose-induced hypertriglyceridemia but without the improvements in hepatic steatosis and hepatic insulin responsiveness [45]. Perilipin-2 (PLIN2) is an abundant lipid droplet protein in the liver. The expression pattern of PLIN2 in NASH livers varies with the size of lipid droplets and is closely associated with oxidative damage [46].
In panel five we illustrated hepatic gene expression and activity of enzymes related to OStress. The findings are homogeneous indicating that most enzymes are reduced in the liver of the fructose animals, but recovered with Metformin, even when the animals were continued to be fed with fructose diet. In this context, SOD is an important antioxidant enzyme, as fructose leads to superoxide anion generation at the complex I of mitochondria and by the activation of NADPH oxidase [8]. It is well known that SOD is the first line of defense against ROS production since it dismutase superoxide anion into hydrogen peroxide and molecular oxygen. Therefore, increased SOD levels may result in a reduction in the oxidative burden caused by a high fructose intake. Our findings agree with the literature that reported reduced SOD activity in the liver of the fructose-fed animals [47]. Catalase (in the liver peroxisome) is also an important antioxidant enzyme. Interestingly, the fructose-fed group had lower catalase gene expression, with the proper activity of this enzyme (compared to the C group). This fact may represent an attempt to attenuate the oxidative stress generated by fructose. A previous study has demonstrated that the activity of catalase varies depending on the substrate in which it is found [48]. Also, we analyzed the GPx and GR, both much diminished in the current study with fructose diet, but restored by Metformin. The main biological role of GPx is to protect the organism from oxidative damage, reducing lipid hydroperoxides to their corresponding alcohols and free hydrogen peroxide to water. GR catalyzes the reduction of glutathione disulfide to the sulfhydryl form glutathione, which is a critical molecule in resisting OStress and maintaining the reducing environment of the cell [49].
In agreement, the fructose diet used in our study induced a rise in hepatic MDA levels, denoting lipid peroxidation. The lipid oxidation in cytoplasmic membranes interferes in their selective permeability, releasing ROS. Chronic imbalance in ROS production may impair the ability of the antioxidant system to reduce the levels of these radicals, reducing their protective function [50]. On the other hand, Metformin attenuates lipid peroxidation in liver, improving the antioxidant system.
5. Conclusion
Our findings are a significant contribution to unraveling the molecular mechanisms involved in the changes caused by fructose diet in the liver and by treatment with Metformin. Our data clearly showed that fructose had increased de novo lipogenesis and lipid peroxidation, reduced the antioxidant defenses, and diminished mitochondrial biogenesis, which has devastating effects on the liver in the long term. After an extended period of fructose intake, Metformin treatment, even in continuing the fructose intake, reversed, at least partially, the liver injury and prevented NAFLD progression to more severe stages of liver disease.
Financial support
The work was supported by the Conselho Nacional de Ciência e Tecnologia (CNPq, grant numbers 302.154/2011-6, and 442673/2014-0), and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Rio de Janeiro (Faperj, grant numbers 201.186/2014, and 010.003093/2014).
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors would like to thank Michele Soares, Aline Penna de Carvalho, Priscila Carapeto, Larissa Santos, Vivian Neves and Wanda Vianna Mury for technical assistance.
Contributor Information
Iara Karise, Email: iarakarise@hotmail.com.
Fernanda Ornellas, Email: cruz.fop@gmail.com.
Sandra Barbosa-da-Silva, Email: sandrabarbosasilva@gmail.com.
Cristiane Matsuura, Email: crismatsuura@gmail.com.
Mariano del Sol, Email: mariano.delsol@ufrontera.cl.
Marcia Barbosa Aguila, Email: marciaguila@gmail.com.
Carlos A. Mandarim-de-Lacerda, Email: mandarim.ca@gmail.com, mandarim@uerj.br, http://www.lmmc.uerj.br.
References
- 1.Zhou J., Li M.L., Zhang D.D., Lin H.Y., Dai X.H., Sun X.L., Li J.T., Song L.Y., Peng H., Wen M.M. The correlation between pancreatic steatosis and metabolic syndrome in a Chinese population. Pancreatology. 2016;16:578–583. doi: 10.1016/j.pan.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Stanhope K.L. Role of fructose-containing sugars in the epidemics of obesity and metabolic syndrome. Annu. Rev. Med. 2012;63:329–343. doi: 10.1146/annurev-med-042010-113026. [DOI] [PubMed] [Google Scholar]
- 3.Le K.A., Ith M., Kreis R., Faeh D., Bortolotti M., Tran C., Boesch C., Tappy L. Fructose overconsumption causes dyslipidemia and ectopic lipid deposition in healthy subjects with and without a family history of type 2 diabetes. Am. J. Clin. Nutr. 2009;89:1760–1765. doi: 10.3945/ajcn.2008.27336. [DOI] [PubMed] [Google Scholar]
- 4.Schultz A., Neil D., Aguila M.B., Mandarim-de-Lacerda C.A. Hepatic adverse effects of fructose consumption independent of overweight/obesity. Int. J. Mol. Sci. 2013;14:21873–21886. doi: 10.3390/ijms141121873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tappy L., Le K.A., Tran C., Paquot N. Fructose and metabolic diseases: new findings, new questions. Nutrition. 2010;26:1044–1049. doi: 10.1016/j.nut.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Michelotti G.A., Machado M.V., Diehl A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013;10:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 7.Tappy L., Le K.A. Does fructose consumption contribute to non-alcoholic fatty liver disease? Clin. Res. Hepatol. Gastroenterol. 2012;36:554–560. doi: 10.1016/j.clinre.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Aronis A., Madar Z., Tirosh O. Mechanism underlying oxidative stress-mediated lipotoxicity: exposure of J774.2 macrophages to triacylglycerols facilitates mitochondrial reactive oxygen species production and cellular necrosis. Free Radic. Biol. Med. 2005;38:1221–1230. doi: 10.1016/j.freeradbiomed.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Schultz A., Barbosa-da-Silva S., Aguila M.B., Mandarim-de-Lacerda C.A. Differences and similarities in hepatic lipogenesis, gluconeogenesis and oxidative imbalance in mice fed diets rich in fructose or sucrose. Food Funct. 2015;6:1684–1691. doi: 10.1039/c5fo00251f. [DOI] [PubMed] [Google Scholar]
- 10.Lim J.S., Mietus-Snyder M., Valente A., Schwarz J.M., Lustig R.H. The role of fructose in the pathogenesis of NAFLD and the metabolic syndrome. Nat. Rev. Gastroenterol. Hepatol. 2010;7:251–264. doi: 10.1038/nrgastro.2010.41. [DOI] [PubMed] [Google Scholar]
- 11.Peverill W., Powell L.W., Skoien R. Evolving concepts in the pathogenesis of NASH: beyond steatosis and inflammation. Int. J. Mol. Sci. 2014;15:8591–8638. doi: 10.3390/ijms15058591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aw D.K., Sinha R.A., Xie S.Y., Yen P.M. Differential AMPK phosphorylation by glucagon and metformin regulates insulin signaling in human hepatic cells. Biochem. Biophys. Res. Commun. 2014;447:569–573. doi: 10.1016/j.bbrc.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 13.Reeves P.G., Nielsen F.H., Fahey G.C., Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 14.Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 15.Katz A., Nambi S.S., Mather K., Baron A.D., Follmann D.A., Sullivan G., Quon M.J. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J. Clin. Endocrinol. Metab. 2000;85:2402–2410. doi: 10.1210/jcem.85.7.6661. [DOI] [PubMed] [Google Scholar]
- 16.Hally A.D. A counting method for measuring the volumes of tissue components in microscopical sections. J. Cell Sci. 1964;3:503–517. [Google Scholar]
- 17.Catta-Preta M., Mendonca L.S., Fraulob-Aquino J., Aguila M.B., Mandarim-de-Lacerda C.A. A critical analysis of three quantitative methods of assessment of hepatic steatosis in liver biopsies. Virchows Arch. 2011;459:477–485. doi: 10.1007/s00428-011-1147-1. [DOI] [PubMed] [Google Scholar]
- 18.Marklund S., Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 19.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 20.Flohe L., Gunzler W.A. Assays of glutathione peroxidase. Methods Enzymol. 1984;105:114–121. doi: 10.1016/s0076-6879(84)05015-1. [DOI] [PubMed] [Google Scholar]
- 21.Draper H.H., Squires E.J., Mahmoodi H., Wu J., Agarwal S., Hadley M. A comparative evaluation of thiobarbituric acid methods for the determination of malondialdehyde in biological materials. Free Radic. Biol. Med. 1993;15:353–363. doi: 10.1016/0891-5849(93)90035-s. [DOI] [PubMed] [Google Scholar]
- 22.Senesi P., Montesano A., Luzi L., Codella R., Benedini S., Terruzzi I. Metformin treatment prevents sedentariness related damages in mice. J. Diabetes Res. 2016;2016 doi: 10.1155/2016/8274689. 8274689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales D.R., Morris A.D. Metformin in cancer treatment and prevention. Annu. Rev. Med. 2015;66:17–29. doi: 10.1146/annurev-med-062613-093128. [DOI] [PubMed] [Google Scholar]
- 24.Willcox B.J., Donlon T.A., He Q., Chen R., Grove J.S., Yano K., Masaki K.H., Willcox D.C., Rodriguez B., Curb J.D. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagul P.K., Middela H., Matapally S., Padiya R., Bastia T., Madhusudana K., Reddy B.R., Chakravarty S., Banerjee S.K. Attenuation of insulin resistance, metabolic syndrome and hepatic oxidative stress by resveratrol in fructose-fed rats. Pharmacol. Res. 2012;66:260–268. doi: 10.1016/j.phrs.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Dai J., Liu M., Ai Q., Lin L., Wu K., Deng X., Jing Y., Jia M., Wan J., Zhang L. Involvement of catalase in the protective benefits of metformin in mice with oxidative liver injury. Chem. Biol. Interact. 2014;216:34–42. doi: 10.1016/j.cbi.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Repiscak P., Erhardt S., Rena G., Paterson M.J. Biomolecular mode of action of metformin in relation to its copper binding properties. Biochemistry. 2014;53:787–795. doi: 10.1021/bi401444n. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Montalvo A., Mercken E.M., Mitchell S.J., Palacios H.H., Mote P.L., Scheibye-Knudsen M., Gomes A.P., Ward T.M., Minor R.K., Blouin M.J., Schwab M., Pollak M., Zhang Y., Yu Y., Becker K.G., Bohr V.A., Ingram D.K., Sinclair D.A., Wolf N.S., Spindler S.R., Bernier M., de Cabo R. Metformin improves healthspan and lifespan in mice. Nat. Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L., Wondisford F.E. Metformin action: concentrations matter. Cell Metab. 2015;21:159–162. doi: 10.1016/j.cmet.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Souza-Mello V., Gregorio B.M., Cardoso-de-Lemos F.S., de Carvalho L., Aguila M.B., Mandarim-de-Lacerda C.A. Comparative effects of telmisartan, sitagliptin and metformin alone or in combination on obesity, insulin resistance, and liver and pancreas remodelling in C57BL/6 mice fed on a very high-fat diet. Clin. Sci. Lond. 2010;119:239–250. doi: 10.1042/CS20100061. [DOI] [PubMed] [Google Scholar]
- 31.Anurag P., Anuradha C.V. Metformin improves lipid metabolism and attenuates lipid peroxidation in high fructose-fed rats. Diabetes Obes. Metab. 2002;4:36–42. doi: 10.1046/j.1463-1326.2002.00178.x. [DOI] [PubMed] [Google Scholar]
- 32.Cleasby M.E., Dzamko N., Hegarty B.D., Cooney G.J., Kraegen E.W., Ye J.M. Metformin prevents the development of acute lipid-induced insulin resistance in the rat through altered hepatic signaling mechanisms. Diabetes. 2004;53:3258–3266. doi: 10.2337/diabetes.53.12.3258. [DOI] [PubMed] [Google Scholar]
- 33.Xu H., Zhou Y., Liu Y., Ping J., Shou Q., Chen F., Ruo R. Metformin improves hepatic IRS2/PI3K/Akt signaling in insulin-resistant rats of NASH and cirrhosis. J. Endocrinol. 2016;229:133–144. doi: 10.1530/JOE-15-0409. [DOI] [PubMed] [Google Scholar]
- 34.Takayama H., Misu H., Iwama H., Chikamoto K., Saito Y., Murao K., Teraguchi A., Lan F., Kikuchi A., Saito R., Tajima N., Shirasaki T., Matsugo S., Miyamoto K., Kaneko S., Takamura T. Metformin suppresses expression of the selenoprotein P gene via an AMP-activated kinase (AMPK)/FoxO3a pathway in H4IIEC3 hepatocytes. J. Biol. Chem. 2014;289:335–345. doi: 10.1074/jbc.M113.479386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spruss A., Kanuri G., Stahl C., Bischoff S.C., Bergheim I. Metformin protects against the development of fructose-induced steatosis in mice: role of the intestinal barrier function. Lab. Investig. 2012;92:1020–1032. doi: 10.1038/labinvest.2012.75. [DOI] [PubMed] [Google Scholar]
- 36.Aharoni-Simon M., Hann-Obercyger M., Pen S., Madar Z., Tirosh O. Fatty liver is associated with impaired activity of PPARγ-coactivator 1α (PGC1α) and mitochondrial biogenesis in mice. Lab. Investig. 2011;91:1018–1028. doi: 10.1038/labinvest.2011.55. [DOI] [PubMed] [Google Scholar]
- 37.Aatsinki S.M., Buler M., Salomaki H., Koulu M., Pavek P., Hakkola J. Metformin induces PGC-1alpha expression and selectively affects hepatic PGC-1alpha functions. Br. J. Pharmacol. 2014;171:2351–2363. doi: 10.1111/bph.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kersten S., Seydoux J., Peters J.M., Gonzalez F.J., Desvergne B., Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Investig. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abd El-Haleim E.A., Bahgat A.K., Saleh S. Effects of combined PPAR-gamma and PPAR-alpha agonist therapy on fructose induced NASH in rats: modulation of gene expression. Eur. J. Pharmacol. 2016;773:59–70. doi: 10.1016/j.ejphar.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Barbosa-da-Silva S., Souza-Mello V., Magliano D.C., Marinho Tde S., Aguila M.B., Mandarim-de-Lacerda C.A. Singular effects of PPAR agonists on nonalcoholic fatty liver disease of diet-induced obese mice. Life Sci. 2015;127:73–81. doi: 10.1016/j.lfs.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Aragno M., Tomasinelli C.E., Vercellinatto I., Catalano M.G., Collino M., Fantozzi R., Danni O., Boccuzzi G. SREBP-1c in nonalcoholic fatty liver disease induced by Western-type high-fat diet plus fructose in rats. Free Radic. Biol. Med. 2009;47:1067–1074. doi: 10.1016/j.freeradbiomed.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Wang Y., Bao C., Xu Y., Shen H., Chen J., Yan J., Chen Y. Metformin interacts with AMPK through binding to γ subunit. Mol. Cell Biochem. 2012;368:69–76. doi: 10.1007/s11010-012-1344-5. [DOI] [PubMed] [Google Scholar]
- 43.He J., Lee J.H., Febbraio M., Xie W. The emerging roles of fatty acid translocase/CD36 and the aryl hydrocarbon receptor in fatty liver disease. Exp. Biol. Med. Maywood. 2011;236:1116–1121. doi: 10.1258/ebm.2011.011128. [DOI] [PubMed] [Google Scholar]
- 44.Miquilena-Colina M.E., Lima-Cabello E., Sanchez-Campos S., Garcia-Mediavilla M.V., Fernandez-Bermejo M., Lozano-Rodriguez T., Vargas-Castrillon J., Buque X., Ochoa B., Aspichueta P., Gonzalez-Gallego J., Garcia-Monzon C. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut. 2011;60:1394–1402. doi: 10.1136/gut.2010.222844. [DOI] [PubMed] [Google Scholar]
- 45.Erion D.M., Popov V., Hsiao J.J., Vatner D., Mitchell K., Yonemitsu S., Nagai Y., Kahn M., Gillum M.P., Dong J., Murray S.F., Manchem V.P., Bhanot S., Cline G.W., Shulman G.I., Samuel V.T. The role of the carbohydrate response element-binding protein in male fructose-fed rats. Endocrinology. 2013;154:36–44. doi: 10.1210/en.2012-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujii H., Ikura Y., Arimoto J., Sugioka K., Iezzoni J.C., Park S.H., Naruko T., Itabe H., Kawada N., Caldwell S.H., Ueda M. Expression of perilipin and adipophilin in nonalcoholic fatty liver disease; relevance to oxidative injury and hepatocyte ballooning. J. Atheroscler. Thromb. 2009;16:893–901. doi: 10.5551/jat.2055. [DOI] [PubMed] [Google Scholar]
- 47.Botezelli J.D., Cambri L.T., Ghezzi A.C., Dalia R.A., Voltarelli F.A., de Mello M.A.R. Fructose-rich diet leads to reduced aerobic capacity and to liver injury in rats. Lipids Health Dis. 2012;11:1. doi: 10.1186/1476-511X-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkman H.N., Gaetani G.F. Mammalian catalase: a venerable enzyme with new mysteries. Trends Biochem. Sci. 2007;32:44–50. doi: 10.1016/j.tibs.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta. 2013;1830:3217–3266. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Girard A., Madani S., Boukortt F., Cherkaoui-Malki M., Belleville J., Prost J. Fructose-enriched diet modifies antioxidant status and lipid metabolism in spontaneously hypertensive rats. Nutrition. 2006;22:758–766. doi: 10.1016/j.nut.2006.05.006. [DOI] [PubMed] [Google Scholar]