Abstract
Background In 2015, the German Federal Ministry of Education and Research initiated a large data integration and data sharing research initiative to improve the reuse of data from patient care and translational research. The Observational Medical Outcomes Partnership (OMOP) common data model and the Observational Health Data Sciences and Informatics (OHDSI) tools could be used as a core element in this initiative for harmonizing the terminologies used as well as facilitating the federation of research analyses across institutions.
Objective To realize an OMOP/OHDSI-based pilot implementation within a consortium of eight German university hospitals, evaluate the applicability to support data harmonization and sharing among them, and identify potential enhancement requirements.
Methods The vocabularies and terminological mapping required for importing the fact data were prepared, and the process for importing the data from the source files was designed. For eight German university hospitals, a virtual machine preconfigured with the OMOP database and the OHDSI tools as well as the jobs to import the data and conduct the analysis was provided. Last, a federated/distributed query to test the approach was executed.
Results While the mapping of ICD-10 German Modification succeeded with a rate of 98.8% of all terms for diagnoses, the procedures could not be mapped and hence an extension to the OMOP standard terminologies had to be made.
Overall, the data of 3 million inpatients with approximately 26 million conditions, 21 million procedures, and 23 million observations have been imported.
A federated query to identify a cohort of colorectal cancer patients was successfully executed and yielded 16,701 patient cases visualized in a Sunburst plot.
Conclusion OMOP/OHDSI is a viable open source solution for data integration in a German research consortium. Once the terminology problems can be solved, researchers can build on an active community for further development.
Keywords: data integration, secondary use, OMOP, OHDSI
Background and Significance
Sharing data and learning from common grounds is at the heart of many medical research projects today. 1 Integrating data within a hospital into a research database or warehouse is an essential step therein.
Adhering to a public common data model (CDM) simplifies the collaboration across institutions. 2 CDMs harmonize data from disparate sources in a standardized way. This allows for easy merging of medical data into larger databases (bringing data to analysis) or for federating research analyzes and aggregating the results (bringing analysis to the data). 3
A preeminent example of a common data model has been provided by the Observational Medical Outcomes Partnership (OMOP), which aims at the evaluation of analytical methods for identifying drug–outcome associations across disparate data sources. 2 For the harmonization of data sources, they developed the OMOP Common Data Model (CDM, OMOP v4.0) for longitudinal patient data associated with standardized medical terminologies.
The Observational Health Data Sciences and Informatics (OHDSI) collaborative continues and advances the work of OMOP. 4 5 It not only provides an updated version of the data model OMOP v5 and a central vocabulary service Athena ( http://athena.ohdsi.org ), but also tools and methods for various types of data analysis. OHDSI defines a workflow for researchers to publish research requests and participate in studies among OHDSI participants fostering an active research community. OMOP/OHDSI is successfully used in several countries with data from hundreds of millions of patients. 6
In Germany, the Federal Ministry of Education and Research (BMBF) commenced a large data integration and data sharing research initiative (BMBF MI-I: Medical Informatics Initiative/Funding Scheme) in 2015 to improve the reuse of data from patient care and translational research. 7 For this purpose, participating German university hospitals are required to establish data integration centers and support collaboration and data exchange within multicenter consortia. Because of the large heterogeneity of clinical documentation across the different hospitals, defining a common data model within a consortium, and even across all consortia is a fundamental prerequisite to achieve the goals of the initiative.
Objective
Since common data models have already been developed and successfully applied in international data sharing projects, one should not try to reinvent, but build upon an existing CDM and assess its applicability in the German initiative. Even though the use of vocabularies or reimbursement schemes in Germany differ from those in the United States, collaborating with a large project consortium, such as OHDSI, and enhancing the OMOP CDM (e.g., with German vocabularies) seem to be more promising, than starting completely from scratch.
Thus, the objectives of our research were:
to pursue an OMOP-based pilot implementation within one large consortium (MIRACUM 8 ) of the BMBF MI-I considering the core data elements currently defined by the interoperability working group of the BMBF MI-I national steering committee, and
to analyze and exemplarily apply the OMOP data model and the OHDSI tools for their applicability to support data harmonization and data sharing between German university hospitals and identify potential enhancement requirements.
Methods
First, the authors got acquainted with the ecosystem of specification, tools, and packages provided by OHDSI on their webpage and via GitHub ( https://www.github.com/OHDSI ). Importing various local datasets provided valuable insight into the structures and inner working of the tools.
For the study, (1) the vocabularies and terminological mapping required for importing the fact data were prepared. Then, (2) a process for importing the data from the source files was designed. For eight German university hospitals of the MIRACUM consortium, 8 (3) a virtual machine preconfigured with the OMOP database and the OHDSI tools as well as the jobs to import the data and conduct the analysis was provided. (4) The eight university hospitals instantiated this virtual machine locally and applied the import jobs to load the data from the standard claims data export set. Last, (5) an exemplary federated/distributed query as proof of the approach was executed.
Dataset
The German BMBF MI-I defines a basic set of data elements comprising patient demographics, visits, diagnoses, procedures, laboratory findings, and medication as the level 1 core dataset that must be provided by each participating hospital.
While laboratory findings and medication data, as of today, cannot be provided in a standardized way by our hospitals, patient demographics, visits, diagnoses, and procedures are based upon a standardized German claims dataset and thus profit from the fact that every hospital is already able to export the data. Such a dataset was available at each participating site; it was anonymized and clearance was received from all local data use and access committees. Although the dataset is limited, it has been used before for research purposes 9 10 11 and suffices the purpose of this study to provide a technically viable proof of concept for a distributed data integration center architecture applying the OMOP common data model.
Terminology
OMOP uses so-called standardized vocabularies for the representation of data in the CDM a such as SNOMED-CT for conditions and observations or RxNorm for drugs. With Athena, OHDSI provides access to the OMOP Vocabulary, a library of more than 70 vocabularies with a complete mapping of terms to the standard concepts. The vocabularies can be extended by local developments, which should map onto standard concepts.
For importing the MI-I core dataset, the authors had to create mappings in three areas of concern:
“International Classification of Diseases, German Modification” (ICD-10-GM) for coding conditions and observations,
“Operationen- und Prozedurenschlüssel” (OPS, surgery and procedure key) for coding procedures, and
individual concept codes for gender, primary and secondary diagnoses in the provided vocabulary.
The general principle for mapping concepts is as follows: Either the concept is available in the library or can be found by translating the concept into English. If not, a concept should be taken from a less granular level (“uphill mapping”) b . An example can be found in Fig. 1 .
Fig. 1.
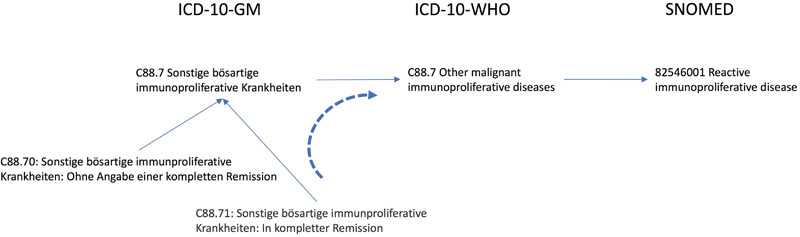
Example of mapping from the German ICD10 via the international ICD by WHO to SNOMED.
The concept of C88.7 in the German ICD-10 has a direct relation to C88.7 in the ICD-10-WHO so that it can be directly mapped to SNOMED 82546001.
The more granular concept of C88.71, which states that the tumor had a complete remission, has no direct relationship to ICD-10-WHO, but it is as well a C88.7 so that it inherits the SNOMED code of its parent.
Diagnoses
For the mapping of ICD-10-GM onto standard diagnosis codes in SNOMED, the presence of ICD-10 WHO proved beneficial. IDC-10 issued by the World Health Organization is modified in Germany for statistical and billing purposes. For that reason, some concepts of the global ICD-10 are omitted (or coarser grained than WHO) or better differentiated (more detailed than WHO) by removing or adding another digit to the code. In any case, the modification is largely compatible for uphill mapping as recommended by OHDSI.
The code of ICD-10-GM was mapped to ICD-10 and then the mapping of ICD-10 to SNOMED as already present in the OMOP Vocabulary (see Fig. 1 ) was used.
ICD-10G-M versions from 2004 to 2016 were mapped including the times of validity ( valid_start/end_date, invalid_reason ).
Procedures
Procedures in Germany are coded using the OPS. Unfortunately, no equivalent terminology is available from Athena : standard concepts for procedures in OHDSI may come e.g., from SNOMED-CT, the Current Procedural Terminology (CPT-4) or the Healthcare Common Procedure Coding System (HCPCS), and ICD-9-Procedures, all of which differ significantly in structure from OPS. Manual mapping of procedures requires extensive medical knowledge and resources that were not available for this preliminary study.
Therefore, OPS vocabulary was imported into OMOP and declared the concepts as “standard.” This breaks the international compatibility, but that was not a requirement for the study.
OPS versions from 2004 to 2016 were imported including the times of validity ( valid_start/end_date, invalid_reason ).
Other Concepts
For individual concepts, such as gender, a direct match using the translated concept was searched manually. In case the target concept found was ambiguous, the mapping was discussed among experts (C.M., L.L., T.G., and M.S.).
Other, larger terminologies were not required for the selected dataset. But other options have to be considered for the import of a much more comprehensive dataset of German university hospitals. Terminologies, such as LOINC, UCUM, and MedDRA, are already provided and could be used. If necessary, a German translation using the same codes could be imported.
Import of Vocabularies
The import of ICD-10-GM and OPS into the vocabulary schema of the OMOP database was modeled using Talend Open Studio for Data Integration (Talend; Redwood City, CA, United States). The import read from the internal data warehouse providing the vocabularies in yearly versions.
Creation of the ETL Jobs
Importing the data into the OMOP database required a mapping of all source attributes and values from various files and columns of the MI-I core dataset onto the OMOP tables and vocabularies. This process of extracting the source data, transforming it, and loading to OMOP (ETL job) requires a design and an implementation phase.
For the design phase, OHDSI recommends using their “Rabbit” tools c . First, WhiteRabbit was used to analyze the source table structure and value domains. The RabbitInAHat reads these results and enables the user to graphically map them onto OMOP target tables and columns and to annotate the transformation with additional information (such as value mapping).
While WhiteRabbit provided input for the first analysis of the source data, the author's departmental wiki for the mapping design as RabbitInAHat does not support collaborative editing in a multiuser environment in the same way.
The mapping of the source data structure onto OMOP was conducted by a medical student (L.L.) supported by a team of computer scientists and physicians with experience in clinical ETL. All mappings were discussed and all decisions were documented using the wiki. The final mapping was entered (manually) to RabbitInAHat.
The implementation of the specification was realized using the Talend Open Studio for Data Integration.
The data import was validated by counting the rows and elements in the source dataset files and the OMOP database to ensure completeness. Additionally, AchillesHeel ( https://github.com/OHDSI/Achilles ) was used to assess the correctness according to OMOP constraints, for example to check the correct import of ICD10 values (diagnoses) from the source dataset into either the condition_occurrence or the observation table.
Provision of a Virtual Machine
A virtual machine (VM) was prepared for the consortium partners. Ubuntu 14.04LTS was used at the base, PostgreSQL 9.6, OpenJDK7, and Apache Tomcat 7. Nginx is the webserver for Atlas (1.4) and AchillesWEB and also serves as a proxy to the WebAPI (v1.5). This allowed us to configure the web tools without using static IP addresses for the REST-services. For running Achilles and other tools, a preconfigured RStudio (RStudio; Boston, MA, United States) as a web service was installed.
The machine was provided preloaded with all vocabularies and distributed as containers for VirtualBox (Oracle Corporation; Redwood Shores, CA, United States) as well as VMware (Dell Technologies Inc., Round Rock, TX, United States) for use with ESX hosts. The ETL-jobs ran outside the VM, expected the data in a predefined directory on the host machine, and uploaded all data in about 3 hours per site, depending on the physical environment and the size of data. The total size of the database including indices was approximately 11GB per site.
Additionally, the SYNPUF1K sample dataset provided by OHDSI d was provided as a “working reference dataset” in the VM template to allow users to explore the functions of Atlas on a wider basis (including drugs and measurements).
Federated Query
To validate the installation and demonstrate the feasibility of federated queries, two distinct queries were specified. As the current dataset is rather limited to general demographics (age, gender, location, visit, primary and secondary diagnoses, and procedures), a federated query was considered to proof the concept rather than trying to generate a medically relevant new finding.
One set of queries was made to get the quantity structure of data, for example the number of patients, procedures, and conditions as well as the unique number of distinct conditions. The queries utilize the pregenerated statistics from the Achilles tables.
A second set of queries was generated in accordance with a medical use case. Based on suggestions from the study group, a cohort of patients with colorectal carcinoma was identified. Depending on the location of the carcinoma, a different treatment path (sequence of operation, radio/chemotherapy) should be expected. Atlas was used to define the following concept_sets and cohort :
diagnose: malignant neoplasm of the colon (ICD-C18), of rectosigmoid junction (C19), and of the rectum (C20)
procedure: “operation” (OPS chapters 5–45*, 5–48*, 5–49)
procedure: “chemotherapy” (OPS chapter 8–52*)
procedure: “radiation therapy” (OPS chapter 8–54*)
procedure: combination of chemotherapy and radiation therapy
The OHDSI tools do not provide direct means for federated query and result aggregation such as i2b2-SHRINE. 12 A direct access to the partner's databases is not allowed for security reasons. Therefore, the queries were prepared centrally, packaged into an easy to use web application in grails 3 ( https://www.grails.org ) and provided as war files to be deployed at each site. The queries generated a local table with the raw results. The raw results data were subsequently collected, merged, and exemplarily visualized as a combined Sunburst plot similar to one of the most recent OHDSI analysis. 6
Results
The core MI-I dataset was mapped onto the OMOP common data model except for laboratory findings and medication, which were not present in the source dataset. The details of the fact data mapping will be described in a future publication. A virtual machine with all required tools for importing and querying the data was configured and provided to the project partners.
Eight university hospitals set up the provided virtual machine, successfully imported their local datasets, and contributed to the federated analysis.
Vocabulary
All required concepts were imported with their relations into the OMOP vocabulary scheme.
The procedure codes from OPS could not be mapped given the available time frame and were not only realized as new vocabulary and hierarchy but also declared “standard” for use in queries. Overall, 58,019 concepts with 225,632 relations have been imported.
The ICD-10-GM terminology was mapped onto SNOMED via the provided mapping of ICD-10 WHO. Not all codes could be directly mapped; out of 19,077 ICD-10-GM codes, there were
12,076 direct mappings to ICD-10 of OMOP (63.3%),
6,530 level 1 uphill mappings to ICD-10 of OMOP (34.2%),
240 level 2 uphill mappings to ICD-10 of OMOP (1.3%),
171 codes not mapped because these do not exist in ICD-10 WHO (0.9%),
60 codes available in ICD-10, but without a relation to SNOMED (0.3%). They belong to U00-U49 “Provisional assignment of new diseases of uncertain etiology or emergency use,” Z75 “Problems related to medical facilities and other healthcare data” and T73 “Effects of other deprivation.”
The concepts have 114,656 relations among ICD-10-GM ( isa/subsumes ) and to SNOMED ( maps_to/maps_from ) standard concepts. None of the codes, that could not be mapped, had data associated with it in our case, so there was no loss of data (all facts were imported as was compared by the line count of the source files with the row counts of the database tables).
Facts
Each site imported data on patient demographics, visits, diagnoses, and procedures according to the MI-I core dataset, which was mostly available for the years 2004 to 2016 (C: 2009–2016, G: 2008–2016). The quantity structure is shown in Table 1 . Overall, the data of almost 3 million patients were imported with 21 million procedures and 50 million diagnoses. While the diagnoses and procedures show a broad range of instances ( unique_ ), only a few observation items were included in this dataset such as the birth weight of infants.
Table 1. Quantity structure of data imported: For each of the eight University Hospitals (A-H), the years of the data available are given as well as the row count of the OMOP database table.
| A | B | C | D | E | F | G | H | Sum | |
|---|---|---|---|---|---|---|---|---|---|
| Years of data available | 2004–2016 | 2004–2016 | 2009–2016 | 2004–2016 | 2004–2016 | 2004–2016 | 2008–2016 | 2004–2016 | |
| condition_occurrence | 4,698,834 | 3,054,937 | 3,871,535 | 3,607,741 | 4,059,270 | 4,054,241 | 3,287,073 | 2,798,597 | 26,145,155 |
| observation | 4,022,012 | 2,874,985 | 3,138,306 | 3,092,768 | 3,389,461 | 4,371,301 | 2,507,428 | 2,772,262 | 23,661,095 |
| procedure_occurrence | 3,993,143 | 2,738,533 | 3,186,826 | 2,716,824 | 2,615,712 | 3,426,254 | 1,925,927 | 2,351,020 | 21,028,312 |
| measurement | 40,351 | 36,535 | 22,012 | 32,268 | 26,897 | 39,117 | 23,929 | 28,881 | 2226,061 |
| person | 393,004 | 453,307 | 426,435 | 495,194 | 319,751 | 409,126 | 357,046 | 478,035 | 22,974,852 |
| visit_occurrence | 912,347 | 628,214 | 612,599 | 711,143 | 742,461 | 895,787 | 573,654 | 661,140 | 55,163,691 |
| death | 12,215 | 11,816 | 8,970 | 12,586 | 10,340 | 11,967 | 10,387 | 9,798 | 777,692 |
| location | 12,845 | 10,894 | 13,245 | 13,803 | 14,774 | 14,285 | 10,355 | 9,497 | |
| care_site | 51 | 51 | 51 | 51 | 51 | 51 | 51 | 51 | |
| unique_procedures | 18,359 | 17,685 | 16,452 | 17,506 | 17,114 | 17,929 | 13,547 | 16,778 | |
| unique_conditions | 7,009 | 6,793 | 6,769 | 6,866 | 6,883 | 6,916 | 6,602 | 6,649 | |
| unique_observations | 3 | 33 | 29 | 31 | 30 | 30 | 32 | 32 |
Abbreviation: OMOP, Observational Medical Outcomes Partnership.
Federated Query
Overall, the paths of 16,701 patients were generated and aggregated ( Fig. 2 ). The innermost ring of the plots shows the distribution of diagnoses, the outer rings visualize the procedure categories per diagnosis. The detailed analysis and interpretation of these results is currently pursued by clinicians of the MIRACUM consortium and shall be part of an additional publication.
Fig. 2.
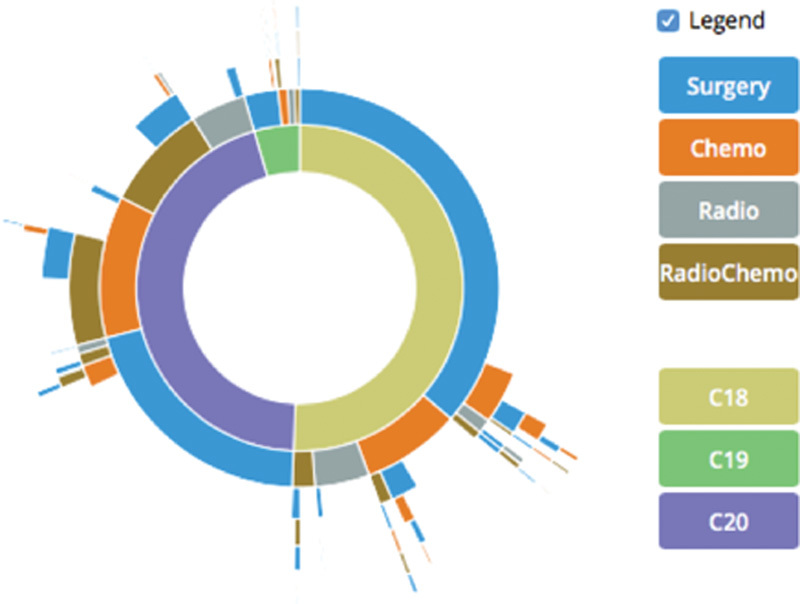
Combined Sunburst plot of 16,701 patients from eight German university hospitals (C18-C20 refer to the location of the tumor in the colon, sigmoid junction, and the rectum).
Discussion
The goal of an OMOP-based pilot implementation within a consortium of the BMBF MI-I based on a subset of the core data elements currently defined by the interoperability working group of the BMBF MI-I national steering committee was accomplished. The authors were able to provide a preconfigured virtual appliance and ETL jobs, including required terminologies and the OHDSI tools to eight university hospitals, import a core dataset locally, distribute queries, and summarize the results.
Overall Experiences
The amount and quality of the tools provided by the OHDSI community for free are impressive. There is a huge momentum and the community is helpful and supportive.
However, the sheer number of different tools developing at various paces requires an attentive management of dependencies. During the few months of the study, the OMOP database specification changed from V5 to V5.01 and V5.1. For example, the definitions of the cost tables have been changed in the table definition. However, the file defining the accompanying indexes was not updated, so the creation failed. It was also not always clear whether e.g., Achilles was updated in accordance as it operates heavily on these tables.
Another tool, Usagi , is available to assist in the mapping of concepts from different vocabularies. The authors tried this on the OPS, but neither the German concepts nor a machine-translated English version (Google translate) seemed to work reliably enough to be pursued in the study.
With regard to vocabularies, Athena already provides access to a plethora of vocabularies to choose from, but neither the ICD-10-GM used for diagnoses nor OPS for procedures are yet available. Mapping ICD-10-GM was possible, because it is primarily based on ICD-10 WHO so that a very similar vocabulary was already available. This is not the case with OPS; therefore, the mapping will require significant resources.
The tools provided to support the ETL process, WhiteRabbit and RabbitInAHat , were helpful to get started. However, the authors preferred their departmental wiki, as it facilitated collaborative editing and commenting. It would be helpful, to provide web-based, collaborative mapping tools in future, especially as mapping is a vital part of any secondary use or CDM project.
For a first look at the data and a check of the quality of the imported data, AchillesWeb and AchillesHeel are valuable tools. AchillesHeel provided insight into the success of import process, especially during the development of the ETL process with reports on the use of missing concepts or data in wrong tables (e.g., if a concept of a condition was used in observation table). AchillesWeb was used to show summary statistics to clinicians (data owners) and helped to break the ice for further data provision. However, at least in Germany, some variables, such as race or ethnicity, are usually not documented, but cannot be hidden in the graphics and thus might distract the users.
Atlas was used to successfully search for terms and define cohorts for the federated query. While its overall usability can be improved, it will become the central entry point not only to search for data, but also for (visual) analytics as more features will be added by the OHDSI community.
The provision of a preconfigured virtual machine enabled the hospitals to quickly set up the environment and import their data. However, in future, a docker-based container infrastructure ( https://www.docker.com ) would allow deploying updates more easily and quickly.
Issues and Details
While the overall experience with OMOP/OHDSI was very positive, some issues exist partly due to lack of features or due to (German) specifics not yet considered by OMOP/OHDSI.
The occurrence of a visit in OMOP is tied to a single care site, whereas in Germany, a patient may be treated by several departments within an institution as part of a single visit (chain of transfers). A possible solution could be to introduce pseudo visit occurrences for each department and chaining them together by a fact_relationship in OMOP. But this could break the assumption of having only a single visit at any time. It was decided to omit the chain of transfers and map all facts onto a single visit. Additionally, the dataset used contains the facts from a patient and the list of treating departments, but not which fact originates from which department. Likewise, Voss et al recommend standardizing organizational concepts such as the definition of a visit across institutions. 13
Some pitfalls are present, although documented in the web. For example, the relationship maps_to is meant from a standard concept to a proprietary concept. Users tend to use local terms for queries in Atlas and just tick maps_to to “include the standard terms required by OMOP.” This implemented behavior makes sense for sharing queries based on standard data, but makes queries on local data harder.
Another example is the determination into which table certain data go. While it may be in a local symptoms database, it may not automatically go into the condition_occurrence table as the table is determined by an attribute domain of the standard concept. This harmonizes the ETL across institutions, but requires local discussions upon disagreement on how the term was used locally.
Regarding security, OMOP/OHDSI does not yet provide any support for limiting access to the data. There is no user management or access control in the web tools and only the database password in the R tools. Beginning with Version 2.0 of the OHDSI tools, this is about to change, as a Java security framework has been introduced to support the development of security-related features.
Other Studies
To best of our knowledge, no publications yet exists describing the explicit application of the OMOP/OHDSI CDM and tools on German datasets and German terminologies.
However, the SALUS project 14 developed a semantic interoperability layer for mapping data from various sources into other, common data models. They tested their approach with data from a German university hospital and the OMOP CDM. Especially the semantic mapping of terminologies 15 could be very interesting to support the required vocabulary mapping.
Schuemie et al 16 replicated the OMOP experiment across several European databases. But they used EU-ADR as CDM and not OMOP while applying the same medical research question to compare, e.g., the incidence rates published by OMOP scientists with one from EU-ADR.
Makadia and Ryan 17 report on the import of the data from a Premier hospital into OMOP v4. They were able to map 91.4% of standard charge codes onto standard concepts in OMOP and import the data except 1% due to bad data quality in the source system. Similarily, Matcho et al 18 imported successfully the British Clinical Practice Research Datalink (CPRD) of more than 11 million patients into OMOP, including medication. They report an accuracy of 99.9% of condition records. While Premier as a US-based hospital can directly benefit from the OMOP vocabularies, an additional mapping was required for diagnoses and conditions (Read code to SNOMED) and medication (Multilex to RxNorm). While the import of the complete database of a university hospital is currently still under development, this study focuses on a small (yet representative) dataset and a common query across hospitals of a consortium.
Fitz Henry et al 19 used OMOP for two institutions, but both based in the US. They report similar challenges in implementing the OMOP CDM: in rare cases an observation is made outside a visit. Also, the “visit within visits” or the chain of referrals within an institution is a challenge to model.
Yoon et al 20 also covered the full dataset of a Korean hospital into the OMOP v4 database and used OHDSI for exploration.
Certainly, common data models other than OMOP, such as EU-ADR and MATRICE, exist and have already been compared, e.g., 21 22 OMOP was chosen mainly because of the broadness of the model, the vocabularies provided, the number of patients and institutions available, and the momentum of the OHDSI collaborative.
Limitations
The dataset chosen for the study is a very special, limited dataset of only inpatient visits. It provides in itself no sufficient data for relevant medical studies. However, it comprises data of major interest such as diagnoses and procedures.
Procedures could not yet be mapped to an OMOP standard terminology. Therefore, OMOP was modified by introducing OPS as a standard terminology. This breaks federated queries with researchers outside Germany, but this was not an issue in the preliminary study. In the long term, a mapping of concepts coded in the German OPS should be generated, validated, and provided to the OMOP vocabulary in Athena .
Tampering with the definition of standard concepts required minor adjustments in the tools, e.g., to allow AchillesWeb to display the hierarchy of concepts.
Conclusion
OMOP is a comprehensive database definition complemented with more than 70 mapped terminologies and a set of tools provided by OHDSI to make use of the database, most important Athena, Atlas , Achilles , and various R packages for analyzing the patient data of cohorts.
The authors were able to successfully implement this tools stack for the first time in a German consortium of university hospitals based on a typical dataset, although in a German flavor due to the coding of procedures. The reception of OMOP/OHDSI by the participating university hospitals was very good and it was agreed to continue using the approach.
However, as the terminologies required and provided by OHDSI are not the ones used in Germany, an additional mapping effort will be required. This can be easy, if compatible or similar terminologies are already provided, but could also lead to major efforts, if this is not the case. Mapping the procedure coding system, OPS will be crucial for the applicability in further use. The terminology mapping efforts achieved (ICD-10 GM, OPS in future) will be shared with OMOP in Athena and the extensions of tools with OHDSI.
Currently, the authors are working on using Kibana ( https://www.elastic.co/de/products/kibana ) for an interactive geovisualization and an integration with the FHIR communication standard ( http://omoponfhir.org ), especially for extension with the SMART-on-FHIR app platform framework. 23
Clinical Relevance Statement
The use of common data models is becoming more and more important for secondary use of clinical data across institutions. This article reports on the experience gathered using OMOP/OHDSI for the first time in Germany. As such, it enables other first timers to build upon the experience and judge what is already possible and where additional research is required.
Multiple Choice Question
Which of the following components is not provided by the OMOP/OHDSI community?
The definition of a common data model to store patient data
A list of vocabularies to annotate data
Ready to use jobs to extract data from various sources and load them into the OMOP CDM
Helper tools to find the right ETL jobs
-
Ready to use tools for visualizing data from the OMOP CDM and define patient cohorts
Correct Answer : The correct answer is c. Because of the vast number of possible source systems, data models, and terminologies, it is not possible to provide general tools centrally. However, with Usagi and the Rabbit tools, the OHDSI community developed tools to support the data engineer during this process.
Protection of Human and Animal Subjects
Neither human nor animal subjects were included in the project.
Conflict of Interest None.
Funding
MIRACUM is funded by the German Federal Ministry of Education and Research (BMBF) within the “Medical Informatics Funding Scheme” (FKZ 01ZZ1606H). The research has been cofunded by the German Federal Ministry of Economics and Technology within the Trusted Cloud initiative (FKZ 01MD11009).
Note
The present work was performed in fulfillment of the requirements for obtaining the degree “Dr. rer. biol. hum.” from the Friedrich-Alexander-Universität Erlangen-Nürnberg.
References
- 1.Safran C, Bloomrosen M, Hammond W E et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J Am Med Inform Assoc. 2007;14(01):1–9. doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overhage J M, Ryan P B, Reich C G, Hartzema A G, Stang P E. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19(01):54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaye A, Marcon Y, Isaeva J et al. DataSHIELD: taking the analysis to the data, not the data to the analysis. Int J Epidemiol. 2014;43(06):1929–1944. doi: 10.1093/ije/dyu188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hripcsak G, Duke J D, Shah N H et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 5.OHDSI Homepage. Available at:https://www.ohdsi.org. Accessed October 14, 2017
- 6.Hripcsak G, Ryan P B, Duke J D et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci U S A. 2016;113(27):7329–7336. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Federal Ministry of Education and Research (BMBF).Medical Informatics Funding Scheme – Networking data – improving healthcare2015. Available at:https://www.gesundheitsforschung-bmbf.de/files/Medical_Informatics_Funding_Scheme.pdf. Accessed December 25, 2017
- 8.MIRACUM Homepage. Available at:https://www.miracum.org. Accessed July 25, 2017
- 9.Mahlke L, Lefering R, Siebert H, Windolf J, Roeder N, Franz D. Description of the severely injured in the DRG system: is treatment of the severely injured still affordable? [Article in German] Chirurg. 2013;84(11):978–986. doi: 10.1007/s00104-013-2490-3. [DOI] [PubMed] [Google Scholar]
- 10.Barufka S, Heller M, Prayon V, Fegert J M. Nonnative guidelines for allocating human resources in child and adolescent psychiatry using average values under convergence conditions instead of price determination - analysis of the data of university hospitals in Germany concerning the costs of calculating day and minute values according to Psych-PV and PEPP-System [Article in German] Z Kinder Jugendpsychiatr Psychother. 2015;43(06):397–409. doi: 10.1024/1422-4917/a000383. [DOI] [PubMed] [Google Scholar]
- 11.Bauer C R, Ganslandt T, Baum B et al. Integrated Data Repository Toolkit (IDRT). A suite of programs to facilitate health analytics on heterogeneous medical data. Methods Inf Med. 2016;55(02):125–135. doi: 10.3414/ME15-01-0082. [DOI] [PubMed] [Google Scholar]
- 12.Klann J G, Abend A, Raghavan V A, Mandl K D, Murphy S N. Data interchange using i2b2. J Am Med Inform Assoc. 2016;23(05):909–915. doi: 10.1093/jamia/ocv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss E A, Ma Q, Ryan P B. The impact of standardizing the definition of visits on the consistency of multi-database observational health research. BMC Medical Research Methodology. BioMed Central. 2015;15:13. doi: 10.1186/s12874-015-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H, Depraetere K, De Roo J et al. Semantic processing of EHR data for clinical research. J Biomed Inform. 2015;58:247–259. doi: 10.1016/j.jbi.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Hussain S, Sun H, Sinaci A et al. A framework for evaluating and utilizing medical terminology mappings. Stud Health Technol Inform. 2014;205:594–598. [PubMed] [Google Scholar]
- 16.Schuemie M J, Gini R, Coloma P M et al. Replication of the OMOP experiment in Europe: evaluating methods for risk identification in electronic health record databases. Drug Saf. 2013;36 01:S159–S169. doi: 10.1007/s40264-013-0109-8. [DOI] [PubMed] [Google Scholar]
- 17.Makadia R, Ryan P B. Transforming the Premier Perspective Hospital Database into the Observational Medical Outcomes Partnership (OMOP) Common Data Model. EGEMS (Wash DC) 2014;2(01):1110. doi: 10.13063/2327-9214.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matcho A, Ryan P, Fife D, Reich C. Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model. Drug Saf. 2014;37(11):945–959. doi: 10.1007/s40264-014-0214-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitz Henry F, Resnic F S, Robbins S L et al. Creating a Common Data Model for Comparative Effectiveness with the Observational Medical Outcomes Partnership. Appl Clin Inform. 2015;6(03):536–547. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon D, Ahn E K, Park M Y et al. Conversion and data quality assessment of electronic health record data at a Korean tertiary teaching hospital to a common data model for distributed network research. Healthc Inform Res. 2016;22(01):54–58. doi: 10.4258/hir.2016.22.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gini R, Schuemie M, Brown J et al. Data Extraction and Management in Networks of Observational Health Care Databases for Scientific Research: A Comparison of EU-ADR, OMOP, Mini-Sentinel and MATRICE Strategies. EGEMS (Wash DC) 2016;4(01):1189. doi: 10.13063/2327-9214.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Zhou X, Suehs B T et al. A comparative assessment of observational medical outcomes partnership and mini-sentinel common data models and analytics: implications for active drug safety surveillance. Drug Saf. 2015;38(08):749–765. doi: 10.1007/s40264-015-0297-5. [DOI] [PubMed] [Google Scholar]
- 23.Mandel J C, Kreda D A, Mandl K D, Kohane I S, Ramoni R B. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc. 2016;23(05):899–908. doi: 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]


