Abstract
β-Galactosidase encoded by the Escherichia coli lacZ gene, is widely used as a reporter molecule in molecular biology in a wide variety of animals. β-Galactosidase retains its enzymatic activity in cells or tissues even after fixation and can degrade X-Gal, a frequently used colormetric substrate, producing a blue color. Therefore, it can be used for the activity staining of fixed tissues. However, the enzymatic activity of the β-galactosidase that is ectopically expressed in the non-fixed tissues of animals has not been extensively studied. Here, we report the characterization of β-galactosidase activity in Drosophila tissues with and without fixation in various experimental conditions comparing the activity of two evolutionarily orthologous β-galactosidases derived from the E. coli lacZ and Drosophila melanogaster DmelGal genes. We performed quantitative analysis of the activity staining of larval imaginal discs and an in vitro assay using larval lysates. Our data showed that both E. coli and Drosophila β-galactosidase can be used for cell-type-specific activity staining, but they have their own preferences in regard to conditions. E. coli β-galactosidase showed a preference for neutral pH but not for acidic pH compared with Drosophila β-galactosidase. Our data suggested that both E. coli and Drosophila β-galactosidase show enzymatic activity in the physiological conditions of living animals when they are ectopically expressed in a desired specific spatial and temporal pattern. This may enable their future application to studies of chemical biology using model animals.
Keywords: β-Galactosidase, lacZ, Drosophila, X-Gal
Abbreviations: CPRG, chlorophenol red-β-d-galactopyranoside; PBS, phosphate-buffered saline; UAS, upstream activation sequence
Highlights
-
•
We created a transgenic fly to express Drosophila endogenous β-galactosidase.
-
•
We compared the properties of β-galactosidase molecules from Escherichia coli and Drosophila.
-
•
Both β-galactosidase molecules were active in both fixed and non-fixed tissues.
-
•
E. coli β-galactosidase showed a preference for neutral pH but not for acidic pH.
1. Introduction
β-Galactosidase encoded by the Escherichia coli lacZ gene, is widely used as a reporter molecule in molecular biology in a wide variety of animals [1]. β-Galactosidase retains its enzymatic activity in cells or tissues even after fixation and can degrade X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside), a widely used colormetric substrate, producing a blue color. Therefore, it can be used for the activity staining of fixed tissues. X-Gal is an indole derivative and is transformed into an indoxyl monomer and galactose when its glycosidic linkage is cleaved by β-galactosidase. Subsequently, two indoxyl monomers form a dimer that is a stable and insoluble blue-colored substance through nonenzymatic oxidation [2], [3]. This process can be enhanced by an oxidation catalyst; for example, it is effectively enhanced by the addition of ferri-ferrocyanide [4]. Without ferri-ferrocyanide, indoxyl monomers become diffused from the original enzymatic sites so that slowly oxidated indoxyl monomers become deposited as blue-colored dimers at the active sites of endogenous peroxidase. Therefore, activity staining is usually performed in a staining buffer that contains ferri-ferrocyanide.
In Drosophila, the Gal4-UAS (upstream activation sequence) system is used to express lacZ in specific tissues or cells. By combining various Gal4 strains that confer a variety of expression patterns of Gal4 with UAS-lacZ, β-galactosidase can be expressed with a desired specificity [5]. Although no background activity of β-galactosidase is detected in frequently examined Drosophila tissues such as the third instar larval imaginal discs in standard experimental conditions, it is reported that there is endogenous activity of β-galactosidase in certain wild-type Drosophila tissues [6]. When whole wild-type third instar larvae were fixed, sectioned by cryostat and stained by X-gal, β-galactosidase activity was detected in tissues including the spiracles, lymph glands and certain parts of the intestine. Additionally, in adult wild-type flies, endogenous β-galactosidase activity was detected in certain tissues. β-Galactosidase activity is also reported to be induced in some Drosophila cultured cells, such as Kc cells, by the administration of ecdysterone [7]. Endogenous β-galactosidase activity was shown to be elevated in association with cellular senescence induced by the ectopic activation of Ras [8]. As a candidate for the origin of this endogenous β-galactosidase activity, the genome of Drosophila melanogaster has an orthologous β-galactosidase-encoding gene called the β-Gal-1, Gal or CG9092 gene [9]. Hereafter, we call this gene DmelGal. This locus encodes an 80 kDa protein. DmelGal protein biochemically purified from adult flies was found to be a 160 kDa homodimer, which showed an optimum enzyme activity at pH 6.0 when it was assayed by the hydrolysis of p-nitrophenyl-β-d-galactopyranoside in vitro [10].
Despite the wide usage of β-galactosidase in fixed tissues, the enzymatic activity of β-galactosidase ectopically expressed in non-fixed tissues of animals has not been extensively studied. Previously, β-galactosidase encoded by lacZ has been suggested to show enzymatic activity in living Drosophila embryos under physiological conditions without fixation [11]. In this study, the activity of β-galactosidase encoded by lacZ was visualized in living embryos by injecting a fluorescent substrate. Another fluorescent substrate of β-galactosidase was also demonstrated for use in the detection of ectopically expressed lacZ activity in imaginal discs without fixation in phosphate-buffered saline (PBS) [12].
Here, we report the characterization of β-galactosidase activity in Drosophila tissues with and without fixation in various experimental conditions, comparing the activity of E. coli lacZ and Drosophila DmelGal. Our data showed that both E. coli and Drosophila β-galactosidase can be used for cell-type-specific activity staining, but they have different preferences in regard to conditions.
2. Materials and methods
2.1. Fly genetics
Flies were reared at 25°C, in 60% humidity and 12 h light/dark cycles. dpp-Gal4 (stock number 1553) was obtained from the Bloomington Drosophila Stock Center. The UAS-DmelGal transgenic fly was created as follows. A full-length cDNA clone (LP09580) of the D. melanogaster DmelGal (CG9092) gene was obtained from the BAC PAC Resources Center at Children's Hospital Oakland Research Institute (Oakland, CA, USA). The 2.3 kb XhoI/EcoRV DNA fragment containing the full-length DmelGal gene was subcloned to the XhoI and EcoRV sites of pBluescriptII SK-(Agilent Technologies). Then the 2.3 kb XhoI/BamHI fragment was subcloned to the XhoI and BglII sites of the pUAST vector plasmid [5]. After the full-length sequence of the Gal gene was checked, it was used to create a transgenic fly by P-element mediated transformation. Six transgenic lines were successfully recovered. Among them, the UAS-DmelGal 2M strain, in which the transgene was inserted into the second chromosome, was chosen for use in the further experiments because it showed the highest expression when driven by ubiquitous actin5C-Gal4, as assayed by staining imaginal discs with X-Gal.
2.2. Staining imaginal discs
The activity staining of imaginal discs with X-Gal was performed essentially as previously described [13]. Wandering third instar larvae were collected and washed in PBS, and leg imaginal discs were dissected. They were fixed in 0.25% glutaraldehyde in 130 mM NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4 for 2 min at 25°C, when necessary. Then, the disc was transferred into 10 μl X-Gal staining solution (0.2% X-Gal in appropriate buffers) on the glass slide between two cover glasses that were used as “pillows”. The cover glass was set on the staining solution containing a sample between two cover glasses above the thickness of the cover glass. X-Gal stock solution (10% in dimethylformamide) was prewarmed at 65°C for 15 min before being dissolved in the appropriate buffers. The buffers used were Fe/NaP buffer (5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 150 mM NaCl, 1 mM MgCl2, 10 mM sodium phosphate buffer, pH 7.0 or 6.0), PBS (4 M NaCl, 27 mM KCl, 97 mM Na2HPO4, 15 mM KH2PO4, pH 7.3) and HL3 (70 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 20 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, 5 mM HEPES-NaOH, pH 7.1). The samples were observed using Axioskop2 (Zeiss), and photos were taken using DP73 camera (Olympus) with a 20× objective lens and Nomarski optics, while incubating at room temperature (25°C). Staining intensities were quantified using CellSens Dimension software (Olympus). 3 to 8 discs were stained for each experimental condition.
2.3. Enzymatic assay using larval lysate
An enzymatic assay of β-galactosidase activity using tissue lysate was performed as described [14], [15]. Wandering third instar larvae were collected and washed with PBS, and each whole larva was collected into a BioMasher II tube (Nippi Inc., Japan). An additional 100 μl appropriate buffer was added and homogenized by 20 strokes with PowerMasher (Nippi Inc., Japan). Additional 100 μl buffer was added and vortexed for 30 s. Then, 2 μl larval lysate was added to 200 μl assay solution (1 mM chlorophenol red-β-d-galactopyranoside; CPRG; Wako pure chemicals, Japan in appropriate buffers) and incubated at 25°C for 30 min. An absorbance of 574 nm was measured. 4 to 10 larvae were assayed for each experimental condition. The background value was obtained from the control assays with no larval lysate. Larvae with no UAS construct were also assayed to examine the activity of endogenous β-galactosidase.
2.4. Statistics
Statistical analyses were performed using SPSS 16.0J.
3. Results
3.1. Creation of a transgenic fly expressing the Drosophila DmelGal gene
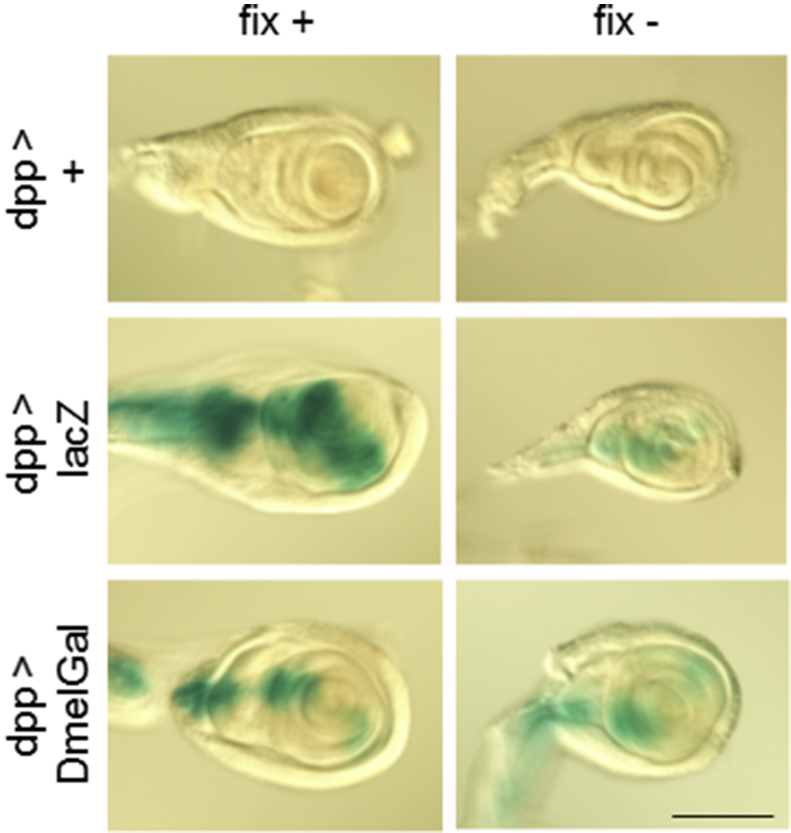
We tested the utility of the DmelGal (CG9092) gene in transgenic flies. For this purpose, we conjugated the full-length DmelGal minigene under the control of the UAS regulatory sequence and used it to create transgenic fly strains. Then, we induced the expression of both lacZ and DmelGal under the control of dpp-Gal4, which drives expression in the boundary of the anterior compartment and the posterior compartment in the larval imaginal discs [16], [17], [18]. We dissected wandering third instar larvae and stained leg imaginal discs with X-Gal using standard techniques [13]. We fixed the discs in a fixative containing 0.25% glutaraldehyde for 2 min, then incubated them in 0.2% X-Gal solution in Fe/NaP buffer (150 mM NaCl, 1 mM MgCl2, 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 10 mM sodium phosphate buffer, pH 7.0) for 20 min. We performed experiments at 25°C, which is a physiologically optimal temperature for Drosophila, not at 37°C that is an optimal temperature of E. coli β-galactosidase. No staining was detected for discs with dpp-Gal4 but no UAS construct (Fig. 1). In dpp > lacZ animals, the anterior/posterior compartment boundary was clearly stained. In dpp > DmelGal animals, the anterior/posterior compartment boundary was also stained, although the intensity of staining was relatively weak. These data indicated that the UAS-DmelGal transgene can be used for the X-Gal activity staining similarly to UAS-lacZ.
Fig. 1.
X-Gal activity staining of leg imaginal discs expressing E. coli and Drosophila genes encoding β-galactosidase driven under the control of dpp-Gal4. dpp-Gal4 is specific to the anterior/posterior compartment boundary. Staining was performed at 25°C in Fe/NaP buffer with or without fixation. While no-UAS control showed no signal, both E-coli lacZ and Drosophila DmelGal showed specific staining both with and without fixation. Scale bar: 100 μm.
Next, we stained leg imaginal discs with X-Gal in Fe/NaP buffer at 25°C for 20 min without fixation. While no staining was observed in the dpp>+ control discs, in the dpp > lacZ and dpp > DmelGal animals, the anterior/posterior compartment boundary was specifically stained (Fig. 1). The intensity of staining was relatively weak compared with the staining after fixation. These data suggested that both E. coli and Drosophila β-galactosidase show cell-type specific enzymatic activity without fixation. We also stained the same series of discs with X-Gal in PBS (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4) and HL3 buffer (70 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 20 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, 5 mM HEPES-NaOH, pH 7.1). HL3 is a buffer used for preserving Drosophila tissues in physiological conditions, for example, for electrophysiological analyses [19]. However, diffuse and non-specific staining was observed for dpp > lacZ and dpp > DmelGal discs, possibly because of the diffusion of indoxyl monomers in the absence of ferri-ferrocyanide as previously reported (data not shown) [4].
3.2. Quantification of X-Gal staining of imaginal discs
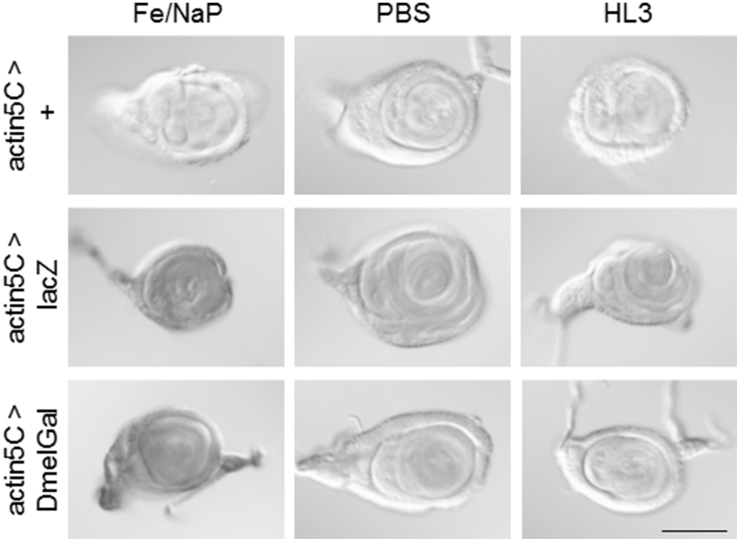
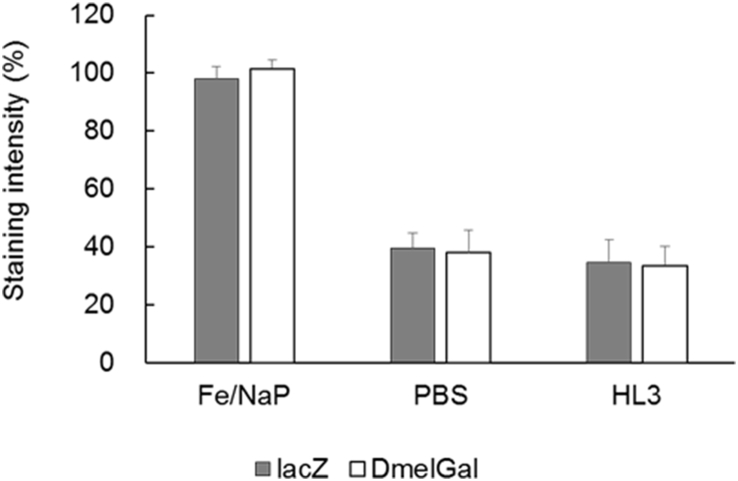
To quantify the intensity of the X-Gal staining, we next used the ubiquitous actin5C-Gal4 driver. We dissected wandering third instar larvae and stained leg imaginal discs with X-Gal without fixation at 25°C for 10 min. In addition to Fe/NaP buffer, which is usually used for X-gal staining, we also tested PBS and HL3 buffer. While no staining was observed in actin5C>+ control animals, whole leg discs were stained in all of the conditions tested in actin5C > lacZ and actin5C > DmelGal animals (Fig. 2). Next, we quantified the staining intensity by imaging analysis (Fig. 3). In Fe/NaP buffer, strong signals were observed, while the signals were relatively weak in PBS and HL3, which are less optimal for colorimetry from X-gal. In all of the buffers tested, there was no significant difference in staining intensity between lacZ and DmelGal.
Fig. 2.
X-Gal activity staining of leg imaginal discs expressing E. coli and Drosophila genes encoding β-galactosidase driven under the control of ubiquitous actin5C-Gal4. Staining was performed at 25°C in Fe/NaP, PBS and HL3 buffers without fixation. While no UAS control showed any signal, both E-coli lacZ and Drosophila DmelGal showed staining throughout the discs. Scale bar: 100 μm.
Fig. 3.
Quantification of the staining intensities of leg imaginal discs expressing E. coli and Drosophila genes encoding β-galactosidase driven under the control of the ubiquitous actin5C-Gal4. Error bars indicate s. e. m.
3.3. Quantification of β-galactosidase activity using larval lysate
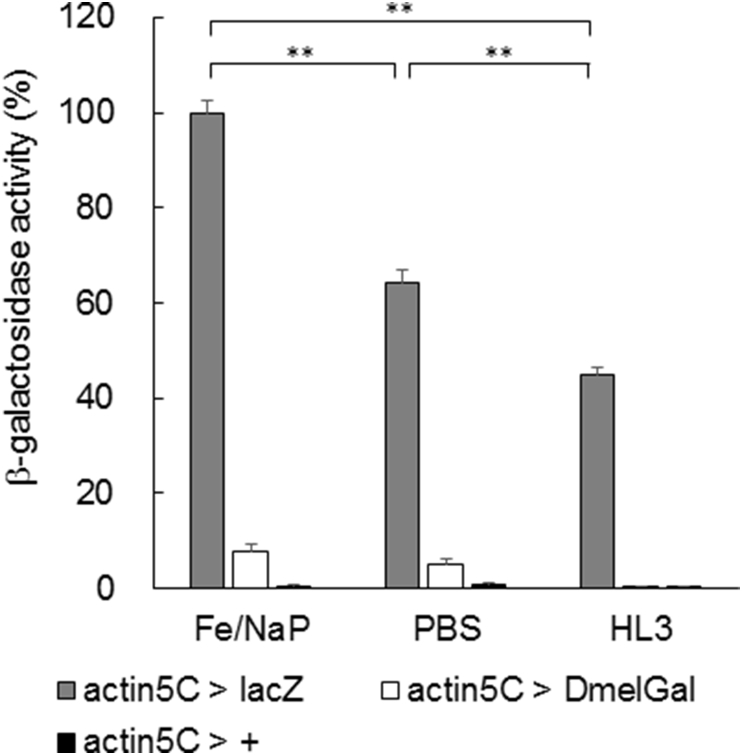
Next, we quantified β-galactosidase activity in vitro in the lysates of whole larvae using the substrate, chlorophenol red-β-d-galactopyranoside (CPRG) by a previously described method [14], [15]. Wandering third instar larvae were collected and homogenized, and then the β-galactosidase activity was assayed by measuring the hydrolyzing activity of CPRG in Fe/NaP, PBS or HL3 buffers. Among the three genotypes tested, actin5C > lacZ, actin5C > DmelGal and actin5C>+, lacZ-expressing animals showed the highest activity compared with the other two genotypes (Fig. 4). They showed the highest activity in Fe/NaP buffer, moderate activity in PBS and the lowest activity in HL3 (one way ANOVA, p < 0.01; Games–Howell test, p < 0.01). DmelGal-expressing animals showed lower activity than lacZ-expressing animals. Finally, almost no β-galactosidase activity was detected in larvae with no UAS transgene.
Fig. 4.
Quantification of the β-galactosidase activity of whole larval lysates expressing E. coli and Drosophila genes encoding β-galactosidase under the control of the ubiquitous actin5C-Gal4. Assays were performed at 25°C in Fe/NaP, PBS and HL3 buffers. Error bars indicate s. e. m. **: p < 0.01 (Games–Howell test followed by one way ANOVA).
3.4. Effect of pH for β-galactosidase activity
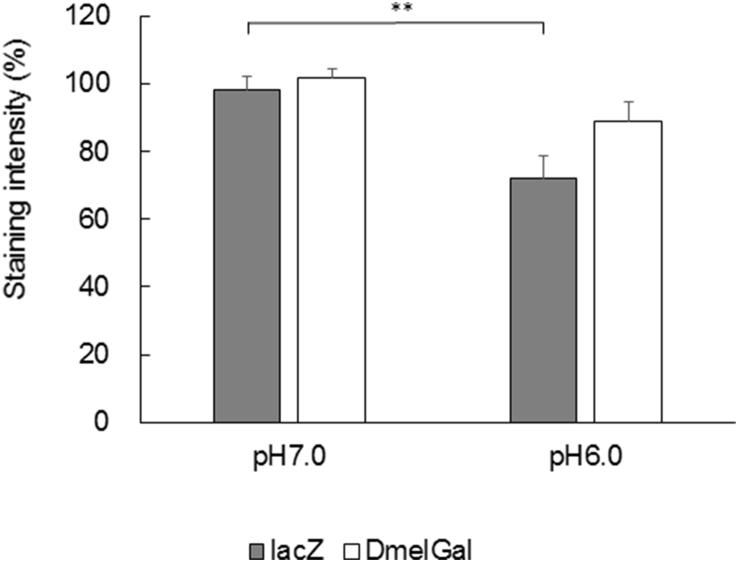
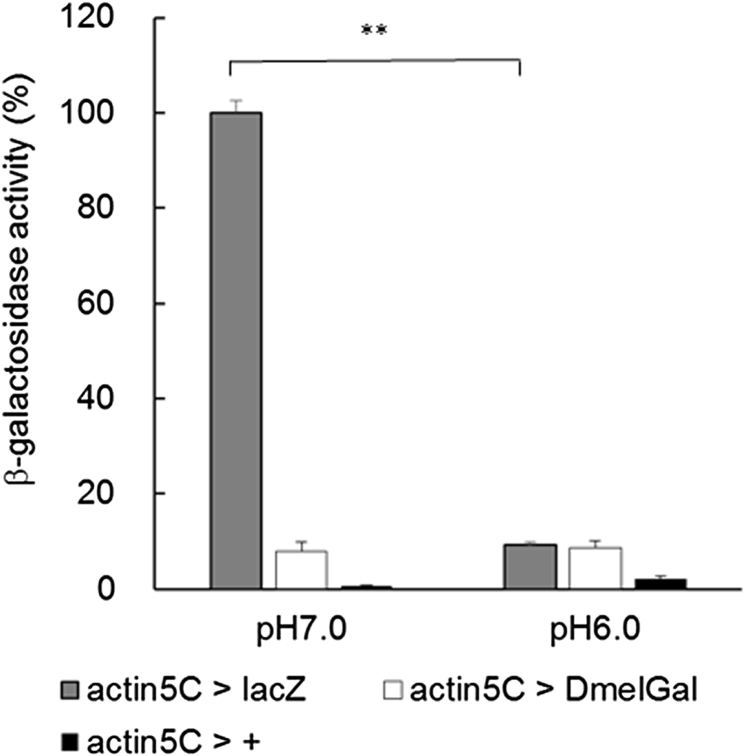
A previous study reported that DmelGal protein purified biochemically from adult flies showed an optimal enzyme activity at pH 6.0 when assayed by the hydrolysis of p-nitrophenyl-β-d-galactopyranoside in vitro [10]. Therefore, we tested the activity of β-galactosidase from E. coli and Drosophila at pH 6.0 by the activity staining of imaginal discs using X-Gal and by an in vitro assay using CPRG without fixation in Fe/NaP buffer. In Fe/NaP buffer at pH 6.0, both actin5C > lacZ and actin5C > DmalGal leg imaginal discs showed staining by X-Gal (Fig. 5). The staining intensity of actin5C > lacZ at pH 6.0 was significantly lower than at pH 7.0, which is a standard experimental condition (Fig. 6; one way ANOVA, p < 0.01; Tukey test, p < 0.01). Next, we examined the in vitro β-galactosidase activity of the larval lysates. Compared with pH 7.0, E. coli β-galactosidase showed remarkably and significantly lower activity at pH 6.0 (Fig. 7; one way ANOVA, p < 0.01; Games–Howell test, p < 0.01). However, the activity of both exogenously expressed and endogenous Drosophila β-galactosidase showed no significant difference between pH 7.0 and pH 6.0.
Fig. 5.
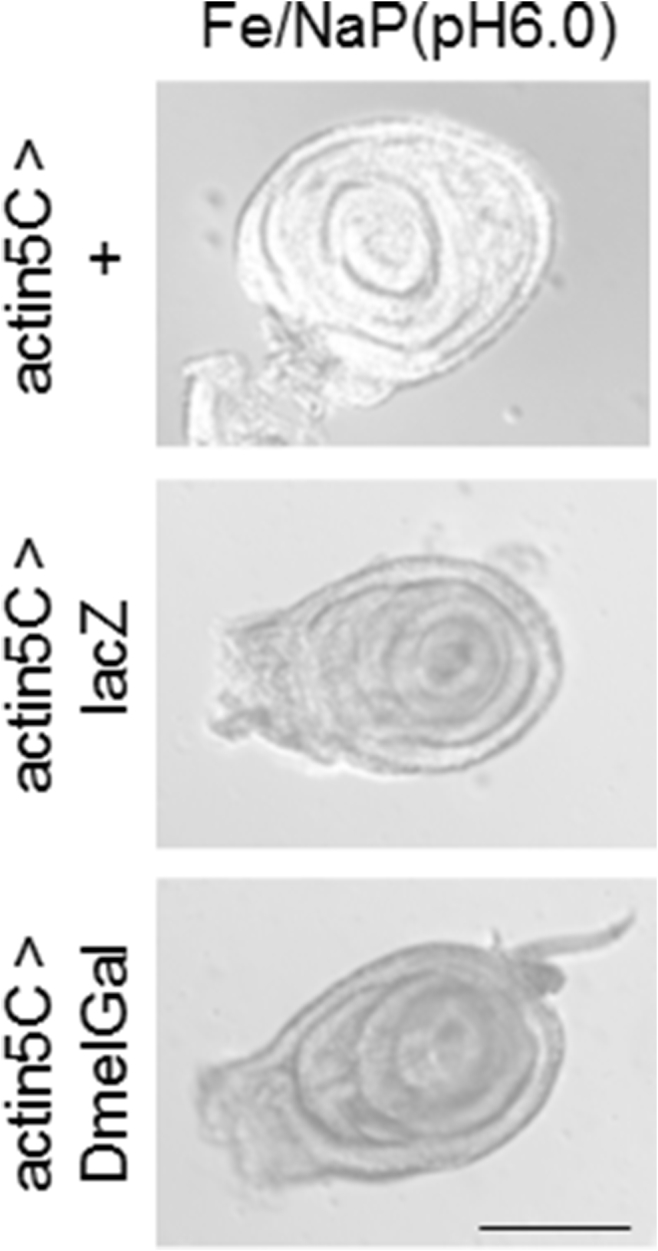
pH dependency of X-Gal activity staining of leg imaginal discs expressing E. coli and Drosophila genes encoding β-galactosidase driven under the control of the ubiquitous actin5C-Gal4. Staining was performed at 25°C in Fe/NaP buffer (pH 6.0) without fixation. While no-UAS control showed no signal, both E-coli lacZ and Drosophila DmelGal showed staining throughout the discs. Scale bar: 100 μm.
Fig. 6.
pH dependency of quantification of the staining intensities of leg imaginal discs expressing E. coli and Drosophila genes encoding β-galactosidase driven under the control of the ubiquitous actin5C-Gal4. Staining was performed at 25°C in Fe/NaP buffer (pH 7.0 and pH 6.0) without fixation. Error bars indicate s. e. m. **: p < 0.01 (Tukey test followed by one way ANOVA).
Fig. 7.
pH dependency of quantification of the β-galactosidase activity of whole larval lysates expressing E. coli and Drosophila genes encoding β-galactosidase driven under the control of the ubiquitous actin5C-Gal4. Assays were performed at 25°C in Fe/NaP buffer (pH 7.0 and pH 6.0). Error bars indicate s. e. m. **: p < 0.01 (Games–Howell test followed by one way ANOVA).
4. Discussion
In this study, we report the characterization of the enzymatic activities of E. coli and Drosophila β-galactosidase that are ectopically expressed through genetic engineering in Drosophila tissues. We showed that the UAS-DmelGal transgenic fly, which we created, can be used for X-Gal activity staining that shows spatial and temporal specificity in combination with Gal4 drivers that confer various specificities in fixed tissues using standard techniques.
We also examined their activities in non-fixed Drosophila tissues. When they were driven by ubiquitous actin5C-Gal4, the intensity of activity staining was not significantly different between E. coli lacZ and Drosophila DmelGal in the Fe/NaP (pH 7.0), PBS and HL3 buffers. Although both lacZ and DmelGal showed weaker staining in PBS and HL3 than in Fe/NaP buffer, this result may reflect the slow oxidation of indoxyl monomers, resulting in the less efficient deposition of blue-colored indoxyl dimers in the absence of an oxidation catalyst such as ferri-ferrocyanide [4]. We also performed in vitro β-galactosidase activity assay using whole larval lysates and the substrate, CPRG. In these assays, E. coli β-galactosidase showed remarkably higher activity than Drosophila β-galactosidase. The activity of E. coli β-galactosidase was dependent on the buffers used. There was a difference in the preferences of experimental conditions for the activity of β-galactosidase between two experimental methods, in the activity staining of imaginal discs and in the in vitro assay using larval lysates. It may reflect the subcellular localization, dimerization or interaction with other molecules of ectopically expressed β-galactosidase. In an in vitro assay, the β-galactosidase molecule exists in the assay buffer that contains homogenously dissolved components of larval lysate. However, in the cells of Drosophila whole-mount imaginal discs, the β-galactosidase proteins are localized in specific subcellular sites that provide appropriate environments for enzymatic reactions, especially in the case of Drosophila β-galactosidase. Such subcellular environments may also enable the cooperative action with other proteins or cofactors. Because mammalian β-galactosidase is known to be localized in the lysosome that provides an acidic environment [20], the lysosome is the candidate subcellular site for the optimal activity of β-galactosidase. Drosophila β-galactosidase is also suggested to be localized in lysosomes [21]. It is noteworthy that there is a remarkable difference in the pH preferences between E. coli and Drosophila β-galactosidase both in the activity staining and the in vitro assay. Although E. coli β-galactosidase is less active at pH 6.0 than at pH 7.0 in both experiments, Drosophila β-galactosidase is as active at pH 6.0 as at pH 7.0. These results suggest that E. coli β-galactosidase prefers neutral pH environments to acidic pH when it was compared with Drosophila β-galactosidase in our experimental conditions.
Although lacZ is widely utilized in fixed tissues, its enzymatic activity has not been extensively examined in non-fixed tissues. In this study, we showed that β-galactosidase that was ectopically expressed from a transgene possesses an enzymatic activity in non-fixed tissues, which means that artificially expressed β-galactosidase can be used to modify chemical compounds that were ectopically introduced into living animals in a tissue or cell-type specific manner. In chemical biology, a variety of chemical compounds are introduced into cells, tissues or animals and their effects will be examined [22]. However, it is difficult to introduce chemical compounds into specific cells in complex multicellular animals such as D. melanogaster. One possible way to solve this problem is to introduce molecules non-specifically and then activate them only in specific cells by means of enzymatic conversion. For this purpose, the chemical compounds to be introduced need to be designed and synthesized to be modified chemically to be inactivated, and then they will be converted to active state by an enzymatic activity that resides in specific cells. In Drosophila, the activity of an enzyme can be triggered in distinct spatial and temporal patterns by sophisticated genetic methods, including the Gal4-UAS system. If the molecule is inactivated by the modification of galactose, it will be able to be activated by an enzymatic activity of β-galactosidase. Drosophila β-galactosidase shows its optimum activity in the lysosomes of living Drosophila cells, which is at 25°C and at acidic pH. It is expected not to be toxic compared with the E. coli molecule. Therefore, for the purpose of the application for the chemical biological experiments using living flies, utilization of Drosophila β-galactosidase transgene may be beneficial because of its properties on optimum temperature, optimum pH and potential toxicity in the living Drosophila cells.
5. Conclusion
Our data suggested that both E. coli and Drosophila β-galactosidase show enzymatic activity at the physiological conditions in living animals when they are ectopically expressed in a desired specific spatial and temporal pattern. Further investigation will be needed to reveal whether it can be applied to the purpose of the cell-specific activation of ectopically introduced chemical compounds.
Conflict of interest
We certify that there is no conflict of interest with any organization regarding the material discussed in the manuscript.
Acknowledgments
We thank Drs. Toshiaki Furuta, Akinobu Suzuki, Eri Oka and Takuya Tamura for their helpful comments and discussion. We thank Dr. Toshiaki Furuta for critically reading the manuscript. This work was supported by the Japan Society for the Promotion of Science KAKENHI Grant Number 26350982 and by a Grant-in-Aid for Scientific Research on Innovative Areas (Comprehensive Brain Science Network) from the Ministry of Education, Science, Sports and Culture of Japan.
References
- 1.Juers D.H., Matthews B.W., Huber R.E. LacZ beta-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Prot. Sci. 2012;21:1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt S.J., Sadler P.W. Studies in enzyme cytochemistry. II. Synthesis of indigogenic substrates for esterases. Proc. R. Soc. Lond. Ser. B, Biol. Sci. 1958;148:481–494. doi: 10.1098/rspb.1958.0040. [DOI] [PubMed] [Google Scholar]
- 3.Coston S., Holt S.J. Studies in enzyme cytochemistry. IV. Kinetics of aerial oxidation of indoxyl and some of its halogen derivatives. Proc. R. Soc. Lond. Ser. B, Biol. Sci. 1958;148:506–519. doi: 10.1098/rspb.1958.0042. [DOI] [PubMed] [Google Scholar]
- 4.Lojda Z. Indigogenic methods for glycosidases. I. An improved method for beta-D-glucosidase and its application to localization studies on intestinal and renal enzymes. Histochemie. 1970;22:347–361. doi: 10.1007/BF00277462. [DOI] [PubMed] [Google Scholar]
- 5.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 6.Schnetzer J.W., Tyler M.S. Endogenous beta-galactosidase activity in the larval, pupal, and adult stages of the fruit fly, Drosophila melanogaster, indicates need for caution in lacZ fusion-gene studies. Biol. Bull. 1996;190:173–187. doi: 10.2307/1542537. [DOI] [PubMed] [Google Scholar]
- 7.Best-Belpomme M., Courgeon A.M., Rambach A. beta-Galactosidase is induced by hormone in Drosophila melanogaster cell cultures. Proc. Natl. Acad. Sci. USA. 1978;75:6102–6106. doi: 10.1073/pnas.75.12.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura M., Ohsawa S., Igaki T. Mitochondrial defects trigger proliferation of neighbouring cells via a senescence-associated secretory phenotype in Drosophila. Nat. Comm. 2014;5:5264. doi: 10.1038/ncomms6264. [DOI] [PubMed] [Google Scholar]
- 9.Knipple D.C., MacIntyre R.J. Cytogenic mapping and isolation of mutations of the beta-Gal-1 locus of Drosophila melanogaster. Mol. Gen. Genet. 1984;198:75–83. doi: 10.1007/BF00328704. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst T.R., Knipple D.C., MacIntyre R.J. Purification and characterization of beta-galactosidase-1 from Drosophila melanogaster. Insect Biochem. 1987;17:1163–1171. [Google Scholar]
- 11.Minden J.S. Synthesis of a new substrate for detection of lacZ gene expression in live Drosophila embryos. BioTechniques. 1996;20:122–129. doi: 10.2144/96201rr02. [DOI] [PubMed] [Google Scholar]
- 12.Kamiya M., Asanuma D., Kuranaga E., Takeishi A., Sakabe M., Miura M., Nagano T., Urano Y. beta-Galactosidase fluorescence probe with improved cellular accumulation based on a spirocyclized rhodol scaffold. J. Am. Chem. Soc. 2011;133:12960–12963. doi: 10.1021/ja204781t. [DOI] [PubMed] [Google Scholar]
- 13.Hama C., Ali Z., Kornberg T.B. Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 1990;4:1079–1093. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- 14.Ashburner M. Cold Spring Harbor Laboratory Press; 1989. Drosophila: a Laboratory Manual. [Google Scholar]
- 15.Simon J.A., Lis J.T. A germline transformation analysis reveals flexibility in the organization of heat shock consensus elements. Nuc. Acids Res. 1987;15:2971–2988. doi: 10.1093/nar/15.7.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staehling-Hampton K., Jackson P.D., Clark M.J., Brand A.H., Hoffmann F.M. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Diff. 1994;5:585–593. [PubMed] [Google Scholar]
- 17.Nellen D., Burke R., Struhl G., Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 18.Wilder E.L., Perrimon N. Dual functions of wingless in the Drosophila leg imaginal disc. Development. 1995;121:477–488. doi: 10.1242/dev.121.2.477. [DOI] [PubMed] [Google Scholar]
- 19.Tamura T., Hou J., Reist N.E., Kidokoro Y. Nerve-evoked synchronous release and high K+-induced quantal events are regulated separately by synaptotagmin I at Drosophila neuromuscular junctions. J. Neurophysiol. 2007;97:540–549. doi: 10.1152/jn.00905.2006. [DOI] [PubMed] [Google Scholar]
- 20.Kurz D.J., Decary S., Hong Y., Erusalimsky J.D. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J. Cell Sci. 2000;113:3613–3622. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 21.Mergliano J., Minden J.S. Caspase-independent cell engulfment mirrors cell death pattern in Drosophila embryos. Development. 2003;130:5779–5789. doi: 10.1242/dev.00824. [DOI] [PubMed] [Google Scholar]
- 22.Narayan P., Ehsani S., Lindquist S. Combating neurodegenerative disease with chemical probes and model systems. Nat. Chem. Biol. 2014;10:911–920. doi: 10.1038/nchembio.1663. [DOI] [PubMed] [Google Scholar]