Abstract
Peptide inter-domain linkers are peptide segments covalently linking two adjacent domains within a protein. Linkers play a variety of structural and functional roles in naturally occurring proteins. In this work we analyze the sequence properties of the predicted linker regions of the bacterial transcriptional regulators belonging to the recently discovered MocR subfamily of the GntR regulators. Analyses were carried out on the MocR sequences taken from the phyla Actinobacteria, Firmicutes, Alpha-, Beta- and Gammaproteobacteria. The results suggest that MocR linkers display phylum-specific characteristics and unique features different from those already described for other classes of inter-domain linkers. They show an average length significantly higher: 31.8 ± 14.3 residues reaching a maximum of about 150 residues. Compositional propensities displayed general and phylum-specific trends. Pro is dominating in all linkers. Dyad propensity analysis indicate Pro–Pro as the most frequent amino acid pair in all linkers. Physicochemical properties of the linker regions were assessed using amino acid indices relative to different features: in general, MocR linkers are flexible, hydrophilic and display propensity for β-turn or coil conformations. Linker sequences are hypervariable: only similarities between MocR linkers from organisms related at the level of species or genus could be found with sequence searches. The results shed light on the properties of the linker regions of the new MocR subfamily of bacterial regulators and may provide knowledge-based rules for designing artificial linkers with desired properties.
Keywords: Linker peptide, Linker length, MocR regulators, Flexibility, Hydrophobicity, Pro–Pro dyad
Graphical abstract
Highlights
-
•
An overview of the structural properties of MocR inter-domain linkers is reported.
-
•
Linker length distribution is heterogeneous in different phyla.
-
•
Linkers are flexible, hydrophilic and have coil conformation propensity.
-
•
Pro and Pro–Pro dyads are very frequent in all the linkers.
-
•
MocR linkers display a few properties different from those reported for other linkers.
1. Introduction
Peptide inter-domain linkers are peptide segments covalently linking two adjacent domains within a protein [1], [2]. Linkers play a variety of structural and functional roles in naturally occurring proteins. For example, they have a role in tuning of biological activities of the connected domains [3], [4], in allosteric coupling [5], in viral replication [6]. They are also of the utmost interest and relevance for applications in protein engineering, for example the alteration of functionality of engineered antibodies [7], [8], [9]. Often, design of efficient and stable linkers with desired properties is hampered by the lack of an adequate knowledge of their structure–function relationship. For that reason, empirical analysis of the characteristics of naturally occurring linkers may provide useful knowledge.
In this work we analyzed the sequence properties of the predicted linker regions of the bacterial transcriptional regulators belonging to the MocR subfamily of GntR regulators [10]. The members of the GntR family of bacterial transcriptional regulators are characterized by the presence of two domains, at the N-terminal and at the C-terminal part of the peptide chain [10]. The N-terminal domain, 60 residue long on average, displays the winged-helix-turn-helix architecture (wHTH) and is responsible for DNA recognition and binding [11]. The C-terminal domain belongs to one of at least four structural families and is essential for oligomerization and effector binding. The two domains are bound to each other by a peptide linker. The MocR subfamily [12], [13] of the GntR regulators is characterized by a large C-terminal domain (350 residue on average) that belongs to the fold type-I pyridoxal 5'-phosphate (PLP) dependent enzymes [14]. Aspartate aminotransferase (AAT) [15] is the archetypal enzyme for this fold. The wHTH and AAT domains are linked to each other by a peptide linker that can have different lengths in different MocRs. The solution of the first three-dimensional structure of GabR from Bacillus subtilis [16], [17] confirmed the presence of a C-terminal fold type-I domain and provided fundamental insights for further investigations aimed at deciphering the mechanism of action of these regulators. Moreover, the same structure suggested that the GabR regulator exists as a domain-swapped dimer and provided an image of the linker segment connecting the two wHTH and AAT domains.
Besides GabR, only a few other MocR proteins have been experimentally characterized: for example, TauR, involved in the regulation of taurine utilization genes in Rhodobacter capsulatus [18]; PdxR, involved in the regulation of the PLP synthesis in several bacteria such as Corynebacterium glutamicum [19], Streptococcus pneumoniae [20], Listeria monocytogenes [21], Streptococcus mutans [22], Bacillus clausii [23]; DdlR from Brevibacillus brevis demonstrated to activate the expression of the gene coding for the enzyme d-alanyl-d-alanine ligase [24].
In this work, we analyzed several structural characteristics of the MocR linkers and suggested that they display a few unique features different from those already described for other classes of peptidic inter-domain linkers.
2. Materials and methods
Only for ease of data processing, analyses were carried out on the MocR sequences taken from the phyla Actinobacteria, Firmicutes, Alpha-, Beta- and Gammaproteobacteria. These phyla are the most populated in the databanks. The sequences of the MocR regulators of each phylum were extracted from the UniProt databank [25] accessed on October, 2015. The regulators were identified using RPSBLAST of the BLAST suite [26] and the CDD databank [27]. The protein sequences containing both the wHTH and AAT domains identified by RPSBLAST were considered genuine MocR regulators. Multiple sequence alignments were calculated with the programs ClustalO [28]. Sequence alignment manipulation and display utilized the software Jalview [29]. Data bank searches utilized BLAST [26] and Hmmer [30] software.
Statistical analyses relied on R statistical package [31]. Python or Perl scripts were written for specific tasks. Physicochemical properties were assigned to the amino acid residues according to the indices provided by the AAindex databank [32] incorporated in the Interpol package [33] of the R project library [31]. Secondary structure prediction utilized the web server Jpred [34] and the program PREDATOR [35]. Sequence redundancy were eliminated with the program CD-HIT [36]. Residue and dipeptide frequency were calculated with the programs “pepstats” or “compseq” of the EMBOSS suite [37].
Residue propensities in the linker region was calculated according to the definition:
| (1) |
where fi,L and fi are the frequencies of the amino acid i in the linker region and in the reference data bank (background frequencies), respectively. In this case, the reference data bank was the bacterial subset of the UniRef50 archive, namely the data bank containing the Uniprot bacterial proteins clustered at 50% sequence identity. Propensities were calculated using the background frequencies of the corresponding phylum. For example, residue propensities for the Actinobacteria linkers were calculated using the background frequencies observed the Actinobacteria subset of UniRef50. Propensity values greater than 1.0 indicates that the corresponding residue is more frequent in the linker region than expected whereas values smaller than 1.0, the opposite. Linker dipeptide (i.e. amino acid pairs or dyads) propensities are the propensity of each of the possible 400 residue pairs to occur preferentially in the linker region [38]. In this case, i in equation (1) refers to each one of these pairs instead of single residues. Phylum-specific dyad background frequencies were used for the propensity calculation as well. Amino acid dyads are obviously not symmetrical: for example, Ala–Arg is not equivalent to the pair Arg–Ala cause of the N- to C- terminal polarity of the peptide chain.
Protein structure display and analysis utilized PyMOL [39] or Chimera [40] software.
3. Results
MocR sequences retrieved from the UniProt data bank were filtered at 75% sequence identity to remove potentially confounding redundancy using the program CD-HIT [36].
Multiple sequence alignments of the entire MocR sequences were calculated within each phylum. Linkers were predicted and extracted from the alignments according to the following criteria: the N-terminal of the linker was the residue immediately following the C-terminal residue of the wHTH domain while its C-terminal was the residue immediately preceding the N-terminal residue of the AAT domain. Domain boundaries were assigned according to the alignment between the MocRs and the two PSSMs (Position Specific Scoring Matrix) of the CDD databank each representing the wHTH or the AAT domains (CDD codes cd07377 and cd00609 or cd01494, respectively). Domain boundaries were “projected” onto all the sequences contained in the multiple sequence alignment and the linker sequences were manually extracted using the editing function of the Jalview software. In the multiple alignment of the Firmicutes bacteria, this procedure was able to correctly identify the boundaries of the linker from the GabR regulator from Bacillus subtilis, independently assigned by the authors of the crystallographic structure [16], [17].
3.1. Length distribution
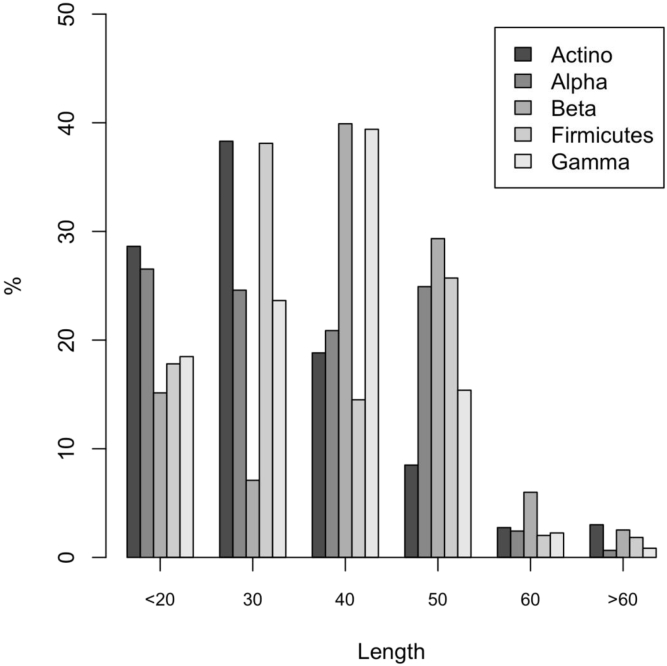
Table 1 reports the number of MocR sequences found in each phylum after filtering at 75% sequence identity. Linker length distributions were calculated within each of the five phyla and are displayed in Fig. 1. The frequency distributions have been calculated for intervals of 10 residue length except for the first and the last frames which have been set to 0–20 and 61–200 residue length, respectively. The observed distributions are rather dispersed around the mean value and show phylum-specific trends (Table 2). In particular, Actinobacteria linkers are on average shorter than those from other phyla while linkers from Betaproteobacteria are longer (Table 2). Moreover, medians of the Beta- and Gammaproteobacteria distributions indicate that their peak frequencies are at higher length intervals. It should be noted that there is a significant number of linkers showing lengths longer than 60 residues. In a few cases, linkers can reach about 150 residue length (see Table 1 in Ref. [41]). At this level, the definition “linker” may not be appropriate anymore. Nonetheless, it will be used throughout the paper to insist that the peptide segments connecting the two main domains of the MocR regulators, wHTH and AAT, are the object of this study.
Table 1.
Number of MocR sequences collected in the databanks for each phylum after redundancy filtering at 75% identity.
| Phylum | MocR numerosity | No. of species |
|---|---|---|
| Actinobacteria | 765 | 129 |
| Alphaproteobacteria | 618 | 105 |
| Betaproteobacteria | 634 | 76 |
| Firmicutes | 1089 | 178 |
| Gammaproteobacteria | 1065 | 180 |
Fig. 1.
Histogram of the linker length distribution in the five phyla considered. Horizontal axis labels indicate length intervals: 20 corresponds to 0–20, 30 (21–30), 40 (31–40), 50 (41–50), 60 (51–60) and >60 (longer than 60 residues). Gray scale corresponds to different phyla as indicated in the box inserted in the plot. Percentage (%) on the vertical axis indicates the fraction of linkers in the length interval.
Table 2.
Linker length distribution parameters.
| Phylum | Mean | Standard deviation | Median |
|---|---|---|---|
| Actinobacteria | 28.9 | 14.8 | 23 |
| Alphaproteobacteria | 30.7 | 12.5 | 30 |
| Betaproteobacteria | 37.0 | 13.4 | 39 |
| Firmicutes | 31.5 | 13.9 | 29 |
| Gammaproteobacteria | 31.8 | 15.1 | 32 |
3.2. Residue propensity
Within each of the five phyla taken into consideration, calculations have been carried out for the entire set of linkers and for length subsets. In particular, linkers were grouped in intervals of 20-residue length (1–20, 21–40, 41–60, 61–200 residues) to assure a sufficient number of residue counts in each subset.
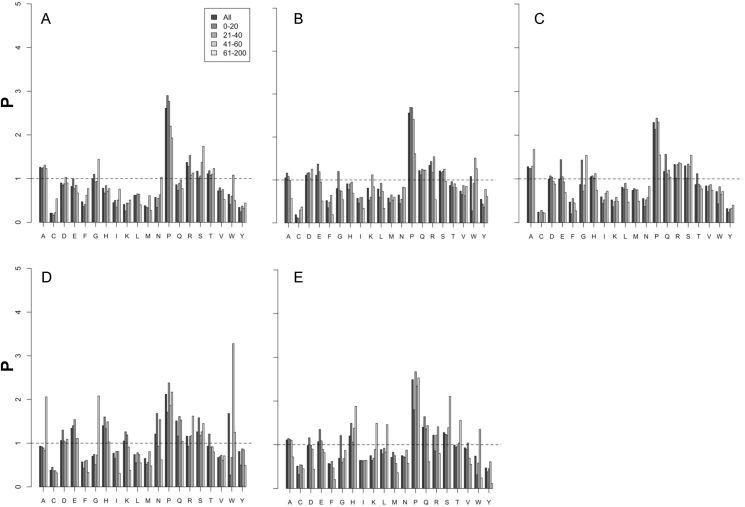
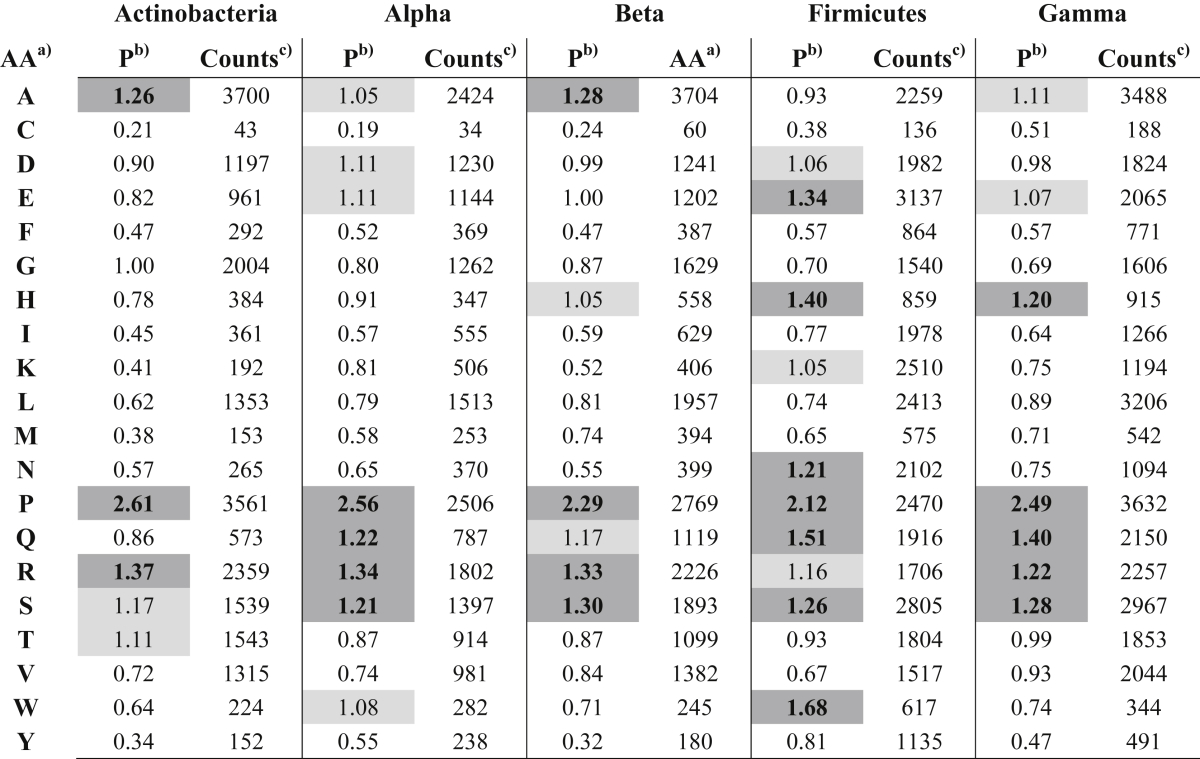
Single residue propensities were first calculated. Results suggest that propensity distributions show phylum- and linker length-specific trends as reported in Fig. 2 and Table 3. In all linkers (Table 3), Pro residue shows a dominating propensity. In Actinobacteria linkers Pro, Ala, Arg are frequent (Fig. 2). Gly displays a neutral propensity (namely 1.0) while tends to be avoided in the linkers of the other phyla (Table 3); in Alphaproteobacteria, Arg and Pro have strong propensities, followed by Gln and Ser with a weaker trend; Betaproteobacteria propensities are similar to those observed in the Alphaproteobacteria except for Ala that displays a stronger propensity and for Gln that has a weaker propensity. Firmicutes possess a relatively “aspecific” distribution: the most represented residues are Glu, His, Asn, Pro, Gln, Ser and Trp. Asp, Lys and Arg have a weaker propensity. Interestingly, Lys appears specific of this phylum since it displays propensities lower than 1.0 in all the others. Gammaproteobacteria linkers are similar to those from Betaproteobacteria except for the higher propensities of His and Gln.
Fig. 2.
Histograms displaying the residue propensity in the linker regions of the MocR from each of the five phyla. Letters indicate: Actinobacteria (A), Alphaproteobacteria (B), Betaproteobacteria (C), Firmicutes (D) and Gammaproteobacteria (E). Single residue propensities is reported for each length interval, with a bar colored according to the grey code shown in the inset of the graph A. X-axis reports the residue one-letter code while the Y-axis indicates residue propensity (P). Horizontal dashed line marks the value 1.0 corresponding to the “neutral” propensity.
Table 3.
Residue propensities in the entire set of linkers.
a) Amino acid one-letter code.
b) Residue propensity; cells containing values ≥1.01 and ≤1.19 or values ≥1.20 are shaded with light or dark grey respectively. In the latter case, numbers are boldfaces.
c) Number of residues in the sample.
Analysis of length-specific propensities highlights several differences (Tables 2–5 in Ref. [41] and Fig. 2) among the phyla considered. Pro is frequent in all the phyla over all the length ranges. At interval 0–20 residues (Table 2 in Ref. [41]) propensity distributions differ from those observed in the overall sample (Table 3). In particular, Asp appears more represented frequent in Firmicutes linkers while Glu has a high propensity in all phyla except in Actinobacteria where it is neutral. Gly now is favored in all phyla except Firmicutes. Lys is frequent in Firmicutes linkers while Arg is underrepresented in Firmicutes and Gammaproteobacteria. The propensities in the 21–40 residue range (Table 3 in Ref. [41]), are very similar to those of the entire set (Table 3). Asn and Trp become less frequent than expected in the Firmicutes. The propensities in the linker range 41–60 are also similar to those reported in Table 3. It should be noted that here Trp displays positive propensity in Alphaproteobacteria, Firmicutes and Gammaproteobacteria. However, Trp is a relatively rare residue and the corresponding counts may be affected by large statistical fluctuation. The last range considered, 61–200 residues, is the least populated and shows marked differences with respect to the propensities of Table 3. Ala is strongly represented in the Firmicutes linkers. Gly is frequent in Actinobacteria, Betaproteobacteria and Firmicutes. Lys and Asn avoid Firmicutes linkers. In Gammaproteobacteria, Leu and Thr become frequent while Gln and Arg relatively rare. Arg and Ser are rare in Alphaproteobateria as well.
Linker dipeptide (i.e. amino acid pairs or dyads) propensities were also calculated separately for each phylum, for all the linkers considered or grouped by length intervals. However, to obtain reliable propensities, each pair should have a sufficient number of counts. This condition is satisfied by the linkers of the five phyla belonging to the subsets “all-linkers”, containing all the considered linkers, and to the 21–40 residue length frame (refer to Table 6 in Ref. [41]). For that reasons, only results from these two sets will be reported here. For completeness, all data are reported in Ref. [41] (Figs. 2–5 therein). Once more, different trends are evident among different phyla and, within each phylum, among length subsets.
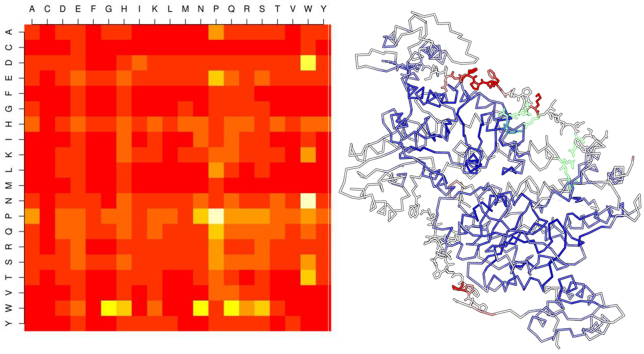
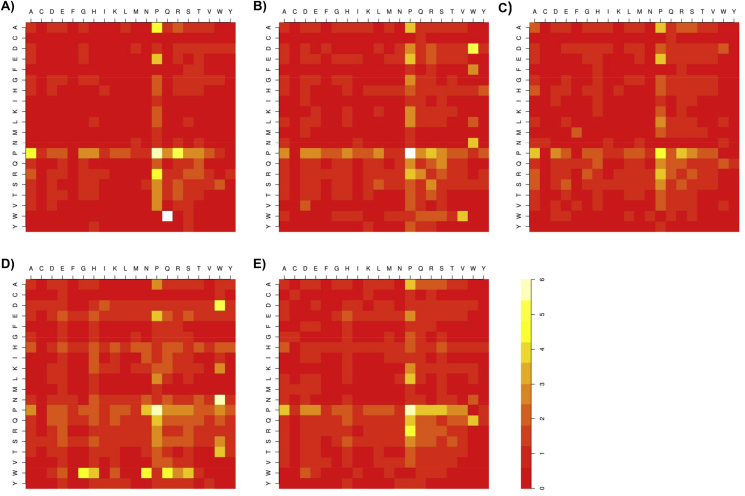
Overall, there is a strong preference for pairs containing Pro at the N- or C-terminal sides of linker dyads in all MocRs (Fig. 3 and refer to Fig. 1 in Ref. [41]). In general, the top most preferred Pro-containing pairs are: Pro–Ala, Pro–Asp, Pro–Glu, Pro–Pro, Pro–Gln, Pro–Arg, Pro–Ser, Ala–Pro, Glu–Pro, Gln–Pro, Arg–Pro, Ser–Pro, Lys–Pro. Among these, Pro–Pro dyad has the strongest propensity. Other dyads are also frequent although not in every phylum. For example: Pro–Gly (Actinobacteria and Betaproteobacteria), Pro–His (Actinobacteria, Alphaprotebacteria and Gammaproteobacteria), Pro–Ile and Pro–Leu (Alphaproteobacteria, Firmicutes and Gammaproteobacteria), Asp–Pro (Alphaproteobacteria), Gly–Pro (Actinobacteria, Alphaproteobacteria, Betaproteobacteria), Lys–Pro (Alphaproteobacteria, Firmicutes, Gammaproteobateria). Interestingly, strong preference for dyads containing Trp can be observed (for example, Trp–Gly, Trp–Gln, Trp–Asn, Asn–Trp in Firmicutes) Trp however is the rarest residue in protein and sampling fluctuations may influence significantly the counts and the reliability of derived frequencies. Firmicutes linkers are distinguished from those of the other phyla because display higher propensity values for dyads containing Glu, His, Lys, Asn and Gln at the N- and/or C-terminal side such as, for example, Glu–Glu, Lys–His, His–Gln, Gln–His Asn–His, Asn–Asn, and the like (Figs. 3 and 1 in Ref. [41]).
Fig. 3.
Heatmaps of the propensity distribution of residue pairs in the “all-linker” sets of the Actinobacteria (A), Alphaproteobacteria (B), Betaproteobacteria (C), Firmicutes (D) and Gammaproteobacteria (E). Vertical and horizontal axes of each map indicate the N-terminal and C-terminal side residues of the dyad using the one-letter code, respectively. Side bar indicates the correspondence between color scale and numerical propensities.
The propensity patterns observed in the 21–40 residue length range reflect largely those seen in the pooled sample (Fig. 3 in Ref. [41]) although their absolute values may differ in the two cases. In particular, propensities of the Trp-containing dyads become lower.
3.3. Physicochemical characteristics
Physicochemical properties of the linker regions were assessed using the AAindex database [42]. This databank contains 544 indices describing many physicochemical characteristics of each of the 20 amino acid residues. Indices were selected as to cover different properties of the peptide chain such as hydrophobicity, flexibility, linker and conformation propensity (Table 4). Moreover, the selected indices map to different groups defined by the cluster analysis based on the correlation coefficient of the AAindex pairs [32], [43]. This assures that the indices in the AAindex set herein used display low cross-correlations. Indices were assigned to each residue of a linker sequence and the average value over the linker length was calculated. The distribution of the index average values for each linker was compared in the form of box plots (Fig. 4). Within each phylum and for each index, the distributions of the average values were contrasted with those calculated in the same way for the wHTH and AAT domains from which the linkers were extracted. As usual, index average distributions were calculated also for different linker interval lengths.
Table 4.
AAindex properties utilized in the linker analysis.
| Property | AAindex code | Interpol code | Reference |
|---|---|---|---|
| Normalized flexibility parameters (B values) average | VINM940101 | 425 | [60] |
| Normalized average hydrophobicity scales | CIDH920105 | 58 | [61] |
| Linker propensity from all dataset | GEOR03010 | 491 | [2] |
| Normalized frequency of β-turn | CHOP780101 | 37 | [62] |
| Normalized frequency of α-helix | CHOP780102 | 38 | [62] |
| Normalized frequency of β-sheet | CHOP780103 | 39 | [62] |
| The Chou-Fasman parameter of coil conformation | CHAM830101 | 24 | [63] |
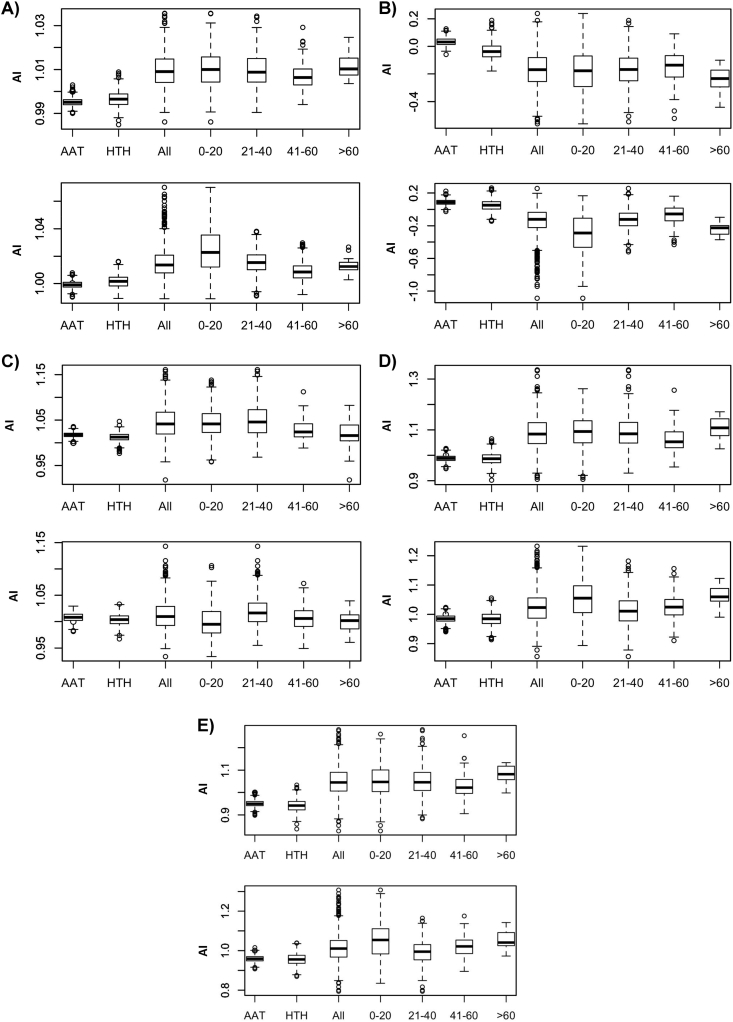
Fig. 4.
Box-plots of the distribution of average AAindices in the Actinobacteria (upper plot of each panel) and Firmicutes (lower plot) phyla. Horizontal axis indicates the average flexibility distribution (A), average hydrophobicity (B), average linker (C), average coil (D), and average β-turn (E) propensities in the wHTH, AAT domains, in all linkers, and in linkers belonging to different length intervals: 0–20, 21–40, 41–60 and > 60 residues. Y-axis reports the corresponding index scale (label AI stands for Average Index).
Linker backbone appears significantly more flexible (in the Actinobacteria, t-test of the null hypothesis of no difference between the mean flexibility of the entire linker set and the wHTH and AAT domain gives a p-value < 10−16) than the AAT and wHTH domains in all phyla and length intervals considered (Actinobacteria and Firmicutes box plots are shown in Fig. 4A while the complete set is reported in Fig. 6 in Ref. [41]). As a general trend, shorter linkers, less than 20 residue long, are more flexible, on average, than the longer ones (Fig. 4A). Likewise, MocR linkers are more hydrophilic (p-value for all-linkers in Actinobacteria <10−16), on average, than the AAT or the HTH domains (Figs. 4B and 7 in Ref. [41]). Also in this case, shorter linker appear to possess a stronger hydrophilic character than the longer ones.
The distribution of the linker residue propensity derived by George and Heringa [2] on their linker set (here referred to as GH-linkers) were also calculated. The distributions should assess the similarity between the composition of the MocR linker sequences and those observed in the GH-linkers (Fig. 4C and Ref [41] Fig. 8). In other words, MocR linker average propensities greater than 1.0 would suggest that they contain residues observed frequently in the GH-linkers, the opposite for propensities lower than 1.0. Results confirm that the MocR linkers shorter than 40 residues possess many of the compositional characteristics observed in the GH-linker set although, interestingly, Firmicutes display different trends (Fig. 4C). Shorter Firmicutes linkers appear indeed to possess residues that show only weak GH-linker like propensities.
Conformational propensity was also assessed using indices related to secondary structure frequency. In general, MocR linkers avoid α-helix and β-strand (Figs. 10 and 11 in Ref. [41]) while display a strong preference for β-turn or coil conformations (Fig. 4D, 9 and 12 in Ref. [41]). To further characterize the linker conformational propensity, secondary structure predictions were carried out. The program PREDATOR was chosen for its ease of use and possibility of computer local installation. Results (Table 7 in Ref. [41]) confirmed that about 80% linkers residues are predicted to be in coil conformation, irrespectively of linker length.
3.4. Sequence similarity
Linker sequences were used as queries in a BLAST search against the entire RefSeq protein databank to verify the presence of significant similarity to any other protein segments. Results suggest that linker sequences are hypervariable: we detected indeed only similarities between MocR linkers from organisms related at the level of species or genus. Only in a few cases, significant similarities were found between MocR linkers from different bacteria phyla (an example is reported in Fig. 5).
Fig. 5.
Sequence alignment between the linkers found in MocR Commensalibacter sp MX01 (Alphaproteobacteria, UniProt: W7E7N5) and Carnobacterium divergens DSM 20623 (Firmicutes, UniProt: A0A0R2I605). Color indicates identical amino acids or with similar physicochemical properties.
3.5. Structure–function relationship
To test whether linker length could be correlated to MocR function, GabR and PdxR regulator sequences were collected from the RegPrecise databank (Table 8 in Ref. [41]) irrespective of the phylum of the source organism. GabR and PdxR have been chosen since they are so far the best characterized MocR regulators. Sequences of the two regulator subfamilies were aligned separately and a Hidden Markov Model profile [44] was calculated for each one of them. Each profile was utilized to search for other putative GabR or PdxR sequences in the reference proteomes data bank available at the Hmmer web server [30]. Data bank sequences to which Hmmer assigned an E-value smaller than 10−120 were retrieved and multiply aligned. The threshold was chosen as to increase the probability of collecting a sufficiently large number of true orthologs while minimizing paralogs. Linker sequences were extracted as described in the Materials and methods section. Linker length distribution in GabR and PdxR were compared (See Fig. 13 in Ref. [41]). Results suggest that the length distribution is rather dispersed around the mean especially in the GabR sample (see Fig. 13 in Ref. [41]): so, it not easy to determine any simple correlation between linker length and regulator function.
4. Discussion
Inter-domain linkers are attracting much interest cause of the functional roles, not yet fully understood, they play in multi-domain proteins and for their potential biotechnological and biomedical applications [45], [46]. For the same reasons, methods for automatic recognition of linker regions in protein structures have also been described in the literature [47], [48], [49]. Scrutiny of new linker systems may provide useful information to the understanding of their structural and functional properties and may help building a set of knowledge-based rules for de-novo design of artificial linkers with novel characteristics.
In this work we report on the analysis of the predicted linkers connecting the wHTH and the AAT domains that constitute the three-dimensional architecture of the recently discovered bacterial family of MocR regulators. The components of this family share the same two-domain asset although they are markedly heterogeneous in terms of sequence similarity and linker structural characteristics [50]. Linkers connecting other multi-domain proteins have already been characterized, for example, the bacterial Q-linkers [51] or GH-linkers [2]. The MocR-linkers share many features with those linkers but appear also to possess a few unique characteristics that, in some cases, are phylum specific. For example, they show an average length significantly higher than the other linkers: 31.8 ± 14.3 residues while Q-linkers are between 15 and 25 residues in length and in the GH-linker average length is 10.0 ± 5.8 residues. Several MocRs of our sample have linkers predicted with a length greater than 60 residues (Table 1 in Ref. [41]) reaching in a few cases the length of about 150 residues. In these cases linkers may represent entire domains rather than a simple peptide stretch connecting two functional domains although they are still predicted to be mostly in extended conformation with only a fraction (about 25%) of putative α-helices (see Table 7 in Ref. [41]). Linker length and composition are parameters influencing the functional properties of the linker itself. For example, in the case of the OmpR regulator (controlling the expression of the porin genes ompF and ompC in E. coli) it has been experimentally demonstrated that linker length and composition influence its function [52]. Other examples are reported in the literature (for a review see Ref. [53]). Therefore, the striking heterogeneity of MocR linker lengths may reflect their functional diversification, the variety of the controlled genes [54], and the different mechanisms of DNA recognition and/or ability to interact with other regulative factors. Interestingly, comparative studies of the predicted MocR binding sites pointed out the lack of any conserved palindromic sequence shared by the whole MocR subfamily [54].
MocR linker residue composition displays both unique and common features compared to the Q-linkers and the GH-linkers. Q-linkers are characterized by the residues Gln, Arg, Glu, Ser and Pro [51] while GH-linkers by Pro, Arg, Phe, Thr, Glu and Gln [2]. High frequency of Pro is a common mark of all the different linker types. In the MocR linkers, Gln has a strong propensity for the Firmicutes and Gammaproteobacteria, weaker for Alphaproteobateria linkers. Arg is instead preferred in all phyla except in Firmicutes where it displays a lower propensity. Notably, Glu is frequent only in the Firmicutes MocR linkers. Interestingly, Lys is represented mainly in Firmicutes linkers, though with a marginal propensity, while it is avoided by all the other phyla. Phe and Thr are not significantly represented in the MocR linkers. Ser has a significant propensity everywhere, although weaker in Actinobacteria. Ala occurs mainly in Actinobacteria and Betaproteobacteria. Within MocR linkers, those from Firmicutes are characterized by the occurrence of Glu, Lys, His and Asn, not observed in the remaining phyla considered except for His that occurs also in Gammaproteobacteria. In general, presence of Arg or Lys suggests that linkers connecting domains of transcriptional regulators may interact with the DNA and play an active role in the regulation and/or recognition mechanism. Conformational role of Pro in the linkers have been clearly discussed by George and Heringa [2]: Pro is an imino acid with no hydrogen-bonding donation potential. For that reason, it generally destabilizes the regular secondary structures α-helix or β-strand and prevents contacts with the neighboring domains. It is therefore well suited for a polypeptide stretch whose function is connecting two domains and permitting their relative movements upon effector binding.
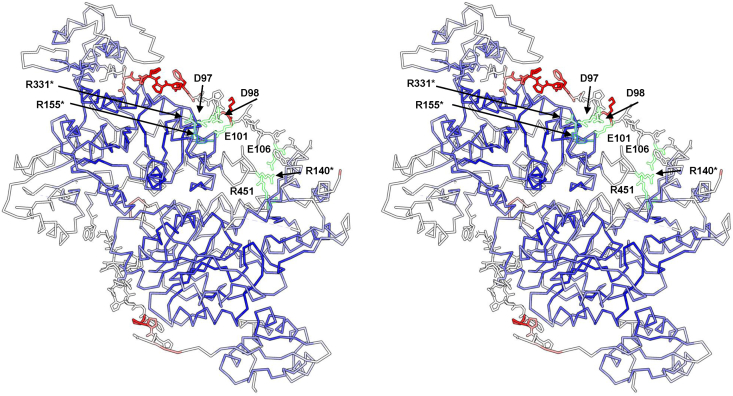
Residue dyad analysis evidences that the Pro–Pro dipeptide is highly represented in linkers from all phyla. Pro–Pro dyad is also the most represented in the non-helical GH-linkers. The helical GH-linkers on the contrary do not show any strong preference for dyads containing Pro either at the N- or C-terminal side. In our linkers, dyads containing Pro at the N- or C-terminal side are also frequent; often, charged or polar residues are associated to the Pro residue within the dyad. Differences can be seen in the different phyla; for example, Firmicutes linkers display a somewhat more dispersed distribution of dyad propensities (Fig. 3D). In the non–helical GH-linkers the dyad Trp–Trp is also very frequent. Interestingly, this dyad is instead rare in all the linkers from all the phyla although, in some cases, dyad containing Trp display a high propensities; for example, Asp–Trp in AlphaProteobacteria and Firmicutes or Asn–Trp in Firmicutes. The high frequency of Pro–Pro sequences supports the notion that the MocR linkers possess an extended conformation [55]. This is corroborated by the analysis of the amino acid properties that indicate a tendency toward hydrophilic character, and flexible extended conformation. These conclusions are also coherent with the properties observed in the linker of GabR from the Firmicutes Bacillus subtilis, the only MocR regulator of which the three dimensional structure has been solved. This linker is 29 residue long and is in an extended, mostly solvent-exposed, conformation. It possesses, among others, 4 and 5 Glu and Asp residues respectively; 3 Pro, one Lys and one Gln residues. No Arg residue is observed. There is also a single Pro–Pro dyad. Two of the Asp and one Glu residue are involved in H-bonds to Arg residues from the other subunit (Table 5). The remaining Asp and Glu residues along with the only Lys are exposed, suggesting a possible interaction with other factors or even with DNA, upon conformational rearrangement of MocR quaternary structure. The linker residues display B-factors higher than in the rest of the dimer (Fig. 6). B-factors magnitude indeed reflects the amount of atom displacement around its average position and may indicate the degree of local flexibility [56], [57], [58]. Flexibility and rigidity are critical parameters for linker mechanical properties and strongly affect, for example, the function of fusion proteins [59]. Current models describing the possible mechanism of action of the MocR regulators predict that linkers allow movement of the wHTH domains to recognize the transcription factor binding sites on the DNA molecule [16], [17], [23].
Table 5.
Residues of GabR linker from Bacillus subtilis (chain D of the PDB structure 4N0B).
| Residue | Solvent exposure | Interactions |
|---|---|---|
| Glu81 | Exposed | |
| Leu82 | Buried | |
| Asp83 | Exposed | |
| Met84 | Exposed | |
| Phe85 | Partly exposed | |
| Ser86 | Exposed | |
| Ala87 | Exposed | |
| Glu88 | Exposed | |
| Glu89 | Exposed | |
| His90 | Exposed | |
| Pro91 | Partly exposed | |
| Pro92 | Exposed | |
| Phe93 | Exposed | |
| Ala94 | Partly exposed | |
| Leu95 | Exposed | |
| Pro96 | Exposed | |
| Asp97 | Exposed | Salt bridge to Arg331 of the other subunit |
| Asp98 | Buried | Salt bridge to Arg331 of the other subunit |
| Leu99 | Exposed | |
| Lys100 | Exposed | |
| Glu101 | Buried | Salt bridge to Arg155 of the other subunit |
| Ile102 | Exposed | |
| His103 | Exposed | |
| Ile104 | Exposed | |
| Asp105 | Exposed | |
| Gln106 | Partly exposed | H-bond to Arg140 of the other subunit; H-bond to Arg451 of the same subunit |
| Ser107 | Exposed | |
| Asp108 | Exposed | |
| Trp109 | Partly exposed |
Fig. 6.
Stereo picture of the trace of the dimer of GabR from Bacillus subtilis (chains C and D in the entry PDB: 4N0B). Side chains are displayed only for residues belonging to the linker region. Atoms of the structure have been colored according to the magnitude of the B-factor; scale ranges from blue (low) to red (high). White and red atoms have the highest B-factors and are the most mobile. Residues mentioned in Table 5 involved in interactions are outlined with green lines and labeled.
This work was meant to study the overall structural properties of the inter-domain peptide linkers in the MocR bacterial regulators. Ideally, possible correlation between linker properties and parent MocR function should be explored to provide insights into the structure–function relationship in these bacterial regulators. Lack of a sufficiently large base of experimental functional characterizations hampers systematic and exhaustive analyses. An attempt to correlate a structural characteristic, namely linker length to function, suggests that a straightforward relation, at least in the GabR and PdxR subfamilies, may not be obvious.
5. Conclusions
This work sheds lights on the properties of the linker regions of the relatively new family of bacterial regulators MocR. The reported results suggest that the MocR linkers may be regarded as a novel linker group. Linkers were grouped according the phylum of source organisms for easy analysis, without any a-priori assumption. The results show that there are statistical trends characteristics of individual phyla, in particular for linkers extracted from the Firmicutes MocRs. Ideally, possible correlation between linker properties and parent MocR function should be tested as more experimental and functional data will accumulate. Even within these limits, the herein reported observations are useful for designing experiments aimed at understanding the role of the linkers within the MocR regulators. The set of linker sequences extracted from the regulators is also useful as a reference library to support the knowledge-based design of novel linkers with desired properties. The entire set of linker sequences will be made available by the authors at the site https://sites.google.com/a/uniroma1.it/pascarella_lab/ and detailed data are reported in Ref. [41].
Authors' contribution
This work was carried out in collaboration among all authors. Authors TM and SP designed the study, carried out the computer programming work and wrote the draft of the manuscript. Authors SA and AT carried out the databank searches and data collection and contributed to write the final version of the manuscript. Authors MLDS and RC read, revised and approved the manuscript and contributed to write the discussion section.
Conflict of interest statement
The authors declare no conflicts of interest pertaining to this work.
Acknowledgments
Authors are grateful to Dr. Cinzia Federici for providing R scripts for AAindex and other statistical analyses. This work was supported by Regione Lazio [grant code FILAS-RU-2014-1020 A/15/2015 to TM], by University of Rome “La Sapienza” and by the Italian Education, University and Research Ministry (MIUR) [grant numbers C26N158EP9; C26A14SY4E].
References
- 1.Bhaskara R.M., de Brevern A.G., Srinivasan N. Understanding the role of domain-domain linkers in the spatial orientation of domains in multi-domain proteins. J. Biomol. Struct. Dyn. 2013;31:1467–1480. doi: 10.1080/07391102.2012.743438. [DOI] [PubMed] [Google Scholar]
- 2.George R.A., Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng. 2002;15:871–879. doi: 10.1093/protein/15.11.871. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q., Cho H., Yeo W.S., Bae T. The extracytoplasmic linker peptide of the sensor protein SaeS tunes the kinase activity required for staphylococcal virulence in response to host signals. PLoS Pathog. 2015;11:e1004799. doi: 10.1371/journal.ppat.1004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller B.R., Sundlov J.A., Drake E.J., Makin T.A., Gulick A.M. Analysis of the linker region joining the adenylation and carrier protein domains of the modular nonribosomal peptide synthetases. Proteins. 2014;82:2691–2702. doi: 10.1002/prot.24635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Register A.C., Leonard S.E., Maly D.J. SH2-catalytic domain linker heterogeneity influences allosteric coupling across the SFK family. Biochemistry. 2014;53:6910–6923. doi: 10.1021/bi5008194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohlway A., Pirakitikulr N., Ding S.C., Yang F., Luo D., Lindenbach B.D., Pyle A.M. The linker region of NS3 plays a critical role in the replication and infectivity of hepatitis C virus. J. Virol. 2014;88:10970–10974. doi: 10.1128/JVI.00745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klement M., Liu C., Loo B.L., Choo A.B., Ow D.S., Lee D.Y. Effect of linker flexibility and length on the functionality of a cytotoxic engineered antibody fragment. J. Biotechnol. 2015;199:90–97. doi: 10.1016/j.jbiotec.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 9.Klein J.S., Jiang S., Galimidi R.P., Keeffe J.R., Bjorkman P.J. Design and characterization of structured protein linkers with differing flexibilities. Protein Eng. 2014;27:325–330. doi: 10.1093/protein/gzu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigali S., Derouaux A., Giannotta F., Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 2002;277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- 11.Hoskisson P.A., Rigali S. Chapter 1: variation in form and function the helix-turn-helix regulators of the GntR superfamily. Adv. Appl. Microbiol. 2009;69:1–22. doi: 10.1016/S0065-2164(09)69001-8. [DOI] [PubMed] [Google Scholar]
- 12.Belitsky B.R., Sonenshein A.L. GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis. Mol. Microbiol. 2002;45:569–583. doi: 10.1046/j.1365-2958.2002.03036.x. [DOI] [PubMed] [Google Scholar]
- 13.Bramucci E., Milano T., Pascarella S. Genomic distribution and heterogeneity of MocR-like transcriptional factors containing a domain belonging to the superfamily of the pyridoxal-5'-phosphate dependent enzymes of fold type I. Biochem. Biophys. Res. Commun. 2011;415:88–93. doi: 10.1016/j.bbrc.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Schneider G., Kack H., Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000;8:R1–R6. doi: 10.1016/s0969-2126(00)00085-x. [DOI] [PubMed] [Google Scholar]
- 15.Kirsch J.F., Eichele G., Ford G.C., Vincent M.G., Jansonius J.N., Gehring H., Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J. Mol. Biol. 1984;174:497–525. doi: 10.1016/0022-2836(84)90333-4. [DOI] [PubMed] [Google Scholar]
- 16.Edayathumangalam R., Wu R., Garcia R., Wang Y., Wang W., Kreinbring C.A., Bach A., Liao J., Stone T.A., Terwilliger T.C., Hoang Q.Q., Belitsky B.R., Petsko G.A., Ringe D., Liu D. Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17820–17825. doi: 10.1073/pnas.1315887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda K., Kato S., Ito T., Shiraki S., Kawase Y., Goto M., Kawashima S., Hemmi H., Fukada H., Yoshimura T. Role of the aminotransferase domain in Bacillus subtilis GabR, a pyridoxal 5'-phosphate-dependent transcriptional regulator. Mol. Microbiol. 2015;95:245–257. doi: 10.1111/mmi.12861. [DOI] [PubMed] [Google Scholar]
- 18.Wiethaus J., Schubert B., Pfander Y., Narberhaus F., Masepohl B. The GntR-like regulator TauR activates expression of taurine utilization genes in Rhodobacter capsulatus. J. Bacteriol. 2008;190:487–493. doi: 10.1128/JB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jochmann N., Gotker S., Tauch A. Positive transcriptional control of the pyridoxal phosphate biosynthesis genes pdxST by the MocR-type regulator PdxR of Corynebacterium glutamicum ATCC 13032. Microbiology. 2011;157:77–88. doi: 10.1099/mic.0.044818-0. [DOI] [PubMed] [Google Scholar]
- 20.El Qaidi S., Yang J., Zhang J.R., Metzger D.W., Bai G. The vitamin B6 biosynthesis pathway in Streptococcus pneumoniae is controlled by pyridoxal 5'-phosphate and the transcription factor PdxR and has an impact on ear infection. J. Bacteriol. 2013;195:2187–2196. doi: 10.1128/JB.00041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belitsky B.R. Role of PdxR in the activation of vitamin B6 biosynthesis in Listeria monocytogenes. Mol. Microbiol. 2014;92:1113–1128. doi: 10.1111/mmi.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao S., Bitoun J.P., Nguyen A.H., Bozner D., Yao X., Wen Z.T. Deficiency of PdxR in Streptococcus mutans affects vitamin B6 metabolism, acid tolerance response and biofilm formation. Mol. Oral Microbiol. 2015;30:255–268. doi: 10.1111/omi.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tramonti A., Fiascarelli A., Milano T., di Salvo M.L., Nogues I., Pascarella S., Contestabile R. Molecular mechanism of PdxR - a transcriptional activator involved in the regulation of vitamin B6 biosynthesis in the probiotic bacterium Bacillus clausii. FEBS J. 2015;282:2966–2984. doi: 10.1111/febs.13338. [DOI] [PubMed] [Google Scholar]
- 24.Takenaka T., Ito T., Miyahara I., Hemmi H., Yoshimura T. A new member of MocR/GabR-type PLP-binding regulator of d-Alanyl-d-Alanine ligase in Brevibacillus brevis. FEBS J. 2015;282:4201–4217. doi: 10.1111/febs.13415. [DOI] [PubMed] [Google Scholar]
- 25.Tatusova T., Ciufo S., Fedorov B., O'Neill K., Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42:D553–D559. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., Lanczycki C.J., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Bryant S.H. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2016. R: a Language and Environment for Statistical Computing. [Google Scholar]
- 32.Kawashima S., Pokarowski P., Pokarowska M., Kolinski A., Katayama T., Kanehisa M. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 2008;36:D202–D205. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heider D. 2012. Interpol: Interpolation of Amino Acid Sequences. R package version 1.3.1. [Google Scholar]
- 34.Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frishman D., Argos P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y., Niu B., Gao Y., Fu L., Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 38.Crasto C.J., Feng J. Sequence codes for extended conformation: a neighbor-dependent sequence analysis of loops in proteins. Proteins. 2001;42:399–413. doi: 10.1002/1097-0134(20010215)42:3<399::aid-prot100>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 39.Schroedinger L.L.C. 2015. The PyMOL Molecular Graphics System. Version 1.8. [Google Scholar]
- 40.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Angelaccio S., Milano T., Tramonti A., Di Salvo M.L., Contestabile R., Pascarella S. Data from computational analysis of the peptide linkers in the MocR bacterial transcriptional regulators. Data Brief. 2016 doi: 10.1016/j.dib.2016.08.064. DIB-D-16-00574; under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawashima S., Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28:374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomii K., Kanehisa M. Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng. 1996;9:27–36. doi: 10.1093/protein/9.1.27. [DOI] [PubMed] [Google Scholar]
- 44.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 45.Li G., Huang Z., Zhang C., Dong B.J., Guo R.H., Yue H.W., Yan L.T., Xing X.H. Construction of a linker library with widely controllable flexibility for fusion protein design. Appl. Microbiol. Biotechnol. 2016;100:215–225. doi: 10.1007/s00253-015-6985-3. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J.H., Yun J., Shang Z.G., Zhang X.H., Pan B.R. Design and optimization of a linker for fusion protein construction. Prog. Nat. Sci. 2009;19:1197–1200. [Google Scholar]
- 47.Bae K., Mallick B.K., Elsik C.G. Prediction of protein interdomain linker regions by a hidden Markov model. Bioinformatics. 2005;21:2264–2270. doi: 10.1093/bioinformatics/bti363. [DOI] [PubMed] [Google Scholar]
- 48.Shatnawi M., Zaki N. Inter-domain linker prediction using amino acid compositional index. Comput. Biol. Chem. 2015;55:23–30. doi: 10.1016/j.compbiolchem.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Liu C., Chin J.X., Lee D.Y. SynLinker: an integrated system for designing linkers and synthetic fusion proteins. Bioinformatics. 2015;31:3700–3702. doi: 10.1093/bioinformatics/btv447. [DOI] [PubMed] [Google Scholar]
- 50.Milano T., Contestabile R., Lo Presti A., Ciccozzi M., Pascarella S. The aspartate aminotransferase-like domain of Firmicutes MocR transcriptional regulators. Comput. Biol. Chem. 2015;58:55–61. doi: 10.1016/j.compbiolchem.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Wootton J.C., Drummond M.H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- 52.Mattison K., Oropeza R., Kenney L.J. The linker region plays an important role in the interdomain communication of the response regulator OmpR. J. Biol. Chem. 2002;277:32714–32721. doi: 10.1074/jbc.M204122200. [DOI] [PubMed] [Google Scholar]
- 53.Gokhale R.S., Khosla C. Role of linkers in communication between protein modules. Curr. Opin. Chem. Biol. 2000;4:22–27. doi: 10.1016/s1367-5931(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 54.Novichkov P.S., Kazakov A.E., Ravcheev D.A., Leyn S.A., Kovaleva G.Y., Sutormin R.A., Kazanov M.D., Riehl W., Arkin A.P., Dubchak I., Rodionov D.A. RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics. 2013;14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saha I., Shamala N. Investigating diproline segments in proteins: occurrences, conformation and classification. Biopolymers. 2012;97:54–64. doi: 10.1002/bip.21703. [DOI] [PubMed] [Google Scholar]
- 56.Ringe D., Petsko G.A. Mapping protein dynamics by X-ray diffraction. Prog. Biophys. Mol. Biol. 1985;45:197–235. doi: 10.1016/0079-6107(85)90002-1. [DOI] [PubMed] [Google Scholar]
- 57.Siglioccolo A., Gerace R., Pascarella S. “Cold spots” in protein cold adaptation: insights from normalized atomic displacement parameters (B'-factors) Biophys. Chem. 2010;153:104–114. doi: 10.1016/j.bpc.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Yuan Z., Zhao J., Wang Z.X. Flexibility analysis of enzyme active sites by crystallographic temperature factors. Protein Eng. 2003;16:109–114. doi: 10.1093/proeng/gzg014. [DOI] [PubMed] [Google Scholar]
- 59.Huang Z., Li G., Zhang C., Xing X.H. A study on the effects of linker flexibility on acid phosphatase PhoC-GFP fusion protein using a novel linker library. Enzyme Microb. Technol. 2016;83:1–6. doi: 10.1016/j.enzmictec.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Vihinen M., Torkkila E., Riikonen P. Accuracy of protein flexibility predictions. Proteins. 1994;19:141–149. doi: 10.1002/prot.340190207. [DOI] [PubMed] [Google Scholar]
- 61.Cid H., Bunster M., Canales M., Gazitua F. Hydrophobicity and structural classes in proteins. Protein Eng. 1992;5:373–375. doi: 10.1093/protein/5.5.373. [DOI] [PubMed] [Google Scholar]
- 62.Chou P.Y., Fasman G.D. Empirical predictions of protein conformation. Annu. Rev. Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 63.Charton M., Charton B.I. The dependence of the Chou-Fasman parameters on amino acid side chain structure. J. Theor. Biol. 1983;102:121–134. doi: 10.1016/0022-5193(83)90265-5. [DOI] [PubMed] [Google Scholar]