Abstract
Glioma is the most aggressive and malignant primary brain tumor in adults. In the present study, we identified a vital oncoprotein, capping actin protein, gelsolin-like (CapG), and investigated its roles in the prognosis, proliferation and metastasis in glioma. The mRNA and protein levels of CapG were significantly increased in human glioma, and higher CapG expression was an independent prognostic factor for predicting unfavorable prognosis. The expression level of CapG was found to be associated with several common molecular features of glioblastoma (GBM; WHO grade IV glioma) in The Cancer Genome Atlas (TCGA) cohort. When analyzing the prognosis of GBM patients according to these molecular features, we observed that the prognostic value of CapG was affected by amplification of CDK6 or EGFR. However, overexpression of CapG markedly promoted cell growth in vitro, while depletion of CapG significantly inhibited cell proliferation by blocking the cell cycle in G1/S transition. Moreover, CapG manipulation in glioma cell lines U87 and U251 showed CapG-dependent cellular migration and invasiveness. These data suggest that CapG may serve as a prognostic biomarker with potentially important therapeutic implications for glioma.
Keywords: CapG, glioma, prognosis, proliferation, migration, invasiveness
Introduction
Glioma, accounting for ~80% of brain malignancies (1), is the most lethal and aggressive type of brain tumor in adults (2). Despite advances in multimodal therapies including surgical resection, radiation and chemotherapy (3), the prognosis of glioma patients remains dismal (4,5), with a 5-year survival rate of 4.3% (6). Due to the fact that most glioma patients have already developed metastasis or malignant diffusion by the time of diagnosis (2), identifying new prognostic biomarkers and molecular targets for glioma is imperative.
To better understand the pathogenesis of glioma, substantial efforts have been made to investigate molecular subtypes (7,8) and potential biomarkers correlated with the clinical prognosis (9–12). For example, The Cancer Genome Atlas (TCGA) (13) database provides researchers an ideal platform to explore tumorigenesis, cancer progression and treatment responses in various types of tumors including GBM (12,14–16).
As proliferation and metastasis play a key role in glioma development (2), we focused on potential genes involved in glioma proliferation and metastasis in the present study. Capping actin protein, gelsolin-like (CapG) was first identified as an actin binding protein of the gelsolin superfamily, which is able to modulate actin length by capping barbed ends in a Ca2+-dependent manner and generate propulsive force (17,18). However, it is involved in the regulation of actin-based cell migration in non-muscle benign cells (19). Moreover, overexpression of CapG has been reported to promote cell motility and increase metastasis in breast cancer and ovarian carcinoma (20). However, the function and prognosis of CapG in human glioma remain unclear.
In the present study, we first investigated levels of CapG expression in a cohort of Chinese glioma patients by tissue microarray (TMA) and estimated its prognostic value. We then tested the differential expression and prognostic value of CapG in an independent cohort from the TCGA dataset. Finally, we assessed the effect of CapG on the proliferation and metastatic ability of glioma cell lines U87 and U251. Our results revealed that CapG is a novel independent prognostic predictor for glioma patients and highlight a key role of CapG in proliferation and metastasis of glioma.
Materials and methods
Patients and tissue samples
For the Chinese TMA cohort, 285 glioma patients of different WHO grades were recruited between January 2000 and December 2010 from the Department of Neurosurgery at Changzheng Hospital, Second Military Medical University (SMMU) (Shanghai, China). Sixteen normal brain tissue samples were taken from trauma outpatients during decompression treatment. Clinical information was obtained from medical records of the patients. Tumor histology was confirmed by two neuropathologists independently. All participants provided written informed consent. The present study was approved by the Ethics Review Committee of the Second Military Medical University and Fudan University.
Immunohistochemistry and tissue microarray analysis
The tissue microarray construction and immunohistochemistry analyses were performed by Shanghai Biochip Co., Ltd. (Shanghai, China), as described previously (21,22). The immunohistochemical staining was read and scored by two independent pathologists in a blinded fashion. The total immunoreactivity score was the product of the staining scores for density and intensity (23). High CapG expression, indicating strongly positive cases (+), had a score of >3, while low expression indicated weakly positive cases (−) with a score of ≤3.
Quantitative real-time PCR
Total RNA was extracted from surgically resected tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. Reverse transcription of total RNA was performed using ReverTra Ace (Toyobo, Tokyo, Japan). The PCR primers are avaiable upon request. Quantitative real-time PCR was run using the ABI Prism 7900HT instruments (Applied Biosystems, Foster City, CA, USA) in the presence of Thunderbird SYBR qPCR Mix (Toyobo). All samples were tested in triplicate and repeated 3 times and melting curve analysis was used to check the specificity of amplification. The 2−ΔΔCt method (24) was used to calculate relative gene expression.
In silico analyses of TCGA data
The Cancer Genome Atlas (TCGA) (13) cohort (N=528; the Affymetrix platform), an independent dataset of GBM, was included in the present study. For the TCGA cohort, multidimensional data of clinical information, molecular subtypes, copy number alterations, common mutations, methylation levels and gene expression were obtained from the TCGA data portal (http://tcga-data.nci.nih.gov/tcga/) and cBioPortal (http://www.cbioportal.org/public-portal/), to analyze the expression profile and prognostic significance of the CapG gene, and their correlations with several molecular alterations. In the analyses of the TCGA cohort, CapG gene levels were dichotomized to low and high according to the ROC curve.
Cell culture and lentivirus infection
U87 and U251 human glioma cell lines were purchased from the Japanese Cancer Research Resources Bank. The glioma cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, Foster City, CA, USA) supplemented with 10% fetal bovine serum (FBS) and penicillin (100 U/ml)/streptomycin (100 U/ml). To generate cell lines stably overexpressing CapG, we cloned CapG from cDNA of the U251 cell line by PCR and cloned it into the CD513B plasmid (System Biosciences, Mountain View, CA, USA) at the EcoRI and BamHI restriction sites. To stably interfere with CapG expression, we selected sequences targeting CapG into the HpaI-XhoI sites plasmid LentiLox 3.7 (pLL3.7) (data available upon request). However, the shLacZ (25) and CD513B empty vector (EV) were used as controls for pLL3.7-shCapG and CD513B-CapG, respectively. Lentiviral production and infection were performed as previously described (26).
Western blotting
The tissue and cell samples were lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail (Sigma, St. Louis, MO, USA). The lysates were separated on 10 or 12% SDS-PAGE gel and electrotransferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk for 1 h at room temperature and incubating with primary antibodies. CapG (Abcam, Cambridge, MA, USA), CDK2, CDK6, Myc, cyclin A1, cyclin D1, cyclin E, CDK4, E2F1, p15, RB and phospho-RB (ProteinTech, Group, Inc., Chicago, IL, USA), the membranes were incubated with the secondary antibody (Epitomics, Burlingame, CA, USA). Anti-GAPDH antibody (Vazyme, Nanjing, China) was used as a loading control.
Cell proliferation assays and cell cycle analysis
Following overexpression or knockdown of CapG, the glioma cells were seeded in 96-well plates (2,000 cells/well) in sextuple, and the cell proliferation assay was performed using Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's instructions. Cell cycle analysis was performed as described previously (27). Glioma cells were washed with 1Χ PBS 3 times and fixed with 75% ice-cold ethanol. Fixed cells were incubated in 1Χ PBS with propidium iodide (PI; 25 µg/ml) and 10 mg/ml RNase for 30 min in the dark before being analyzed by flow cytometry. All assays were performed in 3 independent experiments.
Wound healing assays
Cells were cultured to monolayers in 6-well plates and a reference line was drawn at the back of each well. Three scratches were made from one end to the other end of the well with a sterile p200 pipette tip. The width of the wound distance was measured at baseline. The cells were cultured in serum-free medium for 24 and 48 h, respectively. Wound closure rate was expressed as the distance migrated after 24 or 48 h relative to the basic width at 4 h. Migration experiments were performed in triplicates and repeated at least 3 times.
Transwell invasion assays
Cells were suspended in FBS-free DMEM at a concentration of 1×105 (U87 cells) and 2.5×105 (U251 cells) for Transwell invasion assays using BD BioCoat Matrigel invasion chambers (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's instructions. The suspended cells (500 µl) were pipetted into the upper chamber, while the lower chamber was filled with 750 µl DMEM with EGF (15 ng/ml). After 24 h of incubation at 37°C, the filter side of the upper chamber was scrubbed with cotton swabs to remove non-invading cells and Matrigel matrix. The migrated cells were fixed in methanol and stained with crystal violet. Then, the membranes were cut from the chamber and placed on microscope slides for observation. Each invasion condition was performed in triplicates and repeated at least 3 times.
Statistical analysis
All experiments were conducted in triplicate and are expressed as the means and standard deviation by two-tailed Student's t-test. Analysis of patient survival outcome was conducted by Kaplan-Meier and Cox regression using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA) and SPSS statistics 17.0, respectively (SPSS, Inc., Chicago, IL, USA) (*P<0.05, **P<0.01, ***P<0.001 and ‘ns’, not significant).
Results
CapG is elevated in human glioma tissues
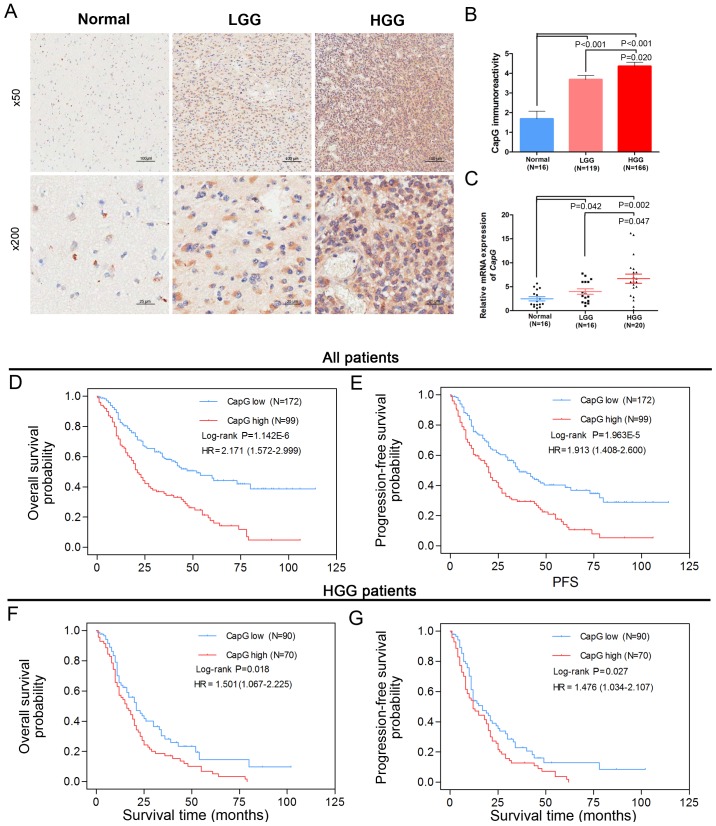
In the present study, a cohort of 285 glioma specimens and 16 normal controls from a Chinese population was enrolled in the TMA. It included 12 grade I (pilocytic astrocytomas), 108 grade II (diffuse astrocytomas), 46 grade III (anaplastic astrocytomas) and 119 grade IV (glioblastomas, GBM) specimens according to the WHO grading schedule. The clinicopathological parameters of the TMA are presented in Table I. The patient median overall survival (OS) was 30 months for all glioma cases, with a 5-year survival rate of 4.3%. Next, we examined the protein level of CapG by immunohistochemical assay performed on the TMA and observed its cytoplasmic expression in most samples. Furthermore, the immunoreactivity of CapG was significantly increased with WHO grade in the human glioma tissues when compared with normal controls: high grade gliomas (HGG; grade III/grade IV) showed stronger immunoreactivity than the low grade gliomas (LGG, grade I/II) (Fig. 1A and B).
Table I.
Clinicopathologic characteristics of the 285 glioma patients in the Chinese TMA cohort.
| WHO grade | |||||
|---|---|---|---|---|---|
| Characteristics | All glioma cases (N=285) | Grade I (N=12) | Grade II (N=108) | Grade III (N=46) | Grade IV (N=119) |
| Sex | |||||
| Male | 188 | 8 (66.7%) | 75 (69.4%) | 27 (58.7%) | 78 (65.5%) |
| Female | 97 | 4 (33.3%) | 33 (30.6%) | 19 (41.3%) | 41 (34.4%) |
| Age at diagnosis (year) | |||||
| <60 | 241 | 11 (91.7%) | 101 (93.5%) | 40 (90.0%) | 89 (74.8%) |
| ≥60 | 44 | 1 (8.3%) | 7 (6.5%) | 6 (10.0%) | 30 (25.2%) |
| Tumor origin | |||||
| Primary | 253 | 12 (100%) | 102 (94.6%) | 35 (76.1%) | 104 (87.4%) |
| Secondary | 32 | 0 (0%) | 7 (6.4%) | 11 (33.9%) | 15 (12.6%) |
| Extent of resection | |||||
| Gross total | 224 | 8 (66.7%) | 89 (82.4%) | 34 (73.9%) | 93 (78.2%) |
| Partial | 61 | 4 (33.3%) | 19 (17.6%) | 12 (26.1%) | 26 (21.8%) |
| CapG expression | |||||
| Low | 180 | 8 (66.7%) | 80 (74.1%) | 30 (65.2%) | 62 (52.1%) |
| High | 105 | 4 (33.3%) | 28 (26.0%) | 16 (34.8%) | 57 (47.9%) |
| OS (months) | 30 (27.4–33.5) | NAa | 45 (41.1–51.4) | 26 (23.9–37.4) | 16 (13.3–18.8) |
| Median (95% CI) | |||||
| PFS (months) | 27 (24.9–30.8) | NAa | 40 (38.2–48.4) | 25 (20.8–33.2) | 14 (11.2–16.4) |
| Median (95% CI) | |||||
TMA, tissue microarray; OS, overall survival; PFS, progression-free survival; CI, confidence interval.
NA, not available due to a large proportion of censored cases.
Figure 1.
Expression profile and prognostic value of CapG in a Chinese glioma cohort. (A) CapG protein expression was analyzed by immunohistochemical staining in different WHO grades of human glioma specimens and normal controls. Scale bars are depicted in figures. (B) Immunoreactivity scores of CapG staining in normal controls, LGG and HGG are represented as mean ± SEM. (C) The relative CapG mRNA expression in 16 LGG and 20 HGG compared with 16 normal controls, and GAPDH was used as an endogenous control. Kaplan-Meier analyses of the associations between differential CapG levels and (D) OS or (E) PFS in 285 glioma patients. Kaplan-Meier analyses of the associations between differential CapG levels and (F) OS or (G) PFS in 165 HGG patients. P-values were determined by log-rank test, while hazard ratio (HR) and 95% confidence interval (CI) were obtained from univariate Cox regression model; *P<0.05; ***P<0.001. LGG, low-grade glioma; HGG, high-grade glioma; OS, overall survival; PFS, progression-free survival.
We then measured the transcriptional level of CapG in a subset of 36 human gliomas (16 LGG and 20 HGG specimens) and 16 normal controls via quantitative RT-PCR assay. These samples were randomly selected from the cohort. Consistent with the TMA data, CapG was considerably increased in human glioma specimens according to WHO grade (Fig. 1C). These results suggest that CapG is frequently elevated in human glioma tissues.
CapG serves as a novel prognostic marker for glioma patients
To further investigate the correlation between CapG expression level and clinical prognosis, we performed Kaplan-Meier analyses and found that glioma patients with higher CapG expression had significantly shorter overall survival (OS, log-rank P=1.142E-06, HR=2.171, 95% CI=1.572–2.999, Fig. 1D) and progression-free survival (PFS, log-rank, P=1.963E-05, HR=1.913, 95% CI=1.408–2.600, Fig. 1E) than those with low expression. Moreover, we observed that HGG patients with higher CapG expression also had an unfavorable OS (log-rank P=0.018, HR=1.501, 95% CI=1.067–2.225, Fig. 1F) and PFS (log-rank P=0.027, HR=1.476, 95% CI=1.034–2.107, Fig. 1G) than those with low CapG expression.
Next, our univariate and multivariate Cox regression analyses revealed that high CapG expression was an independent risk predictor of both OS (P=0.003, HR=1.674, 95% CI=1.198–2.339) and PFS (P=0.008, HR=0.008, 95% CI=1.123–2.12) in our cohort (Table II). In addition, we further identified CapG as an independent prognostic factor for OS (P=0.021, HR=1.541, 95% CI=1.067–2.225) and PFS (P=0.032, HR=1.476, 95% CI=1.034–2.107) of HGG patients; higher levels of CapG predicted poorer survival (Table II).
Table II.
Univariate and multivariate Cox regression of CapG immunoreactivity for OS and PFS in all glioma and HGG patients of the Chinese TMA cohort.
| Univariate Cox regression | Multivariate Cox regression | |||
|---|---|---|---|---|
| Characteristics | P-value | HR (95% CI) | P-value | HR (95% CI) |
| All glioma cases (N=292) | ||||
| OS | ||||
| Sex (male vs. female) | 0.587 | |||
| Age, years (≥60 vs. <60) | 0.004 | 1.844 (1.220–2.786) | 0.037 | 1.580 (1.027–2.429) |
| Tumor origin (secondary vs. primary) | 0.676 | |||
| Resection (gross total vs. patial) | 0.041 | 1.370 (1.013–1.852) | 0.825 | |
| Necrosis (negative vs. positive) | 0.005 | 1.842 (1.198–2.833) | 0.076 | |
| Grade (III+IV vs. I+II) | 1.069E-18 | 6.080 (4.072–9.077) | 1.079E-14 | 5.307 (3.476–8.103) |
| CapG cytoplasm (high vs. low) | 2.553E-06 | 2.171 (1.572–2.999) | 0.003 | 1.674 (1.198–2.339) |
| PFS | ||||
| Sex (male vs. female) | 0.597 | |||
| Age, years (≥60 vs.<60) | 0.001 | 1.976 (1.307–2.986) | 0.003 | 1.806 (1.215–2.684) |
| Tumor origin (secondary vs. primary) | 0.315 | |||
| Resection (gross total vs. patial) | 0.067 | |||
| Necrosis (negative vs. positive) | 0.001 | 2.090 (1.358–3.217) | 0.138 | |
| Grade (III+IV vs. I+II) | 1.911E-19 | 6.409 (4.279–9.594) | 1.424E-14 | 4.456 (3.045–6.519) |
| CapG cytoplasm (high vs. low) | 3.334E-04 | 1.913 (1.408–2.600) | 0.008 | 1.543 (1.123–2.120) |
| HGG (N=174) | ||||
| OS | ||||
| Sex (male vs. female) | 0.522 | |||
| Age, years years (≥60 vs. <60) | 0.303 | |||
| Tumor origin (secondary vs. primary) | 0.055 | |||
| Resection (gross total vs. patial) | 0.408 | |||
| Necrosis (negative vs. positive) | 0.817 | |||
| CapG cytoplasm (high vs. low) | 0.021 | 1.541 (1.067–2.225) | 0.021 | 1.541 (1.067–2.225) |
| PFS | ||||
| Sex (male vs. female) | 0.350 | |||
| Age, years (≥60 vs. <60) | 0.104 | |||
| Tumor origin (secondary vs. primary) | 0.142 | |||
| Resection (gross total vs. patial) | 0.463 | |||
| Necrosis (negative vs. positive) | 0.373 | |||
| CapG cytoplasm (high vs. low) | 0.032 | 1.476 (1.034–2.107) | 0.032 | 1.476 (1.034–2.107) |
OS, overall survival; PFS, progression-free survival; HGG, high-grade gliomas; TMA, tissue microarray; CI, confidence interval; HR, hazard ratio. P-values denoted in bold indicate a significant difference.
CapG gene expression is upregulated in GBM specimens and is correlated with several molecular alterations in the TCGA cohort
We next investigated the expression profile of the CapG gene in an independent cohort from TCGA (13). Consequently, mRNA expression of CapG increased in 97.7% (516/528) of the GBM specimens compared to the normal controls (Fig. 2A). Since TCGA network has introduced a novel molecular classification of GBM according to gene expression, which are proneural, neural, classical and mesenchymal (28), we then assessed CapG expression in each of these subtypes. As shown in Fig. 2B, we observed that CapG expression was drastically decreased in the proneural subtype in comparison with the others. Nevertheless, CapG expression was still elevated in GBM samples.
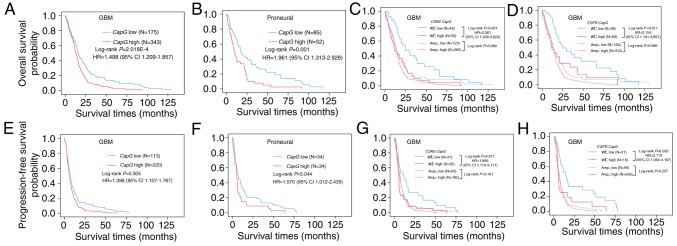
Figure 2.
CapG mRNA expression levels are elevated in glioma (GBM) specimens and are correlated with several common molecular features in the TCGA cohort. (A) CapG mRNA expression levels were detected in 10 cases of normal controls and 528 GBM specimens in the TCGA cohort. The value represents log2 of the CapG mRNA expression level of each GBM sample to the mean of 10 normal controls. (B) CapG mRNA expression levels were compared between normal controls and 4 different molecular subtypes of GBM samples. (C-H) CapG mRNA expression was compared according to several common molecular features in subgroups (proneural and non-proneural) of GBM. ‘NC’, no change; ‘Amp’, amplification; ‘Wt’, wild-type; ‘Mut’, mutation; *P<0.05; **P<0.01; ***P<0.001.
To further explore the expression profile of the CapG gene, we scanned associations between its expression and these common genetic alterations in GBM (13,29) including amplifications of CDK4, CDK6, EGFR, AKT3, PDGFRA, MDM2, MET, MDM4, MYCN, CCND2, PIK3CA, mutations in PTEN, TP53, IDH1, EGFR, NF1, RB1, PIK3R1, ATRX, PIK3CA, CTNND2, SPTA1, GABRA6, LZTR1, and deletions of PTEN, CDKN2A, CDKN2B, CDKN2C, NF1, PARK2, RB1. As a result, we found that CapG expression exhibited a correlation with the amplification of CDK6, EGFR, MET, SYK, ANXA1, MYCN, PDGFRA, and mutations of PTEN, IDH1, RB1.
Since we ascertained that CapG expression is correlated with the GBM subtype, we then explored whether the differential CapG expression with the above-mentioned common genetic alterations was dependent on the specific subtype. Consequently, cases with CDK6 amplification or EGFR amplification or MET amplification had higher levels of CapG expression than those with no copy number alterations in the proneural subtype, while there were no dramatic changes in non-proneural subtypes (Fig. 2C-E). Moreover, cases with SYK amplification had lower CapG expression level than those with no copy number alterations in the proneural subtype, and there was also no obvious change in the non-proneural subtype (Fig. 2F). In contrast, cases with PTEN mutation or RB1 mutation showed a higher level of CapG expression than cases with wild-type only in the non-proneural subtype (Fig. 2G and H). However, cases with ANXA1 amplification, MYCN amplification, PDGFRA amplification, or IDH1 mutation exhibited no significant CapG expression change in the proneural or non-proneural subtypes when compared with those without these genetic alterations (data available on request).
High expression of CapG significantly predicts unfavorable clinical outcome of GBM patients in the TCGA cohort
To further explore the relationship between CapG expression and GBM patient clinical outcome in the TCGA cohort, we compared the OS and PFS of all GBM patients with differential CapG expression and observed statistically significant disadvantage in prognosis for GBM patients with higher CapG expression (OS, log-rank P=2.018E-04, HR=1.498, 95% CI=1.209–1.857, Fig. 3A; PFS, log-rank P=0.005, HR=1.398, 95% CI=1.107–1.767, Fig. 3E). Multivariate Cox regression further validated the CapG expression as an independent predictor for the GBM patient OS (P=0.002, HR=1.546, 95% CI=1.170–2.042) and PFS (P=0.029, HR=1.400, 95% CI=1.035–1.893) in the TCGA cohort (Table III).
Figure 3.
The prognostic value of CapG expression in glioma (GBM) specimens of the TCGA cohort. Kaplan-Meier analyses of differential CapG gene expression for overall survival (upper panels) and progression-free survival (lower panels) of all (A and E) GBM patients or (B and F) proneural subtype or considering the amplification status of (C and G) CDK6 or (D and H) EGFR; P-values were determined by log-rank test, while hazard ratio (HR) and 95% confidence interval (CI) were obtained from the univariate Cox regression model.
Table III.
Univariate and multivariate Cox regression of CapG expression for OS and PFS in GBM patients of the TCGA cohort.
| Univariate Cox regression | Multivariate Cox regression | |||
|---|---|---|---|---|
| Characteristics | P-value | HR (95% CI) | P-value | HR (95% CI) |
| GBM (N=528) | ||||
| OS | ||||
| Sex (male vs. female) | 0.464 | |||
| Age, years (>60 vs. <60) | 1.415E-10 | 1.938 (1.583–2.372) | 1.577E-04 | 1.609 (1.257–2.058) |
| Tumor origin (secondary vs primary) | 4.415E-05 | 0.394 (0.252–0.616) | 0.021 | 0.558 (0.339–0.917) |
| Subtype (proneural vs. non-proneural) | 0.038 | 0.786 (0.626–0.986) | 0.024 | 1.425 (1.047–1.940) |
| EGFR (amplification vs. wild-type) | 1.066E-04 | 1.846 (1.354–2.518) | 0.372 | |
| CDK6 (amplification vs. wild-type) | 0.002 | 1.495 (1.162–1.922) | 0.413 | |
| SYK (amplification vs. wild-type) | 0.831 | |||
| MYCN (amplification vs. wild-type) | 0.087 | |||
| MET (amplification vs. wild-type) | 0.030 | 1.275 (1.023–1.589) | 0.299 | |
| PTEN (mutation vs. wild-type) | 0.863 | |||
| RB1 (mutation vs. wild-type) | 0.363 | |||
| IDH1 (mutation vs. wild-type) | 1.003E-04 | 0.373 (0.227–0.613) | 0.019 | 0.486 (0.266–0.887) |
| CAPG expression (high vs. low) | 2.246E-04 | 1.498 (1.209–1.857) | 0.002 | 1.546 (1.170–2.042) |
| PFS | ||||
| Sex (male vs. female) | 0.350 | |||
| Age, years (>60 vs. <60) | 7.769E-05 | 1.566 (1.254–1.956) | 0.114 | |
| Tumor origin (secondary vs primary) | 1.821E-06 | 0.294 (0.178–0.486) | 3.154E-04 | 0.350 (0.197–0.619) |
| Subtype (proneural vs. non-proneural) | 0.038 | 0.762 (0.589–0.985) | 0.022 | 1.493 (1.060–2.104) |
| EGFR (amplification vs. wild-type) | 4.563E-05 | 2.197 (1.505–3.208) | 0.011 | 2.384 (1.223–4.648) |
| CDK6 (amplification vs. wild-type) | 0.025 | 1.375 (1.041–1.816) | 0.876 | |
| SYK (amplification vs. wild-type) | 0.193 | |||
| MYCN (amplification vs. wild-type) | 0.619 | |||
| MET (amplification vs. wild-type) | 0.184 | |||
| PTEN (mutation vs. wild-type) | 0.780 | |||
| RB1 (mutation vs. wild-type) | 0.816 | |||
| IDH1 (mutation vs. wild-type) | 0.015 | 0.501 (0.287–0.872) | 0.845 | |
| CAPG expression (high vs. low) | 0.005 | 1.398 (1.107–1.767) | 0.029 | 1.400 (1.035–1.893) |
OS, overall survival; PFS, progression-free survival; GBM, glioblastoma; TCGA, The Cancer Genome Atlas; CI, confidence interval; HR, hazard ratio. P-values denoted in bold indicate a significant difference.
Moreover, we next performed survival analysis for each molecular subtype and detected that only the proneural (OS, log-rank P=0.001; HR=1.961, 95% CI=1.313–2.928, Fig. 3B; PFS, log-rank P=0.044; HR=1.570, 95% CI=1.012–2.435, Fig. 3F) subtype retained statistical significance. In addition, we carried out survival analyses stratified by the status of these common genetic alterations in GBM and found the effect of CapG on GBM patient prognosis was associated with EGFR amplification or CDK6 amplification. It is noteworthy that the prognostic value of CapG was highly significant in individuals with wild-type CDK6 (OS, log-rank P=0.001, HR=2.261, 95% CI=1.409–3.628, Fig. 3C; PFS, log-rank P=0.017, HR=1.966, 95% CI=1.119–3.111, Fig. 3G) or EGFR (OS, log-rank P=0.011, HR=2.154, 95% CI=1.191–3.897, Fig. 3D; PFS, log-rank P=0.033, HR=2.113, 95% CI=1.064–4.197, Fig. 3H), but not significant in individuals with amplification of CDK6 or EGFR. Furthermore, patients with low CapG expression and wild-type EGFR or CDK6 had the most favorable prognosis, whereas patients with high CapG expression and amplification of EGFR or CDK6 had the poorest prognosis (Fig. 3C, D, G and H).
CapG regulates glioma cell proliferation in vitro
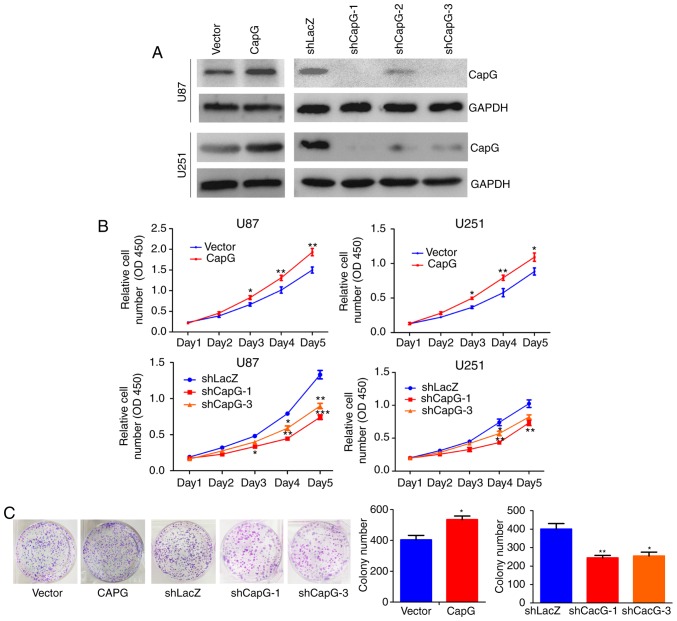
Given that CapG is significantly upregulated in human glioma, we inferred that CapG may promote glioma cell proliferation. To confirm our hypothesis, we performed lentiviral infection to stably overexpress or knockdown CapG in U87 and U251 glioma cells. The efficiency of ectopic expression of CapG was verified by real-time PCR (data not shown) and western blotting (Fig. 4A). Thus shCAPG-1 and shCAPG-3 were chosen for the following study according to their silencing efficiency.
Figure 4.
Overexpression and depletion of CapG in U87 and U251 human glioma cell lines. (A) Protein level of CapG was analyzed by western blot assays. GAPDH served as a loading control. (B) The cell proliferation curve of CapG overexpression and knockdown cells performed by CCK-8 assay. (C) Colony formation assay performed in CapG overexpression and knockdown cells. The assay was determined 3 times with triplicate samples, and bars represent the number of colonies (right panels); *P<0.05; **P<0.01; P<0.001.
We then studied the effect of CapG on cellular proliferation in U87 and U251 cells using CCK-8 assay. The results showed that overexpression of CapG significantly promoted the proliferation in comparison with the control groups, whereas knockdown of endogenous CapG markedly inhibited cellular growth in both cell lines (Fig. 4B). Colony formation assay consistently showed that overexpression of CapG in U251 cells markedly increased the number of colonies, whereas knockdown of CapG significantly decreased the colony numbers compared with the controls (Fig. 4C).
CapG regulates G1/S phase progression of the glioma cell cycle
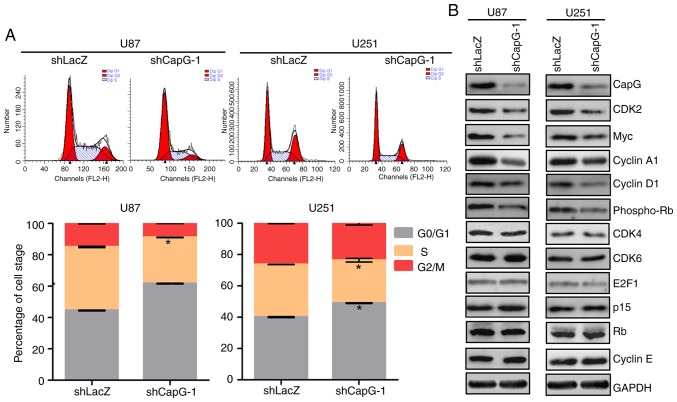
To elucidate the mechanism by which depletion of CapG inhibits cell proliferation in U87 and U251 cells, we performed flow cytometry to analyze whether the inhibition is correlated with specific cell cycle changes. Compared with the shLacZ groups, U87 and U251 cells with CapG depletion showed marked increases in the percentage of cell in the G0/G1 phase and substantial decrease in the S phase (Fig. 5A). To further investigate the molecules involved in the changes in the cell cycle, we determined the expression of several important regulators of the G1 phase by western blot assays (30–32). As shown in Fig. 5B, we observed that depletion of CapG significantly suppressed CDK2, Myc, cyclin A1 and D1, and phospho-RB expression in the U87 and U251 cells. However, the expression levels of cyclin E, CDK4, CDK6, E2F1, p15 and RB had no significant changes after CapG depletion. Collectively, these results suggest that knockdown of endogenous CapG induces G0/G1 cell cycle arrest in glioma cells.
Figure 5.
CapG regulates G1/S phase progression of glioma cell cycle. (A) The cell cycle distribution of U87 and U251 glioma cells after knockdown of endogenous CapG expression. Values are means ± SEM; *P<0.05. (B) The effects of CapG knockdown on cell cycle proteins in the G1/S transition were measured by western blot assays. GAPDH was used as a loading control.
Ectopic CapG expression affects glioma cell migration and invasiveness
Recent research has shown that suppression of CapG expression influences the metastasis of prostate cancer cells (20,29). We then examined the impact of CapG on glioma cell migration and invasiveness in vitro. In wound healing assays, CapG overexpression increased cell migration markedly within 24 or 48 h compared to the control cells, while silencing of CapG resulted in slower wound healing (Fig. 6A-D). However, the effect on cell invasiveness was next assessed by Transwell invasion assays. Consistent with the abovementioned migration analyses, CapG upregulation substantially increased the invasive ability of the glioma cells while CapG depletion resulted in a significant decrease in invasiveness of the cells (Fig. 6E and F). Therefore, glioma cell migration and invasiveness were positively correlated with CapG expression levels.
Figure 6.
CapG expression modulates glioma cell migration and invasiveness in U87 and U251. (A-D) In wound healing assays, CapG depletion cells showed a significant decrease in motility. Consistent with that, CapG overexpression caused a significant earlier wound closure of the scratch. (E and F) In Transwell invasion assay, CapG overexpression had a higher effect on invasion than CapG depletion. Scale bars are depicted in figures; *P<0.05; **P<0.01.
Discussion
In the present study, we demonstrated the prognostic value of CapG as well as its important role in proliferation and metastasis of glioma. In the TCGA data analysis, we attempted to explore the correlation of CapG with common genetic alterations of GBM. Notably, we noted that CapG expression was correlated to several of them (amplifications of CDK6, EGFR, MET and SYK, mutations of PTEN and RB1), and the correlation was only observed in non-proneural or proneural subtype. These data provide a clue for further study of the regulation of CapG expression in GBM patients. Moreover, we investigated the influence of differential CapG expression on GBM patient prognosis stratified by these molecular alterations. Kaplan-Meier analysis suggested that the prognostic significance of CapG was dependent on the status of EGFR or CDK6 alteration.
EGFR is a vital oncogene found in many malignancies, and the majority of GBM show activated EGFR signaling through its amplification or genetic alteration, which drives the proliferation and metastasis of glioma cells (33). However, EGFR, CDK6, a G1 cell cycle regulatory protein, is upregulated in glioma tissues and mediates glioma cell proliferation and migration (34). As the expression of CapG was only correlated with the amplification of EGFR/CDK6 in the proneural subtype, we inferred that the proneural gene expression profile mediated the interaction between EGFR/CDK6 and CapG. Therefore, the change in CapG expression level could further influence the proliferation and metastasis of glioma.
CapG, an actin binding protein, modulates the dynamics of the actin cytoskeleton and aberrant turnover of the actin cytoskeleton, which are closely associated with cell proliferation and motility (35). There is evidence that the CapG expression level is related to tumor cell proliferation and metastasis (36,37), and may be valuable for evaluating tumor progression and prognosis. For example, overexpression of CapG may be associated with tumor progression, as CapG protein is significant increased in pancreatic ductal adenocarcinoma and lung adenocarcinoma (38–40). CapG expression was also found to be upregulated in ovarian cancer and is possibly associated with survival outcome and advanced disease stage (20). In the present study, the mRNA and protein levels of CapG were significantly increased in human glioma, and higher CapG expression was an independent prognostic factor for predicting unfavorable prognosis. In subsequent function validation, CapG promoted cell proliferation and mediated G1/S transition in glioma cell lines U87 and U251. These results suggest that CapG is a putative oncogene in human glioma.
Recent findings have demonstrated that cancer metastasis and diffusion rely on increased tumor cell motility by accelerating cytoplasmic actin turnover (35). In the present study, the wound healing and Transwell invasion assays indicated that CapG plays a critical role in glioma migration and invasiveness. It has been reported that nuclear complexes are significant for migration and invasiveness (41). Furthermore, nuclear complexes carry nuclear actin and are modified by actin binding protein (41). Given that CapG is an actin binding protein, and elevated CapG is found at the tumor invasion interface zone in breast cancer (42), we postulated that CapG expression may directly modulate cell migration and invasion.
In summary, the present study illustrated that CapG expression was significantly elevated in human glioma, and it predicted an unfavorable prognosis of glioma as a novel and independent prognostic factor, which was confirmed in a TMA analyses of a Chinese cohort and the independent TCGA dataset. In addition, the expression profile and prognostic value of CapG were correlated with different subtypes and several molecular features of GBM. Furthermore, CapG plays an important role in the proliferation and metastasis of glioma cells, suggesting its application as a promising therapeutic target of human glioma.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant nos. 81772657, 81372706, 81372235 and 81071739). We thank Bo Zhu (School of Life Sciences, Fudan University) for his help with the Transwell invasion assays. We further thank all participants recruited in the present study.
Glossary
Abbreviations
- CapG
capping actin protein, gelsolin-like
- GBM
glioblastoma
- CCK-8
Cell Counting Kit-8
- TMA
tissue microarray
- TCGA
The Cancer Genome Atlas
- LGG
low-grade glioma
- HGG
high-grade glioma
- OS
overall survival
- PFS
progression-free survival
- HR
hazard ratio
- CI
confidence interval
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westphal M, Lamszus K. The neurobiology of gliomas: From cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12:495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, Van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: No longer a dead end? Nat Rev Clin Oncol. 2013;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Zhang W, Chen D, Lv Y, Zheng J, Lilljebjorn H, Ran L, Bao Z, Soneson C, Sjogren HO, et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA; Proc Natl Acad Sci USA; 2014; pp. 3538–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan W, Zhang W, You G, Zhang J, Han L, Bao Z, Wang Y, Liu Y, Jiang C, Kang C, et al. Molecular classification of gliomas based on whole genome gene expression: A systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neuro Oncol. 2012;14:1432–1440. doi: 10.1093/neuonc/nos263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao YF, Mao XY, Zhu T, Mao CX, Liu ZX, Wang ZB, Li L, Li X, Yin JY, Zhang W, et al. COL3A1 and SNAP91: Novel glioblastoma markers with diagnostic and prognostic value. Oncotarget. 2016;7:70494–70503. doi: 10.18632/oncotarget.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Chen C, Zhang X, Liu Q, Xu JL, Zhang HR, Yao XH, Jiang T, He ZC, Ren Y, et al. Primate-specific miR-663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clin Cancer Res. 2014;20:1803–1813. doi: 10.1158/1078-0432.CCR-13-2284. [DOI] [PubMed] [Google Scholar]
- 11.Meng J, Li P, Zhang Q, Yang Z, Fu S. A radiosensitivity gene signature in predicting glioma prognostic via EMT pathway. Oncotarget. 2014;5:4683–4693. doi: 10.18632/oncotarget.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng D, Chen Y, Yun D, Zhao Y, Wang J, Xu T, Li X, Wang Y, Yuan L, Sun R, et al. High expression of N-myc (and STAT) interactor predicts poor prognosis and promotes tumor growth in human glioblastoma. Oncotarget. 2015;6:4901–4919. doi: 10.18632/oncotarget.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network, corp-author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutledge WC, Kong J, Gao J, Gutman DA, Cooper LA, Appin C, Park Y, Scarpace L, Mikkelsen T, Cohen ML, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res. 2013;19:4951–4960. doi: 10.1158/1078-0432.CCR-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Chen Y, Wang Y, Yang J, Zhu VF, Liu Y, Cui X, Chen L, Yan W, Jiang T, et al. ZIP4 is a novel molecular marker for glioma. Neuro Oncol. 2013;15:1008–1016. doi: 10.1093/neuonc/not042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Meng D, Wang H, Sun R, Wang D, Wang S, Fan J, Zhao Y, Wang J, Yang S, et al. VAMP8 facilitates cellular proliferation and temozolomide resistance in human glioma cells. Neuro Oncol. 2015;17:407–418. doi: 10.1093/neuonc/nou219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: Key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Impe K, De Corte V, Eichinger L, Bruyneel E, Mareel M, Vandekerckhove JL, Gettemans J. The nucleo-cytoplasmic actin-binding protein CapG lacks a nuclear export sequence present in structurally related proteins. J Biol Chem. 2003;278:17945–17952. doi: 10.1074/jbc.M209946200. [DOI] [PubMed] [Google Scholar]
- 19.Witke W, Li W, Kwiatkowski DJ, Southwick FS. Comparisons of CapG and gelsolin-null macrophages: Demonstration of a unique role for CapG in receptor-mediated ruffling, phagocytosis, and vesicle rocketing. J Cell Biol. 2001;154:775–784. doi: 10.1083/jcb.200101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahl E, Sadr-Nabavi A, Klopocki E, Betz B, Grube S, Kreutzfeld R, Himmelfarb M, An HX, Gelling S, Klaman I, et al. Systematic identification and molecular characterization of genes differentially expressed in breast and ovarian cancer. J Pathol. 2005;205:21–28. doi: 10.1002/path.1687. [DOI] [PubMed] [Google Scholar]
- 21.Fan S, Meng D, Xu T, Chen Y, Wang J, Li X, Chen H, Lu D, Chen J, Lan Q. Overexpression of SLC7A7 predicts poor progression-free and overall survival in patients with glioblastoma. Med Oncol. 2013;30:384. doi: 10.1007/s12032-012-0384-8. [DOI] [PubMed] [Google Scholar]
- 22.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 23.Ohuchida K, Mizumoto K, Ishikawa N, Fujii K, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. The role of S100A6 in pancreatic cancer development and its clinical implication as a diagnostic marker and therapeutic target. Clin Cancer Res. 2005;11:7785–7793. doi: 10.1158/1078-0432.CCR-05-0714. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Malliri A, Van Es S, Huveneers S, Collard JG. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J Biol Chem. 2004;279:30092–30098. doi: 10.1074/jbc.M401192200. [DOI] [PubMed] [Google Scholar]
- 26.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Wan XC, Chen HB, Yang S, Liu YY, Mo WJ, Meng DL, Du WT, Huang Y, Wu H, et al. Identification of miR-133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clin Cancer Res. 2014;20:2312–2325. doi: 10.1158/1078-0432.CCR-13-1588. [DOI] [PubMed] [Google Scholar]
- 28.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin-D to the retinoblastoma gene-product (prb) and pRb phosphorylation by the cyclin D-dependent kinase Cdk4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 31.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/MCB.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinberg RA. The retinoblastoma protein and cell-cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 33.Francis JM, Zhang CZ, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, Homer H, Haidar S, Blumenstiel B, Pedamallu CS, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Lei B, Huang S, Zheng M, Liu Z, Li Z, Deng Y. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am J Transl Res. 2015;7:1747–1764. [PMC free article] [PubMed] [Google Scholar]
- 35.Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- 36.Wu JH, Tian XY, Hao CY. The significance of a group of molecular markers and clinicopathological factors in identifying colorectal liver metastasis. Hepatogastroenterology. 2011;58:1182–1188. doi: 10.5754/hge11380. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Hong X, Zhao J, Teng Y, Zheng J, Chen H, Chen H, Li H. Gelsolin-like actin-capping protein is associated with patient prognosis, cellular apoptosis and proliferation in prostate cancer. Biomark Med. 2016;10:1251–1260. doi: 10.2217/bmm-2016-0186. [DOI] [PubMed] [Google Scholar]
- 38.Shao F, Zhang R, Dong L, Ying K. Overexpression of gelsolin-like actin-capping protein is associated with progression of lung adenocarcinoma. Tohoku J Exp Med. 2011;225:95–101. doi: 10.1620/tjem.225.95. [DOI] [PubMed] [Google Scholar]
- 39.Zhu WY, Hunag YY, Liu XG, He JY, Chen DD, Zeng F, Zhou JH, Zhang YK. Prognostic evaluation of CapG, gelsolin, P-gp, GSTP1, and Topo-II proteins in non-small cell lung cancer. Anat Rec. 2012;295:208–214. doi: 10.1002/ar.21523. [DOI] [PubMed] [Google Scholar]
- 40.Thompson CC, Ashcroft FJ, Patel S, Saraga G, Vimalachandran D, Prime W, Campbell F, Dodson A, Jenkins RE, Lemoine NR, et al. Pancreatic cancer cells overexpress gelsolin family-capping proteins, which contribute to their cell motility. Gut. 2007;56:95–106. doi: 10.1136/gut.2005.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubert T, Van Impe K, Vandekerckhove J, Gettemans J. The F-actin filament capping protein CapG is a bona fide nucleolar protein. Biochem Biophys Res Commun. 2008;377:699–704. doi: 10.1016/j.bbrc.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 42.Kang S, Kim MJ, An H, Kim BG, Choi YP, Kang KS, Gao MQ, Park H, Na HJ, Kim HK, et al. Proteomic molecular portrait of interface zone in breast cancer. J Proteome Res. 2010;9:5638–5645. doi: 10.1021/pr1004532. [DOI] [PubMed] [Google Scholar]