Abstract
Increasing evidence has experimentally proved the competitive endogenous RNA (ceRNA) hypothesis that long non-coding RNA (lncRNA) can affect the expression of RNA targets by competitively combining microRNA (miRNA) via miRNA response elements. However, an extensive ceRNA network of thyroid carcinoma in a large cohort has not been evaluated. We analyzed the RNAseq and miRNAseq data of 348 cases of primary papillary thyroid cancer (PTC) patients with clinical information downloaded from The Cancer Genome Atlas (TCGA) project to search for potential biomarkers or therapeutic targets. A computational approach was applied to build an lncRNA-miRNA-mRNA regulatory network of PTC. In total, 780 lncRNAs were detected as collectively dysregulated lncRNAs in all 3 PTC variants compared with normal tissues (fold change >2 and false discovery rate <0.05). The interactions among 45 lncRNAs, 13 miRNAs and 86 mRNAs constituted a ceRNA network of PTC. Nine out of the 45 aberrantly expressed lncRNAs were related to the clinical features of PTC patients. However, the expression levels of 3 lncRNAs (LINC00284, RBMS3-AS1 and ZFX-AS1) were identified to be tightly correlated with the patients overall survival (log-rank, P<0.05). The present study identified a list of specific lncRNAs associated with PTC progression and prognosis. This complex ceRNA interaction network in PTC may provide guidance for better understanding the molecular mechanisms underlying PTC.
Keywords: papillary thyroid cancer, long non-coding RNA, competitive endogenous RNA, microRNA, The Cancer Genome Atlas
Introduction
Thyroid cancer (TC) is the major endocrine malignancy with estimated 298,000 new cases and 40,000 deaths worldwide in 2012 (1). Thyroid carcinoma is the fastest-growing cancer with the increasing morbidity rate >5% due to the advanced diagnostic techniques as well as multiple risk factors. Although the 5-year survival rate for patients with thyroid carcinoma is relatively higher than other malignant neoplasms, the mortality rate of TC has remained ever-increasing over the past decade (2,3). Thyroid cancer is a heterogeneous disease of 3 histological subtypes: papillary TC (PTC), follicular TC (FTC) and anaplastic TC (ATC) (4). As the most primary malignant thyroid tumor, PTC accounts for up to 80% of total cases. Although PTC is generally effectively treated with thyroidectomy and hormone therapy, large numbers of unresponsive patients suffer from progressive disease and metastases (5). Thus, it is urgent to explore the molecular mechanisms underlying PTC for developing effective diagnostic and therapeutic targets.
Non-coding RNAs (ncRNAs) are generally defined as a wide class of RNAs without protein-coding function, but with universal expression in organisms. ncRNAs are loosely split into small ncRNAs and long non-coding RNAs (lncRNAs), both of which have regulatory functions in the various biological processes (6). The well-documented microRNAs (miRNAs; ~22 nucleotides long) belong to small ncRNAs and are regarded as vital regulators of cellular gene expression network (7). lncRNAs are transcripts >200 nucleotides with tissue-specific expression, but neither has significant open reading frames nor translate into proteins (8). Accumulating studies over the past decades have altered the understanding of lncRNA from ‘dark matter’ to important transcriptional and post-transcriptional regulators. Abnormally expressed lncRNAs participate in tumorigenesis by interrupting biological processes of both oncogenic and tumor suppressive pathways (9). In 2011, Pandolfi et al put forward the competitive endogenous RNA (ceRNA) hypothesis that specific RNAs (including lncRNA, mRNA, pseudogene and circular RNA) can impair miRNA activity by sponging miRNA via common miRNA response elements (MREs) and upregulate target RNA expression subsequently (10). This hypothesis introduced a new RNA-RNA crosstalk theory that can generally regulate gene expression. In addition, the promising area of disclosing the landscape of the ceRNA mechanism has evoked much interest. Therefore, the lncRNA-miRNA-mRNA networks that may lead to tumor development and progression have already been constructed in many malignancies, including gastric cancer (11), hepatocellular carcinoma (12), glioblastoma (13) and lung adenocarcinoma (14).
Recent studies have preliminarily revealed several ceRNA regulatory interactions and corresponding mechanisms in thyroid carcinoma. lncRNA H19 was certified to act as ceRNA by sponging miR-17-5p to upregulate the target gene YES1, demonstrating a potential ceRNA crosstalk including H19, miR-17-5p, and YES1 in thyroid cancer pathogenesis (15). A bioinformatic ceRNA analysis revealed that stem cell factor SOX2 could be functionally co-regulated with other genes by crosstalk mediated by several miRNAs in anaplastic thyroid carcinoma (16). Furthermore, both a ceRNA network built by microarray analysis of 5-paired clinical samples and a protein-coding modulatory network that regulated immune responses have been reported in PTC (17,18). However, a whole genome-wide analysis of the lncRNA-miRNA-mRNA regulatory network of papillary thyroid cancer with large-scale sample size is still lacking.
In the present study, we comprehensively investigated both RNA and miRNA sequencing data of 348 PTC primary tumor tissues and 58 non-cancerous thyroid samples from The Cancer Genome Atlas (TCGA) data matrix to obtain the aberrantly expressed lncRNAs, miRNAs, and mRNAs. Next, an lncRNA-associated ceRNA network of papillary thyroid cancer was built based on ceRNA theory. The expression levels of the important lncRNAs involved in ceRNA crosstalk were also assessed for their prognostic values in patients with papillary thyroid cancer.
Materials and methods
Date collection
PTC patients (502) were obtained from the TCGA consortium. Inclusion criteria included: i) patients diagnosed with PTC but with no other malignancies; ii) patients with complete clinical data, including age, sex, race, the American Joint Committee on Cancer (AJCC) TNM and pathologic stage; iii) patients with overall survival time <2,000 days; and iv) patients with complete lncRNA, mRNA and miRNA expression profiles. In total, 348 patients (cohort T) who were pathologically diagnosed as PTC and 58 normal samples (cohort N) were enrolled in downstream analysis. The RNAseq and miRNAseq data (level 3) generated from IlluminaHiseq_RNASeq and IlluminaHiseq_miRNASeq sequencing platform was downloaded from TCGA data portal. The present study followed the publication guidelines of TCGA Research Network (http://cancergenome.nih.gov/publications/publicationguidelines). Thus, no further ethical approvals were required.
Analysis of differentially expressed genes
Differential expression analysis was carried out to identify differentially expressed lncRNAs, mRNAs and miRNAs (DElncRNAs, DEmRNAs, DEmiRNAs) between cohort T and N, by the R/Bioconductor package of edgeR (19) with the cut-off value of |log2FC|>1 and FDR <0.05 (FC, fold change; FDR, false discovery rate).
CeRNA network construction
lncRNA-miRNA interactions were predicted by miRcode (http://www.mircode.org/) (20). TargetScan (http://www.TargetScan.org/mamm_31/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) and miRanda (http://www.microrna.org/microrna/home.do) were cooperatively utilized to predict the mRNAs targeted by miRNAs (21,22). According to the above lncRNA/miRNA and miRNA/mRNA interactions, the Cytoscape (version 3.5.1) was utilized to build and visualize the lncRNA-miRNA-mRNA network (23).
Functional annotation
The Database for Annotation Visualization and Integrated Discovery (DAVID) online tool (https://david.ncifcrf.gov/) was used to conduct the functional and pathway enrichment analyses. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to detect the potential biological functions and pathways of the DEmRNAs involved in the network (P-value <0.05).
Statistical analysis
Unpaired t-test was applied to identify differentially expressed genes between cohort T and N, and to find out DElncRNAs between different pathological subgroups of cohort T. The univariate Cox proportional hazards regression was used to estimate the expression of DElncRNAs in ceRNA network with overall survival. Kaplan-Meier survival analysis was used to analyze the correlation between lncRNA expression and PTC patient prognosis.
Results
Patient characteristics
The clinical and pathological information of patients in cohort T was summarized in Table I. The median age of all patients in cohort T was ~48 years. The distributions of sex and race showed that white female patients accounted for the majority in the male/female ratio of 2.8/1 and white race ratio of 80.2%. The 3 histopathological variants, classical PTC (CPTC), follicular PTC (FPTC) and tall-cell PTC (TCPTC) represented 75.9, 14.3 and 9.8%, respectively. The proportions and distributions of these patients clinical characteristics were consistent with previous studies (24,25).
Table I.
Demographic and clinical characteristics of 348 patients with papillary thyroid cancer in cohort T.
| Parameter/feature | Cohort T (n=348) (%) |
|---|---|
| Age, years (mean ± SD) | 48.1±15.9 |
| Sex | |
| Male | 92 (26.4) |
| Female | 256 (73.6) |
| Race | |
| Asian | 46 (13.2) |
| Black or African American | 22 (6.3) |
| American Indian or | 1 (0.3) |
| Alaska native | |
| White | 279 (80.2) |
| Subtype | |
| CPTC | 264 (75.9) |
| FPTC | 50 (14.3) |
| TCPTC | 34 (9.8) |
| Pathologic stage | |
| I | 192 (55.2) |
| II | 28 (8.0) |
| III | 88 (25.3) |
| IV | 40 (11.5) |
| Tumor size | |
| T1 | 105 (30.2) |
| T2 | 108 (31.0) |
| T3 | 116 (33.3) |
| T4 | 17 (4.9) |
| TX | 2 (0.6) |
| Lymph node | |
| N0 | 164 (47.2) |
| N1 | 157 (45.2) |
| NX | 27 (7.6) |
| Metastasis status | |
| M0 | 231 (66.4) |
| M1 | 6 (1.7) |
| MX | 111 (31.9) |
SD, standard deviation; PTC, papillary thyroid cancer; CPTC, classical PTC; FPTC, follicular PTC; TCPTC, tall-cell PTC.
Differentially expressed lncRNA, mRNA and miRNA
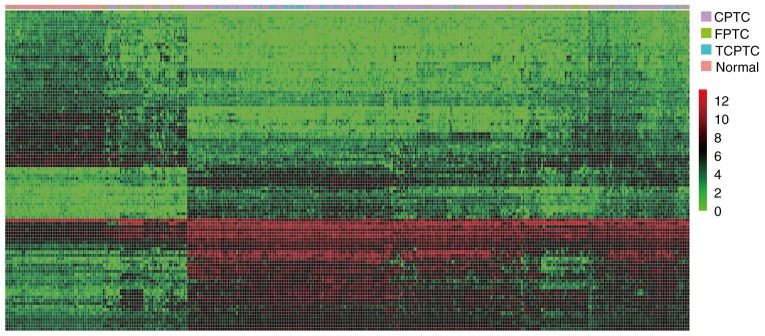
Considering the dissimilarity among the 3 PTC variants, RNA expression levels in the 3 PTC variants (cohort T) vs. normal tissues (cohort N) were investigated, respectively. Genes with an absolute fold change >2 and FDR value <0.05 were considered as discriminatively expressed. As a result, a total of 780 lncRNAs and 1,700 mRNAs were detected to be collectively dysregulated in carcinoma tissues of 3 PTC variants compared with normal tissues. Among them, 440 and 340 lncRNAs were commonly elevated and downregulated, meanwhile, 973 and 727 mRNAs were collectively upregulated and downregulated, respectively. Filtering analysis with the same criteria (|log2FC|>1 and FDR <0.05) also identified 43 miRNAs differentially expressed between 3 PTC variants and normal thyroid tissues, among them 27 upregulated and 16 downregulated (Fig. 1) (data not shown). To visualize the most significant lncRNAs, we selected the top 50 upregulated and top 50 downregulated lncRNAs to construct expression heatmaps (Fig. 2) (data not shown). The data suggested that the expression profiles of DElncRNAs can distinguish PTC tissues from normal tissues.
Figure 1.
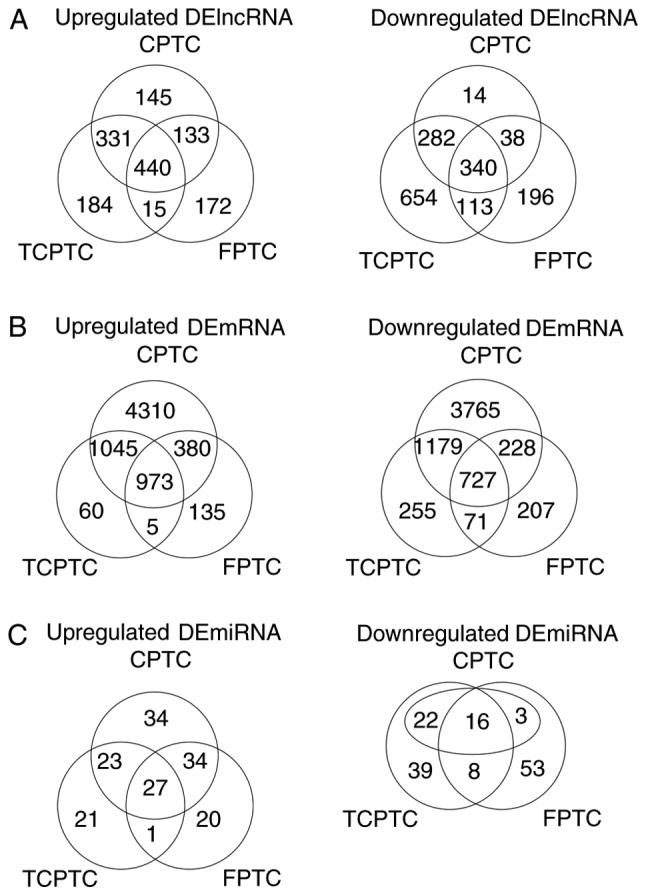
Venn analysis of differentially expressed (DE) lncRNA (A), mRNA (B), and miRNA (C) in 3 PTC variants (CPTC, FPTC and TCPTC), compared with normal tissues.
Figure 2.
Heatmap of the top 50 collectively upregulated and top 50 collectively downregulated DElncRNAs in 3 PTC variants, compared with normal tissues. Each column represents one sample, and each row indicates one lncRNA. Purple, green, blue and red stripes represent CPTC, FPTC, TCPTC and non-tumor samples, respectively. Ascending normalized expression level is colored from green to red.
ceRNA network in PTC
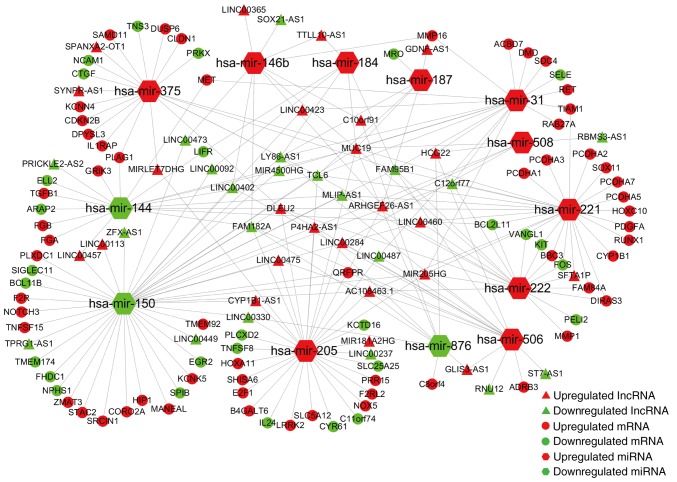
In the next step, we predicted the potential interactions among the above collectively dysregulated genes based on ceRNA hypothesis by computational analysis to further understand the function of DElncRNAs. Thirteen specific DEmiRNAs were identified to interact with 45 DElncRNAs through miRNA response elements by miRcode online tools (Table II). To improve the predictive accuracy, TargetScan, miRTarBase and miRanda databases were combined to predict the candidate mRNA targets of the 11 DEmiRNAs. After excluding the mRNAs that were not involved in the 1,700 DEmRNAs, 86 mRNAs were enrolled to create the ceRNA network (Table III). Finally, the ceRNA network that involved 45 lncRNAs, 13 miRNAs and 86 mRNAs was visualized using Cytoscape software based on the interactions among lncRNAs, miRNAs and mRNAs in Tables II and III (Fig. 3).
Table II.
Putative miRNAs that may target lncRNAs by miRNA response elements.
| lncRNA | miRNAs | lncRNA | miRNAs |
|---|---|---|---|
| AC108463.1 | miR-221, miR-222, miR-876, miR-150, miR-205 | LINC00475 | miR-506, miR-150, miR-205 |
| ARHGEF26-AS1 | miR-146b, miR-187, miR-508, miR-205 | LINC00487 | miR-31, miR-506, miR-205 |
| C10orf91 | miR-146b, miR-876 | LY86-AS1 | miR-375, miR-31, miR-187, miR-506, miR-150, miR-184 |
| C12orf77 | miR-221, miR-222, miR-31, miR-150 | MIR181A2HG | miR-205 |
| CYP1B1-AS1 | miR-150, miR-205 | MIR205HG | miR-221, miR-222, miR-31, miR-506, miR-150, miR-205 |
| DLEU2 | miR-221, miR-222, miR-144, miR-375, miR-506, miR-150, miR-205 | MIR4500HG | miR-144, miR-31, miR-150 |
| FAM182A | miR-146b, miR-144, miR-876, miR-506, miR-150, miR-205 | MIRLET7DHG | miR-375, miR-205 |
| FAM95B1 | miR-221, miR-222, miR-375, miR-31, miR-506, miR-150, miR-184 | MLIP-AS1 | miR-221, miR-222, miR-144, miR-150 |
| GDNF-AS1 | miR-187 | MUC19 | miR-146b, miR-221, miR-222, miR-144, miR-375, miR-31, miR-187, miR-508, miR-876, miR-150, miR-205, miR-184 |
| GLIS3-AS1 | miR-876, miR-506 | P4HA2-AS1 | miR-508, miR-876, miR-150, miR-205 |
| HCG22 | miR-31, miR-508, miR-876, miR-506 | PRICKLE2-AS2 | miR-144 |
| LINC00092 | miR-150, miR-184 | RBMS3-AS1 | miR-508 |
| LINC00113 | miR-150 | RNU12 | miR-506 |
| LINC00237 | miR-205 | SFTA1P | miR-221, miR-222 |
| LINC00284 | miR-508, miR-506, miR-205 | SOX21-AS1 | miR-146b |
| LINC00330 | miR-876, miR-150, miR-205 | SPANXA2-OT1 | miR-375 |
| LINC00365 | miR-146b | ST7-AS1 | miR-506 |
| LINC00402 | miR-146b, miR-506, miR-150 | SYNPR-AS1 | miR-375 |
| LINC00423 | miR-31, miR-150 | TCL6 | miR-221, miR-222, miR-144, miR-375, miR-31, miR-187, miR-150, miR-205 |
| LINC00449 | miR-150 | TPRG1-AS1 | miR-150 |
| LINC00457 | miR-144, miR-150 | TTLL10-AS1 | miR-146b, miR-184 |
| LINC00460 | miR-221, miR-222, miR-150 | ZFX-AS1 | miR-150 |
| LINC00473 | miR-146b, miR-150 |
Table III.
miRNAs targeting mRNAs in papillary thyroid carcinoma.
| miRNA | mRNAs targeted by miRNA |
|---|---|
| miR-144 | FGA, ARAP2, PLAG1, FGB, MET, TGFB1, LIFR, BCL2L11, ELL2, GRIK3 |
| miR-146b | MMP16 |
| miR-150 | BCL11B, STAC2, CORO2A, TMEM174, SPIB, F2R, PLXDC1, KCNK5, HIP1, NOTCH3, FHDC1, SRCIN1, TNFSF15, TMEM92, EGR2, NPHS1, MANEAL, QRFPR, ZMAT3, SIGLEC11 |
| miR-184 | LIFR |
| miR-187 | MRO |
| miR-205 | NOX5, TNFSF8, KCTD16, CYR61, SLC25A25, PLCXD2, SLC5A12, C11orf74, F2RL2, LRRK2, E2F1, HOXA11, IL24, B4GALT6, PRR15, SHISA6 |
| miR-221 | PCDHA2, SOX11, PCDHA1, FOS, PCDHA3, PCDHA7, FAM84A, RUNX1, PDGFA, KIT, PCDHA5, TIAM1, DIRAS3, BBC3, BCL2L11, VANGL1, HOXC10, CYP1B1 |
| miR-222 | BBC3, DIRAS3, FOS, VANGL1, KIT, PELI2, FAM84A, MMP1, BCL2L11 |
| miR-31 | DMD, RET, ACBD7, SELE, MMP16, RAB27A, TIAM1, MET, SDC4 |
| miR-375 | KCNN4, CTGF, DUSP6, TNS3, IL1RAP, PLAG1, DPYSL3, NCAM1, PRKX, SAMD11, CLDN1, CDKN2B |
| miR-506 | ADRB3, QRFPR |
| miR-876 | C8orf4 |
Figure 3.
The lncRNA-miRNA-mRNA ceRNA network of PTC. Different colors represent expression levels and different shapes represent different RNA types.
Functional analysis of DEmRNAs in the ceRNA network
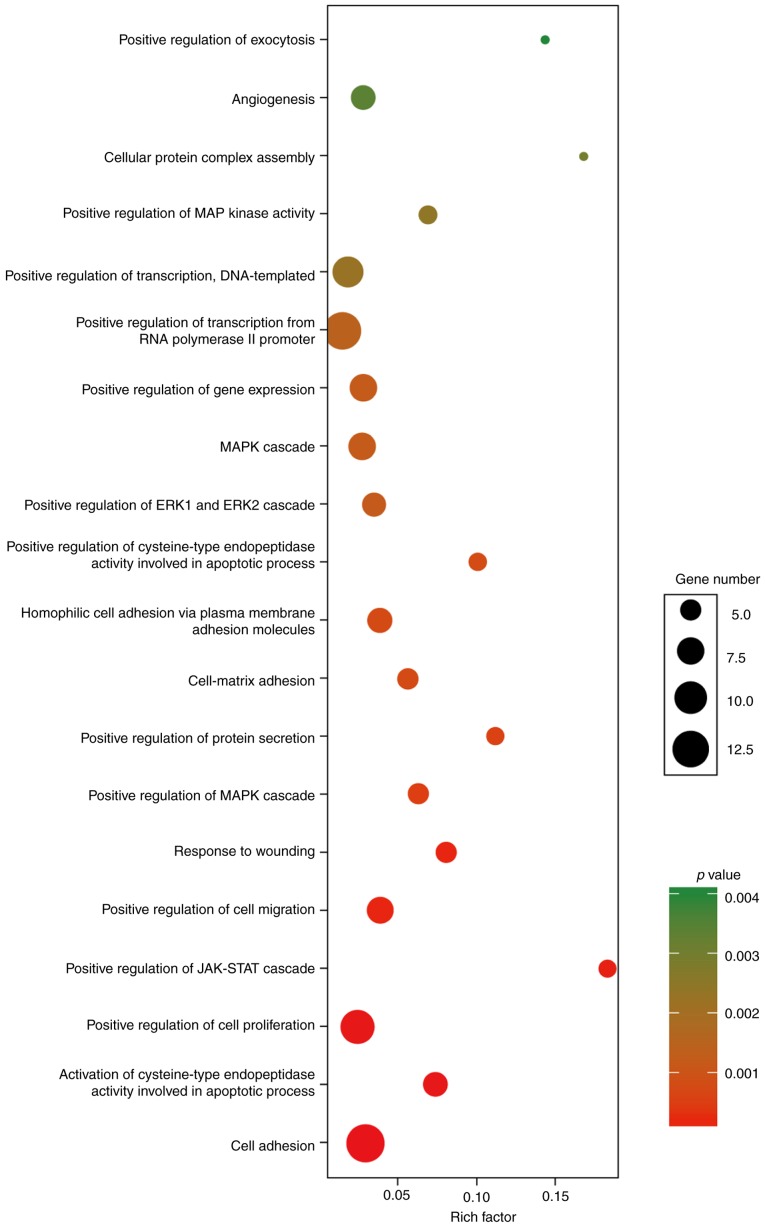
The function analysis revealed that the 86 DEmRNAs in the above ceRNA network were enriched in 60 GO biological process categories and 7 KEGG categories (P<0.05). The significant GO biological processes of dysregulated genes were 3 cell adhesion terms (GO:0007155, GO:0007160 and GO:007156) (Fig. 4). Table IV contains the significantly enriched pathways of these DEmRNAs by KEGG analysis. There were two cancer-related pathways, including pathways in cancer and miRNAs in cancer. Notably, transcriptional factor E2F1 had an elevated expression in PTC tissues and was involved as the key component in both cancer-related pathways.
Figure 4.
Top 20 enrichment Gene Ontology (GO) biological process terms of DEmRNAs involved in the ceRNA network. The size of balls represents gene number and color represents P-value.
Table IV.
KEGG pathways enriched by DEmRNA involved in the ceRNA network.
| Pathway ID | Description | P-value | Numbers of DEmRNAs |
|---|---|---|---|
| hsa05200 | Pathways in cancer | 8.99E-05 | E2F1, FOS, RET, CDKN2B, PDGFA, MET, KIT, RUNX1, TGFB1, MMP1, F2R |
| hsa04060 | Cytokine-cytokine receptor interaction | 4.12E-04 | PDGFA, IL1RAP, MET, TNFSF15, LIFR, KIT, TGFB1, TNFSF8 |
| hsa04144 | Endocytosis | 0.0043302 | ADRB3, RET, MET, KIT, ARAP2, TGFB1, F2R |
| hsa05206 | MicroRNAs in cancer | 0.0070069 | NOTCH3, E2F1, CYP1B1, PDGFA, MET, MMP16, BCL2L11 |
| hsa05166 | HTLV–I infection | 0.018641 | E2F1, FOS, EGR2, CDKN2B, PDGFA, TGFB1 |
| hsa05144 | Malaria | 0.0354242 | MET, SELE, TGFB1 |
| hsa04015 | Rap1 signaling pathway | 0.0377107 | TIAM1, PDGFA, MET, KIT, F2R |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
lncRNAs in relation to clinical outcome
To further understand whether dysregulated lncRNAs were correlated with patient outcome, the 45 DElncRNAs involved in the network were analyzed according to clinical characteristics including sex, race, TNM staging system and pathologic stage. The expression levels of 9 lncRNAs were aberrantly expressed in clinical feature comparisons of cohort T (|log2FC|>1 and FDR<0.05) (Table V). Among them, we identified 3 lncRNAs (FAM182A, LINC00402 and LINC00473) differentially expressed in race subgroup. Four upregulated lncRNAs (MIR205HG, C12orf7, TCL6 and LINC00460) and 4 downregulated lncRNAs (GDNF-AS1, LINC00402, LINC00473 and LINC00237) were markedly related to tumor progression. LINC00473 may not only inhibit tumor growth (T3+T4 vs. T1+T2), but also decrease in individuals with poor pathologic stage, suggesting its potential negative role in tumor development of PTC. Overexpression lncRNA LINC00460 was identified to be correlated with the poor pathologic stage.
Table V.
The correlation between lncRNA in the ceRNA network and patient characteristics.
| Comparisons | Downregulated | Upregulated |
|---|---|---|
| Race | FAM182A, LINC00402 LINC00473 | |
| (Asian vs. White) | ||
| Tumor size | GDNF-AS1, LINC00402, LINC00473 | |
| (T3+T4 vs. T1+T2) | ||
| Lymph node | LINC00237 | MIR205HG, C12orf77, TCL6 |
| (N1 vs. N0) | ||
| Pathologic stage | LINC00473 | LINC00460 |
| (Stage III+IV vs. stage I+II) |
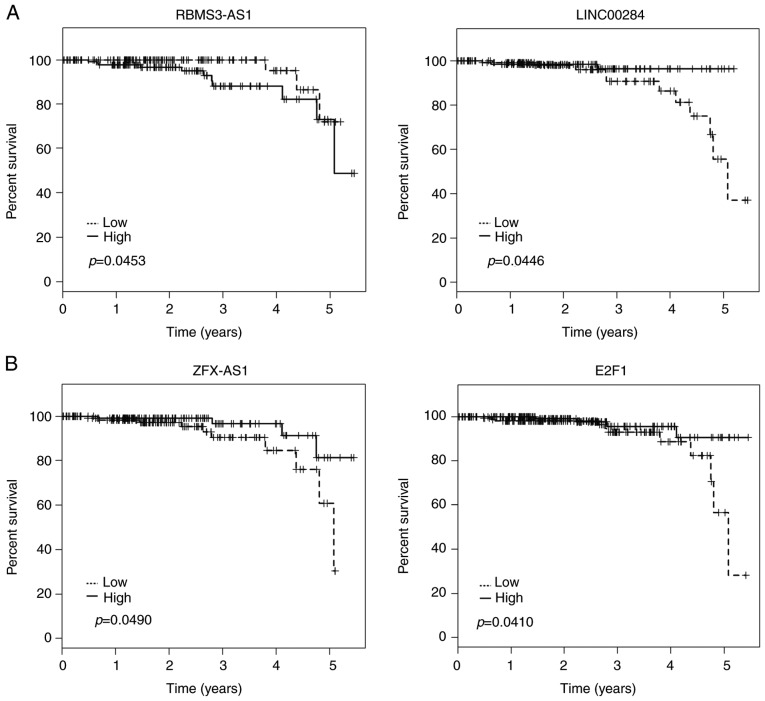
Kaplan-Meier analysis was applied to investigate overall survival for DElncRNAs in cohort T. Among the 45 DElncRNAs in the ceRNA network, 3 lncRNAs (LINC00284, RBMS3-AS1 and ZFX-AS1) were identified to be associated with overall survival by univariate Cox regression analysis (log-rank, P<0.05). Kaplan-Meier plots indicated that individuals with relatively higher RBMS3-AS1 expression tended to have shorter overall survival time. On the contrary, LINC00284 and ZFX-AS1 could prolong patient survival time (Fig. 5A). Kaplan-Meier survival analysis was also assessed to estimate overall survival for the potentially associated DEmRNAs of these lncRNAs. Notably, high expression of E2F1 was also predicted to be positively associated with patient survival time (Fig. 5B).
Figure 5.
Kaplan-Meier curves for (A) 3 DElncRNAs and (B) one mRNA associated with overall survival. Horizontal axis, overall survival time (years); Vertical axis, survival function. Tumor samples were divided into ‘high’ group (solid lines, ≥median) and ‘low’ group (dotted lines, <median) according to expression levels.
Discussion
PTC is the most frequent endocrine neoplasia characterized by a variety of aberrant molecular events driving tumori-genesis (26). Currently, much effort has been made to elucidate the roles of ncRNAs involved in tumorigenesis, disease progression and metastasis of PTC (17,27,28). According to the ceRNA hypothesis, increasing evidence has demonstrated that lncRNAs regulated target genes through microRNAs (miRNAs) competition (29). To comprehensively identify the landscape regarding how lncRNA-associated ceRNA network affects PTC, the present study analyzed the large-scale sequencing data of PTC patient cohort in TCGA database. We identified the aberrantly expressed lncRNAs, mRNAs and miRNAs, and further successfully constructed the dysregulated lncRNA-associated ceRNA network in PTC by biological prediction. We also identified several lncRNAs and their related mRNAs to be potential prognostic indicators for PTC patients.
In previous studies, several groups have identified numerous abnormally expressed lncRNAs in PTC compared with non-tumor tissues. Two small cohort studies based on microarray sequencing of 3 and 5 pairs of PTC samples, respectively, have identified thousands of significantly dysregulated lncRNAs and validated several DElncRNAs by real-time PCR (18,30). Xie et al reported a ceRNA network in PTC containing 29 lncRNAs, 9 miRNAs and 67 mRNAs by microarray analysis of 5-paired clinical samples (31). However, lncRNA-associated ceRNA network based on whole-genome gene expression profiling with large study population in PTC has not been described.
In the present study, 780 lncRNAs were detected as commonly dysregulated lncRNAs in 3 PTC variants (CPTC, FPTC and TCPTC) compared with normal tissues, including 440 upregulated and 340 downregulated. Among them, 45 aberrantly expressed lncRNAs were identified in the ceRNA network, and 8 out of them were related to tumor progression. The low expression of an intergenic lncRNA LINC00473 in PTC tumor tissues (~10-folds) may have potential capacity to promote tumor growth and pathologic stage. It has been identified to have prognostic value for lung cancer in a cyclic AMP-dependent manner (32). However, whether LINC00473 can utilize similar mechanism to lung cancer in PTC needs further research. LINC00460, a human gene transcribed from chromosome 13, is upregulated in various cancers, including thyroid cancer (33). The upregulation of LINC00460 was also observed in the present study and was positively correlated with the pathologic stage of PTC. It was reported that LINC00460 participated in a series of biological processes (e.g., metabolic process, chromatin modification, cell cycle and cell death) and showed prognostic prediction power for head and neck squamous cell carcinoma (33,34). These findings implied that LINC00460 may functionally be involved in the tumorigenesis of PTC. Moreover, RBMS3-AS1 and ZFX-AS1 also had potential values for PTC patients prognosis. Since these lncRNAs are reported for the first time as prognostic predictors, much effort deserves to be made to elucidate their biological function in neoplasms. Although the lncRNAs listed in Table V had no statistical significance in Kaplan Meier analysis (data not shown), both the lnRNAs that were associated with the progression of PTC and with patients' overall survival may have potential roles in PTC and are worth studying in the future.
However, having identified several specific lncRNAs for PTC, the present study drew a comprehensive ceRNA network, which revealed the complexity of the regulatory relationships among lncRNAs, miRNAs and mRNAs. E2F1, the best-studied E2F member, was significantly upregulated in PTC tumor tissues, which was consistent with a previous study (35). E2F1 was involved in both cancer-related pathways: pathways in cancer and miRNAs in cancer. Although previously E2F1 was found to promote thyroid carcinogenesis (35), our results demonstrated that high expression of E2F1 could prolong survival time of PTC patients, which strongly suggested its tumor-suppressive activity in thyroid cancer. Thus, we supposed that E2F1 may exhibit both oncogenic and tumor suppressive properties in thyroid cancer resulting from its capacity to induce tumor cell proliferation and apoptosis (36–38). The ceRNA network predicted that E2F1 was a target gene of miR-205. It was consistent with the previous study that miR-205 induced cancer senescence by regulating E2F1 and ultimately suppressed melanoma development (39). Our bioinformatic analysis predicted that miR-205 may interact with LINC00284, the increased expression of which was also positively correlated with the prognosis of PTC patients. Although the function of LINC00284 is poorly annotated, it is conceivable that LINC00284 may act as a ceRNA to interact with miR-205 and E2F1 in PTC.
There are several limitations in the present study. First, ceRNA hypothesis indicates that lncRNAs can serve as ceRNAs to regulate expression of the target genes by merging miRNAs. Therefore, the complex ceRNA network of PTC was built based on this hypothesis using biological prediction. Second, the ceRNA network with hundreds of connections suggested that one lncRNA connected with a number of miRNAs and relevant mRNAs. In this regard, our further study may focus on conducting experiments to validate several crucial interactions, particularly to verify the key interactions we discussed above and determine the roles of these lncRNAs as ceRNA in PTC. Third, the 3 lncRNAs that were aberrantly expressed in race need extensive validation studies, which must be based on a large-scale multiracial cohort.
In conclusion, we identified various dysregulated lncRNAs, miRNAs and mRNAs between tumor and non-tumor samples by genome-wide analysis of a large PTC patient cohort from TCGA. The lncRNA-related ceRNA network will be useful to guide further investigation concerning the involved lncRNAs of their definite mechanisms and functions in the tumorigenesis of PTC.
Acknowledgements
We thank The Cancer Genome Atlas (TCGA) Research Network for providing data and publication permission (http://cancergenome.nih.gov/). The present study was supported by grants from the Chinese National Natural Science Foundation Grant (nos. 81671541, 81273202 and 31400773), the Clinical Medicine Science and Technology Project of Jiangsu Province of China (BL2013024), the Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX17_1820) and the China Scholarship Council. The present study was also supported by the Program of Innovative Research Team of Jiangsu Province, the Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions and the Project of the Key Academic Program Development of Jiangsu University (1291270019).
Glossary
Abbreviations
- TC
thyroid cancer
- PTC
papillary thyroid cancer
- FTC
follicular thyroid cancer
- ATC
anaplastic thyroid cancer
- CPTC
classical PTC
- FPTC
follicular PTC
- TCPTC
tall-cell PTC
- ncRNA
non-coding RNA
- lncRNA
long non-coding RNA
- miRNA
microRNA
- ceRNA
competitive endogenous RNA
- TCGA
The Cancer Genome Atlas
- MREs
miRNA response elements
- DAVID
The Database for Annotation Visualization and Integrated Discovery
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AJCC
American Joint Committee on Cancer
- FC
fold change
- FDR
false discovery rate
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R. Worldwide increasing incidence of thyroid cancer: Update on epidemiology and risk factors. J Cancer Epidemiol. 2013;2013:965212. doi: 10.1155/2013/965212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider DF, Chen H. New developments in the diagnosis and treatment of thyroid cancer. CA Cancer J Clin. 2013;63:374–394. doi: 10.3322/caac.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum MA, McHenry CR. Contemporary management of papillary carcinoma of the thyroid gland. Expert Rev Anticancer Ther. 2009;9:317–329. doi: 10.1586/14737140.9.3.317. [DOI] [PubMed] [Google Scholar]
- 6.Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 9.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Fan D, Jian Z, Chen GG, Lai P. Cancer specific long noncoding RNAs show differential expression patterns and competing endogenous RNA potential in hepatocellular carcinoma. PLoS One. 2015;10:e0141042. doi: 10.1371/journal.pone.0141042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K, Li Q, Kang X, Wang Y, Wang S. Identification and functional characterization of lncRNAs acting as ceRNA involved in the malignant progression of glioblastoma multiforme. Oncol Rep. 2016;36:2911–2925. doi: 10.3892/or.2016.5070. [DOI] [PubMed] [Google Scholar]
- 14.Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao WZ, Peng H, Hong WW, Yin LH, Pu YP, et al. Integrated analysis of long non-coding RNA-associated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49:2023–2036. doi: 10.3892/ijo.2016.3716. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283:2326–2339. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 16.Arancio W, Carina V, Pizzolanti G, Tomasello L, Pitrone M, Baiamonte C, Amato MC, Giordano C. Anaplastic thyroid carcinoma: A ceRNA analysis pointed to a crosstalk between SOX2, TP53, and microRNA biogenesis. Int J Endocrinol. 2015;2015:439370. doi: 10.1155/2015/439370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CT, Oyang YJ, Huang HC, Juan HF. MicroRNA-mediated networks underlie immune response regulation in papillary thyroid carcinoma. Sci Rep. 2015;4:6495. doi: 10.1038/srep06495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan X, Zhang H, Wang Z, Dong W, Sun W, Shao L, Zhang T, Zhang D. Genome-wide analysis of long noncoding RNA expression profile in papillary thyroid carcinoma. Gene. 2015;569:109–117. doi: 10.1016/j.gene.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeggari A, Marks DS, Larsson E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riffo-Campos ÁL, Riquelme I, Brebi-Mieville P. Tools for sequence-based miRNA target prediction: What to choose? Int J Mol Sci. 2016;17(pii):E1987. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LiVolsi VA. Papillary thyroid carcinoma: An update. Mod Pathol. 2011;24(Suppl 2):S1–S9. doi: 10.1038/modpathol.2010.129. [DOI] [PubMed] [Google Scholar]
- 25.Tunca F, Sormaz IC, Iscan Y, Senyurek YG, Terzioglu T. Comparison of histopathological features and prognosis of classical and follicular variant papillary thyroid carcinoma. J Endocrinol Invest. 2015;38:1327–1334. doi: 10.1007/s40618-015-0376-6. [DOI] [PubMed] [Google Scholar]
- 26.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aragon Han P, Weng CH, Khawaja HT, Nagarajan N, Schneider EB, Umbricht CB, Witwer KW, Zeiger MA. MicroRNA expression and association with clinicopathologic features in papillary thyroid cancer: A systematic review. Thyroid. 2015;25:1322–1329. doi: 10.1089/thy.2015.0193. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Li H, Zhang L, Zhang C, Yan W, Wang C. Identification of novel long non-coding RNA biomarkers for prognosis prediction of papillary thyroid cancer. Oncotarget. 2017;8:46136–46144. doi: 10.18632/oncotarget.17556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumor Biol. 2015;36:3129–3136. doi: 10.1007/s13277-015-3346-x. [DOI] [PubMed] [Google Scholar]
- 30.Yang M, Tian J, Guo X, Yang Y, Guan R, Qiu M, Li Y, Sun X, Zhen Y, Zhang Y, et al. Long noncoding RNA are aberrantly expressed in human papillary thyroid carcinoma. Oncol Lett. 2016;12:544–552. doi: 10.3892/ol.2016.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J, Guo B, Ding Z, Kang J, Deng X, Wu B, Fan Y. Microarray analysis of lncRNAs and mRNAs co-expression network and lncRNA function as ceRNA in papillary thyroid carcinoma. J Biomater Tiss Eng. 2015;5:872–880. doi: 10.1166/jbt.2015.1389. [DOI] [Google Scholar]
- 32.Chen Z, Li J, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126:2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Tao Y, Liao Q. Long noncoding RNA: A crosslink in biological regulatory network. Brief Bioinform. 2017 Apr 24; doi: 10.1093/bib/bbx042. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 34.Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T, Chen WT, Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi: 10.1016/j.oraloncology.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 35.Onda M, Nagai H, Yoshida A, Miyamoto S, Asaka S, Akaishi J, Takatsu K, Nagahama M, Ito K, Shimizu K, Emi M. Up-regulation of transcriptional factor E2F1 in papillary and anaplastic thyroid cancers. J Hum Genet. 2004;49:312–318. doi: 10.1007/s10038-004-0146-3. [DOI] [PubMed] [Google Scholar]
- 36.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian W, Cui F, Esteban MA. E2F1 in renal cancer: Mr Hyde disguised as Dr Jekyll? J Pathol. 2013;231:143–146. doi: 10.1002/path.4238. [DOI] [PubMed] [Google Scholar]
- 38.Zhan L, Zhang Y, Wang W, Song E, Fan Y, Wei B. E2F1: A promising regulator in ovarian carcinoma. Tumour Biol. 2016;37:2823–2831. doi: 10.1007/s13277-015-4770-7. [DOI] [PubMed] [Google Scholar]
- 39.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286:16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]