Abstract
Oral squamous cell carcinoma (OSCC) is the most frequent oral cancer in the world, accounting for more than 90% of all oral cancer diagnosis. Circular RNAs (circRNAs) are large types of non-coding RNAs, demonstrating a great capacity of regulating the expression of genes. However, most of the functions of circRNAs are still unknown. Recent research revealed that circRNAs could serve as a miRNA-sponge, consequently regulating the expression of target genes indirectly, including oncogenes. In this study, we built an apoptotic model with TNF-α, and then we confirmed a circRNA associated with the apoptosis of OSCC cells, circDOCK1 by comparing the expression profile of circRNAs in an apoptotic model with that in untreated OSCC cells. We ascertained the presence of circDOCK1 with qRT-PCR and circRNA sequencing. The knockdown of the expression of circDOCK1 led to the increase of apoptosis. Utilizing multiple bioinformatics methods, we predicted the interactions among circRNAs, miRNAs and genes, and built the circDOCK1/miR-196a-5p/BIRC3 axis. Both the silencing of circDOCK1 with small interfering RNA and the upregulation of the expression of miR-196a-5p with mimics led OSCC cells to increase apoptosis and decrease BIRC3 formation. We further confirmed this outcome by comparing the expression of circDOCK1, miR-196a-5p and BIRC3 in oral squamous carcinoma tissue with those in para-carcinoma tissue, and examining the expression profile of circRNAs in oral squamous carcinoma tissue and para-carcinoma tissue with microarray. Our results demonstrated that circDOCK1 regulated BIRC3 expression by functioning as a competing endogenous RNA (ceRNA) and participated in the process of OSCC apoptosis. Thus, we propose that circDOCK1 could represent a novel potential biomarker and therapeutic target of OSCC.
Keywords: circDOCK1, miR-196a-5p, BIRC3, apoptosis, sponge, oral squamous cell carcinoma
Introduction
Cancer of the oral cavity (oral cancer) is the 11th most frequent malignancy in the world, especially in developing countries, ranking ninth in the total number of incidences (1). Ninety-percent of oral cancers histologically originate in squamous cells (2). The incidence and mortality of this cancer exhibits variability resulting from: geographic location, gender, as well as occurrence site (lips, tongue, gingiva, mouth floor, parotid and saliva glands) (http://globocan.iarc.fr/Pages/summary_table_pop_sel.aspx). Although with the development of iatrotechniques, the mortality of oral squamous cell carcinoma (OSCC) has remained relatively unchanged. Worse still, the age of oral cancer patients is trending younger and younger. The overall prognosis of this cancer is poor; the survival rate is roughly 55–65% (3). This is probably due to late diagnosis. An early diagnosis is key in decreasing the mortality of oral cancer, which subsequently improves the overall survival and prognosis. Therefore, seeking a novel biomarker and therapeutic target is of the utmost importance.
Circular RNAs (circRNAs) are special types of non-coding RNAs characterized by their circular shape resulting from covalently closed continuous loops, widely expressed in cells that regulate the expression of relative genes in multiple pathways (4,5). CircRNAs were initially thought to originate due to a failure in alternative splicing owing to their low expression levels (6). Since next-generation sequencing technologies and bioinformatics analysis have developped, the regulatory function of circRNAs in eukaryotic cells is beginning to increasingly attract emphasis. As particular non-coding RNAs, circRNAs are widely expressed, highly conserved and stable in the cytoplasm (RNase R resistant) (6). With the deepening of research, two circRNAs (CDR1as and Sry) have been revealed to be miRNA sponges regulating the expression of relevant genes (7,8). Moreover, they were also found to be regulators of alternative splicing (9), and transcription factors (10) and to encode for proteins (11,12). CircRNAs participate in several pathological states, including neurological diseases (13,14), hematopathy (15), cartilage degradation (16), Hirschsprung's disease (17) and multiple cancers, which may serve as promising diagnostic biomarkers and therapeutic targets. It is known that circRNAs could compete for binding to cancer-related miRNAs. Multiple circRNA/miRNA/mRNA axes in cancer-associated pathways have been investigated in colorectal cancer (18,19), pancreatic cancer (20), bladder carcinoma (21), esophageal squamous cell carcinoma (ESCC) (22), glioblastoma (23), cervical cancer (24), chronic lymphocytic leukemia (CLL) (25), non-small cell lung cancer (NSCLC) (26), cholangiocarcinoma (27), ovarian cancer (28) and hepatocellular carcinoma (HCC) (29,30), with both agonist and antagonist effects on carcinogenesis. However, whether circRNAs harbor miRNAs with regulatory roles in OSCC is still unknown.
MicroRNAs (miRNAs) are one of the large families of non-coding RNAs with small, endogenous, and evolutionarily conserved features, (approximately 18–24 nucleotides) which are involved in regulating several essential, cellular and functional processes such as cell differentiation, migration, growth, proliferation, apoptosis, metabolism and tumorigenesis (31–33). It is well known that miRNAs are known to play key roles in most types of cancer and function as oncogenes or tumor suppressors in upregulation or downregulation processes respectively (31). miRNAs have the potential to be used as prognostic and diagnostic biomarkers. Generally, to be a negative regulator, miRNAs bind to the 3′-untranslated region (3′-UTR) of their target mRNA based on the principle of complementary base pairing and inhibit translation (34). miRNAs play crucial roles in OSCC initiation and development (35).
In this study, we investigated the molecular mechanisms underlying the circRNAs in OSCC. We built an apoptosis model by stimulating CAL-27 cells with TNF-α and acquired a differentially expressed circRNA profile from the apoptosis model and normal CAL-27 cells using high-throughput microarray. Several significant differentially expressed circRNAs were further confirmed with qRT-RCR and we found that the circRNA has_circ_100721 (circDOCK1) was markedly overexpressed and the knockdown of the expression of circDOCK1 led to the increase of apoptosis. According to a set of bioinformatics approaches, we constructed a circRNA/miRNA/gene network and determined that circDOCK1 interacted with miR-196a-5p by competing with BIRC3, eventually regulating cell apoptosis. Finally, we explored the functions of circDOCK1 in OSCC in vivo and in vitro. Hence, we concluded that circDOCK1 suppressed cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC.
Materials and methods
Ethics statement and tissue samples
This study was approved and supervised by The Ethics Committee of Guangzhou Medical University. All the experiments were carried out in accordance with relevant guidelines and regulations. All oral carcinoma tissues and adjacent normal tissues were collected from patients who had undergone surgery at the Cancer Center of Guangzhou Medical University and The First Affiliated Hospital of Guangzhou Medical University from 2015 to 2016. All carcinoma and paired normal tissues were ten groups each, which were confirmed by pathological detection. The clinical and pathological characteristics for each patient were noted before and after surgery. The patients were made aware of the research purposes and informed consent was given.
Cell culture
The human OSCC cell lines CAL-27, SCC-9 and SCC-25 were purchased from ATCC (American Type Culture Collection, Manassas, VA, USA). Human normal oral epithelial keratinocytes (HOK) were purchased from ScienCell Research Laboratories (San Diego, CA, USA). HOK was cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Carlsbad, CA, USA) containing with 10% fetal bovine serum (FBS) (Gibco). The other cell lines were cultured in Dulbecco's modified Eagle's medium: nutrient Mixture F-12 (DMEM/F12) (Gibco) with 10% FBS. All cell lines were incubated at 37°C in a humidified chamber containing 5% CO2.
Apoptosis model and flow cytometry (FCM)
OSCC cell line CAL-27 cells were stimulated with 100 ng/ml recombinant human TNF-α (PeproTech, Rocky Hill, NJ, USA) for 48 h. The FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, Franklin Lakes, NJ, USA) was used according to the FITC Annexin V Staining Protocol to detect apoptotic cells 48 h after being treating with TNF-α. First, the cells were collected and washed twice with cold PBS and then resuspended in 1X binding buffer at a concentration of 1×106 cells/ml. One hundred microliters of the solution (1×105 cells) was transferred to a 5 ml culture tube. FITC Annexin V (5 µl) and propidium iodide (PI; 5 µl) staining solutions were added to the cells. The cells were gently cycled in a vortex and incubated for 15 min at RT (25°C) in the dark. Then 400 µl of 1X binding buffer were added to each tube and the cells were analyzed by flow cytometry (BD Biosciences) within 1 h.
CircRNA Microarray
An apoptosis model and a negative control group, oral carcinoma tissues and adjacent normal tissues were used to detect the expression of circRNAs by CircRNA Microarray. Total RNA from each sample was quantified using the NanoDrop ND-1000. The sample preparation and microarray hybridization were performed based on the standard protocols of Arraystar Inc. (Rockville, MD, USA). Briefly, total RNA from each sample was amplified and transcribed into fluorescent cRNA utilizing a random primer according to Arraystar's Super RNA Labeling protocol (Arraystar Inc.). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Microarray (8×15K, Arraystar Inc.). After having washed the slides, the arrays were scanned using the Agilent DNA Microarray Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images while quantile normalization and subsequent data processing were performed using the R software package. Differentially expressed circRNAs with statistical significance between two groups were identified through volcano plot filtering and these two groups were also identified through fold change filtering. Hierarchical clustering was performed to reveal the distinguishable circRNA expression patterns among samples. The microarray hybridization and the collection of data were performed by KangChen Bio-tech, Shanghai, China.
Quantitative real-time PCR and circRNA sequencing
Total RNA was extracted from OSCC cell lines and oral cancer samples using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and quantified using NanoDrop 2000 (Thermo Scientific, Carlsbad, CA, USA). After RNA extraction, SuperScript® III Reverse Transcriptase (Invitrogen) was used for reverse-transcription to cDNA following the manufacturer's protocol. The RT products were used as templates for amplification using the SYBR® Premix Ex Taq™ II PCR amplification reagent (Takara, Shiga, Japan). Specific divergent primers were designed to amplify the circular transcripts. The reaction system of qRT-PCR was performed in 25 µl of reaction volume, including 2 µl of cDNA, 12.5 µl 2X SYBR Premix Ex Taq II, 0.5 µl of forward primer (10 µM), 0.5 µl of reverse primer (10 µM) and 9.5 µl of double distilled water. The reaction was set at 95°C for 30 sec for pre-denaturation, then at 95°C for 5 sec, at 60°C for 30 sec and at 72°C for 30 sec for 39 cycles. Reactions for each sample were performed in triplicate and the relative levels of target genes were calculated using the 2−∆∆Ct method and normalized to GAPDH. The products of qRT-PCR were purified using Agencourt AMPure XP PCR purification kit from Beckman Coulter Inc. (Brea, CA, USA). Direct PCR product Sanger sequencing was performed by LGC Genomics (Berlin, Germany) Ready2 Run services. The forward primer for circDOCK1 was provided for sequencing the product of circDOCK1. All the primer sequences of the genes used in this study are shown in Table I.
Table I.
All the primer sequences of the genes used for this study.
| Genes | Primer sequence (5′-3′) |
|---|---|
| GAPDH | F: GCACCGTCAAGGCTGAGAAC |
| R: TGGTGAAGACGCCAGTGGA | |
| circDOCK1 | F: GAAATCGTCCACAGTGACCT |
| R: CACAGTGTCTCCGATCTGTAAA | |
| circRNA_101531 | F: TGCAGGGCGTCATCAAAACC |
| R: CACACACCTGTTCTCCTGTGG | |
| circRNA_101315 | F: AGATGCGCAGGCTCAACATA |
| R: CAATGGATTGTTCCTTGGCTGT | |
| circRNA_104313 | F: GCTACCATTTTGGCTGTTTGGA |
| R: TCTTCAGCCTTGGGCTTCTTTT | |
| circRNA_101226 | F: GCCTGTGTATATCCCAGTTCCT |
| R: TTTGGTCTGCATGTGAGGTTT | |
| circRNA_104838 | F: TTCCATAAATGGGGTCGAG |
| R: TTGAGTGGTCCAAAATCTGC | |
| circRNA_102765 | F: AAGCCTTCACCTGCATCCAA |
| R: AGACGTCTCTCAGTGACAAAG | |
| circRNA_100790 | F: CTGCGTCTCAGCCTCAAGTAT |
| R: TGGTTCCTTCTGTGGTCGTT | |
| circRNA_102367 | F: TGGAAGAATTGCTGTGCTGTT |
| R: TGCACCAAGATTTCCTGTTCG | |
| circRNA_102956 | F: GTTTTGGGCGTGCTAGAGGT |
| R: GCTCGAATTCCTCCATGTCCA | |
| BIRC3 | F: CAACAGATCTGGCAAAAGCA |
| R: TTGCTCAATTTTCCACCACA | |
| FOXO1 | F: AAGAGCGTGCCCTACTTCAA |
| R: CTGTTGTTGTCCATGGATGC | |
| MDM2 | F: CATTGTCCATGGCAAAACAG |
| R: GGCAGGGCTTATTCCTTTT | |
| DOCK1 | F: TCGGACGCCGTACTCTTTCTC |
| R: CTGATTGCTTGGCAGATTCCT | |
| miR-196a-5p | F: TAGGTAGTTTCATGTTGTTGG |
| miR-138-2-3p | F: CTATTTCACGACACCAGGGTT |
F, forward; R, reverse.
RNA interference, miRNA mimic and transfection
The small interfering RNAs (siRNAs) (RiboBio, Guangzhou, China) were designed for the junction of circDOCK1 and the mimic was designed for miR-196a-5p. The concentration of the mimic for miR-196a-5p, the siRNA for circDOCK1 and the siRNA for the negative control was 50 nM. Transfection of the siRNA and mimic was conducted with GenMute™ siRNA Transfection Reagent (SignaGen Laboratories, Rockville, MD, USA) according to the standard protocol. Before transfection, the complex which included 100 µl 1X GenMute™ Transfection Buffer, 5 µl siRNA or mimic and 4 µl GenMute™ Reagent was incubated for 30 min at room temperature, and then the above mixture was added to OSCC cell lines with 1 ml DMEM/F12 growth medium in 6-well plates. The cells were cultured in a humidified atmosphere with 5% CO2 at 37°C. After transfection for 48 h, the levels of circDOCK1 and miR-196a-5p were assessed by qRT-PCR. All the sequence of siRNAs and mimics are shown in Tables II and III.
Table II.
The sequence of siRNAs for circDOCK1.
| siRNA | Target sequence (5′-3′) |
|---|---|
| siRNA 1 | CTTCACACAGCATGCTTTT |
| siRNA 2 | TCACACAGCATGCTTTTTA |
| siRNA 3 | CACACAGCATGCTTTTTAT |
Table III.
The sequence of miR-196a-5p mimics.
| miRNA mimics | Primer sequence (5′-3′) |
|---|---|
| miR-196a-5p | F: UAGGUAGUUUCAUGUUGUUGGG |
| R: CCCAACAACAUGAAACUACCUA |
F, forward; R, reverse.
Prediction for circRNA/miRNA/mRNA pathways
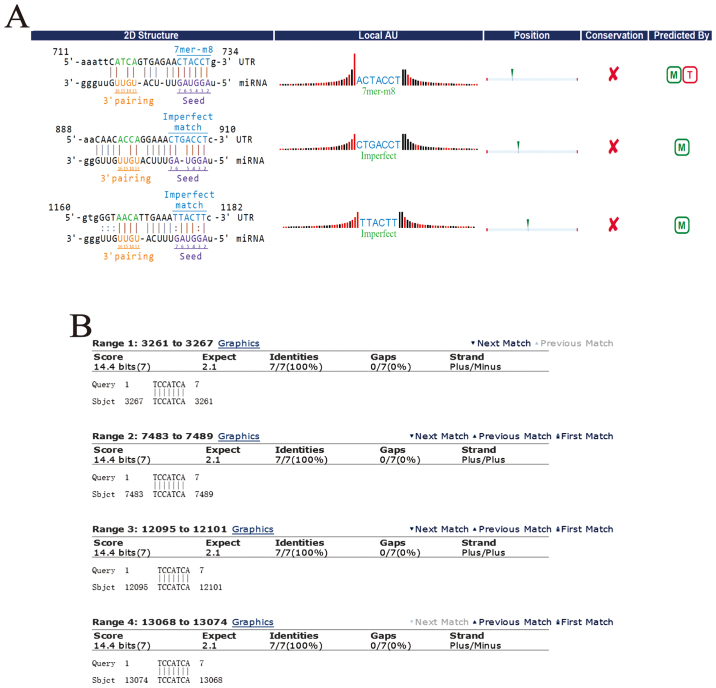
The relationship between circRNAs and miRNAs was predicted by miRNA target prediction software based on miRanda and TargetScan. Also, the circRNA/microRNA interaction was speculated using Circinteractome according to the name of circRNAs and miRDB according to the sequence of circRNAs. Through combination of the three methods the capability of seed match sequences was ranked. Using the Cytoscape 3.4.0, the graph of circRNA/miRNA network was drawn. The top 15 miRNA pathways were predicted based on DIANA TOOLS-miRPath v.3 and the cancer-related pathways and apoptosis-related pathways of the top 15 miRNAs were selected. The target genes were acquired based on Database for Annotation, Visualization and Integrated Discovery (DAVID). Finally, several cancer-related pathways and apoptosis-related pathways associating with circRNAs, miRNAs and target genes were constructed.
Western blotting
The total protein of samples was extracted using RIPA Lysis Buffer (Beyotime, China, which contained PMSF (100:1) according to the standard procedure. The protein samples were loaded onto 10% SDS PAGE and separated. Then, they were transferred to a PVDF membrane. The membrane was blocked for 2 h with 0.5% skim milk powder. The target proteins on the membranes were incubated with the relative primary antibodies β-actin (1:4,000; cat. no. ab8227) and BIRC3 (1:1,000; cat no. ab32059) (both from Abcam, Cambridge, UK) for 2 h on table concentrator at 4°C. After being washed three times, the membranes were sequentially incubated with the HRP conjugated secondary antibody anti-Rabbit IgG H&L (HRP) (1:4,000; cat. no. ab205718) (Abcam) for 1 h at RT and ECL detection followed.
Statistical analysis
All of the experiments were repeated three times independently. Data are expressed as the mean ± standard deviation (SD). Student's t-test (two-tailed) was applied for data analysis. The values P<0.05 indicated a statistically significant difference.
Results
Construction of a cell apoptosis model and its differentially expressed circRNA profiles
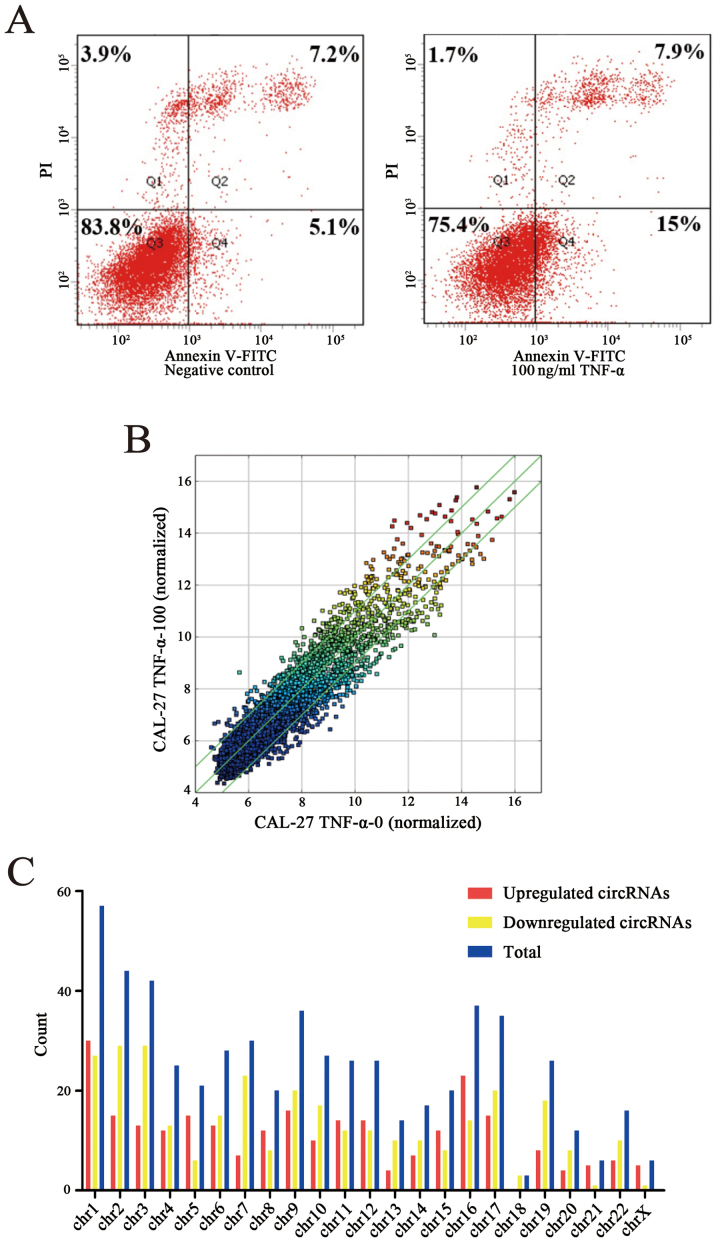
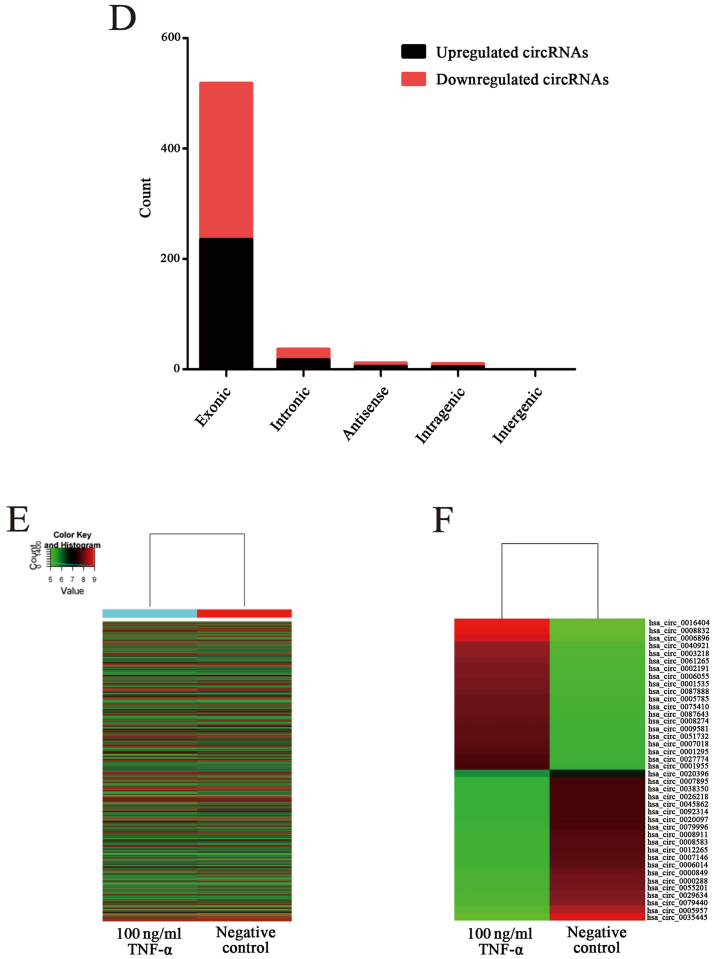
OSCC cell line CAL-27 was stimulated with 100 ng/ml recombinant human TNF-α and cultured for 48 h. The apoptosis rate of the apoptosis group and the negative group were detected through FCM and the apoptosis rate of the apoptosis model was increased by 10.6% compared to the negative control (Fig. 1A). Following construction of the apoptosis model the circRNA profile of the model was detected by means of high throughput microarray assay. According to the fold change (≥2.0), 628 circRNAs were differentially expressed, among which 287 circRNAs were upregulated while 341 circRNAs were downregulated (Fig. 1B). By analyzing the data of differentially expressed circRNAs, we classified their chromosomal origin (Fig. 1C) and determined that most of the circRNAs came from the protein coding exons (Fig. 1D). Furthermore, we listed the top twenty upregulated and downregulated circRNAs in line with the fold changes of the expression of circRNAs (Table IV). Moreover, hierarchical clustering was drawn to reveal the differentially expressed circRNAs between the apoptosis model and the negative control (Fig. 1E and F). All of the data revealed that the expression of circRNAs in the apoptosis group was different from that which was matched in the negative group.
Figure 1.
Construction of an OSCC cell apoptosis model and depiction of differentially expressed circRNA profiles. (A) FCM detected the apoptosis rate of the apoptosis model compared to the negative control and the apoptosis rate was increased by 10.6%. (B) A scatter diagram was used to evaluate the differentially expressed circRNAs. The x-axis represents the relative expression of circRNAs in the negative control, and the y-axis represents the relative expression of circRNAs in the apoptosis model. The spots above the top green line and below the bottom green line represent more than 2.0-fold changes between the two groups. (C) The distribution of differentially expressed circRNAs in human chromosomes. (D) The bar diagram shows the origin of differentially expressed circRNAs. Most of them originated from exons. Some originated from introns, while, a few were from other sources. (E and F) Heat map and hierarchical clustering revealed expression values of all differentially expressed circRNAs and the top 20 upregulated and downregulated circRNAs. OSCC, oral squamous cell carcinoma; FCM, flow cytometry; circRNAs, circular RNAs.
Table IV.
Top 20 upregulated and downregulated circRNAs were chosen in line with the fold changes of the expression of circRNAs.
| Upregulated circRNAs | Downregulated circRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CircRNA ID | CircRNA type | Chrom | Fold change | GeneSymbol | CircRNA ID | CircRNA type | Chrom | Fold change | GeneSymbol |
| hsa_circRNA_100445 | Exonic | chr1 | 8.0656065 | TATDN3 | hsa_circRNA_101531 | Exonic | chr15 | −7.5435655 | ALDH1A2 |
| hsa_circRNA_103205 | Exonic | chr22 | 7.8829886 | FBXO7 | hsa_circRNA_101315 | Exonic | chr14 | −6.1496856 | METTL3 |
| hsa_circRNA_104650 | Exonic | chr8 | 7.2938337 | KIAA1429 | hsa_circRNA_104313 | Exonic | chr7 | −5.1219124 | PHF14 |
| hsa_circRNA_101906 | Exonic | chr16 | 5.4328383 | ANKRD11 | hsa_circRNA_101226 | Exonic | chr13 | −4.9973297 | ZMYM2 |
| hsa_circRNA_102895 | Exonic | chr2 | 5.3716472 | BMPR2 | hsa_circRNA_102765 | Exonic | chr2 | −4.7427653 | SFXN5 |
| hsa_circRNA_103108 | Exonic | chr21 | 5.0335912 | BC048201 | hsa_circRNA_100790 | Exonic | chr11 | −4.7006624 | CAPRIN1 |
| hsa_circRNA_104833 | Exonic | chr9 | 4.8887767 | C9orf3 | hsa_circRNA_102367 | Exonic | chr18 | −4.6127982 | DYM |
| hsa_circRNA_102991 | Exonic | chr20 | 4.875669 | TMEM230 | hsa_circRNA_102956 | Exonic | chr2 | −4.2670317 | ILKAP |
| hsa_circRNA_103951 | Exonic | chr5 | 4.7518169 | FAM13B | hsa_circRNA_100721 | Exonic | chr10 | −4.1949433 | DOCK1 |
| hsa_circRNA_104855 | Exonic | chr9 | 4.7120789 | TMEM245 | hsa_circRNA_104838 | Exonic | chr9 | −4.1273329 | CDC14B |
| hsa_circRNA_101141 | Exonic | chr12 | 4.5303581 | ANAPC7 | hsa_circRNA_103563 | Exonic | chr3 | −4.1234249 | DLG1 |
| hsa_circRNA_104040 | Exonic | chr6 | 4.5210355 | DUSP22 | hsa_circRNA_102651 | Exonic | chr2 | −4.0871929 | ZNF512 |
| hsa_circRNA_104839 | Exonic | chr9 | 4.4748769 | CDC14B | hsa_circRNA_101695 | Exonic | chr16 | −4.0480481 | NAGPA |
| hsa_circRNA_101287 | Exonic | chr13 | 4.3717281 | UGGT2 | hsa_circRNA_104356 | Exonic | chr7 | −3.9404514 | POLR2J4 |
| hsa_circRNA_100033 | Exonic | chr1 | 4.3075694 | RERE | hsa_circRNA_100688 | Exonic | chr10 | −3.9125353 | ATRNL1 |
| hsa_circRNA_102587 | Exonic | chr19 | 4.30598 | LIG1 | hsa_circRNA_400068 | intronic | chr22 | −3.8772576 | RANBP1 |
| hsa_circRNA_101850 | Exonic | chr16 | 4.1911027 | CIRH1A | hsa_circRNA_102205 | Exonic | chr17 | −3.8669576 | TNRC6C |
| hsa_circRNA_103360 | Exonic | chr3 | 4.0908172 | SMARCC1 | hsa_circRNA_101055 | Exonic | chr12 | −3.8643985 | CERS5 |
| hsa_circRNA_101115 | Exonic | chr12 | 3.9659427 | METAP2 | hsa_circRNA_101747 | Exonic | chr16 | −3.8581905 | C16orf62 |
| hsa_circRNA_101555 | Exonic | chr15 | 3.897013 | CSNK1G1 | hsa_circRNA_100117 | Exonic | chr1 | −3.8552525 | EYA3 |
Construction of the circRNA/miRNA connection network
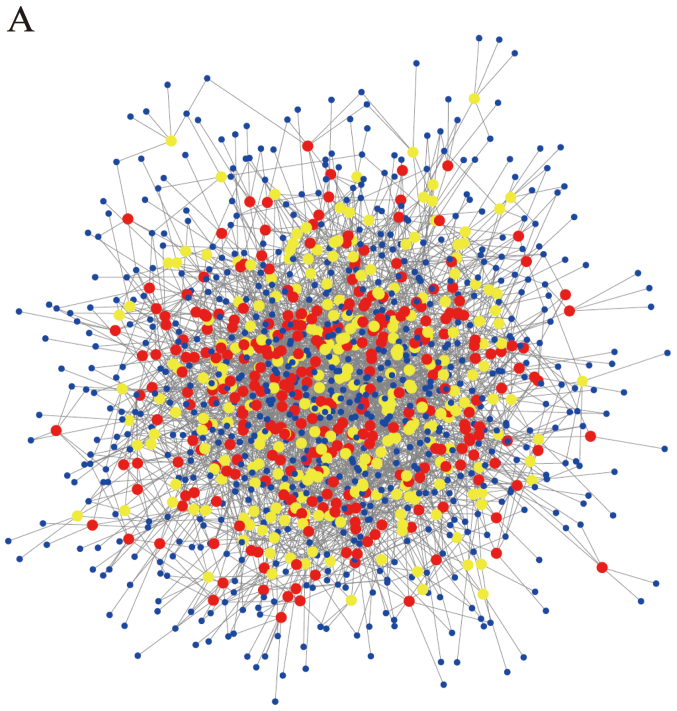
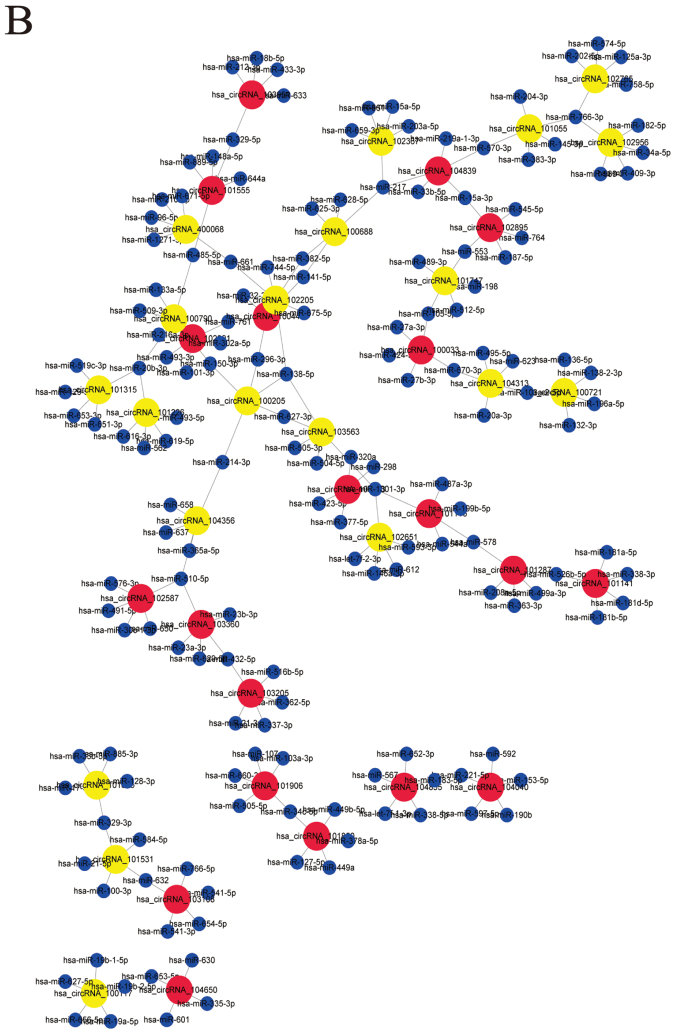
According to the principle of complementary base pairing and using the prediction software based on miRanda, TargetScan and websites Circinteractome and miRDB, we found hundreds of relative miRNAs targeting the differentially expressed circRNAs. The top five candidate miRNAs out of a total of 628 circRNAs were selected in accordance with the strength of their connection. The circRNA/miRNA network was depicted using Cytoscape (Fig. 2A). In addition to the whole data, the network of top 20 upregulated and downregulated circRNAs and their candidate miRNAs was also depicted (Fig. 2B).
Figure 2.
Differentially expressed circRNA/miRNA connection network. (A) All of the differentially expressed circRNAs and their candidate miRNAs constitute a complex network. (B) The top 20 upregulated and downregulated circRNAs and their candidate miRNAs could also constitute a connective network. Red nodes, upregulated circRNAs. Yellow nodes, downregulated circRNAs. Blue nodes, target miRNAs. CircRNAs, circular RNAs.
Validation of the differentially expressed circRNAs in the apoptosis model and OSCC tissue by qRT-PCR and sequencing
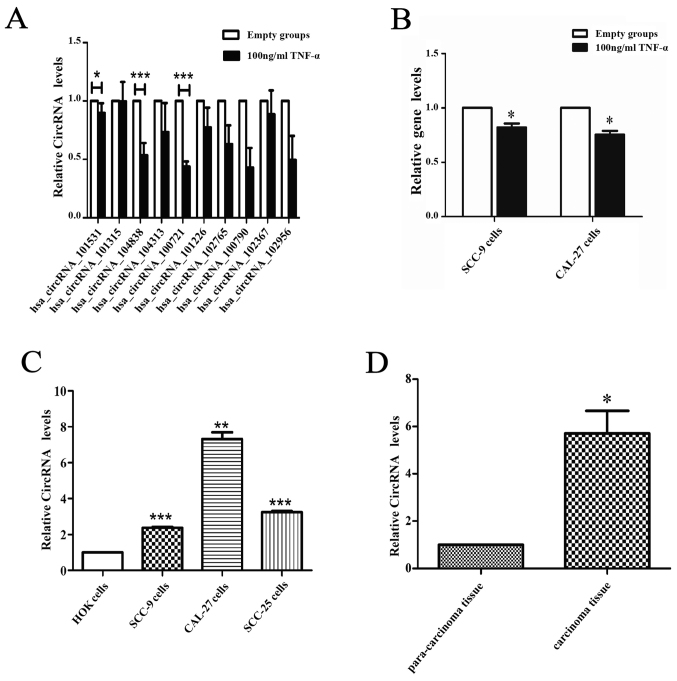
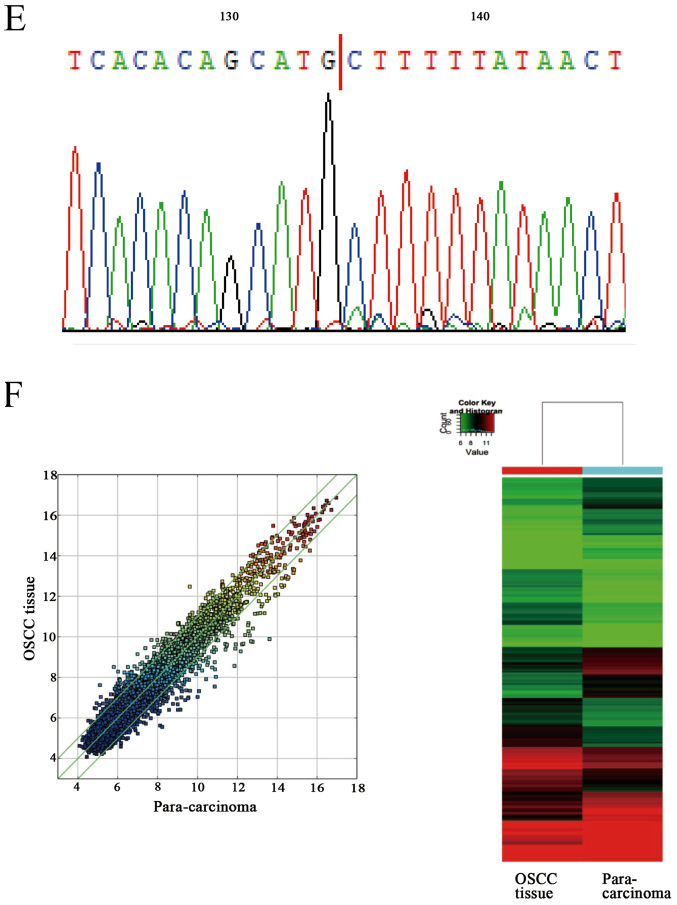
Ten circRNAs were selected to confirm the differential expression between the apoptosis model and the negative group via qRT-PCR. The results revealed, two circRNAs, circCDC14B (hsa_circ_104838) and circDOCK1 (hsa_circ_100721) that were significantly downregulated in the apoptosis group (Fig. 3A). However, the expression levels of linear DOCK1 were slightly affected by TNF-α in SCC-9 and CAL-27 cells (Fig. 3B). To further confirm the significance of these two circRNAs, their expression in OSCC cell lines (CAL-27, SCC-9, SCC-25) and normal oral epithelial cells (HOK), OSCC tissue and para-carcinoma tissue (ten groups) were ascertained by qRT-PCR. Furthermore, the circRNA profiles of one of the OSCC tissue groups compared with the para-carcinoma tissue was detected via high throughput microarray assay. The results revealed that circDOCK1 was distinctly upregulated in OSCC cell lines and OSCC tissue (Fig. 3C and D). The sequencing of circDOCK1 confirmed its presence in OSCC (Fig. 3E). The data indicated that microarray analysis was consistent with the qRT-PCR results regarding the expression levels of circDOCK1 (Fig. 3F). Consequently, circDOCK1 was chosen to be studied due to its mechanism of action.
Figure 3.
Confirmation of the presence of circDOCK1 and its differentially expressed levels in the apoptosis model and OSCC cell lines. (A) The expression level of circDOCK1 (circ_100721) was significantly downregulated in the apoptosis model compared to the negative control. (B) The expression level of linear DOCK1 was slighlty affected by TNF-α in SCC-9 and CAL-27 cells. (C) The expression level of circDOCK1 (circ_100721) was significantly upregulated in OSCC cell lines compared to HOK cells. (D) The expression level of circDOCK1 (circ_100721) was significantly upregulated in OSCC tissue compared to para-carcinoma tissue. (E) The qRT-PCR product of circDOCK1 was sequenced and the red line represents the junction, which confirmed the presence of circDOCK1 in OSCC. (F) The circRNA profiles of OSCC tissue were detected via high throughput microarray assay and the outcome of the data was consistent with the qRT-PCR results mentioned above. All data are expressed as the mean ± SE of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the control. OSCC, oral squamous cell carcinoma; circRNAs, circular RNAs.
Confirmation of circDOCK1 apoptosis in OSCC cell lines
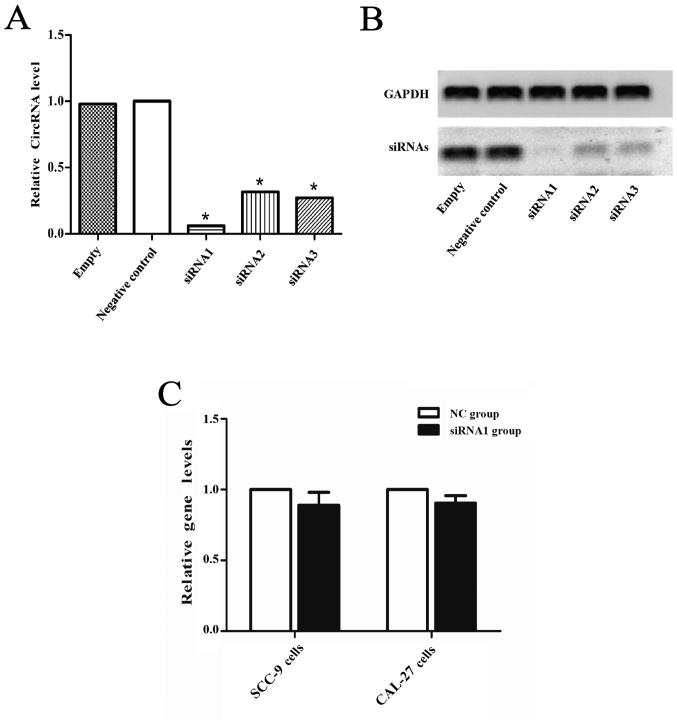
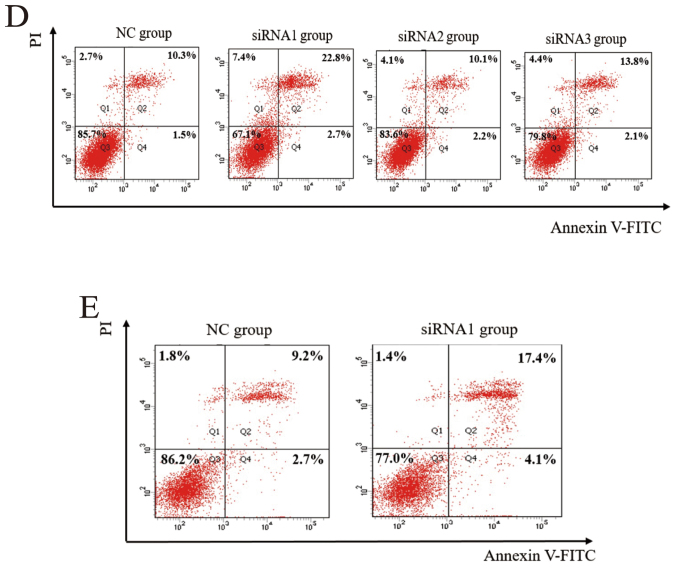
CircDOCK1 was significantly downregulated in the apoptosis model compared with the negative group and markedly upregulated in the OSCC tissue compared with the para-carcinoma tissue, however whether it could regulate the apoptosis rate in OSCC cell lines was still unclear. Hence, a low expression model of circDOCK1 was constructed with three siRNAs for circDOCK1 in CAL-27 cells. After transfection for 48 h, the effect of the three different siRNAs against circDOCK1 (si-circDOCK1) was analyzed by qRT-PCR and nucleic acid electrophoresis. The most marked inhibitory result was achieved with si-circDOCK1-1 (siRNA1) (Fig. 4A and B). However, the expression levels of linear DOCK1 were not affected by siRNA1 in SCC-9 and CAL-27 cells (Fig. 4C). FCM detected the apoptosis rate of the low expression model in CAL-27 and SCC-9 cells. Following transfection with si-circDOCK1-1 for 48 h, the apoptosis rate was increased by 13.7% and 9.6% in CAL-27 and SCC-9 cells, respectively (Fig. 4D and E). Consequently, circDOCK1 regulated the apoptosis of OSCC cell lines.
Figure 4.
Confirmation of circDOCK1 apoptosis in OSCC cell lines. (A and B) Three siRNAs were used to downregulate the expression of circDOCK1. The results of qRT-PCR and nucleic acid electrophoresis demonstrated that the most marked inhibitory effect was achieved with si-circDOCK1-1 (siRNA1). (C) The expression levels of linear DOCK1 were not affected by siRNA1 in SCC-9 and CAL-27 cells, which exhibited no statistical significance. (D and E) Following transfection of siRNA1 in circDOCK1, FCM detected the apoptosis rate which was increased by 13.7% and 9.6% in CAL-27 and SCC-9 cells, respectively. All data are expressed as the mean ± SE of three independent experiments. *P<0.05 vs. the control. OSCC, oral squamous cell carcinoma; FCM, flow cytometry.
Forecasting the circRNA/miRNA/mRNA axis of circDOCK1
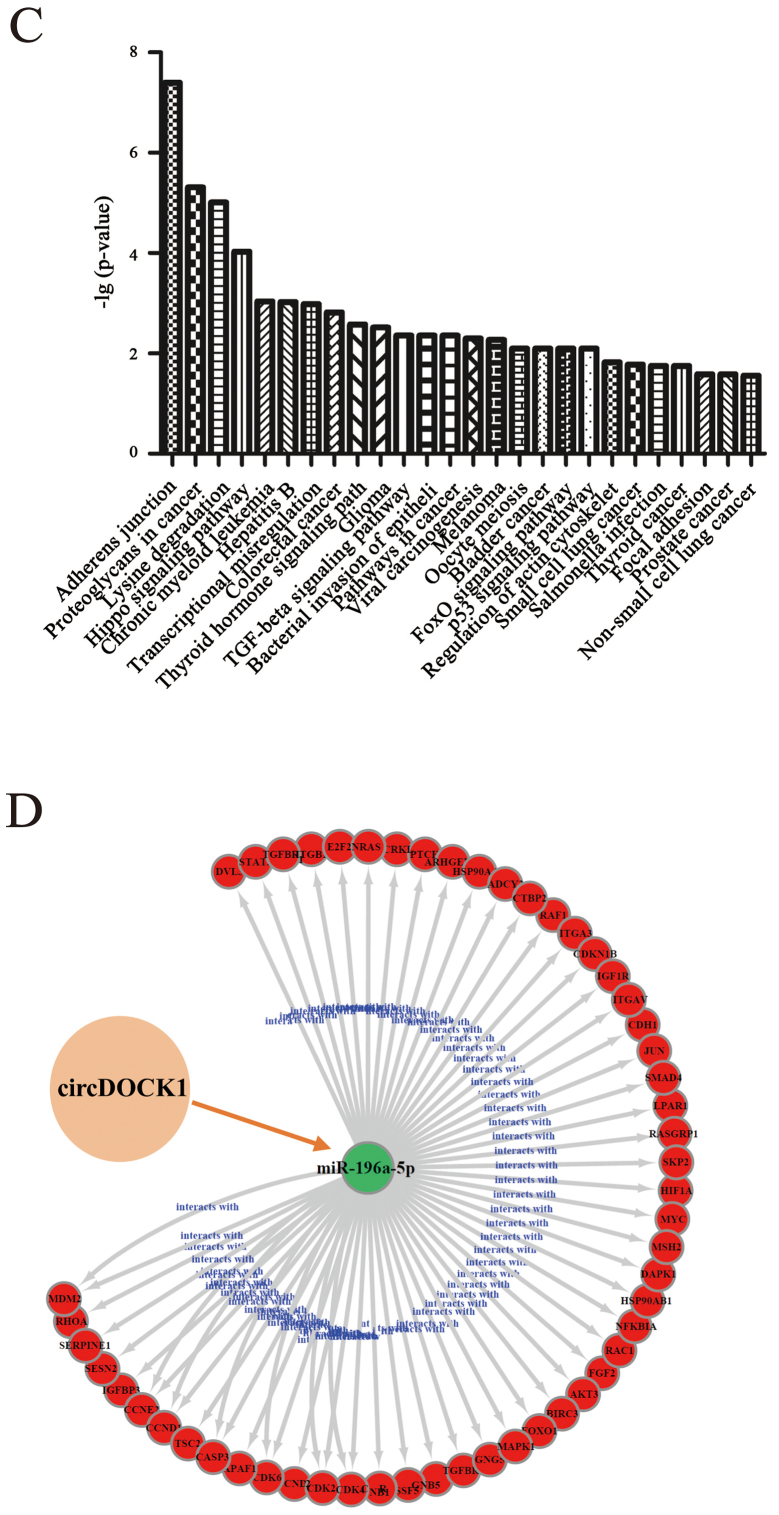
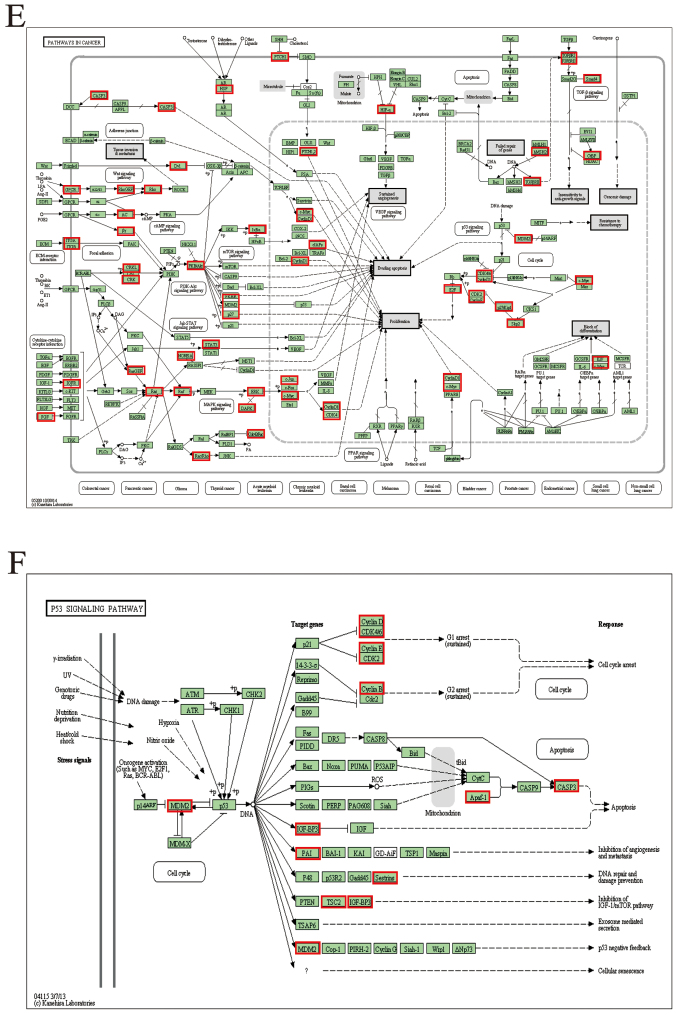
It is well known that circRNAs could serve as miRNA sponges, indirectly regulating the expression of downstream mRNAs. The combination of circRNAs and miRNAs as well as miRNAs and mRNAs were based on complementary base pairing (Fig. 5A and B). To study the apoptosis mechanism of circDOCK1, circDOCK1/miRNA/mRNA was predicted. DIANA TOOLS-mirPath v.3 ascertained all the candidate miRNAs connecting with circDOCK1, which participated in cancer-related pathways and apoptosis-related pathways. According to the P-value (P<0.05), the data revealed that the candidate miRNAs for circDOCK1 hsa-miR-138-2-3p and hsa-miR-196a-5p were involved in cancer-related pathways. Further analysis of these two pathways revealed that only hsa-miR-196a-5p was associated with apoptosis-related pathways, including the p53 signaling pathway (Fig. 5C); while 48 genes and 14 genes related to hsa-miR-196a-5p were involved in the cancer-related pathways and the p53 signaling pathway, respectively (Fig. 5D). In order to further construct the circDOCK1/miR-196a-5p/mRNA axis which was associated with the apoptosis pathway, DAVID functional annotation was used. A total of 55 genes (7 genes were repetitive in cancer-related pathways and the p53 signaling pathway) were analyzed (Fig. 5E and F). The analysis revealed that some of these genes were significantly correlated with cell apoptosis. Three genes were chosen: BIRC3, FOXO1, MDM2, which participated in apoptosis-related pathways. Furthermore, it is well known that Cyclin D1 and CDK4 could affect the cell cycle process and cell proliferation. In addition E2F and c-Myc may block cell differentiation. Ras, Rac1, ITGA, ITGB and HSP90 were involved in the PI3K-Akt signaling pathway which regulated multiple cancer-related phenotypes. In addition, FOXO could serve as a tumor-inhibiting factor. This indicated that most of the genes correlated to circDOCK1 were closely related to anti-apoptosis, tumorigenesis and metastasis.
Figure 5.
Prediction of the circDOCK1/miR-196a-5p apoptosis gene axis. (A and B) According to the principle of complementary base pairing, circDOCK1 and BIRC3 targeted the seed sequence of miR-196a-5p. (C) The signaling pathways related to miR-196a-5p were listed, which were based on P-values. (D) According to the bioinformatics prediction, the circDOCK1/miRNA gene network was depicted, including 55 target genes, which were associated with cancer-related pathways and the p53 signaling pathway. (E and F) These two pathway maps indicate cancer-related pathways and the p53 signaling pathway related to miR-196a-5p. The red frames represent the genes were regulated by the circDOCK1/miR-196a-5p axis.
OSCC cell apoptosis is regulated via the circDOCK1/miR-196a-5p/BIRC3 pathway
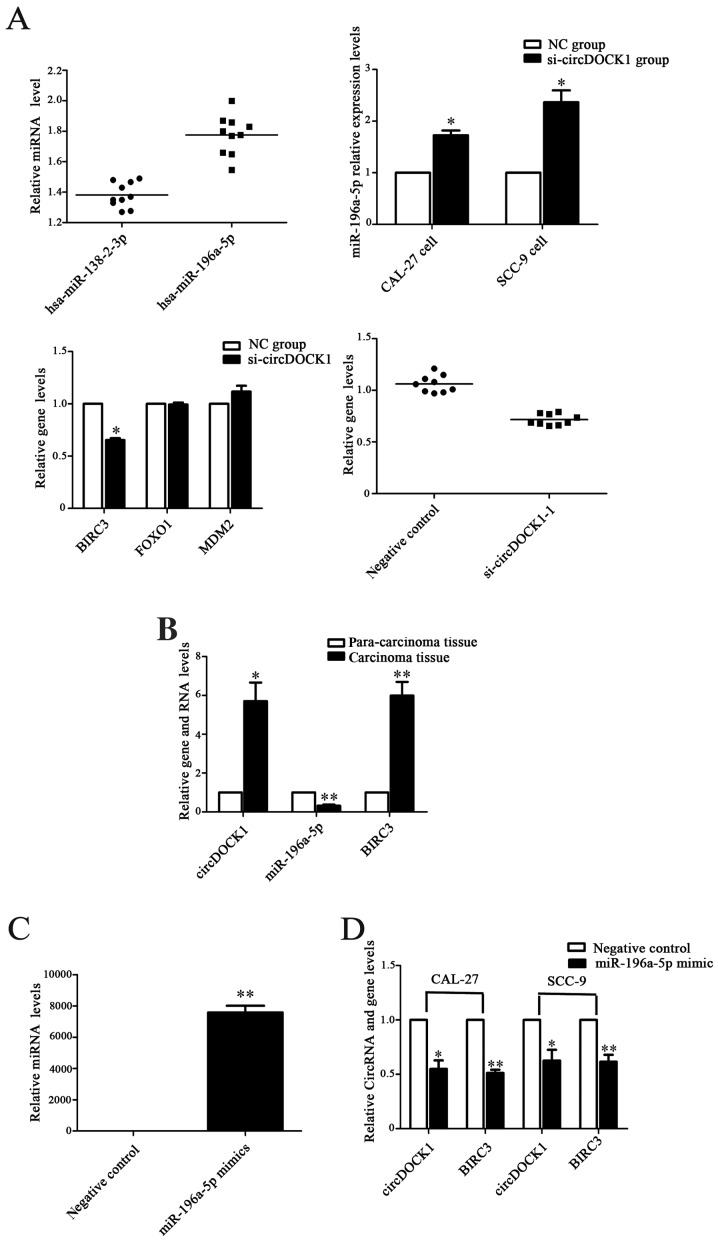
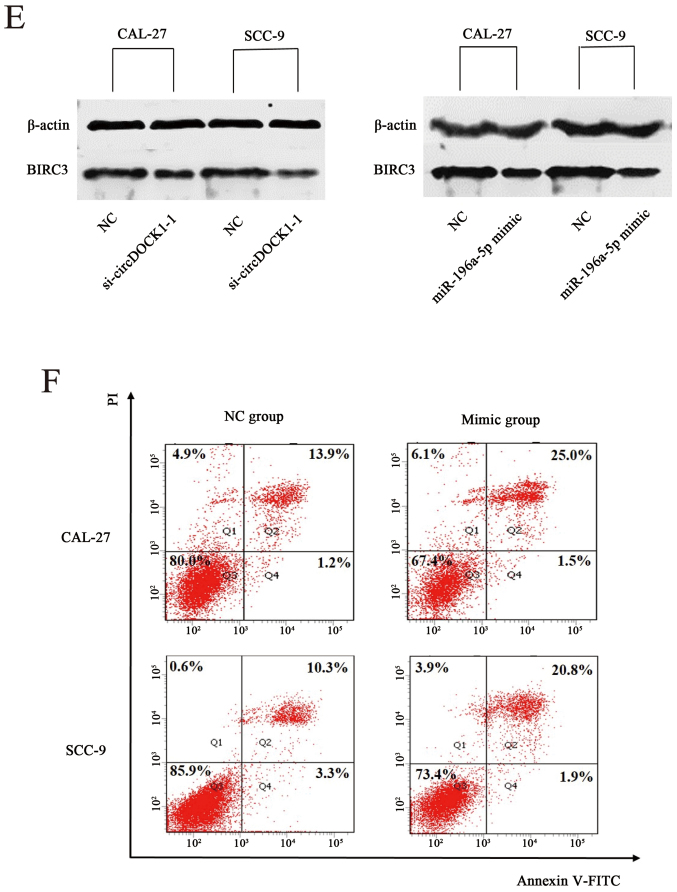
The anti-apoptosis effect of circDOCK1 was previously demonstrated. Moreover, a low expression model of circDOCK1 was also constructed. Bioinformatic prediction indicated that miR-196-5p may be one of the target miRNAs of circDOCK1. Therefore, the low expression model of circDOCK1 was used to detect the expression of miR-196a-5p. In addition, a negative group served as a control. Following transfection for 48 h, the expression levels of miR-196a-5p were upregulated distinctly in the low-expression circDOCK1 group compared with the negative control in CAL-27 and SCC-9 cells, while the expression levels of BIRC3 in the low-expression circDOCK1 group were downregulated. However, FOXO1 and MDM2 expressed no significant differences in expression (Fig. 6A). Moreover, the expression levels of miR-196a-5p in the OSCC tissue were lower than those in the para-carcinoma tissue as determined by qRT-PCR, while higher expression of BIRC3 was consistently detected in OSCC tissue (ten groups) (Fig. 6B). Then, the overexpression model of miR-196a-5p was constructed with mimics of miR-196a-5p in CAL-27 cells. Following transfection with miR-196a-5p mimics for 48 h, the expression levels increased nearly seven 7,000-fold (Fig. 6C). Accordingly, the expression levels of circDOCK1 and BIRC3 were downregulated in CAL-27 and SCC-9 cells as determined by qRT-PCR and western blotting (Fig. 6D and E). Furthermore, FCM revealed that the cell apoptosis rate after transfection with miR-196a-5p mimics increased by 11.4% and 9.1% in CAL-27 and SCC-9 cells, respectively (Fig. 6F). These results revealed that OSCC cell apoptosis could be regulated through the circDOCK1/miR-196a-5p/BIRC3 pathway.
Figure 6.
The circDOCK1/miR-196a-5p/BIRC3 pathway regulated OSCC cell apoptosis. (A) Following transfection with si-circDOCK1-1 for 48 h, the expression levels of miR-196-5p were distinctly upregulated, while the expression level of BIRC3 in the low-expression circDOCK1 group was downregulated. However, FOXO1 and MDM2 exhibited no significant difference in expression. (B) The expression level of miR-196a-5p was significantly downregulated in OSCC tissue compared to para-carcinoma tissue, while the expression levels of BIRC3 were upregulated. (C) Following transfection with miR-196a-5p mimics, the expression level was increased nearly 7,000-fold. (D) The expression levels of circDOCK1 and BIRC3 were downregulated in CAL-27 and SCC-9 cells after transfection with miR-196a-5p mimics as determined by qRT-PCR. (E) The expression levels of circDOCK1 and BIRC3 were downregulated in CAL-27 and SCC-9 cells after transfection with miR-196a-5p mimics as determined by western blotting. (F) FCM revealed that the cell apoptosis rate following transfection with miR-196a-5p mimics was increased by 11.4% and 9.1% in CAL-27 and SCC-9 cells, respectively. All data are expressed as the mean ± SE of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the control. OSCC, oral squamous cell carcinoma; FCM, flow cytometry.
Discussion
As early as 1976, circRNAs have been found in viroids (36) however they were often considered as splicing byproducts with low abundance (6). With the emergence of new generation sequencing technology and the progress of bioinformatics, numerous circRNAs were found in eukaryotic organisms and viruses (37–40). The regulatory effect of circRNAs in eukaryotic cells was gradually recognized (4,10,11). Sanger et al (36) found that viroids were single-stranded covalently closed circRNA molecules, which could infect certain higher plants. Unlike the linear RNAs, the 3- and 5-ends of circRNAs were found to be joined together to form covalently closed loop structures, which indicated their potential important function (8). Salzman et al found that numerous circRNAs existed in most normal cells and cancer cells. The function of circRNAs were confirmed in succession (41). In addition to the potential regulatory function, circRNAs may become new biomarkers of cancer diagnosis and targeted therapy.
TNF-α could induce cancer cell apoptosis, which is an accepted fact. The apoptosis effect in multiple types of cancer have been demonstrated (42). The construction of an apoptosis model in OSCC cell lines was successful, which revealed that TNF-α could affect apoptosis in OSCC.
In the present study, we first constructed an OSCC cell apoptosis model and corresponding differentially expressed circRNA profiles. The circRNA profiles revealed that hundreds of circRNAs were aberrantly expressed in the OSCC cell apoptosis model compared with the negative control, suggesting that circRNAs were involved in the regulation of cell phenotype. We also detected the differentially expressed circRNA profiles in OSCC tissue and para-carcinoma tissue. Upon comparison of these two sets of data, we determined that dozens of circRNAs overlapped, such as circDOCK1, circDLG1, circFBXL5, circPLCB1, circLOC401320, circDHDDS, circZFYVE27, circPRDX3, circPPAPDC1A, circQSER1, circCAPRIN1, circGAPDH, circRNFT2, circMETTL3, circEML1, circHERC2P3, circCIB2, circABCC1, circCLN3, circABCC3, circPGPEP1, circCACNG7, circLMF2, circRAD54L2, circABHD6, circLRIG1, circYEATS2, circSENP5, circDEPDC1B, circG3BP1, circNUP153, circLRRC16A, circBVES, circFAM120B, circSND1, circCREB3L2, circTRIM24, circDOCK5, circKAT6A, circC9orf5, circSUSD1, circZMIZ1, circRPL27A, circEXOC7, circMCM5, circQRICH1, circTJAP1, circCOL26A1, which indicated that these overlapped differentially expressed circRNAs may be significant in the regulation of OSCC cell apoptosis. CircDOCK1 was confirmed to be markedly dysregulated in the apoptosis model, OSCC cell lines and OSCC tissue compared with the negative control, HOK and para-carcinoma tissue, respectively. A previous study revealed that circRNAs from DOCK1 were strongly decreased by TGF-β treatment, while the DOCK1 mRNA was increased 2-fold. Furthermore TGF-β treatment could induce epithelial mesenchymal transformation (EMT). This indicated that one of the functions of circRNAs from DOCK1 may induce the downregulation of mRNAs in epithelial cells, keeping cellular stability (43). However, numerous circRNAs from DOCK1, may possess other functions in biological regulation. Therefore, circDOCK1 may be a promising biomarker and therapeutic target for OSCC, and its other functions in OSCC will be detected continuously in our laboratory.
In addition, it is the first time that differentially expressed circRNA profiles in OSCC were constructed. We also constructed the circRNA/miRNA networks of OSCC, which enriched competing endogenous RNAs (ceRNAs). As negative regulatory factors, miRNAs could degrade or suppress their target mRNAs (32,44,45). However, in the past two years, circRNAs have been demonstrated to serve as miRNA sponges, such as the circRNAs CDR1as (7), SRY(7) and circ-ITCH (46). They could interact with relevant miRNAs based on conserved seed sequence matches, which suppress the negative effect of miRNAs, thus upregulating the target mRNAs. For instance, miR-7, which could be adsorbed by CDR1as, and could inhibit the EGFR signaling pathway by targeting EGFR, Raf1, PSME3, PLEC1, CKAP4, CNOT8, CNN3, CAPZA1, PFN2 and ARF4 (47). Furthermore, the EGFR signaling pathway exhibited a significant connection with cancer development (48). Therefore, circRNAs play a crucial role in the ceRNA system. As an upstream regulator, the change in the expression levels of circRNA may lead to the change of miRNAs and mRNAs. Eventually the balance of cells would lead to abnormity, including cancers. According to recent research, apart from their function as miRNA-sponges, circRNAs could serve as regulators of alternative splicing (9), transcription factors (10) and encode for proteins (11). The regulatory mechanism of circRNAs is complex and it is not clear how many circRNAs could function as miRNA-sponges. To date, there has been no report on circRNAs in OSCC.
Improving the apoptosis rate is a great approach to hinder the development of cancers. As a result, we found that circDOCK1 was significantly downregulated in the apoptosis model compared to the control. Moreover, our results revealed that circDOCK1 was highly expressed in OSCC tissue and cell lines compared to para-carcinoma and HOK cells, respectively. In order to further confirm the biological function of circDOCK1 in OSCC, we downregulated its expression using siRNAs and the apoptosis rate increased, which was consistent with our previous results. With the use of bioinformatics analysis, we surmised that circDOCK1 could serve as miR-196a-5p sponge. DIANA-miRPath analysis revealed that miR-196a-5p may be implicated in cancer-associated pathways and regulate apoptosis-related genes. Based on diverse bioinformatics data, we depicted a circRNA/miRNA/mRNA regulatory pathway in OSCC. The data revealed that 61 genes, which were related to miR-196a-5p, were involved in cancer-related pathways and the p53 signaling pathway. Following the use of DAVID functional annotations, we found that BIRC3, FOXO1 and MDM2 participated in apoptosis-related pathways. In order to validate the circDOCK1/miR-196a-5p/mRNA axis, our results revealed that when the expression level of circDOCK1 was downregulated, miR-196-5p was upregulated while BIRC3 was downregulated accordingly. Moreover, when the expression level of miR-196-5p was upregulated, circDOCK1 and BIRC3 were downregulated accordingly, too. We also detected the expression level of miR-196a-5p and BIRC3 in OSCC tissue and cell lines compared to para-carcinoma and HOK cells, respectively and found that miR-196a-5p was expressed at a low level while BIRC3 was highly expressed in OSCC tissue and cell lines. Both the RNA level and protein level exhibited the same outcome. A recent study demonstrated that both in vivo and in vitro BIRC3 upregulation led to apoptosis evasion and therapeutic resistance in genome-wide analysis of glioblastoma (GBM) (49). Inhibition of IAP protein expression by small molecule inhibitors or molecular biological assays could promote tumor cell apoptosis and improve sensitivity to chemotherapy (50), which was consistent with our results. However, we noted that the apoptotic rate increased predominately in quadrant 2 rather than quadrant 4 [compared figs. 4D and E, 6F and 1A (apoptosis model)] in the siRNA experiment, which may signify that more late apoptosis/necrosis occurred.
In conclusion, our results demonstrated that circDOCK1 was upregulated and revealed its apoptosis-regulating function in OSCC. Using multiple bioinformatic methods, we constructed the circRNA/miRNA/mRNA regulatory network in OSCC. Finally, the apoptosis-regulating function of the circDOCK1/miR-196a-5p/BIRC3 axis was further confirmed in OSCC. In the future, other functions of circDOCK1 may be detected in our laboratory however our present results revealed that circDOCK1 may serve as a promising diagnostic biomarker and therapeutic target in OSCC.
References
- 1.Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, et al. Global Burden of Disease Cancer Collaboration The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lingen MW, Kalmar JR, Karrison T, Speight PM. Critical evaluation of diagnostic aids for the detection of oral cancer. Oral Oncol. 2008;44:10–22. doi: 10.1016/j.oraloncology.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghantous Y, Yaffi V, Abu-Elnaaj I, Oral cavity cancer epidemiology and early diagnosis. Refuat Hapeh Vehashinayim (1993) 2015;32(71):55–63. (In Hebrew) [PubMed] [Google Scholar]
- 4.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasda E, Parker R. Circular RNAs: Diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30:2243–2246. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar L, Shamsuzzama, Haque R, Baghel T, Nazir A. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol. 2016 Oct 29; doi: 10.1007/s12035-016-0213-8. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 14.Nan A, Chen L, Zhang N, Liu Z, Yang T, Wang Z, Yang C, Jiang Y. A novel regulatory network among LncRpa, CircRar1, miR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch Toxicol. 2017;91:1671–1684. doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonizzato A, Gaffo E, Te Kronnie G, Bortoluzzi S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016;6:e483. doi: 10.1038/bcj.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a miR-136 ‘sponge’ in human cartilage degradation. Sci Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R, Su Y, Xie H, Li H, Xu X, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 19.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Hao X, Wang H, Liu Z, He Y, Pu M, Zhang H, Yu H, Duan J, Qu S. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem. 2016;40:1334–1344. doi: 10.1159/000453186. [DOI] [PubMed] [Google Scholar]
- 21.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, Li G, Zhu Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget. 2016;7:63449–63455. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao Z, Yang J, Wang C, Li Y, Zhang Y, Dong X, Zhou L, Liu J, Zhang Y, Qian J. MicroRNA-7 inhibits metastasis and invasion through targeting focal adhesion kinase in cervical cancer. Int J Clin Exp Med. 2015;8:480–487. [PMC free article] [PubMed] [Google Scholar]
- 25.Berg V, Rusch M, Vartak N, Jüngst C, Schauss A, Waldmann H, Hedberg C, Pallasch CP, Bastiaens PI, Hallek M, et al. miRs-138 and −424 control palmitoylation-dependent CD95-mediated cell death by targeting acyl protein thioesterases 1 and 2 in CLL. Blood. 2015;125:2948–2957. doi: 10.1182/blood-2014-07-586511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R, Liu X, Zheng Y, Gu J, Xiong S, Jiang P, Jiang X, Huang E, Yang Y, Ge D, et al. MicroRNA-7 sensitizes non-small cell lung cancer cells to paclitaxel. Oncol Lett. 2014;8:2193–2200. doi: 10.3892/ol.2014.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Tang H, Yin S, Dong C. Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep. 2013;29:2046–2052. doi: 10.3892/or.2013.2304. [DOI] [PubMed] [Google Scholar]
- 28.Ayaz L, Çayan F, Balci Ş, Görür A, Akbayir S, Yıldırım Yaroğlu H, Doğruer Unal N, Tamer L. Circulating microRNA expression profiles in ovarian cancer. J Obstet Gynaecol. 2014;34:620–624. doi: 10.3109/01443615.2014.919998. [DOI] [PubMed] [Google Scholar]
- 29.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 31.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 35.Gomes CC, de Sousa SF, Gomez RS. MicroRNAs: Small molecules with a potentially role in oral squamous cell carcinoma. Curr Pharm Des. 2013;19:1285–1291. doi: 10.2174/1381612811319070012. [DOI] [PubMed] [Google Scholar]
- 36.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures; Proc Natl Acad Sci USA; 1976; pp. 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313–319. doi: 10.1016/0092-8674(80)90505-X. [DOI] [PubMed] [Google Scholar]
- 38.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 39.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-Y. [DOI] [PubMed] [Google Scholar]
- 40.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: Correlation with exon skipping; Proc Natl Acad Sci USA; 1996; pp. 6536–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 43.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 44.Mallory AC, Vaucheret H. MicroRNAs: Something important between the genes. Curr Opin Plant Biol. 2004;7:120–125. doi: 10.1016/j.pbi.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 46.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YJ, Chien PH, Chen WS, Chien YF, Hsu YY, Wang LY, Chen JY, Lin CW, Huang TC, Yu YL, et al. Hepatitis B virus-encoded X protein downregulates EGFR expression via inducing microRNA-7 in hepatocellular carcinoma cells. Evid Based Complement Alternat Med. 2013;2013:682380. doi: 10.1155/2013/682380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W, Jing H, Zhang F. Epidermal growth factor receptor-targeted therapy in colorectal cancer. Front Biosci (Landmark Ed) 2016;21:410–418. doi: 10.2741/4397. [DOI] [PubMed] [Google Scholar]
- 49.Wang D, Berglund A, Kenchappa RS, Forsyth PA, Mulé JJ, Etame AB. BIRC3 is a novel driver of therapeutic resistance in Glioblastoma. Sci Rep. 2016;6:21710. doi: 10.1038/srep21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hird AW, Aquila BM, Hennessy EJ, Vasbinder MM, Yang B. Small molecule inhibitor of apoptosis proteins antagonists: A patent review. Expert Opin Ther Pat. 2015;25:755–774. doi: 10.1517/13543776.2015.1041922. [DOI] [PubMed] [Google Scholar]