Abstract
Background
The rs2282679 A>C polymorphism in the vitamin D binding protein gene is associated with lower circulating levels of vitamin D. We investigated associations of this SNP with colorectal cancer (CRC) risk and survival and whether the associations vary by dietary vitamin D intake and tumor molecular phenotype.
Methods
A population-based case-control study identified 637 incident CRC cases (including 489 participants with follow-up data on mortality end-points) and 489 matched controls. Germline DNA samples were genotyped with the Illumina Omni-Quad 1 Million chip in cases and the Affymetrix Axiom® myDesign™ Array in controls. Logistic regression examined the association between the rs2282679 polymorphism and CRC risk with inclusion of potential confounders. Kaplan-Meier curves and multivariable Cox models assessed the polymorphism relative to overall survival (OS) and disease-free survival (DFS).
Results
The rs2282679 polymorphism was not associated with overall CRC risk; there was evidence, however, of effect modification by total vitamin D intake (Pinteraction = 0.019). Survival analyses showed that the C allele was correlated with poor DFS (per-allele HR, 1.36; 95%CI, 1.05–1.77). The association of rs2282679 on DFS was limited to BRAF wild-type tumors (HR, 1.58; 95%CI, 1.12–2.23). For OS, the C allele was associated with higher all-cause mortality among patients with higher levels of dietary vitamin D (HR, 2.11; 95%CI, 1.29–3.74), calcium (HR, 1.93; 95%CI, 1.08–3.46), milk (HR, 2.36; 95%CI, 1.26–4.44), and total dairy product intakes (HR, 2.03; 95%CI, 1.11–3.72).
Conclusion
The rs2282679 SNP was not associated with overall CRC risk, but may be associated with survival after cancer diagnosis. The association of this SNP on survival among CRC patients may differ according to dietary vitamin D and calcium intakes and according to tumor BRAF mutation status.
Keywords: Vitamin D binding protein, Genetic polymorphism, Colorectal cancer, Dietary vitamin D, Microsatellite instability, BRAF mutations
Background
Colorectal cancer (CRC) is a complex, multifactorial disease resulting from multiple genetic and environmental factors [1]. Vitamin D from diet, supplements, and cutaneous synthesis from sunlight, is associated with lower risks of CRC incidence [2–5] and mortality [6–11]. The anti-carcinogenic effects of vitamin D might vary by the vitamin D-binding protein (DBP) [12]. As the major carrier protein in systemic circulation, DBP reversibly binds and transports vitamin D metabolites to different target organs, including the colorectum, thereby influencing the bioavailability of active 25-hydroxyvitamin D (25(OH)D) [12, 13]. Additionally, DBP is the precursor molecule of a potent macrophage-activating factor (GcMAF) [14], which is highly tumoricidal against various malignancies through its ability to inhibit endothelial angiogenesis [15, 16] and stimulate the inflammation-primed phagocytic activity of tumoricidal macrophages [17]. Therefore, DBP would be hypothesized to play an important role in CRC initiation and progression, either alone or in combination with vitamin D [18].
The gene encoding DBP, GC gene, is highly polymorphic. The single nucleotide polymorphism (SNP) rs2282679 A>C is one of the most commonly studied variants in this gene, which has been shown to be robustly correlated with serum levels of 25(OH)D in recent genome-wide association studies (GWAS) [19, 20]; specifically, the C allele of this SNP is associated with lower levels of 25(OH)D [20]. Prior studies on GC variants have been performed on melanoma [21], prostate [22] and breast cancers [23, 24]; however, we found only two studies that evaluated the specific association of the GC rs2282679 polymorphism with CRC risk, with both studies reporting no evidence of association [25, 26]. Another study reported that the GC rs2282679 SNP was associated with prognosis for patients diagnosed with stages II and III colon cancer [27].
Microsatellite instability (MSI) and BRAF V600E hotspot mutation are important molecular classifiers in CRC, which define distinct CRC subgroups arising from different oncogenic pathways. Microsatellite unstable (MSI-H) tumors are generally associated with superior prognosis [28] whereas BRAF-mutated cancers are related to inferior survival [29, 30]. Therefore, it is plausible that factors associated with CRC risk and survival differ across tumor molecular subtypes defined by MSI and BRAF mutation status. Prior studies have reported that the associations between genetic variations in vitamin D and calcium metabolic pathway and CRC vary according to MSI status, with significant associations for microsatellite unstable CRC only [31, 32]. However, no study has yet evaluated the relationship between the GC rs2282679 polymorphism and CRC by these tumor molecular alternations.
In this analysis, we assessed the associations of the GC rs2282679 variant with CRC risk and survival. We additionally evaluated the potential influence of this SNP according to dietary vitamin D, calcium, milk, and total dairy product intake and whether associations varied by tumor microsatellite instability (MSI) or BRAF Val600Glu mutation status.
Methods
Study participants
Study data and biologic specimens were drawn from the Newfoundland and Ontario Familial Colorectal Cancer Study (NFCCS), a large population-based case-control study designed to identify genetic and environmental risk and prognostic factors for CRC [33, 34]. The detailed rationale and methodology of NFCCS has been described elsewhere [4, 33, 35–37]. For the current study, only the participants from the NL portion were analyzed. Briefly, men and women with pathologically confirmed CRC were identified through the Newfoundland Familial Colorectal Cancer Registry (NFCCR). Eligibility criteria included patients newly diagnosed with CRC from 1999 to 2003 and aged 20–75 years at the time of diagnosis. Controls were selected by random digit dialing and matched on age (±5 years) and sex with cases at baseline [38]. All consenting participants were sent self-administered risk factor questionnaires (a Food Frequency Questionnaire (FFQ), a Family History Questionnaire (FHQ), and a Personal History Questionnaire (PHQ)), and were asked to provide blood samples and for permission to access their tumor specimens and medical records (for cases). A total of 656 cases and 696 controls completed detailed questionnaires and donated a blood sample. Of the 656 cases, 490 were followed for mortality and recurrence from the date of cancer diagnosis to April 2010. Vital status (i.e., death, recurrence, and metastasis) was ascertained through periodic follow-up questionnaires (e.g., FHQ), local newspapers, death certificates, pathology records, autopsy records, physicians’ notes, surgical reports, and from records at the Dr. H. Bliss Murphy Cancer Care Foundation. The main study survival outcomes were death from all-causes (i.e., overall survival (OS)) and disease-free survival (DFS), defined as death, recurrence, or metastasis (whichever came first). Follow-up time began at CRC diagnosis, and individuals who were lost to follow-up or did not die, had a recurrence or had a metastasis were censored at the time of their last contact.
Exclusions were made if patients had equivocal genotype or clinical outcome, or failed to provide sufficient information on other critical predictors. Thus, 637 cases and 489 controls for risk analyses and 489 patients for survival analysis were included in the final study.
Diet assessment and baseline information collection
Information on diet and other lifestyle, medical and demographic characteristics was gathered with self-administered questionnaires. The dietary questionnaire was an adaptation of the Hawaii semi-quantitative FFQ to assess the dietary habits of participants from a year prior to disease diagnosis (cases) or interview (controls), which has been validated in a prior study [39]. The FFQ contained questions regarding the brand and frequency of consumption of 170 foods and beverages plus multivitamin and individual vitamin supplements [40]. The nutrient intakes from diet were calculated by multiplying the frequency of consumption of each food item by the nutrient content per average unit [4]. Total daily nutrient intakes were computed by incorporating supplement use in addition to intakes from diet. The PHQ collected information from each participant on socio-demographics (e.g., age, gender, ethnicity, and education attainment), medical conditions, bowel screening history, aspirin use, physical activity, and recent or prior alcohol and tobacco use. The FHQ gathered baseline and follow-up family history data from the participants.
Genotyping
Genotyping for the GC rs2282679 allele was conducted using the Illumina Human Omni-Quad Beadchip that contains about 1.1 million SNPs at Centrillion Biosciences (USA). Control individuals were genotyped in the Laboratory of Dr. Stephen Gruber (Director, USC Norris Comprehensive Cancer Center, Los Angeles) using the Affymetrix Axiom® myDesign™ GW Array Plate, which contains 1.3 million probes. To monitor quality and consistency between the two platforms, DNA samples from 200 CRC patients were typed on both platforms. As the DNA from cases and controls were genotyped on different platforms, a genotype imputation strategy was implemented to integrate the two datasets using IMPUTE2 [41] with multi-population reference panels from 1000 Genomes (Phase 1). The imputation approach was validated based on the overlapping SNPs between the two platforms and the genotypes from 200 CRC samples that were typed on both platforms. SNPs with genotype concordance < 97% across the two platforms were removed from further analysis. For the purpose of the current study, directly measured data from both arrays on rs2282679 were retrieved from the genome-wide SNP genotype database of the NFCCR.
Our protocol for MSI and BRAF V600E mutation analyses in tumor DNA has been described previously [42–44]. MSI status was evaluated with 5 to 10 microsatellite markers. Tumors were deemed MSI-high if ≥30% of the repeats were unstable and MS-stable/MSI-low if < 30% of the repeats were unstable. The c.1799 T > A variant (Val600Glu mutation) region of the BRAF gene was amplified by BRAF allele-specific polymerase chain reaction technique [44].
Statistical analysis
Group comparisons between cases and controls were performed with two-sample t test for continuous variables and Chi-square (χ2) test for categorical variables. The Hardy-Weinberg Equilibrium for rs2282679 genotype was evaluated using χ2 goodness-of-fit test. Unconditional logistic regression was used to estimate the association between the rs2282679 GC SNP and risk for CRC as odds ratio (OR) with 95% confidence interval (CI). Initially, logistic regression models only included genotype, age and sex. More complex models also included family history of CRC, screening procedure, multivitamin use, folic acid intake, smoking history, and education attainment. These covariates were retained in the final models because they entered the model at P < 0.1, altered the parameter estimates by > 10%, and/or improved the model fit.
In survival analysis, survival curves were constructed with the Kaplan–Meier method. The log-rank test and the Cox regression models were used for univariable and multivariable survival analyses to assess the association between the SNP of interest and OS and DFS of CRC. The assumption of proportional hazards for each Cox model was verified by testing the statistical significance of time-dependent covariates in the model. The hazard rate ratio (HR) and 95% CI were calculated from the Cox models. As the true inheritance mode of the rs2282679 polymorphism has not yet been established, the SNP was analyzed for risk and survival under dominant, additive, and recessive models. Given the limited sample size in some subgroups, we combined those who carried at least one of the minor C alleles in stratified analysis by selected tumor molecular phenotype. Linear trend for gene dose effect was tested by modeling ordinal variables of allele dose (0, 1, and 2) as a continuous variable. Gene-environment interactions were tested by introducing a multiplicative interaction term into the model and assessing its significance with the Wald method. Two-sided exact P < 0.05 was considered statistically significant. We did not adjust for multiple comparisons because the sub-tests in the current study are not independent of each other since the stratified variables are highly correlated (i.e., vitamin D, calcium, and dairy products). Although adjustment for multiple testing reduces type I error, it increases type II error and errors of interpretation [45]. All data management and analyses were performed using SAS software, Version 9.3.
Results
The rs2282679 polymorphism was in Hardy-Weinberg equilibrium (P > 0.05). Among controls, the genotype frequency was 7.4% homozygous (CC), 42.5% heterozygous (AC) and 50.1% wild-type homozygous (AA); the observed minor allele frequencies in the controls were comparable to that previously reported [25]. During a maximum follow-up of 10.9 years (mean: 6.3 years), 150 deaths occurred among the 489 patients included in the survival analysis. The cause of death defined by ICD codes was obtained for 105 of 150 deceased patients; thereof the majority (90.5%) was due to CRC.
Baseline characteristics of cases and controls
Cases and controls had similar sex and ethnicity distributions, and the majority reported their race as White (Table 1). Relative to cases, controls were slightly younger, leaner (lower body mass index), better educated, less likely to smoke, and more likely to have had a colorectal cancer screening/early detection procedure. Among those who completed the FFQ, total vitamin D and calcium intakes were significantly higher in controls than in cases (P = 0.001). Family history of CRC (first-degree relatives affected only) was reported by 9.8% of the patients and 7.5% of the controls. MSI-high was identified in 61 of 507 (12.0%) tumors, and BRAF Val600Glu mutation was detected in 10.8% of tumors.
Table 1.
Selected demographical and clinicopathological characteristics of study population (cases and controls)
| Characteristics | Cases | Controls | P-valuea |
|---|---|---|---|
| No.(%) | No.(%) | ||
| Age (year)b | 63.1 ± 8.6 | 61.2 ± 9.0 | 0.001 |
| BMI (kg/m2)b | 28.1 ± 5.0 | 27.3 ± 4.5 | 0.003 |
| Sex | |||
| Men | 399 (62.9) | 272 (58.5) | |
| Women | 235 (37.1) | 193 (41.5) | 0.136 |
| Race | |||
| White | 615 (97.0) | 444 (95.5) | |
| Other | 19 (3.0) | 21 (5.0) | 0.184 |
| Family history of CRC | |||
| Yes | 62 (9.8) | 35 (7.5) | |
| No | 572 (90.2) | 430 (92.5) | 0.194 |
| Level of education | |||
| Lower than high school | 302 (47.8) | 140 (30.2) | |
| High-school graduate | 100 (15.8) | 74 (16.0) | |
| College | 177 (28.0) | 175 (37.7) | |
| Bachelor or higher | 53 (8.4) | 75 (16.1) | < 0.001 |
| Smoking history | |||
| Current | 129 (20.3) | 58 (12.5) | |
| Former | 325 (51.3) | 228 (49.4) | |
| Never | 180 (28.4) | 176 (38.1) | < 0.001 |
| Reported screening procedure | |||
| Yes | 66 (10.4) | 107 (23.0) | |
| No | 568 (89.6) | 358 (77.0) | < 0.001 |
| Fruit intake (servings c/wk)b | 9.7 ± 8.3 | 10.9 ± 8.1 | 0.027 |
| Total vitamin D intake (μg/d)b | 8.7 ± 6.4 | 10.2 ± 7.9 | 0.001 |
| Total calcium intake (mg/d)b | 1019.8 ± 488.8 | 1097.1 ± 567.7 | 0.018 |
| Milk (g/d)b | 287.1 ± 278.0 | 299.9 ± 295.5 | 0.465 |
| Total dairy products (g/d)b | 364.5 ± 303.3 | 389.9 ± 340.0 | 0.200 |
| Tumor location | |||
| Colon | 417 (65.8) | – | – |
| Rectum | 217 (34.2) | – | – |
| Stage at diagnosis | |||
| I/II | 288 (58.9) | – | – |
| III/IV | 201 (41.1) | – | – |
| Chemoradiotherapy | |||
| Yes | 106 (19.9) | – | – |
| No | 427 (80.1) | – | – |
| MSI status | |||
| MSS/MSI-L | 446 (88.0) | – | – |
| MSI-H | 61 (12.0) | – | – |
| BRAF mutation status | |||
| Wild type | 445 (89.2) | – | – |
| BRAF mutant | 54 (10.8) | – | – |
Abbreviations: BMI body mass index, CRC colorectal cancer, MSI microsatellite instability, MSI-H microsatellite instability-high, MSS/MSI-L microsatellite stable/microsatellite instability-low
aPvalues are for the significance of the two-sample t test for continuous variables and of the chi-square test for categorical variables
bContinuous variables presented as mean ± s.d. (standard deviation)
cA serving of fruit is: 1 medium-sized fresh fruit; ½ cup of chopped, cooked or canned fruit; ¼ cup of dried fruit; 6 oz of fruit juice (50%–100% pure juice)
Subjects with missing values are not reported in the table
Association of rs2282679 genotype with CRC risk
The rs2282679 SNP was not associated with risk of CRC overall or when stratified by MSI or BRAF-mutation subtypes (Table 2). Specifically, the odds ratio was 1.10 (95% CI, 0.88–1.37) per variant C allele and 1.21 (95% CI, 0.70–2.09) in CC homozygotes compared with AA homozygotes. The ORs were similar for men and women and did not differ according to tumor anatomical sub-site (data available upon request).
Table 2.
Frequency distribution and associations of GC SNP rs2282679 with CRC risk (overall and by molecularly defined subtypes)
| rs2282679 Genotype/Allele | Cases N(%) |
Controls N(%) |
OR (95% CI)a | OR (95% CI)b |
|---|---|---|---|---|
| All cases vs. controls | ||||
| AA | 309 (48.5) | 245 (50.1) | 1.00 | 1.00 |
| AC | 282 (44.3) | 208 (42.5) | 1.08 (0.82–1.41) | 1.10 (0.83–1.47) |
| CC | 46 (7.2) | 36 (7.4) | 1.06 (0.63–1.77) | 1.21 (0.70–2.09) |
| P trendc | 0.647 | 0.403 | ||
| CC + AC (vs. AA) | 328 (51.5) | 244 (49.9) | 1.07 (0.83–1.39) | 1.12 (0.85–1.47) |
| CC (vs. AC + AA) | 46 (7.2) | 36 (7.4) | 1.02 (0.62–1.68) | 1.15 (0.68–1.97) |
| Per C allele | 1.05 (0.85–1.29) | 1.10 (0.88–1.37) | ||
| BRAF V600E mutant cases vs. controls | ||||
| AA | 25 (46.3) | 245 (50.1) | 1.00 | 1.00 |
| CC + ACd | 29 (53.7) | 244 (49.9) | 1.03 (0.54–1.96) | 1.10 (0.56–2.16) |
| BRAF wild-type cases vs. controls | ||||
| AA | 223 (50.1) | 245 (50.1) | 1.00 | 1.00 |
| CC + ACd | 222 (49.9) | 244 (49.9) | 1.08 (0.82–1.42) | 1.10 (0.82–1.47) |
| MSI-H cases vs. controls | ||||
| AA | 31 (50.8) | 245 (50.1) | 1.00 | 1.00 |
| CC + ACd | 30 (49.2) | 244 (49.9) | 0.99 (0.56–1.80) | 1.03 (0.56–1.90) |
| MSS/MSI-Low cases vs. controls | ||||
| AA | 223 (50.0) | 245 (50.1) | 1.00 | 1.00 |
| CC + ACd | 223 (50.0) | 244 (49.9) | 1.09 (0.83–1.44) | 1.15 (0.86–1.53) |
Abbreviations: CRC colorectal cancer, OR odds ratio, MSI-H microsatellite instability-high, MSS/MSI-L microsatellite stable/microsatellite instability-low
aCrude model adjusted for age and sex
bMultivariable model additionally adjusted for family history of CRC, screening procedure, multivitamin use, folic acid intake, smoking history, and education attainment where applicable
cLinear trend tested by modeling the ordinal variables of genotype dose as a continuous variable
dCC and AC genotypes were analyzed jointly because of limited sample size in some subgroups
Interactions of rs2282679 genotype with dietary characteristics in relation to CRC risk
A previous NFCCS study demonstrated that total vitamin D intake was inversely associated with CRC incidence [4]. We therefore cross classified subjects on total vitamin D intake and other related dietary factors with rs2282679 genotype for CRC risk (Table 3). Among participants with the more common AA genotype, we confirmed the association, observing a 2.5-fold increased risk of CRC in individuals consuming total vitamin D in the lowest tertile than in those in the highest tertile (95% CI, 1.52–4.09). Among carriers of the C allele, however, no appreciable difference in ORs between strata of total vitamin D intake was observed. Intriguingly, carrying the risk allele (i.e. the heterozygous/homozygous genotypes) conferred an enhanced risk for this malignancy in the presence of high level of vitamin D intake (OR, 1.65; 95% CI, 1.01–2.71; P interaction = 0.019). Stratification on consumption of milk and total dairy products suggested borderline significant effect modifications (P interaction = 0.056 for milk and 0.079 for total dairy products). Additionally, there was no evidence that the associations of this SNP with CRC risk were modified by body mass index (obese vs. not obese), drinking status (non-drinkers vs. drinkers) or smoking history (non-smokers vs. smokers) (data available upon request).
Table 3.
Risk estimates for interactions between GC rs2282679 genotypes and non-genetic dietary factors
| GC rs2282679 genotype | P interaction b | ||
|---|---|---|---|
| AA OR (95% CI)a |
AC + CC OR (95% CI)a |
||
| Total vitamin D intake (μg/d) | |||
| Highest tertile (≥10.1) | 1.00 | 1.65 (1.01–2.71) | |
| Lowest tertile (< 5.0) | 2.50 (1.52–4.09) | 1.72 (1.03–2.90) | 0.019 |
| Total calcium intake (mg/d) | |||
| Highest tertile (≥1195.4) | 1.00 | 1.45 (0.89–2.36) | |
| Lowest tertile (< 761.4) | 1.21 (0.73–2.00) | 1.63 (0.95–2.81) | 0.827 |
| Milk (g/d) | |||
| Highest tertile (≥321.3) | 1.00 | 1.61 (0.99–2.61) | |
| Lowest tertile (< 146.5) | 1.46 (0.90–2.38) | 1.21 (0.72–2.04) | 0.056 |
| Total dairy products (g/d) | |||
| Highest tertile (≥432.0) | 1.00 | 1.61 (0.99–2.61) | |
| Lowest tertile (< 210.6) | 1.44 (0.90–2.32) | 1.27 (0.75–2.14) | 0.079 |
Abbreviations: CRC colorectal cancer, OR odds ratio
aAdjusted for age, sex, family history of CRC, screening procedure, folic acid intake, smoking history, and education attainment where applicable
bP for interaction is computed with Wald method testing significance of multiplicative interaction term between GC SNP rs2282679 genotype and respective stratified variable
Those with P < 0.05 are in bold
Association of rs2282679 genotype and survival outcome
Survival analysis showed a positive association between rs2282679 polymorphism and reduced DFS of CRC, with the co-dominant CC and the dominant CC + AC vs. AA model exhibiting 1.93- and 1.40- fold increases in the risk for DFS, respectively (Table 4, Fig. 1). The per-allele HR was 1.36 (95% CI, 1.05–1.77; P trend = 0.020). The adverse prognosis in relation to the risk allele was limited to BRAF wild-type tumors (HR, 1.58; 95% CI, 1.12–2.23). Among patients harboring the BRAF Val600Glu mutation, the DFS was essentially the same for individuals with and without the C allele (HR, 0.95; 95% CI, 0.25–3.62; P interaction = 0.043). Although no evidence existed for a differential prognostic role of rs2282679 according to MSI status, patients with the AA genotype/MSI-high tumors experienced the most favorable DFS whereas patients with AC + CC genotypes/MSS + MSI-low tumors had the worst DFS (Fig. 2). The 5-year DFS of CRC was 92% for AA/MSI-high, 82% for AC + CC genotypes/MSI-high, 68% for AA genotype/MSS + MSI-low, and 65% for AC + CC genotypes/MSS + MSI-low tumors (Log-rank P = 0.0013, Fig. 2). Our results did not confirm a prognostic relevance of rs2282679 in OS of CRC.
Table 4.
Association between GC SNP rs2282679 genotypes and overall and disease free colorectal cancer survival (overall and stratified by tumor molecular phenotype)
| rs2282679 Genotype/Allele | Disease-Free Survival | P interaction c | Overall Survival | P interaction c | ||
|---|---|---|---|---|---|---|
| No. of Eventsa /At Risk |
HR (95% CI)b | No. of Eventsa /At Risk |
HR (95% CI)b | |||
| Total colorectal cancer | ||||||
| AA | 83/245 | 1.00 | 69/245 | 1.00 | ||
| AC | 80/207 | 1.33 (0.94–1.88) | 68/208 | 1.24 (0.85–1.81) | ||
| CC | 16/36 | 1.93 (1.06–3.52) | 13/36 | 1.60 (0.85–3.02) | ||
| P trendd | 0.020 | 0.107 | ||||
| CC + AC (vs. AA) | 96/243 | 1.40 (1.00–1.95) | 81/244 | 1.29 (0.90–1.85) | ||
| CC (vs. AC + AA) | 16/36 | 1.69 (0.95–2.99) | 13/36 | 1.44 (0.79–2.65) | ||
| Per C allele | 1.36 (1.05–1.77) | 1.26 (0.95–1.66) | ||||
| BRAF Val600Glu mutant | ||||||
| AA | 8/20 | 1.00 | 5/20 | 1.00 | ||
| CC + ACe | 8/26 | 0.95 (0.25–3.62) | 7/26 | 1.53 (0.47–4.95) | ||
| BRAF wild-type | ||||||
| AA | 65/200 | 1.00 | 55/200 | 1.00 | ||
| CC + AC e | 83/195 | 1.58 (1.12–2.23) | 0.043 | 70/196 | 1.33 (0.93–1.91) | 0.892 |
| MSI-H | ||||||
| AA | 3/25 | 1.00 | 0/26 | 1.00 | ||
| CC + ACe | 5/28 | 1.26 (0.18–8.96) | 3/28 | NCf | ||
| MSS/MSI-L | ||||||
| AA | 76/204 | 1.00 | 65/204 | 1.00 | ||
| CC + ACe | 87/203 | 1.34 (0.97–1.87) | 0.702 | 74/204 | 1.26 (0.89–1.77) | 0.210 |
Abbreviations: HR hazard rate ratios, CI confidence interval, MSI-H microsatellite instability-high, MSS/MSI-L microsatellite stable/microsatellite instability-low
aEvents are defined as deaths for overall survival and death, recurrence, or metastasis (whichever occurred earliest) for disease-free survival
bMultivariable Cox model adjusted for sex, age at diagnosis, tumor stage at diagnosis, marital status, race, reported chemoradiotherapy, MSI status, BRAF mutation status, tumor location, fruit intake, and body mass index where applicable
cP for interaction is computed with Wald method testing significance of multiplicative interaction term between GC SNP rs2282679 genotype and molecular subtype
dLinear trend tested by modeling the ordinal variables of genotype dose as a continuous variable
eCC and AC genotypes were analyzed jointly because of limited sample size in some subgroups
fNC: not calculated
Those with P < 0.05 are in bold
Fig. 1.
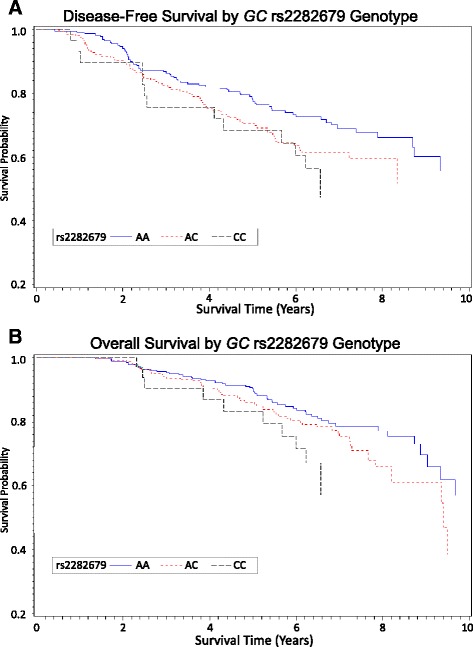
Survival curves for (a) disease-free survival and (b) overall survival by GC rs2282679 genotypes. Adjusted for sex, age at diagnosis, tumor stage at diagnosis, marital status, race, reported chemoradiotherapy, MSI status, BRAF mutation status, tumor location, fruit intake, and body mass index
Fig. 2.

Kaplan-Meier survival curves for disease-free survival according to GC rs2282679 genotypes and MSI status
Interactions of rs2282679 genotype with dietary characteristics in relation to CRC survival
The GC rs2282679 genotype interacted with dietary factors to influence OS after CRC diagnosis (Table 5). Specifically, the positive association between carriage of the C allele and poor OS seemed limited to patients in higher categories of dietary vitamin D, calcium, milk, and total dairy product intakes; the HRs associated with the AC + CC genotypes were 2.11 (95% CI, 1.29–3.74; P interaction = 0.040), 1.93 (95% CI, 1.08–3.46; P interaction = 0.043), 2.36 (95% CI, 1.26–4.44; P interaction = 0.004), and 2.03 (95% CI, 1.11–3.72; P interaction = 0.024), respectively. Low intake of pre-diagnostic milk was associated with worse OS among wild-type homozygotes (AA) only, with the HR equaled to 2.09 (95% CI, 1.09–4.02). For DFS, effect-modification analyses yielded comparable but nonsignificant results. Analyses using total vitamin D or total calcium intakes (diet+supplements) did not show any associations (patterns) more significant or different. Therefore, we only present results stratified by dietary intakes. Stratification on tumor subsite (colon vs. rectum), reported chemoradiation therapy (yes vs. no) or smoking history (non-smokers vs. smokers) detected no significant interaction (data available upon request).
Table 5.
Association between GC rs2282679 genotypes and overall and disease free colorectal cancer survival stratified by non-genetic dietary factors
| GC rs2282679 genotype | P interaction b | ||
|---|---|---|---|
| AA HR (95% CI)a |
AC + CC HR (95% CI)a |
||
| Disease-Free Survival | |||
| Dietary vitamin D (μg/d) | |||
| Highest tertile (≥7.1) | 1.00 | 1.65 (0.97–2.82) | |
| Lowest tertile (< 4.6) | 1.03 (0.58–1.83) | 1.08 (0.56–2.08) | 0.191 |
| Dietary calcium (mg/d) | |||
| Highest tertile (≥1120.2) | 1.00 | 1.58 (0.92–2.71) | |
| Lowest tertile (< 702.1) | 1.06 (0.59–1.91) | 1.14 (0.62–2.11) | 0.355 |
| Milk (g/d) | |||
| Highest tertile (≥274.6) | 1.00 | 1.63 (0.92–1.84) | |
| Lowest tertile (< 128.1) | 1.21 (0.68–2.16) | 0.97 (0.51–1.84) | 0.083 |
| Total dairy products (g/d) | |||
| Highest tertile (≥411.8) | 1.00 | 1.51 (0.87–2.60) | |
| Lowest tertile (< 199.2) | 1.00 (0.57–1.77) | 0.99 (0.53–1.83) | 0.289 |
| Overall Survival | |||
| Dietary vitamin D (μg/d) | |||
| Highest tertile (≥7.1) | 1.00 | 2.11 (1.29–3.74) | |
| Lowest tertile (< 4.6) | 1.28 (0.68–2.42) | 1.11 (0.54–2.27) | 0.040 |
| Dietary calcium (mg/d) | |||
| Highest tertile (≥1120.2) | 1.00 | 1.93 (1.08–3.46) | |
| Lowest tertile (< 702.1) | 1.55 (0.82–2.91) | 1.18 (0.59–2.35) | 0.043 |
| Milk (g/d) | |||
| Highest tertile (≥274.6) | 1.00 | 2.36 (1.26–4.44) | |
| Lowest tertile (< 128.1) | 2.09 (1.09–4.02) | 1.26 (0.61–2.62) | 0.004 |
| Total dairy products (g/d) | |||
| Highest tertile (≥411.8) | 1.00 | 2.03 (1.11–3.72) | |
| Lowest tertile (< 199.2) | 1.77 (0.94–3.33) | 1.27 (0.63–2.58) | 0.024 |
Abbreviations: HR hazard rate ratios, CI confidence interval
aAdjusted for sex, age at diagnosis, stage at diagnosis, marital status, race, reported chemoradiotherapy, MSI status, BRAF mutation status, tumor location, body mass index where applicable
bP for interaction is computed with Wald method testing significance of multiplicative interaction term between GC SNP rs2282679 genotype and respective stratified variable
Those with P < 0.05 are in bold
Discussion
In this study, we observed no clear association of rs2282679 SNP with overall CRC risk, but noted a suggestive association of the CC genotype with DFS. A previous multicenter case-control study of 10,061 CRC cases and 12,768 controls of European ancestry found no evidence for associations between GC rs2282679 and the risk of CRC overall or for colon or rectal tumor separately [26]. In a Mendelian Randomization study in Scotland, Theodoratou et al. [25] reported a nonsignificant association of rs2282679 wild-type A allele with CRC risk (Per A allele: OR, 0.97; 95% CI, 0.90–1.06). The only available previous study investigating the prognostic effect of the SNP on CRC reported consistent results that GC rs2282679 polymorphism was significantly associated with reduced time to recurrence (HR, 3.30; 95% CI, 1.09–9.97, P = 0.034) in stages II and III colon cancer patients treated with surgery alone [27].
Referring to two recent GWAS studies [19, 20], the GC rs2282679 has been identified as the strongest genomic predictor of serum vitamin D level (P = 2.0 × 10− 30). Per copy of the risk C allele was associated with an approximately 50% elevated risk for hypovitaminosis among Caucasians [20]. In another study by Zhang et al. [46], the C allele of this SNP was associated with lower circulating DBP concentrations and thus lower 25(OH)D bioavailability to target organs. Together with the fact that vitamin D has been shown to reduce the growth of CRC xenografts by influencing cell growth, differentiation, apoptosis, as well as immune-modulation, the elevated risk of C allele carriers may be attributed to their low circulating DBP and 25(OH)D concentrations relative to noncarriers [47–49]. Alternatively, DBP can be converted to GcMAF, an activator of macrophages, by stepwise incubation of β-galactosidase and sialidase [17]. GcMAF could activate phagocytosis of macrophages during inflammation, reduce tumor growth and stimulate cell apoptosis [15, 16]. In addition, GcMAF has been demonstrated to have the potential utility as an antitumorigenic drug for metastatic breast cancer [50]. Therefore, genetic variation in GC may alternatively influence cancer outcome via GcMAF, a biological mechanism independent of vitamin D levels.
We found that carriage of the risk C allele was associated with an increased likelihood of CRC incidence in patients with high vitamin D intake (P interaction = 0.019). No prior studies were found that specifically examined the interaction between rs2282679 polymorphism and vitamin D on CRC; yet, two other GC SNPs, rs17467825 and rs7041, which are in strong linkage disequilibrium with GC rs2282679 (γ2 = 1.0 and 0.6 respectively) [20], have been associated with a slightly greater risk of CRC among individuals who consumed total vitamin D above the median in a multicenter case–unaffected sibling control study, though the interactions were not significant [32]. In addition, low vitamin D intake conferred higher risk of CRC among wild-type AA carriers but less obvious effect among AC or CC carriers; therefore, subjects with the AC/CC genotype might derive little benefit from high vitamin D intake, which may be due to their low affinity and abundance of DBP that might influence the function of vitamin D. Previous research [51] found that serum 25(OH)D level had greater effect on colorectal adenoma among patients with high total calcium intake; it is therefore unsurprising that we also observed a particularly strong association between the variation and all-cause mortality in patients at the higher calcium category. Based on these observations, we may speculate that the influence of rs2282679 polymorphism on either carcinogenesis or progression of CRC was strengthened by a metabolically permissive environmental condition characterized by high levels of dietary vitamin D, calcium, or foods rich in vitamin D and calcium [52].
In this study, the associations between rs2282679 SNP and DFS and OS were of similar patterns but stronger with DFS than OS. The difference in results may be explained by several reasons. It is plausible that many deaths among CRC patients are preceded by tumor metastasis or recurrence. Thus, the DFS end point may be dominated by metastasis and recurrence rather than deaths from all causes [53]; the difference in outcomes may have affected the results. A second possible explanation is that the power to detect an association for OS is less than that for DFS as the OS end point requires extended follow-up [53]. Therefore, the non-significant P values for OS might reflect inadequate power rather than a true lack of effect.
Our data suggest that the GC rs2282679 variation may be associated with poor DFS among patients with BRAF wild-type tumors, but not among BRAF mutant tumors (P interaction = 0.043). Although intriguing, the interaction of the SNP with tumor BRAF mutation status should be interpreted with caution because of a limited statistical power caused by low number of patients with BRAF mutant tumors, as well as the lack (at least to date) of exact mechanism of action underlying the prognostic value of this gene only in BRAF mutated CRC. Additionally, we observed an additive effect of the rs2282679 genotype combined with MSI status; unsurprisingly, the most favorable prognosis as determined by DFS was seen among patients with AA/MSI-high tumors (vs. AC + CC/MSS + MSI-low). MSI has been established as a prognostic biomarker that confers survival advantage to CRC due to increased apoptosis rate and high lymphocytic infiltration [54–57]. Our observations suggest that the prediction model of CRC outcome should additionally integrate the rs2282679 genotype. These results may provide relevant information for identification of patients with increased susceptibility to CRC incidence and mortality and for patient assignment to interventions that are tailored to the individual. Additional studies should be addressed to investigate the role of rs2282679/MSI classification in predicting the response to therapeutic lifestyle interventions.
One limitation of our study is that only one genetic variant of the GC gene was evaluated, thereby providing incomplete coverage of this gene; and we cannot exclude that genetic polymorphisms in other genes in the vitamin D metabolism pathway (e.g., vitamin D receptor) may also influence overall CRC initiation and progression. It is also possible that rs2282679 is not the true causal variant in itself but acts as a proxy through linkage disequilibrium (LD). Moreover, plasma 25(OH)D levels were not measured in this study. The lack of 25(OH)D measurements impeded us to test the relations of GC rs2282679 polymorphisms with plasma vitamin D concentration and to evaluate the extent to which the high risk of CRC mortality associated with the C allele is mediated through low 25(OH)D levels. Furthermore, dietary vitamin D intake may not accurately reflect each participant’s vitamin D status since dietary history as measured by the FFQ is imprecise, and neither dermatic synthesis of vitamin D from sun exposure nor long-term dietary vitamin D intake was taken into account. Additionally, individuals were asked to report dietary exposures from one year prior to diagnosis for cases and one year prior to recruitment for controls; therefore, cases recalled dietary intakes from years earlier than controls. The longer recall period increases the rate of recall error resulting in higher likelihood of exposure misclassification in the case group.
Among the strengths of the study is the careful data collection, with a combination of results from genotyping and epidemiologic questionnaires. The availability of information on known environmental and genetic risk factors of CRC allowed us to investigate potential gene-gene or gene-environment interactions. The relatively large sample size with up to 10 years of follow-up permitted enough power to discern the significant gene-gene and gene-environment interactions in modifying CRC risk and survival using stratified analyses, which could be missed in smaller investigations. Finally, we were able to link the GC rs2282679 genotype to both risk and survival of CRC to recapitulate the entire spectrum of the disease from initiation through progression [58].
Conclusions
Our data demonstrate that the GC rs2282679 polymorphism is not associated with CRC risk overall, but suggest a possible reduced DFS after CRC diagnosis. These results identified an association between the GC SNP rs2282679 and DFS of CRC and effect modifications by vitamin D intake and BRAF mutation status. The genotype at the CG rs2282679 locus, along with vitamin D and BRAF mutation status, has potential utility as a susceptibility and prognostic biomarker of CRC. Future studies should verify these findings in other populations as well as clarify the molecular mechanisms behind the differential effects of the SNP on CRC outcomes according to vitamin D and BRAF mutation status.
Acknowledgments
The authors wish to thank Dr. Meera Jain and all those who contributed to data collection for the Newfoundland Familial Colorectal Cancer Study.
Funding
This work was supported by the Canadian Institutes of Health Research Team Grant [CIHR-CPT79845] and Canadian Institutes of Health Research Team in Interdisciplinary Research on Colorectal Cancer Studentship [205835]. Yun Zhu was awarded by the Beatrice Hunter Cancer Research Institute (BHCRI) through a CRTP Traineeship Award, the Newfoundland and Labrador Centre for Applied Health Research (NLCAHR) through a Doctoral Thesis Research Grant, and the Faculty of Medicine, Memorial University through a Dean’s Fellowship.
Availability of data and materials
The datasets analyzed during this study are available from the corresponding author upon reasonable request.
Abbreviations
- CI
Confidence Interval
- CRC
Colorectal Cancer
- DBP
Vitamin D-Binding Protein
- DFS
Disease-Free Survival
- FFQ
Food Frequency Questionnaire
- FHQ
Family History Questionnaire
- GcMAF
Macrophage-Activating Factor
- GWAS
Genome-Wide Association Study
- HR
Hazard Ratio
- MSI
Microsatellite Instability
- NFCCR
Newfoundland Familial Colorectal Cancer Registry
- NFCCS
Newfoundland and Ontario Familial Colorectal Cancer Study
- OR
Odds Ratio
- OS
Overall Survival
- PHQ
Personal History Questionnaire
- SNP
Single Nucleotide Polymorphism
Authors’ contributions
YZ led the data analysis and findings interpretation, and wrote the first version of the manuscript. JRW, SS, YL, PTC, XZ, and NY consequently revised the manuscript. PPW, GZ, BB conceptualized, devised, and commented on the overall research design and the results interpretation. ED, JRM, PSP critically commented, and oversaw the scientific implementation of this study. All authors read and approved the final version.
Ethics approval and consent to participate
The study protocol was approved by the Health Research Ethics Authority of Memorial University of Newfoundland, and all participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huhn S, Pardini B, Naccarati A, Vodicka P, Hemminki K, Forsti A. Ancestral susceptibility to colorectal cancer. Mutagenesis. 2012;27(2):197–204. doi: 10.1093/mutage/ger061. [DOI] [PubMed] [Google Scholar]
- 2.Emerson JC, Weiss NS. Colorectal cancer and solar radiation. Cancer Causes Control. 1992;3(1):95–99. doi: 10.1007/BF00051919. [DOI] [PubMed] [Google Scholar]
- 3.Bostick RM, Potter JD, Sellers TA, DR MK, Kushi LH, Folsom AR. Relation of calcium, vitamin D, and dairy food intake to incidence of colon cancer among older women. The Iowa Women's health study. Am J Epidemiol. 1993;137(12):1302–1317. doi: 10.1093/oxfordjournals.aje.a116640. [DOI] [PubMed] [Google Scholar]
- 4.Sun Z, Wang PP, Roebothan B, Cotterchio M, Green R, Buehler S, Zhao J, Squires J, Zhu Y, Dicks E, et al. Calcium and vitamin D and risk of colorectal cancer: results from a large population-based case-control study in Newfoundland and Labrador and Ontario. Can J Public Health. 2011;102(5):382–389. doi: 10.1007/BF03404181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garland CF, Comstock GW, Garland FC, Helsing KJ, Shaw EK, Gorham ED. Serum 25-hydroxyvitamin D and colon cancer: eight-year prospective study. Lancet. 1989;2(8673):1176–1178. doi: 10.1016/S0140-6736(89)91789-3. [DOI] [PubMed] [Google Scholar]
- 6.Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014;14(5):342–357. doi: 10.1038/nrc3691. [DOI] [PubMed] [Google Scholar]
- 7.Garland CF, Garland FC. Do sunlight and vitamin D reduce the likelihood of colon cancer? Int J Epidemiol. 1980;9(3):227–231. doi: 10.1093/ije/9.3.227. [DOI] [PubMed] [Google Scholar]
- 8.Ng K, Meyerhardt JA, Wu K, Feskanich D, Hollis BW, Giovannucci EL, Fuchs CS. Circulating 25-hydroxyvitamin d levels and survival in patients with colorectal cancer. J Clin Oncol. 2008;26(18):2984–2991. doi: 10.1200/JCO.2007.15.1027. [DOI] [PubMed] [Google Scholar]
- 9.Zgaga L, Theodoratou E, Farrington SM, Din FV, Ooi LY, Glodzik D, Johnston S, Tenesa A, Campbell H, Dunlop MG. Plasma vitamin D concentration influences survival outcome after a diagnosis of colorectal cancer. J Clin Oncol. 2014;32(23):2430–2439. doi: 10.1200/JCO.2013.54.5947. [DOI] [PubMed] [Google Scholar]
- 10.Maalmi H, Ordonez-Mena JM, Schottker B, Brenner H. Serum 25-hydroxyvitamin D levels and survival in colorectal and breast cancer patients: systematic review and meta-analysis of prospective cohort studies. Eur J Cancer. 2014;50(8):1510–1521. doi: 10.1016/j.ejca.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Ng K, Wolpin BM, Meyerhardt JA, Wu K, Chan AT, Hollis BW, Giovannucci EL, Stampfer MJ, Willett WC, Fuchs CS. Prospective study of predictors of vitamin D status and survival in patients with colorectal cancer. Br J Cancer. 2009;101(6):916–923. doi: 10.1038/sj.bjc.6605262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speeckaert M, Huang G, Delanghe JR, Taes YE. Biological and clinical aspects of the vitamin D binding protein (Gc-globulin) and its polymorphism. Clin Chim Acta. 2006;372(1–2):33–42. doi: 10.1016/j.cca.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Chun RF, Peercy BE, Adams JS, Hewison M. Vitamin D binding protein and monocyte response to 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D: analysis by mathematical modeling. PLoS One. 2012;7(1):e30773. doi: 10.1371/journal.pone.0030773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto N, Naraparaju VR. Vitamin D3-binding protein as a precursor for macrophage activating factor in the inflammation-primed macrophage activation cascade in rats. Cell Immunol. 1996;170(2):161–167. doi: 10.1006/cimm.1996.0148. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto N, Naraparaju VR. Role of vitamin D3-binding protein in activation of mouse macrophages. J Immunol. 1996;157(4):1744–1749. [PubMed] [Google Scholar]
- 16.Kanda S, Mochizuki Y, Miyata Y, Kanetake H, Yamamoto N. Effects of vitamin D(3)-binding protein-derived macrophage activating factor (GcMAF) on angiogenesis. J Natl Cancer Inst. 2002;94(17):1311–1319. doi: 10.1093/jnci/94.17.1311. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto N, Kumashiro R. Conversion of vitamin D3 binding protein (group-specific component) to a macrophage activating factor by the stepwise action of beta-galactosidase of B cells and sialidase of T cells. J Immunol. 1993;151(5):2794–2802. [PubMed] [Google Scholar]
- 18.Zhou L, Zhang X, Chen X, Liu L, Lu C, Tang X, Shi J, Li M, Zhou M, Zhang Z, et al. GC Glu416Asp and Thr420Lys polymorphisms contribute to gastrointestinal cancer susceptibility in a Chinese population. Int J Clin Exp Med. 2012;5(1):72–79. [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, ML MC, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs KB, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19(13):2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies JR, Field S, Randerson-Moor J, Harland M, Kumar R, Anic GM, Nagore E, Hansson J, Hoiom V, Jonsson G, et al. An inherited variant in the gene coding for vitamin D-binding protein and survival from cutaneous melanoma: a BioGenoMEL study. Pigment Cell Melanoma Res. 2014;27(2):234–243. doi: 10.1111/pcmr.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidd LC, Paltoo DN, Wang S, Chen W, Akereyeni F, Isaacs W, Ahaghotu C, Kittles R. Sequence variation within the 5′ regulatory regions of the vitamin D binding protein and receptor genes and prostate cancer risk. Prostate. 2005;64(3):272–282. doi: 10.1002/pros.20204. [DOI] [PubMed] [Google Scholar]
- 23.McCullough ML, Stevens VL, Diver WR, Feigelson HS, Rodriguez C, Bostick RM, Thun MJ, Calle EE. Vitamin D pathway gene polymorphisms, diet, and risk of postmenopausal breast cancer: a nested case-control study. Breast Cancer Res. 2007;9(1):R9. doi: 10.1186/bcr1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abbas S, Linseisen J, Slanger T, Kropp S, Mutschelknauss EJ, Flesch-Janys D, Chang-Claude J. The Gc2 allele of the vitamin D binding protein is associated with a decreased postmenopausal breast cancer risk, independent of the vitamin D status. Cancer Epidemiol Biomark Prev. 2008;17(6):1339–1343. doi: 10.1158/1055-9965.EPI-08-0162. [DOI] [PubMed] [Google Scholar]
- 25.Theodoratou E, Palmer T, Zgaga L, Farrington SM, McKeigue P, Din FV, Tenesa A, Davey-Smith G, Dunlop MG, Campbell H. Instrumental variable estimation of the causal effect of plasma 25-hydroxy-vitamin D on colorectal cancer risk: a mendelian randomization analysis. PLoS One. 2012;7(6):e37662. doi: 10.1371/journal.pone.0037662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiraki LT, Qu C, Hutter CM, Baron JA, Berndt SI, Bezieau S, Brenner H, Caan BJ, Casey G, Chang-Claude J, et al. Genetic predictors of circulating 25-hydroxyvitamin d and risk of colorectal cancer. Cancer Epidemiol Biomark Prev. 2013;22(11):2037–2046. doi: 10.1158/1055-9965.EPI-13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szkandera J, Absenger G, Pichler M, Stotz M, Langsenlehner T, Samonigg H, Renner W, Gerger A. Association of common gene variants in vitamin D modulating genes and colon cancer recurrence. J Cancer Res Clin Oncol. 2013;139(9):1457–1464. doi: 10.1007/s00432-013-1461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilar E, Tabernero J. Molecular dissection of microsatellite instable colorectal cancer. Cancer Discov. 2013;3(5):502–511. doi: 10.1158/2159-8290.CD-12-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seppala TT, Bohm JP, Friman M, Lahtinen L, Vayrynen VM, Liipo TK, Ristimaki AP, Kairaluoma MV, Kellokumpu IH, Kuopio TH, et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112(12):1966–1975. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogino S, Shima K, Meyerhardt JA, NJ MC, Ng K, Hollis D, Saltz LB, Mayer RJ, Schaefer P, Whittom R, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18(3):890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs ET, Martinez ME, Campbell PT, Conti DV, Duggan D, Figueiredo JC, Haile RW, EC LR, Poynter JN, Thompson PA, et al. Genetic variation in the retinoid X receptor and calcium-sensing receptor and risk of colorectal cancer in the colon cancer family registry. Carcinogenesis. 2010;31(8):1412–1416. doi: 10.1093/carcin/bgq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poynter JN, Jacobs ET, Figueiredo JC, Lee WH, Conti DV, Campbell PT, Levine AJ, Limburg P, Le Marchand L, Cotterchio M, et al. Genetic variation in the vitamin D receptor (VDR) and the vitamin D-binding protein (GC) and risk for colorectal cancer: results from the colon cancer family registry. Cancer Epidemiol Biomark Prev. 2010;19(2):525–536. doi: 10.1158/1055-9965.EPI-09-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green RC, Green JS, Buehler SK, Robb JD, Daftary D, Gallinger S, JR ML, Parfrey PS, Younghusband HB. Very high incidence of familial colorectal cancer in Newfoundland: a comparison with Ontario and 13 other population-based studies. Familial Cancer. 2007;6(1):53–62. doi: 10.1007/s10689-006-9104-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J, Zhu Y, Wang PP, West R, Buehler S, Sun Z, Squires J, Roebothan B, JR ML, Campbell PT, et al. Interaction between alcohol drinking and obesity in relation to colorectal cancer risk: a case-control study in Newfoundland and Labrador, Canada. BMC Public Health. 2012;12:94. doi: 10.1186/1471-2458-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y, Wang PP, Zhao J, Green R, Sun Z, Roebothan B, Squires J, Buehler S, Dicks E, Cotterchio M, et al. Dietary N-nitroso compounds and risk of colorectal cancer: a case-control study in Newfoundland and Labrador and Ontario, Canada. Br J Nutr. 2014;111(6):1109–17. [DOI] [PMC free article] [PubMed]
- 36.Sun Z, Zhu Y, Wang PP, Roebothan B, Zhao J, Dicks E, Cotterchio M, Buehler S, Campbell PT, JR ML, et al. Reported intake of selected micronutrients and risk of colorectal cancer: results from a large population-based case-control study in Newfoundland, Labrador and Ontario, Canada. Anticancer Res. 2012;32(2):687–696. [PubMed] [Google Scholar]
- 37.Zhu Y, Yang SR, Wang PP, Savas S, Wish T, Zhao J, Green R, Woods M, Sun Z, Roebothan B, et al. Influence of pre-diagnostic cigarette smoking on colorectal cancer survival: overall and by tumour molecular phenotype. Br J Cancer. 2014;110(5):1359–66. [DOI] [PMC free article] [PubMed]
- 38.Wang PP, Dicks E, Gong X, Buehler S, Zhao J, Squires J, Younghusband B, JR ML, Parfrey PS. Validity of random-digit-dialing in recruiting controls in a case-control study. Am J Health Behav. 2009;33(5):513–520. doi: 10.5993/AJHB.33.5.4. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Wang PP, Roebothan B, Ryan A, Tucker CS, Colbourne J, Baker N, Cotterchio M, Yi Y, Sun G. Assessing the validity of a self-administered food-frequency questionnaire (FFQ) in the adult population of Newfoundland and Labrador, Canada. Nutr J. 2013;12:49. doi: 10.1186/1475-2891-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hankin JH, Wilkens LR, Kolonel LN, Yoshizawa CN. Validation of a quantitative diet history method in Hawaii. Am J Epidemiol. 1991;133(6):616–628. doi: 10.1093/oxfordjournals.aje.a115934. [DOI] [PubMed] [Google Scholar]
- 41.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods MO, Younghusband HB, Parfrey PS, Gallinger S, McLaughlin J, Dicks E, Stuckless S, Pollett A, Bapat B, Mrkonjic M, et al. The genetic basis of colorectal cancer in a population-based incident cohort with a high rate of familial disease. Gut. 2010;59(10):1369–1377. doi: 10.1136/gut.2010.208462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woods MO, Hyde AJ, Curtis FK, Stuckless S, Green JS, Pollett AF, Robb JD, Green RC, Croitoru ME, Careen A, et al. High frequency of hereditary colorectal cancer in Newfoundland likely involves novel susceptibility genes. Clin Cancer Res. 2005;11(19 Pt 1):6853–6861. doi: 10.1158/1078-0432.CCR-05-0726. [DOI] [PubMed] [Google Scholar]
- 44.Loughrey MB, Waring PM, Tan A, Trivett M, Kovalenko S, Beshay V, Young MA, McArthur G, Boussioutas A, Dobrovic A. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Familial Cancer. 2007;6(3):301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 45.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1(1):43–46. doi: 10.1097/00001648-199001000-00010. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, He JW, Fu WZ, Zhang CQ, Zhang ZL. An analysis of the association between the vitamin D pathway and serum 25-hydroxyvitamin D levels in a healthy Chinese population. J Bone Miner Res. 2013;28(8):1784–1792. doi: 10.1002/jbmr.1926. [DOI] [PubMed] [Google Scholar]
- 47.Christakos S, Dhawan P, Liu Y, Peng X, Porta A. New insights into the mechanisms of vitamin D action. J Cell Biochem. 2003;88(4):695–705. doi: 10.1002/jcb.10423. [DOI] [PubMed] [Google Scholar]
- 48.Bouillon R, Eelen G, Verlinden L, Mathieu C, Carmeliet G, Verstuyf A. Vitamin D and cancer. J Steroid Biochem Mol Biol. 2006;102(1–5):156–162. doi: 10.1016/j.jsbmb.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 49.Ordonez-Moran P, Larriba MJ, Pendas-Franco N, Aguilera O, Gonzalez-Sancho JM, Munoz A. Vitamin D and cancer: an update of in vitro and in vivo data. Front Biosci. 2005;10:2723–2749. doi: 10.2741/1731. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto N, Suyama H, Ushijima N. Immunotherapy of metastatic breast cancer patients with vitamin D-binding protein-derived macrophage activating factor (GcMAF) Int J Cancer. 2008;122(2):461–467. doi: 10.1002/ijc.23107. [DOI] [PubMed] [Google Scholar]
- 51.Peters U, McGlynn KA, Chatterjee N, Gunter E, Garcia-Closas M, Rothman N, Sinha R. Vitamin D, calcium, and vitamin D receptor polymorphism in colorectal adenomas. Cancer Epidemiol Biomark Prev. 2001;10(12):1267–1274. [PubMed] [Google Scholar]
- 52.Nguyen PL, Ma J, Chavarro JE, Freedman ML, Lis R, Fedele G, Fiore C, Qiu W, Fiorentino M, Finn S, et al. Fatty acid synthase polymorphisms, tumor expression, body mass index, prostate cancer risk, and survival. J Clin Oncol. 2010;28(25):3958–3964. doi: 10.1200/JCO.2009.27.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sargent DJ, Wieand HS, Haller DG, Gray R, Benedetti JK, Buyse M, Labianca R, Seitz JF, O'Callaghan CJ, Francini G, et al. Disease-free survival versus overall survival as a primary end point for adjuvant colon cancer studies: individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2005;23(34):8664–8670. doi: 10.1200/JCO.2005.01.6071. [DOI] [PubMed] [Google Scholar]
- 54.Roth AD, Delorenzi M, Tejpar S, Yan P, Klingbiel D, Fiocca R, d'Ario G, Cisar L, Labianca R, Cunningham D, et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst. 2012;104(21):1635–1646. doi: 10.1093/jnci/djs427. [DOI] [PubMed] [Google Scholar]
- 55.Bertagnolli MM, Redston M, Compton CC, Niedzwiecki D, Mayer RJ, Goldberg RM, Colacchio TA, Saltz LB, Warren RS. Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III colon cancer--a study of CALGB 9581 and 89803. J Clin Oncol. 2011;29(23):3153–3162. doi: 10.1200/JCO.2010.33.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim GP, Colangelo LH, Wieand HS, Paik S, Kirsch IR, Wolmark N, Allegra CJ. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and bowel project collaborative study. J Clin Oncol. 2007;25(7):767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 57.Benatti P, Gafa R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11(23):8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 58.Mucci LA, Stark JR, Figg WD, Schumacher F, Li H, Abe M, Hennessy K, Stampfer MJ, Gaziano JM, Ma J, et al. Polymorphism in endostatin, an angiogenesis inhibitor, and prostate cancer risk and survival: a prospective study. Int J Cancer. 2009;125(5):1143–1146. doi: 10.1002/ijc.24423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during this study are available from the corresponding author upon reasonable request.


