Abstract
Background
Breast cancer is the most common malignancy and the leading cause of cancer-related deaths among Jordanian women. With a median age of 50 years at diagnosis, a higher prevalence of hereditary breast cancer may be expected. The objective of this pilot study is to evaluate, for the first time, the contribution of germline mutations in BRCA1/2 to breast cancer among Jordanian patients.
Methods
Jordanian breast cancer women with a selected high risk profile were invited to participate. Peripheral blood samples were obtained for DNA extraction. A detailed 3-generation family history was also collected. BRCA sequencing was performed at a reference laboratory. Mutations were classified as deleterious, suspected deleterious, variant of uncertain significance or favor polymorphisms. Patients’ medical records were reviewed for extraction of clinical and tumor pathology data.
Results
One hundred patients were enrolled to the study. Median age was 40 (22–75) years. In total, 20 patients had deleterious and 7 suspected deleterious mutations in BRCA1 or BRCA2 genes. Seven variants of uncertain significance were also detected. After excluding patients tested subsequent to the index case in their families, highest mutation rates were observed among triple negatives (9/16, 56.3%) especially among those with positive family history of breast and/or ovarian cancer (9/13, 69.2%), patients with bilateral or second primary breast cancer (10/15, 66.7%) and those with family history of male breast cancer (2/5, 40.0%).
Conclusions
BRCA1/2 mutations are not uncommon among selected Jordanian females with breast cancer. The contribution of these findings to much younger age at diagnosis is debatable.
Although small, our selected patient cohort shows an important incidence of deleterious and suspected deleterious BRCA1/2 mutations suggesting that genetic testing should be offered to patients with certain high risk features.
Keywords: Breast cancer, BRCA1, BRCA2, Jordan, Hereditary breast cancer
Background
Breast cancer is the most common cancer and the leading cause of cancer-related deaths among Jordanian women. The latest annual report of the Jordan Cancer Registry stated a total of 1067 breast cancer cases, accounting for 19.7% of all cancer cases diagnosed in Jordan [1].
Like many neighboring countries, breast cancer in Jordan presents with many peculiar features. The median age at presentation is 50 years; 10 years younger than western societies. Additionally more than a third of patients present with locally-advanced or metastatic disease highlighting the importance of early detection programs [2, 3].
Given the limited resources and recent debates about the value of national screening mammography [4–6], identifying higher risk group(s) of patients to which preventive and early detection efforts can be directed is extremely important.
Hereditary breast cancer is well-described; around 5–10% of breast cancer patients carry high risk gene mutations like BRCA1 and BRCA2 [7, 8]. Given the high penetrance rates among such mutation carriers [9, 10], it will be important to identify those patients to whom many additional risk-reduction clinical interventions, like bilateral mastectomies and oophorectomies can be performed. The Frequency of BRCA1 and BRCA2 carrier rates varies from 1/400 in the general Caucasian population to as high as 1/40 among the Ashkenazi Jewish population [11].
Data related to hereditary breast cancer among the Arab countries is very scarce; none reported from Jordan. In a recent study that included 250 high risk Lebanese patients, 14 (5.6%) were found to carry a deleterious BRCA mutation (7 BRCA1, 7 BRCA2) and 31 others (12.4%) carried a variant of uncertain significance (VUS) [12]. High risk patients were defined as those diagnosed at young age (≤40 years), those ≤50 years old with positive family history of breast or ovarian cancer and those with personal history of ovarian cancer. However, an earlier study from the same country that included 72 unrelated patients with positive family history of breast and/or ovarian cancers or with an early onset breast cancer reported higher carrier rates; deleterious BRCA1 and BRCA2 mutations were reported in 12.5% [13].
BRCA1 gene analysis was also performed in 121 Moroccan women diagnosed with breast cancer; 31.6% (6/19) of familial and 1% (1/102) of early-onset sporadic cases (< 45 years) were found to be associated with BRCA1 mutations [14]. In Egypt, 60 breast cancer patients, derived from 60 families, were selected for molecular genetic testing of BRCA1 and BRCA2 genes. The study also included 120 healthy first degree female relatives of the patients, either sisters and/or daughters, for early detection of presymptomatic breast cancer mutation carriers. Mutations were detected in 86.7% of the families; 60% were BRCA1, while 26.7% were attributable to BRCA2 mutations [15]. Few other smaller regional studies had reported variable rates [16–18]. The variability of results from the above-mentioned studies might be related to patient selection criteria, referral patterns, small number of patients enrolled and different methods of testing.
The aim of our study is to evaluate, and for the first time, the contribution of germline mutations in BRCA1/2 to breast cancer among Jordanian patients with a selected high risk profile.
Methods
Patient population
Jordanian breast cancer patients with a selected high risk profile; as per the National Comprehensive Cancer Network (NCCN) guidelines [19] were invited to participate. This includes patients 40 years or younger, triple negative patients (i.e. negative for estrogen receptors ER, progesterone receptors PR, and HER2 receptors) age ≤ 50 years, patients diagnosed at any age with ≥2 close relatives (any age) with breast, epithelial ovarian, fallopian tube or primary peritoneal cancer, patients with family history of male breast cancer and patients with two breast cancer primaries, or breast and ovarian/fallopian tube/primary peritoneal cancer. Eligible patients were identified by review of the King Hussein Cancer Center Tumor Registry and medical records, and approached during routine clinic visits. Patients were interviewed for 30 min for proper consent and were given full autonomy to decide whether they want to know their test result, want to inform their treating physician or place a copy of the test result in their medical record. A detailed 3-generation family history was also obtained by one of the investigators.
Patients were made aware of all clinical and psychosocial consequences of positive test results. When needed and requested by the patient, such meeting and discussion were also carried out with the spouse and/or family members.
Patients’ medical records were reviewed for extraction of clinical data and tumor pathology.
The study was funded by a competitive grant from the King Hussein Cancer Center/MD Anderson Cancer Center Sister Institution Network Fund (SINF). The study was approved by King Hussein Cancer Center Institutional Review Board (IRB) under project number 11KHCC63. All patients signed informed consent.
BRCA1/2 testing
BRCA1/2 testing was done at no-cost to participants. Ten mL peripheral blood samples were obtained for DNA extraction. BRCA sequencing was performed at Myriad Genetics laboratory (Myriad Genetics, Salt Lake City, UT) utilizing the Comprehensive BRACAnalysis® and BRACAnalysis® Rearrangement Test (BART). Analysis consists of sequencing of all translated exons and immediately adjacent intronic regions of the BRCA1 and BRCA2 genes and a comprehensive rearrangement test of both BRCA1 and BRCA2 by quantitative PCR analysis.
A disease-causing mutation, also called deleterious mutation, pathogenic variant, predisposing mutation, and susceptibility gene, is a genetic alteration that increases an individual’s susceptibility or predisposition to a certain disease or disorder. When such a variant (or mutation) is inherited, development of symptoms is more likely, but not certain. BRCA mutations were classified as deleterious, suspected deleterious, variant of uncertain significance (VUS) or favor polymorphism based on established criteria [20].
Statistical analysis
Patient characteristics were tabulated and described by their medians, ranges or percentages (%). Relatives tested later to the index case in the family were excluded from subsequent analyses. χ2 test or Fisher exact test were used to compare the proportion of positive BRCA1/2 deleterious/suspected deleterious mutations according to age (cut-off ≤40), triple negative status, first and/or second-degree family history of breast and/or ovarian cancer, number of first and/or second-degree relatives with breast and/or ovarian cancer (cut-off ≥2), bilateral or second primary breast cancer and family history of male breast cancer. Multivariate analysis using a logistic regression model adjusting for age, triple negative status, number of first and/or second-degree relatives with breast and/or ovarian cancer and bilateral or second primary breast cancer was performed. Odds ratios and their related 95% confidence intervals were calculated.
A significance level of p ≤ 0.05 was used in the analysis. All analyses were performed using SAS version 9.4 (SAS Institute Inc., Cary, NC).
Results
Between July 2012 and April 2015, a total of 100 eligible patients were included. Only two patients fulfilling the eligibility criteria and approached for the study declined to participate. Median age of participants was 40 (22–75 years). Fifty one (51%) were ≤40 years. Majority (91; 91%) had infiltrating ductal carcinoma (IDC) and most patients presented with early stage disease. Eighty nine (89%) patients had positive first and/or second-degree family history of breast and/or ovarian cancers. Majority (77; 77%) of the patients had hormone-receptor (ER and/or PR) positive disease. Among the 93 patients with known HER-2 status, 13 (14%) were positive by immunohistochemistry and/or FISH (Fluorescent In Situ Hybridization). Most of the patients had grade II and III disease, Table 1.
Table 1.
Patients characteristics, N = 100
| Characteristics | Number (%) |
|---|---|
| Age | |
| Median (years) | 40 |
| Range (years) | 22–75 |
| Pathology | |
| IDC | 91 (91%) |
| ILC and others | 9 (9%) |
| Stage | |
| I | 16 (16%) |
| II | 51 (51%) |
| III | 22 (22%) |
| IV | 2 (2%) |
| Unknown | 9 (9%) |
| Grade | |
| I | 10 (10%) |
| II | 37 (37%) |
| III | 45 (45%) |
| Unknown | 8 (8%) |
| Hormone Receptor Status | |
| ER and/or PR Positive | 77 (77%) |
| ER Positive | 77 (77%) |
| ER Negative | 22 (22%) |
| ER Unknown | 1 (1%) |
| PR Positive | 77 (77%) |
| PR Negative | 22 (22%) |
| PR Unknown | 1 (1%) |
| HER-2 Status | |
| Positive | 13 (13%) |
| Negative | 80 (80%) |
| Unknown | 7 (7%) |
| Triple-Negative | 17 (17%) |
IDC Infiltrating Ductal Carcinoma, ILC Infiltrating Lobular Carcinoma
Overall 20 (20%) patients had deleterious BRCA1 or BRCA2 mutations (7 BRCA1, 13 BRCA2). Seven (7%) patients had suspected deleterious mutations; all were in the BRCA2 gene. Seven (7.0%) variants of uncertain significance (VUS) were detected, one in BRCA1 and six in BRCA2. Table 2 summarizes the genetic and histopathologic characteristics of patients with BRCA1 and BRCA2 variants.
Table 2.
Genetic and histopathologic characteristic of Jordanian breast cancer patients with BRCA1 and BRCA2 genetic variants
| Test Result | Patients | Base change | AA change | Variant type | Interpretation | Age at diagnosis (Years) | Tumor histopathology | ER | PR | HER2 | Family history (Breast/ Ovarian cancer) (n) 1st deg. 2nd deg. 3rd deg |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 genetic variants | |||||||||||||
| 3450del4 | BRACA 029 | del4 | Stop 1115 | Deletion/Frame shift | Deleterious | 50 | IDC/GIII | -VE | -VE | -VE | 3 | 0 | 0 |
| 3954delG | BRACA 073 | delG | Stop 1306 | Deletion/Frame shift | Deleterious | 41 | IDC/GII | +VE | +VE | -VE | 2 | 0 | 0 |
| 3954delG | BRACA 096 | delG | Stop 1306 | Deletion/Frame shift | Deleterious | 37 | IDC/GIII | -VE | -VE | -VE | 0 | 1 | 0 |
| 3555del4 | BRACA 078 | del4 | Stop 1153 | Deletion/Frame shift | Deleterious | 29 | IDC/GIII | -VE | -VE | -VE | 1 | 0 | 0 |
| E1373X (4236G > T) | BRACA 091 | G > T | E1373X | Nonsense | Deleterious | 44 | IDC/GIII | -VE | -VE | -VE | 1 | 1 | 0 |
| IVS17 + 3 A > G | BRACA 086 | A > G | – | Intronic | Deleterious | 34 | IDC/GIII | -VE | -VE | -VE | 1 | 1 | 0 |
| 5149del4 | BRACA 094 | del4 | Stop 1678 | Deletion Frame shift | Deleterious | 33 | IDC/GIII | -VE | -VE | -VE | 1 | 1 | 0 |
| E1478D (4553G > C)b | BRACA 066 | G > C | E1478D | Missense | VUS | 35 | IDC/GII | +VE | +VE | -VE | 1 | 2 | 2 |
| E445Q (1452G > C) | BRACA 010 | G > C | E445Q | Missense | FP | 35 | IDC/GIII | +VE | +VE | +VE | 1 | 0 | 0 |
| BRCA2 genetic variants | |||||||||||||
| 999del5 | BRACA 082 | del5 | Stop 273 | Deletion/Frame shift | Deleterious | 56 | IDC/GIII | -VE | -VE | -VE | 0 | 3 | 1 |
| 1461insA | BRACA 060 d | insA | Stop 420 | Insertion/Frame shift | Deleterious | 33 | IDC/GIII | -VE | -VE | -VE | 1 | 2 | 0 |
| 1461insA | BRACA 063 d | insA | Stop 420 | Insertion/Frame shift | Deleterious | 34 | ILC/GII | +VE | +VE | -VE | 1 | 2 | 0 |
| 2482del4 | BRACA 018 d | del4 | Stop 770 | Deletion/Frame shift | Deleterious | 48 | IDC/G UNK | +VE | +VE | -VE | 2 | 2 | 2 |
| 2482del4 | BRACA 070 d | del4 | Stop 770 | Deletion/Frame shift | Deleterious | 46 | IDC/GII | +VE | +VE | -VE | 1 | 2 | 4 |
| 2482del4 | BRACA 071 d | del4 | Stop 770 | Deletion/Frame shift | Deleterious | 46 | IDC/GIII | -VE | -VE | -VE | 1 | 2 | 4 |
| 2482del4 | BRACA 084 C | del4 | Stop 770 | Deletion/Frame shift | Deleterious | 30 | IDC/GII | +VE | +VE | -VE | 1 | 1 | 0 |
| L2039X (6344 T > A) | BRACA 064 | T > A | L2039X | Nonsense | Deleterious | 44 | IDC/GII | +VE | +VE | -VE | 1 | 3 | 0 |
| 6855del8 | BRACA 049 | del8 | Stop 2221 | Deletion/Frame shift | Deleterious | 33 | IDC/GIII | +VE | +VE | -VE | 1 | 2 | 0 |
| 6862del4 | BRACA 041 | del4 | Stop 2227 | Deletion Frame shift | Deleterious | 42 | IDC/GIII | +VE | +VE | -VE | 2 | 0 | 0 |
| E2229X (6913G > T) | BRACA 059 | G > T | E2229X | Nonsense | Deleterious | 37 | IDC/GIII | -VE | -VE | -VE | 1 | 2 | 1 |
| E2229X (6913G > T) | BRACA 080 | G > T | E2229X | Nonsense | Deleterious | 29 | IDC/GIII | +VE | +VE | -VE | 1 | 0 | 0 |
| IVS23-1G > A | BRACA 057 | G > A | – | Intronic | Deleterious | 51 | IDC/GIII | +VE | +VE | -VE | 3 | 2 | 0 |
| IVS24-1G > A | BRACA 055 C, d | G > A | – | Intronic | Suspected Deleterious | 42 | IDC/GIII | +VE | +VE | -VE | 2 | 2 | 0 |
| IVS24-1G > A | BRACA 061 C, d | G > A | – | Intronic | Suspected Deleterious | 55 | IDC/GIII | +VE | +VE | UNK | 2 | 2 | 0 |
| IVS24-1G > A | BRACA 067 | G > A | – | Intronic | Suspected Deleterious | 32 | IDC/GIII | +VE | +VE | -VE | 0 | 2 | 0 |
| dup exons 5–11(5′) a | BRACA 008 d | – | – | – | Suspected Deleterious | 25 | IDC/GIII | +VE | +VE | -VE | 1 | 1 | 0 |
| dup exons 5–11(5′) a | BRACA 020 d | – | – | – | Suspected Deleterious | 50 | IDC/GII | +VE | +VE | -VE | 1 | 1 | 1 |
| dup exons 5–11(5′) a | BRACA 047 | – | – | – | Suspected Deleterious | 37 | UNK | +VE | +VE | UNK | 3 | 1 | 0 |
| dup exons 5–11(5′) a | BRACA 089 | – | – | – | Suspected Deleterious | 36 | IDC/GIII | +VE | +VE | -VE | 1 | 0 | 0 |
| P168A (730C > G) | BRACA 093 | C > G | P168A | Missense | VUS | 36 | IDC/GI | +VE | +VE | -VE | 1 | 0 | 0 |
| T251R (980C > G) | BRACA084 C | C > G | T251R | Missense | VUS | 30 | IDC/GII | +VE | +VE | -VE | 1 | 1 | 0 |
| A2306P (7144G > C) | BRACA 035 | G > C | A2306P | Missense | VUS | 35 | IDC/GII | +VE | +VE | -VE | 0 | 1 | 0 |
| Q2925R (9002A > G) | BRACA 053 | A > G | Q2925R | Missense | VUS | 52 | IDC/GI | +VE | +VE | -VE | 1 | 2 | 0 |
| Q2925R (9002A > G) | BRACA 098 | A > G | Q2925R | Missense | VUS | 42 | IDC/GIII | -VE | -VE | +VE | 1 | 0 | 3 |
| E2193K (6805G > A) | BRACA 099 | G > A | E2193K | Missense | VUS | 36 | IDC/GIII | -VE | -VE | -VE | 2 | 0 | 0 |
| K21R (290A > G) | BRACA 043 | A > G | K21R | Missense | FP | 35 | IDC/GIII | +VE | +VE | -VE | 0 | 3 | 2 |
| K3416E (10474A > G) | BRACA 055 C, d | A > G | K3416E | Missense | FP | 42 | IDC/GIII | +VE | +VE | -VE | 2 | 2 | 0 |
| K3416E (10474A > G) | BRACA 061 C, d | A > G | K3416E | Missense | FP | 55 | IDC/GII | +VE | +VE | UNK | 2 | 2 | 0 |
aThis mutation consist of a duplication of exons 5–10 of the BRCA2 gene; the 5′ end of BRCA2 exon 11 is also duplicated
bAccording to Myriad Genetic Laboratories- variant information sheet, this is the first observation for this variant
CPatients 055 & 061 had both a suspected deleterious variant and a favor polymorphism variant; patient 084 had both a deleterious variant and variant of uncertain significance
dThe following patients are relatives: Patients 055 & 061 (sisters), patients 060 & 063 (sisters), patients 008 & 020 (second degree relatives) and patients 18, 70 & 71 (first and third degree relatives)
AA Amino Acid, VUS variants of uncertain significance, FP favor polymorphism, IDC invasive ductal carcinoma, ILC invasive lobular carcinoma
Excluding 5 relatives tested subsequent to the index case in their families (patients 061, 063, 020, 070 and 071), 10 (45.5%) of the 22 patients with deleterious/suspected deleterious mutations had bilateral or contralateral breast cancer, developed 2–9 years after the initial diagnosis, compared to only 5 (6.8%) out of the other 73 patients with either no known mutations, VUS or favor polymorphisms, p-value< 0.001, Table 3.
Table 3.
Association of different variables with BRCA1/2 mutation status, N=95a
| Variable | Level | Total | BRCA1/2 mutation status | P-value | |
|---|---|---|---|---|---|
| Positive (deleterious and suspected deleterious) | Others (No variant, FP, VUS) | ||||
| Age N = 95 | age < =40 | 50 | 13(26.0%) | 37(74.0%) | NS |
| age > 40 | 45 | 9(20.0%) | 36 (80.0%) | ||
| Triple negative N = 95 | No | 79 | 13 (16.5%) | 66 (83.5%) | 0.001 |
| Yes | 16 | 9 (56.3%) | 7 (43.8%) | ||
| Triple negative (age < =50 (N = 75)) | No | 60 | 12 (20.0%) | 48 (80.0%) | |
| Yes | 15 | 8 (53.3%) | 7 (46.7%) | 0.009 | |
| Triple negative (age < =40 (N = 50)) | No | 38 | 7 (18.4%) | 31 (81.6%) | |
| Yes | 12 | 6 (50.0%) | 6 (50.0%) | 0.030 | |
| Triple negative (family history = Yes (N = 84)) | No | 71 | 13 (18.3%) | 58 (81.7%) | |
| Yes | 13 | 9 (69.2%) | 4 (30.8%) | 0.000 | |
| Number of relatives with breast and/or ovarian cancer (first and second degree) | Relatives < 2 | 31 | 4 (12.9%) | 27 (87.1%) | NS |
| Relatives > = 2 | 64 | 18 (28.1%) | 46 (71.9%) | ||
| Family history of breast cancer and/or ovarian cancer (first and second degree) | No | 11 | 11(100%) | 0.063 | |
| Yes | 84 | 22 (26.2%) | 62 (73.8%) | ||
| Bilateral or second primary breast cancer | no | 80 | 12 (15.0%) | 68 (85.0%) | 0.000 |
| yes | 15 | 10 (66.7%) | 5 (33.3%) | ||
| Family history of male breast cancer | no | 90 | 20 (22.2%) | 70 (77.8%) | NS |
| yes | 5 | 2 (40.0%) | 3 (60.0%) | ||
| Family history (age < =40 (N = 50)) | No | 10 | 10(100%) | 0.046 | |
| Yes | 40 | 13 (32.5%) | 27 (67.5%) | ||
FP favor polymorphism, VUS variant of uncertain significance, NS non-significant
aFive patients (patients 061, 063, 020, 070 and 071) were relatives to the index case tested in their families, therefore they were excluded from this analysis, see footnote to Table 2
Young patients
Fifty one young patients (40 years or younger, range: 22–40, Median: 35 years) were included; 10 (19.6%) had deleterious mutations (4 (7.8%) BRCA1, 6 (11.8%) BRCA2). Four (7.8%) others had suspected deleterious BRCA2 mutations while 5 (9.8%) had VUS; 4 of them where in BRCA2.
Among the 40 (80.0%) young patients with positive first or second-degree family history of breast and/or ovarian cancer, 13 (32.5%) had deleterious/suspected deleterious BRCA1 or BRCA2 mutations, while no known mutations were found in the 10 other patients without a significant family history, p-value = 0.046. Twelve (24.0%) young patients had triple-negative disease, 6 (50.0%) had positive deleterious/suspected deleterious BRCA1/2 mutations compared to 7 (18.4%) out of the 38 none-triple negative patients (p-value = 0.030), Table 3.
Triple-negative patients
Sixteen patients had triple-negative disease. Nine (56.3%) had deleterious mutations in BRCA1 or BRCA2, compared to 13 (16.5%) out of 79 patients with non-triple negative disease, p-value = 0.001 (Table 3). Six (37.5%) of these triple-negative patients had BRCA1 deleterious mutations while 3 (18.75%) had BRCA2 deleterious mutations. One triple-negative patient had a VUS in BRCA2.
Patients with family history
Of eighty four patients (84/95; 88.4%) with first and/or second-degree family history of breast and/or ovarian cancer; 22 (26.2%) had deleterious/suspected deleterious mutations in either BRCA1 (7; 8.3%) or BRCA2 (15; 17.9%). None of the other 11 patients were positive for a deleterious/suspected deleterious mutation in BRCA 1 or 2, p-value = 0.063. Among the 13 patients who also had a triple-negative disease, 9 (69.2%) had deleterious BRCA1 or BRCA2 mutations, while 13 (18.3%) out of the other 71 patients who had family history but were not triple-negative harbored deleterious/suspected deleterious mutations in BRCA 1 or 2 (p-value< 0.001), Table 3.
Other patients
Five patients had family history of male breast cancer, two (40.0%) of them had deleterious/suspected deleterious mutations in BRCA2, another patient harbored a VUS in BRCA2.
Among the 15 patients with bilateral or second primary breast cancer; 10 (66.7%) had deleterious or suspected deleterious BRCA1/2 mutations; 5 (33.3%) were in BRCA2 and 5 (33.3) in BRCA1. Fig. 1 summarizes positive test results among different patients’ risk groups.
Fig. 1.
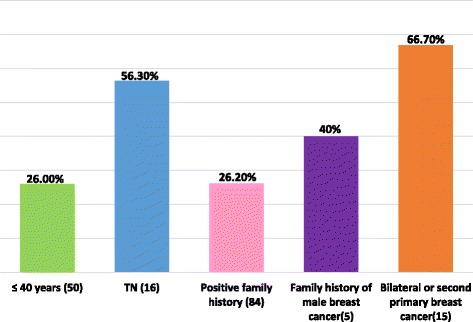
Percentage of BRCA1/2 positivity among different patients’ risk groups, N = 95*. Patients with BRCA1/2 deleterious or suspected deleterious mutations were considered BRCA1/2 positive. * Five patients (patients 061, 063, 020, 070 and 071) were relatives to the index case tested in their families, therefore they were excluded from this analysis, see footnote to Table 2. TN: Triple Negative breast cancer
Using a multivariate logistic regression model, adjusting for age, triple negative status, number of first and/or second degree relatives with breast and/or ovarian cancer and bilateral or second primary breast cancer; the later three variables were significantly associated with the incidence of BRCA1/2 deleterious/suspected deleterious mutations. Odds ratios and 95% confidence intervals for triple negative, number of relatives and bilateral or second breast primary were 7.46 (1.66–33.62), 13.21 (2.20–79.30) and 19.30 (3.97–93.88), and p-values = 0.0089, 0.0048 and 0.0002, respectively, Table 4.
Table 4.
Multivariate logistic regression, N=95a
| Variable | Reference | OR | 95% CI | P- value | |
|---|---|---|---|---|---|
| Age | age > 40 vs age ≤ 40 | 0.315 | 0.080 | 1.234 | 0.0972 |
| Triple negative | Yes vs No | 7.460 | 1.655 | 33.624 | 0.0089 |
| Number of 1st or 2nd degree relatives | Relatives ≥2 vs Relatives < 2 | 13.212 | 2.201 | 79.296 | 0.0048 |
| Bilateral or second primary breast cancer | Yes vs No | 19.304 | 3.969 | 93.882 | 0.0002 |
OR Odds Ratio estimates, CI Wald Confidence Interval
aFive patients (patients 061, 063, 020, 070 and 071) were relatives to the index case tested in their families, therefore they were excluded from this analysis, see footnote to Table 2
Discussion
This is the first BRCA mutation study from Jordan. Our data showed that such mutations are not uncommon among highly selected Jordanian females with breast cancer. Using a multivariate logistic regression model, adjusting for age, triple negative status, number of first and/or second degree relatives with breast and/or ovarian cancer and bilateral or second primary breast cancer; the later three variables were significantly associated with the incidence of BRCA1/2 deleterious/suspected deleterious mutations while age was not an independent predictor of carrier status. The contribution of these findings to much younger age at diagnosis among Jordanian females is debatable. Considering the young population structure of Jordan, with around 80% of the population below the age of 40 [21], a larger fraction of breast cancer cases is expected to be younger. Nonetheless, our findings suggest that BRCA1/2 screening should be offered to patients with certain high risk features.
BRCA1/2 penetrance rates are high; results from prospective analysis of EMBRACE trial were recently reported and showed that the average cumulative risks, by age 70 years, for BRCA1 carriers were estimated to be 60% for breast cancer, 59% for ovarian cancer, and 83% for contralateral breast cancer. For BRCA2 carriers, the corresponding risks were 55% for breast cancer, 16.5% for ovarian cancer, and 62% for contralateral breast cancer [10].
Given that BRCA mutations are not uncommon and given their high penetrance rate, risk-reduction strategies including bilateral mastectomy and salpingo-oophorectomy are becoming standard of care and are widely accepted by patients and family-at-risk [22]. Most of our patients with positive deleterious/suspected deleterious mutations who were offered such risk-reduction surgeries had accepted and many already had undergone the recommended procedure(s).
International guidelines had identified specific patients with high-risk profile for which genetic counselling and testing are recommended [19, 23]. Depending on the specific ethnicity and the population studied, this group of patients can be large enough to put significant pressure on health care budgets especially in low or middle income countries, like ours, where the cost of testing is still relatively high. Identifying smaller subgroups of patients with “higher” probability of positive mutations can improve implementation of the genetic testing guidelines.
In our study, we identified subgroups of patients with significantly higher risk of having deleterious mutations. Even after excluding relatives tested subsequent to the index case in their families, 9(56.3%) patients were positive for BRCA1/2 deleterious/suspected deleterious mutations among 16 triple-negative patients. Moreover, in 12 patients with early onset triple negative breast cancer (age ≤ 40), 6 patients (50.0%) reported deleterious mutations in BRCA1/2. Such positive mutation rate was even higher (69.2%) among the 13 triple negative patients with positive first and/or second-degree family history of breast and/or ovarian cancers. The association of BRCA1 mutations with triple-negative breast cancer is well-described [24] and in our study 6 out of the 10 deleterious mutations in this subgroup were in BRCA1, Table 2.
An interesting spectrum of mutations were identified in both BRCA1 and BRCA2, Table 2. Of note, there were many recurrent mutations with more than one carrier found to harbor the identical BRCA1 or BRCA2 mutation. However, most of these carriers were either first or second-degree relatives (see footnote to Table 2) rendering this an expected finding. The small sample size of this pilot study and the fact that genetic analysis was performed at Myriad Genetics laboratories did not allow for haplotype and founder mutation analyses which will be sought in future studies. Most of the detected mutations were reported previously in the Breast Cancer Information Core (BIC) [25] among Caucasian and Western populations, possibly due to similarity of genetic makeup between Middle Eastern population and Western population [26]. Only one variant of uncertain significance (VUS) in BRCA1, E1478D (4553G > C), was reported by Myriad Genetics variant information sheet as the first observation for this variant (personal communication). The 3450del4 deleterious mutation in BRCA1 was also previously reported in patients from Egypt [25] and Tunisia [27]. Also, BRCA1 E1373X (4236G > T) was originally described in a Palestinian family [28] and was recently reported again in a Palestinian patient [29]. Similarly, the 2482del4 deleterious mutation in BRCA2 was reported among Palestinian Arabs in BIC, and the BRCA2 E2229X seems to be recurrent among Arabs [25]. It is not unexpected to find BRCA1/2 mutations among Jordanians that were previously reported in Palestinians, knowing the Palestinian-Jordanian blended nature of families in Jordan. The BRCA2 VUS Q2925R (9002A > G) was also reported in Near Eastern and Middle Eastern populations [25]. Interestingly, the Icelandic founder mutation, BRCA2 999del5 [30], was also detected in one of our patients, but we do not have data to explain this finding.
The mutation rates we are reporting are similar to what Fostira et al. had reported among 403 Greek triple-negative patients; BRCA1 mutation was found in 47.6% among a subgroup of 105 triple-negative patients with family history of breast or ovarian cancers. A rate of 35.9% was reported among a subgroup of 106 women with early-onset (< 40 years) triple-negative breast cancer [31].
In a recent study, researchers at MD Anderson Cancer Center (MDACC) reported a similar incidence of BRCA1/2 mutations in patients with ER low-positive/PR low-positive/HER-2 neu negative tumors and patients with triple-negative breast cancer, suggesting that genetic counseling and BRCA testing should also be offered to patients who have hormone receptor–low-positive breast cancers [32]. Moreover, in an earlier publication Gonzalez-Angulo et al. reported a 19.5% incidence rate of BRCA mutations among an unselected cohort of triple negative breast cancer patients and patients with mutations had a significantly lower risk of relapse [33].
Our results support the conclusion that our ethnic group is not different and as such, women with early-onset triple-negative breast cancer, and ideally all triple-negative breast cancer patients, are candidates for BRCA genetic testing especially if they have family history of breast and/or ovarian cancers.
Among the other patients’ risk groups recruited to the study, 2 out of five (40.0%) patients with family history of male breast cancer and 10 out of 15 (66.7%) patients with bilateral or second primary breast cancer reported deleterious/suspected deleterious BRCA1/2 mutations (Fig. 1). Therefore, if cost is an issue for full adaption and implementation of international guidelines in low and middle-income countries, then testing patients with these “higher” risk features can be an option, at least in the initial phases of adaptations. Since recurrent mutations in our cohort occurred mostly among first and second-degree relatives, then initial testing for these recurrent mutations cannot be recommended for cost-saving before large-scale sequencing analyses are pursed to determine BRCA1/2 mutation status. Future larger studies aiming on haplotype and founder mutation detection may help in this regard.
Our positive rates, however, are significantly higher than what have been recently reported among neighboring Lebanese women where deleterious BRCA mutations were found in only 5.6%, and an additional 12.4% with VUS [12]. The difference in mutation rates may be explained by the different testing methodology, but more importantly this difference can be justified by the different inclusion criteria. However, when comparing similar groups of enrolled patients, significant differences were still observed. Among a subgroup of 148 young Lebanese patients (≤ 40 years at diagnosis) only 9 (6.1%) harbored deleterious mutations [12], while in our cohort of 50 young patients 26.0% reported deleterious/suspected deleterious mutations. Additionally, our rate was significantly higher (32.5%) in the 40 tested young patients with positive family history compared to 10.8% in 74 similar Lebanese patients. Such differences in closely related ethnic groups are difficult to explain, but the highly selective criteria we used to include patients may still be a confounder since many of our patients satisfied more than one inclusion criterion. In addition, differences in the methodology and techniques used in BRCA testing might be a contributing factor. Moreover, considering the small number of highly selected patients included in our study, the reported BRCA1/2 mutation rates should be interpreted with caution and within context.
Conducting a culturally sensitive genetic testing research in a developing country with limited resources is a challenge. Many ethical and cultural difficulties were encountered during the course of our study. Ensuring confidentiality and privacy were major issues in a tribal-based closely-related community and culture like the Jordanian population. Many patients expressed their concerns about labeling and stigmatization. Preserving other family members’ confidentiality when documenting family history was also addressed with the patients and occasionally with the relatives. Many concerns were related to the scope of physician-patient confidentiality when relatives are at genetic risk of cancer. Sharing information with at-risk relatives was not an issue despite our IRB concerns. Except for very few (3 patients), all our patients with deleterious/suspected deleterious mutations shared results with their at-risk relatives without major issues.
Potential insurance, employment and social discrimination were also addressed with the patients prior to testing and in more detail after receiving positive mutation results. These issues are expected to be a challenge once genetic testing is made routinely available to eligible patients as a standard clinical practice, especially that most insurance agencies don’t cover risk-reduction procedures including contralateral mastectomies and oophorectomies.
Following this exploratory pilot study, BRCA testing has started to be routinely offered at our institution, initially for the “higher” risk groups (discussed above) with the intention to gradually expand to include a wider patient population as suggested by the ASCO (American Society of Clinical Oncology) [23] and the NCCN (National Comprehensive Cancer Network) guidelines [19]. This process should enhance our understanding of the prevalence of BRCA1/2 mutations in our patient population. An ongoing project is currently collecting this information prospectively on all patients tested for BRCA1/2 mutations. The results of this project will help to assess whether a founder effect exists in the Jordanian population and whether a subset of mutations can be tested for cost-saving. Future recommendations for establishing a Clinical Cancer Genetics program are envisioned, where unaffected family members can also benefit from early screening and take appropriate risk-reduction measures.
Our study is limited by the small sample size and the highly selective criteria used for patient accrual. We were conscious to these limitations from the onset of the study. The small sample size was due to the limited funds available and to the high cost of BRCA testing at an accredited and reliable laboratory. Therefore we opted for highly selective inclusion criteria to test the more high risk patients in order for the results to have relevance in the clinical setting, especially that colleagues at other academic institutions in Jordan were reporting lack of BRCA1/2 mutations among Jordanians based on scholarly research performed in-house in their laboratories (personal communication). Our selected patient cohort shows an important incidence of deleterious and suspected deleterious BRCA mutations suggesting that genetic testing should be discussed with and offered to patients with such a high risk profile. Further studies are needed to confirm the results of this pilot study. Moreover, since many of the recruited high risk patients tested negative for BRCA1/2 mutations, it is plausible to take advantage of the collected DNA samples and test for mutations in other breast cancer susceptibility genes, e.g. CHEK2, PALB2 and BRIP1. Using next-generation sequencing will enable simultaneous testing for mutations in these and other genes, and multigene panels are now commercially available and are increasingly being used [34–36].
Conclusions
In summary, our results support the conclusion that BRCA1/2 mutations are common among Jordanian breast cancer patients with a highly selected risk profile and may contribute to the pathogenesis of disease in this patient population. This has significant clinical implications, both for the management and prevention of breast cancer. Therefore, full BRCA1/2 screening should be offered to patients with characteristic high risk features.
Acknowledgements
The authors sincerely thank Mrs. Dalia Al-Rimawi and Mrs. Ayat Taqash for their help in the statistical analysis of results.
Funding
The study was funded by a competitive grant from the King Hussein Cancer Center/MD Anderson Cancer Center Sister Institution Network Fund (SINF). There was no role for the funding body in the design of the study, the collection, analysis, and interpretation of data or in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article. Raw data sets are not publically available due to the culturally sensitive nature of the study and to protect the confidentiality of patients, but are available from the corresponding author on reasonable request.
Abbreviations
- ASCO
American Society of Clinical Oncology
- BIC
Breast cancer Information Core
- BRCA1
BReast CAncer susceptibility gene 1
- BRCA2
BReast CAncer susceptibility gene 2
- ER
Estrogen Receptor
- FISH
Fluorescent In Situ Hybridization
- FP
Favor Polymorphism
- HER2
Human Epidermal growth factor Receptor 2
- IDC
Infiltrating Ductal Carcinoma
- ILC
Infiltrating Lobular Carcinoma
- IRB
Institutional Review Board
- NCCN
National Comprehensive Cancer Network
- PCR
Polymerase Chain Reaction
- PR
Progesterone Receptor
- SINF
Sister Institution Network Fund
- VUS
Variant of Uncertain Significance
Authors’ contributions
HAR conceptualized the research idea, contributed to patient recruitment and drafted the manuscript. AAO contributed to the conceptualization and development of research methodology, funding acquisition, patient recruitment and critical review and editing of the manuscript. FZ was involved in patient recruitment, data collection and critical review and editing of the manuscript. BA critically reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the King Hussein Cancer Center, Amman, Jordan. IRB approval number 11KHCC63. All patients provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hikmat Abdel-Razeq, Email: habdelrazeq@khcc.jo.
Amal Al-Omari, Email: asomari@khcc.jo.
Farah Zahran, Email: fzahran@khcc.jo.
Banu Arun, Email: barun@mdanderson.org.
References
- 1.Cancer incidence in Jordan 2013. Jordan cancer registry. Non-Communicable Diseases Directorate Ministry of Health, Jordan Available at http://www.moh.gov.jo/Echobusv3.0/SystemAssets/7c73e811-df26-411d-8ed4-de050373aea0.pdf Accessed 18 Dec 2016.
- 2.El Saghir NS, Khalil MK, Eid T, El Kinge AR, Charafeddine M, Geara F, et al. Trends in epidemiology and management of breast cancer in developing Arab countries: a literature and registry analysis. Int J Surg. 2007;5:225–233. doi: 10.1016/j.ijsu.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Chouchane L, Boussen H, Sastry KS. Breast cancer in Arab populations: molecular characteristics and disease management implications. Lancet Oncol. 2013;14:e417–e424. doi: 10.1016/S1470-2045(13)70165-7. [DOI] [PubMed] [Google Scholar]
- 4.Gonzales FA, Taplin SH, Yu M, Breen N, Cronin KA. Receipt of mammography recommendations among white and non-white women before and after the 2009 United States preventive services task force recommendation change. Cancer Causes Control. 2016;27:977–987. doi: 10.1007/s10552-016-0775-9. [DOI] [PubMed] [Google Scholar]
- 5.Pace LE, He Y, Keating NL. Trends in mammography screening rates after publication of the 2009 US preventive services task force recommendations. Cancer. 2013;119:2518–2523. doi: 10.1002/cncr.28105. [DOI] [PubMed] [Google Scholar]
- 6.Finney Rutten LJ, Ebbert JO, Jacobson DJ, Squiers LB, Fan C, Breitkopf CR, et al. Changes in U.S. preventive services task force recommendations: effect on mammography screening in Olmsted County, MN 2004-2013. Prev Med. 2014;69:235–238. doi: 10.1016/j.ypmed.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med. 2008;359:2143–2153. doi: 10.1056/NEJMra0802968. [DOI] [PubMed] [Google Scholar]
- 8.Peto J, Collins N, Batfoot R, Seal S, Warren W, Rahman N, et al. Prevalence of BRCA1 and BRCA2 gene mutations in patients with early onset breast cancer. J Natl Cancer Inst. 1999;91:943–949. doi: 10.1093/jnci/91.11.943. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 2013;105:812–822. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 11.Karami F, Mehdipour P. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int. 2013;2013:1–21. doi: 10.1155/2013/928562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Saghir NS, Zgheib NK, Assi HA, Khoury KE, Bidet Y, Jaber SM, et al. BRCA1 and BRCA2 mutations in ethnic Lebanese Arab women with high hereditary risk breast cancer. Oncologist. 2015;20:357–364. doi: 10.1634/theoncologist.2014-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jalkh N, Nassar-Slaba J, Chouery E, Salem N, Uhrchammer N, Golmard L, et al. Prevalance of BRCA1 and BRCA2 mutations in familial breast cancer patients in Lebanon. Hered Cancer Clin Pract. 2012;10:7. doi: 10.1186/1897-4287-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laraqui A, Uhrhammer N, Lahlou-Amine I, EL Rhaffouli H, El Baghdadi J, Dehayni M, et al. Mutation screening of the BRCA1 gene in early onset and familial breast/ovarian cancer in Moroccan population. Int J Med Sci. 2013;10:60–67. doi: 10.7150/ijms.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim SS, Hafez EE, Hashishe MM. Presymptomatic breast cancer in Egypt: role of BRCA1 and BRCA2 tumor suppressor genes mutations detection. J Exp Clin Cancer Res. 2010;29:82–91. doi: 10.1186/1756-9966-29-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awadelkarim KD, AcetoG VS, Elhaj A, Morgano A, Mohamedani AA, et al. BRCA1 and BRCA2 status in a central Sudanese series of breast cancer patients: interactions with genetic, ethnic and reproductive factors. Breast Cancer Res Treat. 2007;102:189–199. doi: 10.1007/s10549-006-9303-z. [DOI] [PubMed] [Google Scholar]
- 17.Hasan TN, Shafi G, Syed NA, Alsaif MA, Alsaif AA, Alshatwi AA. Lack of association of BRCA1 and BRCA2 variants with breast cancer in an ethnic population of Saudi Arabia, an emerging high-risk area. Asian Pac J Cancer Prev. 2013;14:5671–5674. doi: 10.7314/APJCP.2013.14.10.5671. [DOI] [PubMed] [Google Scholar]
- 18.Troudi W, Uhrhammer N, Romdhane KB, Sibille C, Amor MB, El Khil HK, et al. Complete mutation screening and haplotype characterization of BRCA1 gene in Tunisian patients with familial breast cancer. Cancer Biomark. 2008;4:11–18. doi: 10.3233/CBM-2008-4102. [DOI] [PubMed] [Google Scholar]
- 19.NCCN Clinical Practice Guidelines in Oncology, Genetic/Familial High-Risk Assessment: Breast and Ovarian. Version 2.2017. https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf Accessed 18 Dec. 2016.
- 20.Beaudet AL, Tsui LC. A suggested nomenclature for designating mutations. Hum Mutat. 1993;2:245–248. doi: 10.1002/humu.1380020402. [DOI] [PubMed] [Google Scholar]
- 21.PopulationPyramid.net available at https://www.populationpyramid.net/jordan/2017/. Accessed 3 Dec. 2017.
- 22.Glassey R, Ives A, Saunders C, Musiello T. Decision making, psychological wellbeing and psychosocial outcomes for high risk women who choose to undergo bilateral prophylactic mastectomy - a review of the literature. Breast. 2016;28:130–135. doi: 10.1016/j.breast.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, et al. American Society of Clinical Oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2015;33:3660–3667. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 24.Greenup R, Buchanan A, Lorizio W, Rhoads K, Chan S, Leedom T, et al. Prevalence of BRCA mutations among women with triple-negative breast cancer (TNBC) in a genetic counseling cohort. Ann Surg Oncol. 2013;20:3254–3258. doi: 10.1245/s10434-013-3205-1. [DOI] [PubMed] [Google Scholar]
- 25.Breast Cancer Information Core: An open access on-line breast cancer mutation data base. Available at http://research.nhgri.nih.gov/bic/. Accessed 2 Dec. 2017.
- 26.Bu R, Siraj AK, Al-Obaisi KA, Beg S, Al Hazmi M, Ajarim D, et al. Identification of novel BRCA founder mutations in middle eastern breast cancer patients using capture and sanger sequencing analysis. Int J Cancer. 2016;139(5):1091–1097. doi: 10.1002/ijc.30143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahfoudh W, Bouaouina N, Ahmed SB, Gabbouj S, Shan J, Mathew R, et al. Hereditary breast cancer in middle eastern and north African (MENA) populations: identification of novel, recurrent and founder BRCA1 mutations in the Tunisian population. Mol Biol Rep. 2012;39(2):1037–1046. doi: 10.1007/s11033-011-0829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadouri L, Bercovich D, Elimelech A, Lerer I, Sagi M, Glusman G, et al. A novel BRCA-1 mutation in Arab kindred from east Jerusalem with breast and ovarian cancer. BMC Cancer. 2007;7:14. doi: 10.1186/1471-2407-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lolas Hamameh S, Renbaum P, Kamal L, Dweik D, Salahat M, Jaraysa T, et al. Genomic analysis of inherited breast cancer among Palestinian women: genetic heterogeneity and a founder mutation in TP53. Int J Cancer. 2017;141(4):750–756. doi: 10.1002/ijc.30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barkardottir RB, Sarantaus L, Arason A, Vehmanen P, Bendahl PO, Kainu T, et al. Haplotype analysis in Icelandic and Finnish BRCA2 999del5 breast cancer families. Eur J Hum Genet. 2001;9(10):773–779. doi: 10.1038/sj.ejhg.5200717. [DOI] [PubMed] [Google Scholar]
- 31.Fostira F, Tsitlaidou M, Papadimitriou C, Pertesi M, Timotheadou E, Stavropoulou AV, et al. Prevalence of BRCA1 mutations among 403 women with triple-negative breast cancer: implications for genetic screening selection criteria: a Hellenic cooperative oncology group study. Breast Cancer Res Treat. 2012;134:353–362. doi: 10.1007/s10549-012-2021-9. [DOI] [PubMed] [Google Scholar]
- 32.Sanford RA, Song J, Gutierrez-Barrera AM, Profato J, Woodson A, Litton JK, et al. High incidence of germline BRCA mutation in patients with ER low-positive/PR low-positive/HER-2 neu negative tumors. Cancer. 2015;121:3422–3427. doi: 10.1002/cncr.29572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17:1082–1089. doi: 10.1158/1078-0432.CCR-10-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narod SA. Testing For CHEK2 in the cancer genetics clinic: ready for prime time? Clin Genet. 2010;78:1–7. doi: 10.1111/j.1399-0004.2010.01402.x. [DOI] [PubMed] [Google Scholar]
- 35.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. Raw data sets are not publically available due to the culturally sensitive nature of the study and to protect the confidentiality of patients, but are available from the corresponding author on reasonable request.


