Abstract
Thousands of long noncoding RNAs (lncRNAs) have been reported and represent an important subset of pervasive genes associated with a broad range of biological functions. Abnormal expression levels of lncRNAs have been demonstrated in multiple types of human disease. However, the role of lncRNAs in systemic lupus erythematosus (SLE) remains poorly understood. In the present study, the expression patterns of lncRNAs and messenger RNAs (mRNAs) were investigated in peripheral blood mononuclear cells (PBMCs) in SLE using Human lncRNA Array v3.0 (8×60 K; Arraystar, Inc., Rockville, MD, USA). The microarray results indicated that 8,868 lncRNAs (3,657 upregulated and 5,211 downregulated) and 6,876 mRNAs (2,862 upregulated and 4,014 downregulated) were highly differentially expressed in SLE samples compared with the healthy group. Gene ontology (GO) analysis of lncRNA target prediction indicated the presence of 474 matched lncRNA-mRNA pairs for 293 differentially expressed lncRNAs (fold change, ≥3.0) and 381 differentially expressed mRNAs (fold change, ≥3.0). The most enriched pathways were ‘Transcriptional misregulation in cancer’ and ‘Valine, leucine and isoleucine degradation’. Furthermore, reverse transcription-quantitative polymerase chain reaction data verified six abnormal lncRNAs and mRNAs in SLE. The results indicate that the lncRNA expression profile in SLE was significantly changed. In addition, a range of SLE-associated lncRNAs were identified. Thus, the present results provide important insights regarding lncRNAs in the pathogenesis of SLE.
Keywords: systemic lupus erythematosus, long non-coding RNAs, messenger RNA, peripheral blood mononuclear cells
Introduction
Systemic lupus erythematosus (SLE), a complex autoimmune disease, is characterized by multiple immunologic abnormalities. It has previously been reported that ~7.4–159.4/100,000 people suffer from SLE worldwide, and SLE predominantly affects women of childbearing age (1–3). The presence of high titres of autoantibodies against nuclear components, elevated circulating immune complexes and complement deficiency are the predominant characteristics of the disease. The etiology of SLE is incompletely understood; however, genetic factors are important in the susceptibility to the disease.
Long noncoding RNAs (lncRNAs) are transcript RNA molecules, longer than 200 nucleotides, which do not encode a protein and reside in the nucleus or cytoplasm (4). lncRNAs are classified by their position relative to protein-coding messenger RNAs (mRNAs), and comprise the long intergenic ncRNA (lincRNA), intronic lncRNA, antisense lncRNA, transcribed pseudogene lncRNAs and enhancer RNA (eRNA). Although dysregulation of lncRNA expression has been characterized predominantly in cancer, it has recently been evaluated in autoimmune diseases, such as autoimmune thyroid disease (AITD) and rheumatoid arthritis (RA) (5,6). Specifically, lncRNAs have been proposed as a regulator of immune response (7–9) and contribute to the inflammatory response (10–13). Thus, it is hypothesized that lncRNAs, in combination with mRNAs, are also involved in the germination and development of SLE. However, until now, knowledge regarding the possible association between abnormal lncRNAs and SLE remains limited.
In the present study, integrative lncRNA-mRNA microarray analysis was performed to determine the expression profiles of lncRNAs in SLE samples and normal samples. In addition, three lncRNAs and three mRNAs were confirmed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were used to predict the function and signaling pathways affected by the differentially expressed mRNAs, which were target genes of aberrantly expressed lncRNAs. The results of the present study suggest that lncRNA expression patterns may provide further insight into the pathogenesis of SLE.
Materials and methods
SLE patients and healthy control subjects
Whole blood was collected from 29 SLE patients and 34 age- and sex-matched healthy control subjects (HCs), who were enrolled between July 2015 and May 2016, in the Department of Rheumatology and Immunology, The First Affiliated Hospital of Nanchang University (Nanchang, China). HCs were selected based on no history of autoimmune disease and all SLE patients were diagnosed according to the American College of Rheumatology classification criteria for SLE (14). A total of 10 of the already enrolled 29 patients with SLE and 10 of the already enrolled 34 age- and sex-matched HCs were recruited to isolate the peripheral blood mononuclear cells (PBMCs) and perform microarray analysis. Other samples (19 patients with SLE and 24 HCs) were used to verify the results of microarray by RT-qPCR assay. The study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (approval no. 2014003) and was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all the participants before commencing the study.
Blood sample collection and RNA isolation
Venous blood samples were obtained from 29 SLE patients and 34 HCs and stored in EDTA tubes. PBMCs were isolated from the venous blood by Ficoll-Histopaque (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) density gradient centrifugation (1,000 × g, 20 min, 22°C). Total RNA was extracted from PBMCs from each specimen using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. The concentration and quality of the RNA were assessed by absorbance spectrometry (260 nm/280 nm) using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.).
Microarray and data analysis
RNA samples were analyzed using Arraystar Human LncRNA Array v3.0 (Arraystar, Inc., Rockville, MD, USA), according to the manufacturer's instructions, which includes 33,045 lncRNAs and 30,215 coding transcripts. Briefly, rRNA was removed from total RNA and the mRNAs were obtained using an mRNA-ONLY™ Eukaryotic mRNA Isolation kit (Epicentre; Illumina, Inc., San Diego, CA, USA). The random priming method was utilized to amplify each sample and mRNA was transcribed into fluorescent complementary RNAs (cRNAs) without 3′bias. Labeled cRNAs were hybridized to the Human LncRNA Microarray. After washing the slides using Gene Expression Wash Buffer 1 (Agilent Technologies, Inc., Santa Clara, CA, USA), and Gene Expression Wash Buffer 2 (Agilent Technologies, Inc.), the arrays were scanned using an Agilent G2505C Scanner. Raw data were extracted using GeneSpring GX v12.0 software package (Agilent Technologies, Inc.). The microarray work was performed by Kangcheng Biology Engineering Co., Ltd. (Shanghai, China).
Functional group analysis
GO (www.geneontology.org) and KEGG (www.genome.ad.jp/kegg) databases were used to analyze the biological functions and signaling pathways affected by disexpression of mRNAs (the cut-off P-value was 0.05).
RT-qPCR assay
The expression levels of differentially expressed lncRNAs and mRNAs were confirmed by RT-qPCR. Briefly, total RNA was extracted from PBMCs using TRIzol reagent (Sigma-Aldrich; Merck KGaA), and 5 µg samples were used for the synthesis of first strand cDNA using a PrimeScript™ RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. The RT reaction was performed in a 10 µl reaction containing 5X PrimeScript™ Buffer, 1 µl RT specific primer, 0.5 µl PrimeScript™ RT Enzyme Mix and 5 µg of total RNA. The RT assay was set at an initial denaturation step at 37°C for 15 min, followed by 85°C for 5 sec. Following first strand cDNA synthesis, the PCR reaction was performed in a 10 µl reaction containing 1X SYBR-Green PCR Master mix (Takara Biotechnology Co., Ltd.), 0.4 µM of each specific forward and reverse primer and 0.5 µl of cDNA template. The PCR assay was set at an initial denaturation step at 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min with an ABI 7500 Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers are detailed in Table I. Gene expression levels were normalized to GAPDH in cDNA samples, and all experiments were performed in triplicate. The relative expression levels of the genes were determined using the ∆Cq method, where ∆Cq=Cq median gene-Cq median GAPDH (15,16).
Table I.
Primer sequence.
| Gene name | Primer sequence | Product (bp) length |
|---|---|---|
| AK130076 | F: CCAACATGCTGACTCACCCTTCC | 185 |
| R: ATGGAGTCTCGCTCTGTCACCCA | ||
| CTC-471J1.2 | F: ACAAATCTGAAAATACCACCTTG | 106 |
| R: TTTCCTAGAAATCATTTAACCCA | ||
| RP11-875O11.1 | F: CCCGATGGAATCTTACTCTGTTG | 129 |
| R: CATGCCTGTAATCCCAGCTACTC | ||
| PGLYRP1 | F: CACATGAAGACACTGGGCTGGT | 147 |
| R: CATGAAGCTGATGCCAATGGAC | ||
| PDCD4 | F: GGATGAAAGGGCATTTGAGAAGAC | 152 |
| R: CCAATGCTAAGGATACTGCCAACA | ||
| PTEN | F: ATCATTTCTTCATAGTGCTCCCC | 125 |
| R: CAATAGTAGTTGTACTCCGCTTA | ||
| GAPDH | F: GCACCGTCAAGGCTGAGAAC | 138 |
| R: TGGTGAAGACGCCAGTGGA |
Long none-coding RNAs included AK130076, CTC-471J1.2 and RP11-875O11.1. F, forward; R, reverse; PGLYRP1, peptidoglycan recognition protein 1; PDCD4, programmed cell death 4; PTEN, phosphatase and tensin homolog; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Statistical analysis
Data are presented as mean ± standard deviation. The Mann-Whitney test or Student's t-test was used to perform statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Aberrant lncRNA expression in PBMCs of SLE
To investigate the possible biological functions of lncRNAs in SLE, the lncRNA and mRNA expression profiles were estimated in PBMCs of SLE using microarray analysis (Figs. 1 and 2). The expression profiles of 8,868 (28.9%) lncRNAs indicated that they were differentially expressed (more than two-fold) between two groups (data no shown). Among these, 3,657 were upregulated while 5,211 were downregulated in the SLE group, compared with those in the healthy group. The most significantly deregulated lncRNAs were ENST00000523884 (fold change up, 303.1127021) and ENST00000559539 (fold change down, 4868.2903529).
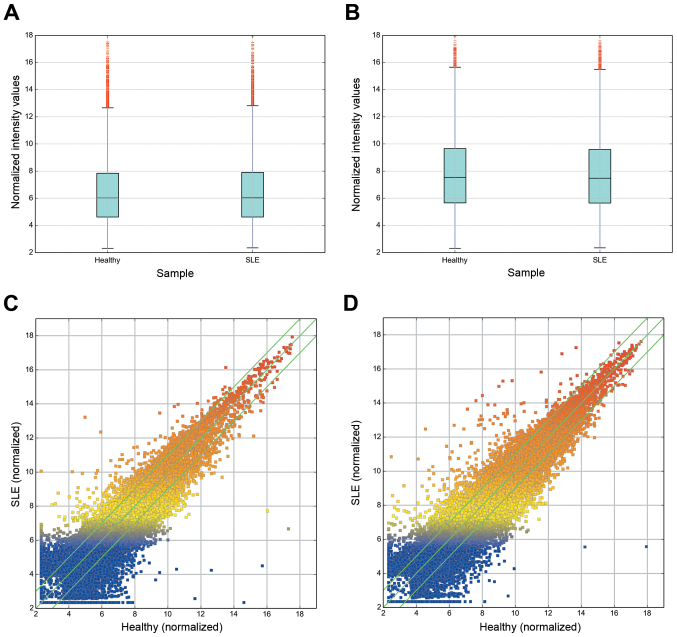
Figure 1.
Box plots and scatter plots demonstrating the variation in lncRNA and mRNA expression levels between the SLE and healthy group arrays. Box plots representing the distribution of the (A) lncRNA and (B) mRNA profiles. Following normalization, the distributions of the log2-ratios among the tested samples were nearly the same. The scatter plots of (C) lncRNA and (D) mRNA assess the variation between the SLE and healthy group arrays. The x- and y-axes in the scatter plot represent the normalized signal values of each group (log2 scale). The lncRNAs and mRNAs above the top green line and below the bottom green line are those with a two-fold change in expression between groups. lncRNA, long none-coding RNA; mRNA, messenger RNA; SLE, systemic lupus erythematosus.
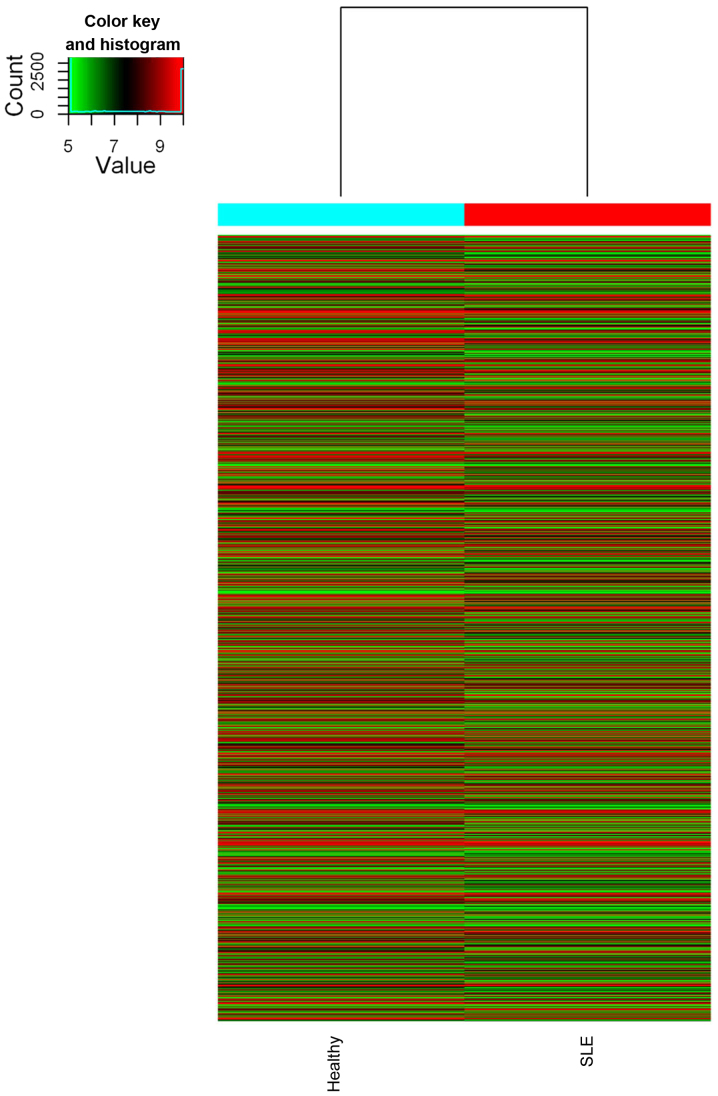
Figure 2.
Hierarchical clustering analysis of lncRNA and mRNA profile comparison between the SLE and healthy groups (n=10 per group). SLE, systemic lupus erythematosus.
Aberrant mRNA expression in PBMCs of SLE
To evaluate the involvement of lncRNAs in transcriptional, epigenetic or posttranscriptional regulation of gene expression, the expression levels of potential target mRNAs of the differentially expressed lncRNA data were predicted. In total, compared with the healthy group, 6,876 mRNAs (2,862 upregulated and 4,014 downregulated) were identified to be differentially expressed (P<0.05). The most markedly deregulated mRNAs were thyroglobulin (fold change up, 329.7330982) and potassium calcium-activated channel subfamily M regulatory beta subunit 3 (fold change down, 5246.5342205).
lncRNA classification and subgroup analysis
According to previous reports, lncRNAs are classified into different subgroups, such as lncRNAs with enhancer-like function (lncRNA-a), antisense lncRNA and lincRNAs (17). Previous studies demonstrated that subgroups, such as lincRNA and enhancer-like lncRNAs are involved in numerous types of disease (18,19). lncRNA-a were identified using GENCODE annotation (16). Further analysis was performed in the current study by classifying and stratifying the lncRNAs into the aforementioned subgroups (17). A total of 847 enhancer-like lncRNAs were identified to be significantly differentially expressed (fold change ≥2.0; P<0.05, data not shown), among which 430 lncRNAs were upregulated and 417 were downregulated in the SLE group, compared with the healthy group. The expression profiles of 1,911 lincRNAs (1,225 of which were upregulated) indicated that they were significantly differentially expressed (fold change ≥2.0; P<0.05, data not shown) between the SLE and healthy groups. Among these, 1,225 were upregulated and 686 were downregulated. It was also identified in antisense lncRNA profiling that 449 antisense lncRNAs (147 upregulated and 302 downregulated) were differentially expressed (fold change ≥2.0; P<0.05, data not shown) between the groups. In addition, certain nearby coding genes may be regulated by these subgroup lncRNAs. Furthermore, 474 matched lncRNA-mRNA pairs were identified for 293 differentially expressed lncRNAs (fold change ≥3.0) and 381 differentially expressed mRNAs (fold change ≥3.0). Among them, 310 pairs were differentially expressed unidirectionally (up or down), while 164 pairs were differentially expressed bidirectionally.
GO and KEGG signaling pathway analysis
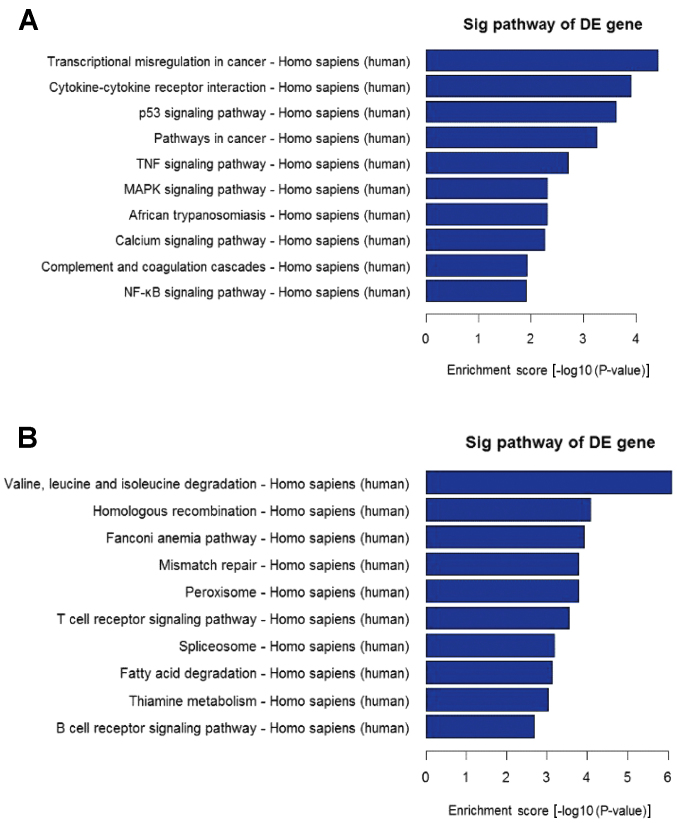
Through GO analysis, the downregulated and upregulated transcripts of lncRNAs were identified to be associated with biological processes, cellular components and molecular function. Additionally, the differentially expressed mRNAs between the SLE and healthy groups were significantly enriched in the cellular macromolecule metabolic process, intracellular part, nucleic acid binding, single-multicellular organism process, cell periphery and sequence-specific DNA binding, amongst others (data no shown). Using the latest version of the KEGG database, KEGG pathway analysis was performed to evaluate differentially expressed mRNAs. Pathway analysis determined that these lncRNAs may target 65 gene pathways, including 26 upregulated pathways, for example, ‘Transcriptional misregulation in cancer’ and 39 downregulated pathways, for example, ‘Valine, leucine and isoleucine degradation’ (Fig. 3).
Figure 3.
Pathway analysis of (A) upregulated and (B) downregulated mRNAs in SLE. (A) Twenty-six upregulated pathways were identified, including ‘Transcriptional misregulation in cancer’ and ‘Cytokine-cytokine receptor interaction’. (B) Thirty-nine downregulated pathways were identified, including ‘Valine, leucine and isoleucine degradation’ and ‘Homologous recombination’. mRNA, messenger RNA; SLE, systemic lupus erythematosus; DE, differently expressed.
RT-qPCR validation
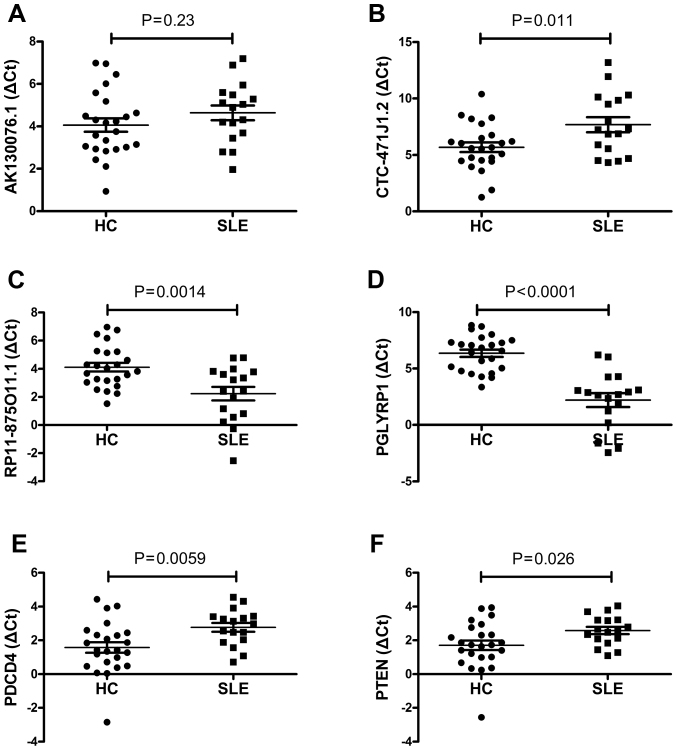
In order to verify the reliability of the microarray data, a number of interesting candidate lncRNAs (AK130076.1, CTC-471J1.2 and RP11-875O11.1) and mRNAs [peptidoglycan recognition protein 1 (PGLYRP1), programmed cell death 4 (PDCD4) and phosphatase and tensin homolog (PTEN)] were initially identified for further analysis. The expression levels of these lncRNAs and mRNAs were confirmed by RT-qPCR. The primers are presented in Table I. For the lncRNAs, the results demonstrated that RP11-875O11.1 were upregulated and that CTC-471J1.2 were downregulated in 17 SLE patients relative to the healthy group (all P<0.05; Fig. 4B and C). The expression level of AK130076.1 was not significantly different between SLE patients and healthy control subjects (P>0.05; Fig. 4A). For the mRNAs, the RT-qPCR analysis indicated that the expression levels of PGLYRP1, PDCD4 and PTEN changed significantly between the two groups (P<0.05 for each mRNA; Fig. 4). The results demonstrated that the expression level patterns of RP11-875O11.1, CTC-471J1.2, PGLYRP1, PDCD4 and PTEN were consistent with those obtained by microarray analysis.
Figure 4.
Relative expression levels of lncRNA and mRNA in SLE patients and healthy controls determined by reverse transcription-quantitative polymerase chain reaction. (A) AK130076.1 tends to be downregulated in SLE patients when compared with the healthy group, although the difference was not statistically significant (P=0.23). (B) CTC-471J1.2 was found to be significantly downregulated in SLE patients (P=0.011). (C) RP11-875O11.1 was found to be significantly upregulated in SLE patients (P=0.0014). (D) PGLYRP1 was found to be significantly upregulated in SLE patients (P<0.00010). (E) PDCD4 was found to be significantly downregulated in SLE patients (P=0.0059). (F) PTEN was found to be significantly downregulated in SLE patients (P=0.032). The expression levels of lncRNAs and mRNA were normalized against GAPDH, which served an internal control. The relative expression levels of the genes were calculated using the ΔCq method.
Discussion
SLE, an autoimmune disease, is characterized by aberrant lymphocyte activation. It has a prevalence of 40 cases per 100,000 individuals, with onset typically occurring in women of childbearing age (female:male ratio, 9:1) (20). Current treatment strategies using immunosuppressive drugs and other medications for SLE are effective at reducing morbidity and mortality, but fail to effectively cure the disease (21). However, the pathogenic mechanisms underlying SLE remain largely unknown; therefore, further investigations of SLE are considered to be of great importance.
Evidence from previous research indicated the aberrant expression of lncRNAs in the pathogenesis of SLE (22,23); however, fewer studies have examined the expression profile of lncRNAs in SLE. PBMCs, which include numerous immune cells, are key in host defense, and are used to identify novel disease mediators, disease variants and treatment responses (24–26). PBMCs have been used to discriminate the differences in non-coding RNA (microRNAs) profiles in SLE using microarray technology (27,28). Therefore, in an attempt to obtain the expression pattern of lncRNAs in SLE, lncRNA microarray technology was used to investigate the lncRNA signatures of 10 SLE patients.
In the present study, a number of differentially expressed mRNAs and lncRNAs were detected in PBMCs of SLE patients to investigate the potential functions of lncRNAs in its pathogenesis. A total of 8,868 lncRNAs (3,657 upregulated and 5,211 downregulated) and 6,876 mRNAs (2,862 upregulated and 4,014 downregulated) were significantly expressed in the SLE group compared with the healthy group. lncRNAs include lincRNAs, intronic lncRNAs, antisense lncRNAs, transcribed pseudogene lncRNAs and eRNAs. lncRNAs function via a variety of mechanisms; however, a common and important function of lncRNAs is to alter nearby encoding gene expression by affecting the process of transcription (29) or directly performing an enhancer-like role (17,30). Wu et al (22) demonstrated that lincRNAs were associated with the pathogenesis of SLE. Furthermore, the results demonstrated that enhancer-like lncRNAs (430 upregulated and 417 downregulated), lincRNAs (1,225 upregulated and 686 downregulated) and antisense lncRNAs (147 upregulated and 302 downregulated) were aberrantly expressed. In addition, results from microarray analysis revealed that these subgroup lncRNAs regulate certain nearby coding genes.
To investigate the potential regulatory roles of lncRNAs, GO category and KEGG pathway annotation were used to analyze the target gene pool. GO and KEGG pathway analyses demonstrated that downregulated and upregulated transcripts of lncRNAs were associated with biological process, cellular components and molecular function, which were associated with 65 gene pathways that corresponded to transcripts, for example, ‘Cytokine-cytokine receptor interaction’, ‘TNF signaling pathway’, ‘MAPK signaling pathway’ and ‘NF-κB signaling pathways’, which indicated that the pathology of SLE is predominantly associated with the regulation of multiple genes. Furthermore, these pathways are associated with the initiation and development of SLE (31,32).
Subsequently, six altered lncRNAs and mRNAs were selected, and their expression levels and the microarray results were assessed via RT-qPCR. The change in mRNA expression level was confirmed to be concordant with the microarray data, while the expression tendency of lncRNA demonstrated a similar trend, although it was not exactly the same. This may be due to the heterogeneity of SLE, differing sample sizes and variation in the sensitivity of the methods performed. Generally, the data from microarray analysis requires confirmation by RT-qPCR, which is considered to be more accurate.
In conclusion, lncRNA and mRNA expression levels were analyzed in PBMCs from SLE samples using microarrays, which revealed a novel and interesting foundation for improving the understanding of the association between lncRNA homeostasis in PBMCs and SLE. However, the findings described in the present study represent a starting point for the investigation of lncRNAs in SLE. Further investigations are required to evaluate the signaling pathways identified in the GO and KEGG analyzes with regard to their role in the development and progression of SLE.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81360459) and the Jiangxi Provincial Natural Science Foundation of China (grant no. 20151BAB215031). The authors would like to thank Dr Rui Wu at the Department of Rheumatology, The First Affiliated Hospital of Nanchang University (Nanchang, China).
References
- 1.Bertsias GK, Salmon JE, Boumpas DT. Therapeutic opportunities in systemic lupus erythematosus: State of the art and prospects for the new decade. Ann Rheum Dis. 2010;69:1603–1611. doi: 10.1136/ard.2010.135186. [DOI] [PubMed] [Google Scholar]
- 2.Kow NY, Mak A. Costimulatory pathways: Physiology and potential therapeutic manipulation in systemic lupus erythematosus. Clin Dev Immunol. 2013;2013:245928. doi: 10.1155/2013/245928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirou KA, Gkrouzman E. Anti-interferon alpha treatment in SLE. Clin Immunol. 2013;148:303–312. doi: 10.1016/j.clim.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature. 2012;482:310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- 5.Shirasawa S, Harada H, Furugaki K, Akamizu T, Ishikawa N, Ito K, Ito K, Tamai H, Kuma K, Kubota S, et al. SNPs in the promoter of a B cell-specific antisense transcript, SAS-ZFAT, determine susceptibility to autoimmune thyroid disease. Hum Mol Genet. 2004;13:2221–2231. doi: 10.1093/hmg/ddh245. [DOI] [PubMed] [Google Scholar]
- 6.Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. 2015;15:121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 7.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 9.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: Influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL; Proc Natl Acad Sci USA; 2014; pp. 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, Kochkodan JJ, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L, Zhou C, Ao Y. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66:969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- 13.Müller N, Döring F, Klapper M, Neumann K, Schulte DM, Türk K, Schröder JO, Zeuner RA, Freitag-Wolf S, Schreiber S, Laudes M. Interleukin-6 and tumour necrosis factor-α differentially regulate lincRNA transcripts in cells of the innate immune system in vivo in human subjects with rheumatoid arthritis. Cytokine. 2014;68:65–68. doi: 10.1016/j.cyto.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 15.Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2017 Jun 15; doi: 10.1002/jcla.22281. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Chen J, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One. 2014;9:e104044. doi: 10.1371/journal.pone.0104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Wan ZY, Song F, Sun Z, Chen YF, Zhang WL, Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: A microarray related study. Arthritis Res Ther. 2014;16:465. doi: 10.1186/s13075-014-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurevitz SL, Snyder JA, Wessel EK, Frey J, Williamson BA. Systemic lupus erythematosus: A review of the disease and treatment options. Consult Pharm. 2013;28:110–121. doi: 10.4140/TCP.n.2013.110. [DOI] [PubMed] [Google Scholar]
- 21.Xiong W, Lahita RG. Pragmatic approaches to therapy for systemic lupus erythematosus. Nat Rev Rheumatol. 2014;10:97–107. doi: 10.1038/nrrheum.2013.157. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, Xia S, Chen S, Tang Y, Shen N. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. 2015;17:131. doi: 10.1186/s13075-015-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, Wu L, Qian J, Qu B, Xia S, La T, Wu Y, Ma J, Zeng J, Guo Q, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96–104. doi: 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Toonen EJ, Barrera P, Radstake TR, van Riel PL, Scheffer H, Franke B, Coenen MJ. Gene expression profiling in rheumatoid arthritis: Current concepts and future directions. Ann Rheum Dis. 2008;67:1663–1669. doi: 10.1136/ard.2007.076588. [DOI] [PubMed] [Google Scholar]
- 25.Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, Lam LT, Davis RE, Staudt LM. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 26.Yamagata T, Benoist C, Mathis DA. Shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- 27.Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA, Bruner GR, Harley JB, Ojwang JO. Identification of unique microRNA signature associated with lupus nephritis. PLoS One. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Zhao H, Zhao S, Wang X. MicroRNA expression profiles of peripheral blood mononuclear cells in patients with systemic lupus erythematosus. Acta Histochem. 2014;116:891–897. doi: 10.1016/j.acthis.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: The role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- 30.Mattick JS. Lincing long noncoding RNAs and enhancer function. Dev Cell. 2010;19:485–486. doi: 10.1016/j.devcel.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Yougbaré I, Boire G, Roy M, Lugnier C, Rouseau E. NCS 613 exhibits anti-inflammatory effects on PBMCs from lupus patients by inhibiting p38 MAPK and NF-κB signalling pathways while reducing proinflammatory cytokine production. Can J Physiol Pharmacol. 2013;91:353–361. doi: 10.1139/cjpp-2012-0233. [DOI] [PubMed] [Google Scholar]
- 32.Zhu LJ, Landolt-Marticorena C, Li T, Yang X, Yu XQ, Gladman DD, Urowitz MB, Fortin PR, Wither JE. Altered expression of TNF-alpha signaling pathway proteins in systemic lupus erythematosus. J Rheumatol. 2010;37:1658–1666. doi: 10.3899/jrheum.091123. [DOI] [PubMed] [Google Scholar]