Abstract
Lethal-7 (let-7) microRNA (miRNA) serves a pivotal role in a number of physiological processes and is associated with the occurrence and development of multiple disorders such as cancer. The present study aimed to use a newly developed stem-loop strategy for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) to distinguish let-7 miRNA family members that differ by as little as a single nucleotide. For the miRNAs comprising 16 identical nucleotides at the 5′-end, different stem-loop RT primers were designed and used in RT-qPCR to assess the expression profiles of a panel of let-7 family member miRNAs in human glioblastoma U87 cells. Amplification efficiency was evaluated through correlation analysis between total RNA input and the quantification threshold values. Melting curve profiles were measured to estimate the amplification specificity of the improved stem-loop RT-qPCR compared with those of the poly(A)-tailing method. In addition, the discrimination ability of the modified stem-loop method was examined. Compared with poly(A) tailing, the modified stem-loop RT method was able to specifically reverse transcribe the diverse let-7 miRNA family members followed by accurate quantification, with a theoretical amplification efficiency of ~100%. This modified stem-loop method was able to distinguish miRNAs with a single base difference. This innovative method may be used in the clinical detection of let-7 expression levels in a variety of tumour samples, and may provide valuable data for disease diagnosis and prognostic evaluation. In addition, this method may offer a new avenue for developing particular stem-loop approaches in measuring other miRNAs with little discrepancy.
Keywords: microRNA, lethal-7 family, stem-loop primer, reverse transcription-quantitative polymerase chain reaction
Introduction
MicroRNAs (miRNAs) are short (20–22 nucleotides in length), single-stranded, noncoding regulatory ribonucleic acids that participate in gene expression modulation (1–5) by complementary base paring to its target mRNA. miRNAs have been implicated as having a role in a number of cellular events, including proliferation, differentiation, communication and apoptosis (6), as well as in the regulation of multiple physiological and pathological processes, such as DNA methylation, chromatin modification and oncogenesis (7,8). A large majority of miRNAs exist as members of a large family with similar nucleotide sequences, therefore it may be hard to specifically discriminate one individual family member from others using conventional polymerase chain reaction (PCR).
Originally identified in Caenorhabditis elegans in 2000, a homolog of lethal-7 (let-7) was subsequently identified in humans and other mammalian species that exhibited numerous functions (9). The let-7 family is the largest miRNA family discovered to date, and it is ubiquitously expressed. In Homo sapiens, at least nine let-7 family members have been identified that are highly evolutionarily conserved, including let-7a-g, let-7i and microRNA (miR)-98 (10,11). The let-7 family not only regulates a series of crucial physiological functions, such as growth, development and homeostasis, but also acts as a suppressor that impedes tumour generation and growth (12). Indeed, plenty of evidence revealed that let-7 was downregulated in malignant tumours (13,14). For example, the expression level of let-7 was previously correlated with the degree of tumour malignancy, which suggested a significant role for let-7 expression signatures in cancer diagnosis and prognosis evaluation (15). However, owing to their short length and sequence similarity, the identification of an individual let-7 gene from the other family members, and the subsequent accurate quantitative profiling for each mature let-7 miRNA in neoplasms remains challenging.
Current methods that have been extensively used for the detection and quantification of miRNAs largely depend on poly(A) tailing and stem-loop reverse transcription-quantitative PCR (RT-qPCR). A previous study reported a novel stem-loop RT-qPCR in 2005 (16), in which a specially designed stem-loop RT primer that hybridized to 6–8 nucleotides at the 3′-end of mature miRNAs and was able to reverse transcribe them. Subsequently, the products were subjected to TaqMan-based conventional qPCR using specific forward primers and the aforementioned stem-loop reverse primers. Although high sensitivity and specificity were observed using this method compared with conventional qPCR, it may not be used extensively on account of the expensive TaqMan probes and the low amplification efficiency attained using TaqMan miRNA assays coupled with qPCR. Furthermore, the linear primers used in the poly (A) tailing approach may not distinguish between mature miRNAs and primary miRNA precursors, which may lead to relatively poor amplification specificity.
The present study provided a modified stem-loop RT-based qPCR strategy for the specific and sensitive measurement of individual let-7 miRNA family members using a SYBR green-based miRNA qPCR assay. This approach was able to specifically detect and quantify individual let-7 genes, of which the expression signatures may serve as potential biomarkers for various disorders.
Materials and methods
Cell culture and total RNA extraction
The U87 human glioblastoma cell line was purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% (v/v) foetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml of penicillin and 100 µg/ml of streptomycin (both from Invitrogen; Thermo Fisher Scientific, Inc.). Cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C. Total RNA was isolated from 2×106 U87 cells using 1 ml TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RNA was quantified and evaluated for purity with a spectrophotometer based on A260 and A280 values, followed by visualization on a 1.0% (w/v) agarose gel stained with ethidium bromide.
Stem-loop RT-qPCR
Stem-loop RT and RT-qPCR primers were synthesized by Genewindows Biotech. Co. Ltd. (Guangzhou, China) and are listed in Table I. cDNA was generated by reverse transcription using 1 µg RNA as template with ReverTra Ace-α-Transcriptase (Toyobo Life Science, Osaka, Japan). For amplification synthetic DNA, ~0.1 ng of each let-7 member were mixed and used as template. Briefly, stem-loop and U6 RT primers were combined to transcribe the total RNA. cDNA was prepared in a series of dilutions and thus a concentration gradient was generated prior to RT-qPCR assay. RT-qPCR was performed using the SYBR Premix Ex Taq II kit (Tli RNaseH Plus; Takara Bio, Inc., Otsu, Japan), following the manufacturer's protocol. qPCR was performed with a LightCycler 480 Real-Time PCR system (Roche Applied Science, Rotkreuz, Switzerland) and quantitation cycle (Cq) values were normalized to U6 small nuclear RNA (16), which was used as an internal control. The amplification profile was: 1 cycle of denaturation at 95°C for 30 sec; followed by 45 cycles of denaturation at 95°C for 5 sec, annealing and extension at 60°C for 1 min; and fluorescence intensity was measured at 75°C. For melting curve analysis, the following parameters were used: Denaturation at 95°C for 5 sec, annealing at a rate of −2.8°C/sec until 60°C, fluorescence intensity were measured throughout the process. All experiments were performed in triplicate. Analysis was performed using Microsoft Excel 2013 (Microsoft Corporation, Redmond, WA, USA) and Origin 8.0 (OriginLab, Wellesley, MA, USA) software.
Table I.
Stem-loop RT and RT-quantitative polymerase chain reaction primer sequences.
| Gene | Sequence (5′→3′) |
|---|---|
| U6 (RT) | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATATG |
| U6 (F) | CTCGCTTCGGCAGCACA |
| U6 (R) | AACGCTTCACGAATTTGCGT |
| Let-7a/e/f-5p (RT) | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACTATAC |
| Let-7b-5p (RT) | CTCAACTGGAGCTAGTTTCGTCGTAGGGCAGTTGAGAACCACAC |
| Let-7c (RT) | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCAT |
| Let-7d-5p (RT) | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACTATGC |
| Let-7g-5p (RT) | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACTGTAC |
| Let-7i-5p (RT) | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACAGC |
| miR-98 (RT) | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACAATAC |
| Let-7a/b/c-5p (F) | CCAGCTGGGTGAGGTAGTAGGTTGT |
| Let-7d-5p (F) | CCAGCTGGGAGAGGTAGTAGGTTGC |
| Let-7e-5p (F) | CCAGCTGGGTGAGGTAGGAGGTTGT |
| Let-7f-5p (F) | CCAGCTGGGTGAGGTAGTAGATTGT |
| Let-7g/i-5p (F) | CCAGCTGGGTGAGGTAGTAGTTTGT |
| miR-98-5p (F) | CCAGCTGGGTGAGGTAGTAAGTTGT |
| Uni (R1)a | TCCAGTGCAGGGTCCGAGGTA |
| Uni (R2)b | CTGGTGTCGTGGAGTCGGCAATT |
| Uni (R3)c | CTGGAGCTAGTTTCGTCGTAGGG |
| Poly(A) (RT) | GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTTTTTTTVN |
| Poly(A) (Uni R) | GCGAGCACAGAATTAATACGACTCA |
Uni (R1) was used as the reverse primer for the detection of let-7c and let-7i.
Uni (R2) was used as a common reverse primer.
Uni (R3) was used as the reverse primer for the detection of let-7b. F, forward; miR, microRNA; N, any nucleotide; R, reverse; RT, reverse transcription; Uni, universal; V, any nucleotide but T.
Poly(A) tailing and RT
Poly(A) tails were added to total RNA 3′-ends using the miRNA First-Strand cDNA Synthesis kit (ComWin Biotech Co., Ltd., Beijing, China). Subsequently, RNA was reverse transcribed with anchor primer consisting of oligo(dT) for binding poly(A), a specific sequence for the combination with the reverse primer used in subsequent RT-qPCR, and a binucleotide VN (V indicates any nucleotide except for dT whereas N refers to any nucleotide). qPCR forward primers and thermocycling profiles were similar to the aforementioned stem-loop RT-qPCR.
Data analysis
Cq values and melting curves were obtained using the LightCycler 480 software V1.5, and Origin 8.0 software (OriginLab) was exploited to draw standard curves. The Cq values were plotted against the log2 of total RNA input, and the amplification efficiency was evaluated through correlation analysis between these two values using Origin 8.0 software. Amplification efficiency (E) was calculated as: E=2−1/a-1; where a is the slope of the standard curve line. The Clustal algorithm (Bio-Edit software V7.0.5) was applied in multiple sequence alignment, whereas primer designation schemes were produced using Adobe Illustrator CS4 software (Adobe Systems Incorporated, San Jose, CA, USA).
Statistical analysis
All data were presented as the mean ± standard deviation. Significant differences between samples were calculated by Student's t-test. All tests were performed at least in triplicate. All statistical analyses were carried out using the Origin 8.0 (Additive GmbH, Friedrichsdorf, Germany) and Excel software (Microsoft Corporation, Redmond, WA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
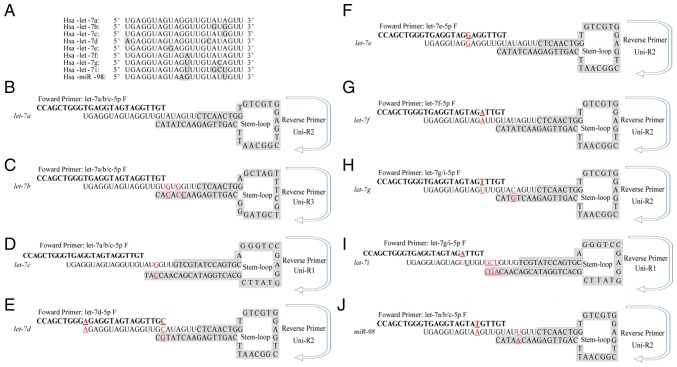
Sequence alignment identified no differences in the 16 nucleotide base sequences at the 5′-ends of let-7a, let-7b and let-7c; therefore, the same forward primers was used for these genes. Similarly, let-7i and let-7g shared a common forward primer. However, distinct stem-loop structures and different reverse primers were designed for used in SYBR Green-based RT-qPCR, which would ensure amplification specificity. The specific stem-loop RT primers designed for each let-7 family member is illustrated in Fig. 1.
Figure 1.
Different stem-loop primers were designed to distinguish let-7 family members. (A) Multiple sequence alignment of nine let-7 family members was performed to analyse the conserved nature of let-7 family. (B-J) Sequences of the specific stem-loop primers for let-7 family members, including (B) let-7a, (C) let-7b, (D) let-7d, (E) let-7d, (F) let-7e, (G) let-7f, (H) let-7g, (I) let-7i and (J) miR-98. Red underscored characters indicated variant nucleotides that differ from let-7a. F, forward; Hsa, Homo sapiens; R, Reverse; uni, universal.
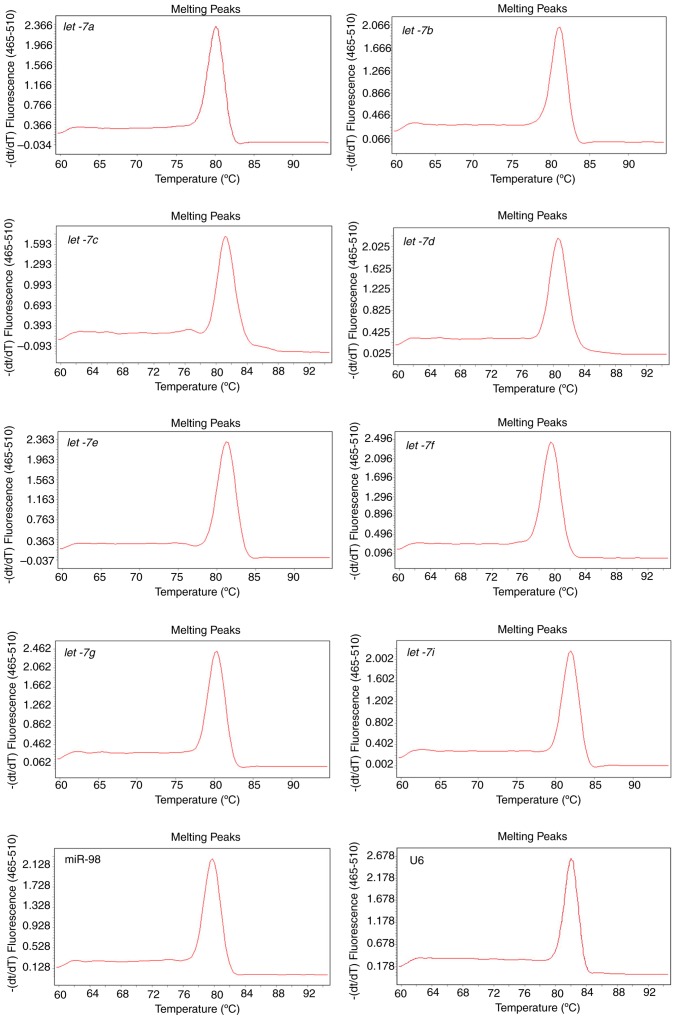
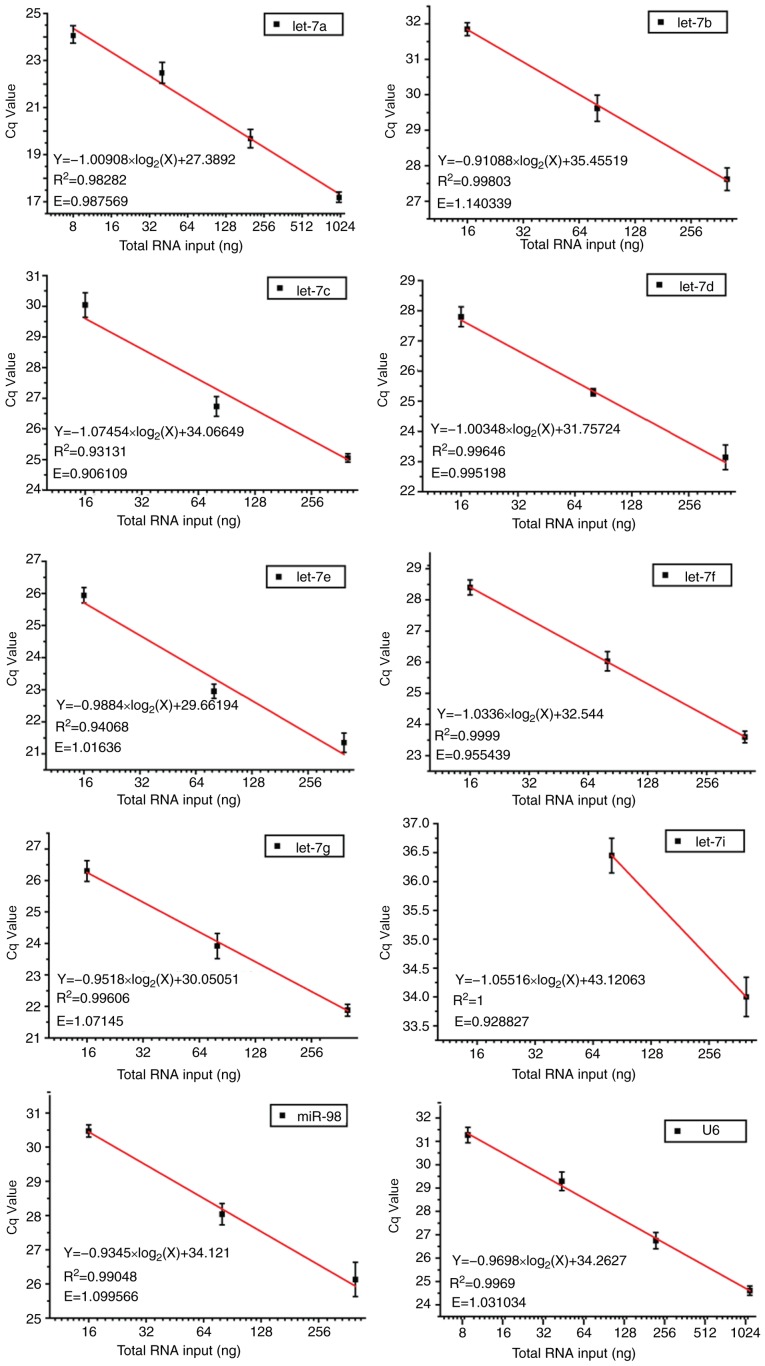
Robust amplification efficiency and specificity are achieved
Total RNA was extracted from U87 cells and reverse transcribed with a mixture of distinct stem-loop RT primers specifically designed for let-7 family members and U6 primers. Melting curves that are representative of amplification specificity combined with amplification efficiency of let-7 and U6 were also measured. Melting curve analysis of the amplified products demonstrated a single, sharp peak, which indicated good specificity (Fig. 2). Except for let-7i, which had a low concentration and therefore cannot be detected at a high dilution ratio, other amplification efficiencies ranged between 0.9288 and 1.1403. In addition, good linear relationships were indicated between PCR Cq values and log2 of RNA input (R2>0.9; Fig. 3). These data suggested that by performing only one miRNA qPCR reaction, it was feasible to simultaneously detect and quantify all nine let-7 family members, and to further discriminate between let-7a, let-7b and let-7c, as well as between let-7i and let-7g. Notably, the mixture containing our modified RT primers and U6 primer was still able to efficiently and specifically quantify each of the let-7 congeners and U6 internal control, respectively, contributing to a considerable decrease in time and cost.
Figure 2.
Melting curves of the let-7 family members and U6 amplification assay.
Figure 3.
Correlation analysis of total RNA input and the Cq values for let-7 miRNA family members and U6 from reverse transcription-quantitative polymerase chain reaction. Cq, quantification cycle; E, amplification efficiency.
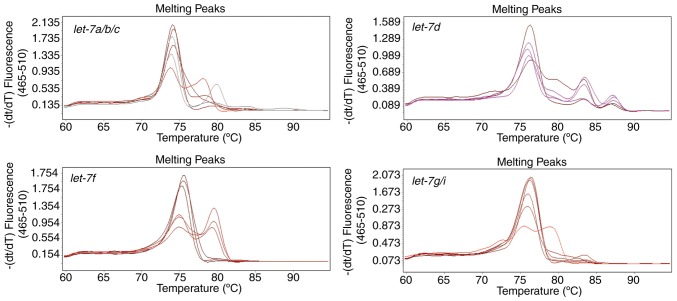
Stem-loop RT-qPCR is superior to Poly(A) tailing-based RT-qPCR in amplifying the mature let-7 family
The expression profiles of let-7 family members in U87 cells were also assessed by the poly(A) tailing method (Fig. 4). This method was initially established to detect miRNAs for large-scale and high-throughput screening, so the forward primers in the stem-loop method and the poly(A) tailing method designed in the present study were the same. There is only a one base difference between mature let-7 miRNA family members, therefore, if the stem-loop section of primers used to reverse transcribe in different let-7 members were identical, the melting temperature values of these distinct let-7 members were alike (Fig. 2). However, in the poly(A) method, cross-reactions between let-7a, let-7b and let-7c or between let-7g and let-7i cannot result in a difference of >5°C in the melting temperature. Therefore, multiple peaks that occurred in the melting curves in the poly(A) tailing method were caused by other reasons.
Figure 4.
Melting curves were obtained from quantitative polymerase chain reaction amplification products of representative let-7 family members using the poly(A) tailing method. For each plot, six curves were produced that represent the quantitative polymerase chain reactions that was performed twice at different concentrations of template (a high concentration and a low concentration), and each reaction using a specific concentration of template was repeated in triplicate.
However, the melting curves of each amplification reaction that were generated using the poly(A) method contained multiple peaks representing different products (Fig. 4), regardless of the specific forward primers of let-7d and let-7f. These may because miRNA precursors were also amplified, since the linear primers used in poly (A) tailing method lack specificity.
Modified stem-loop RT-qPCR discriminates distinct mature let-7 family members that differ by as little as a single nucleotide
The ability of the modified stem-loop RT-qPCR assay to discriminate between miRNAs that differ by as little as a single nucleotide was examined using synthetic templates that contained a certain amount of each let-7 member and the differential Cq values were analysed. Different concentrations of synthetic let-7a, let-7b and let-7c were mixed and reverse transcribed with a mixture of corresponding primers and RT-qPCR was performed with let-7a primers alone. The results demonstrated that the addition of equal concentrations of let-7b or let-7c did not affect the detection of let-7a (Fig. 5). qPCR reactions devoid of let-7a had significantly higher Cq values compared with those that also contained synthetic let-7a. Although 5-fold excess concentration of let-7b or let-7c slightly interfered with let-7a detection, this interference was not statistically significant.
Figure 5.
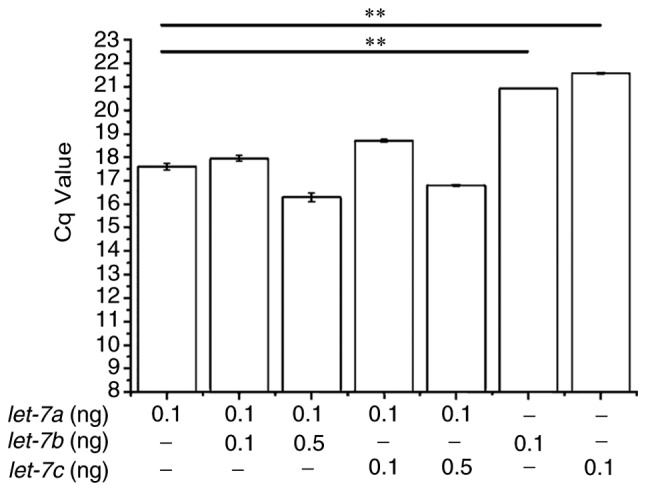
Specificity of the modified stem-loop RT-PCR assay. Mature let-7a specific qPCR was conducted by adding synthesized analogues of let-7b and let-7c as interference. The indicated amounts of synthetic let-7a, let-7b and let-7c were mixed together and reverse transcribed with mixed RT primers prior to RT-qPCR **P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
The let-7 family of miRNAs are highly conserved, differing by only 1–4 nucleotides among its members, which enables these congeners to share common target mRNAs that are downregulated by complementary sequences within their 3′-untranslated regions. Let-7 miRNA was first discovered in C. elegans as a switch gene that controlled cell fate transitions between larva and adult, and was later documented in humans in repressing several cancers, including lung cancer (17), colon cancer (18) and melanoma (19). Underexpression of let-7 in lung cancer was reported to be associated with shorter survival following tumorectomy (17), whereas overexpression of let-7 suppressed cancer cell proliferation and tumourigenicity (20–22). Accordingly, let-7 may be regarded as a miRNA candidate for cancer detection and monitoring.
However, small discrepancies among the let-7 family members generally escape detection by current routine techniques, except for high-throughput sequencing (23) or stem-loop RT-qPCR combined with TaqMan-based real-time quantification. Nevertheless, their clinical utilization in detection and intensive research for miRNAs was still hindered, due to the relatively high cost. Conversely, despite the low cost of SYBR Green dyes, it cannot be used as a substitute for TaqMan probes to simultaneously detect miRNAs with few differences. However, the present study offered a newly developed method based on the method previously described by Chen et al (16). The initial stem-loop method described by Chen et al was able to differentiate between let-7a and let-7b, but not between let-7a, let-7c and let-7d (16). This was likely to the fact that the TaqMan probe used in their system restricted the amplification efficiency. Notably, the modified method used in the present study was able to adequately discriminate let-7a and let-7c, which indicated that this method is able to differentiate miRNAs that differ by as little as a single nucleotide and therefore render it capable of being used in numerous of clinical applications.
Using an identical upstream primer, the present method enabled specific RT and quantification of several distinct miRNAs without a major loss of specificity of downstream primers that were mixed together, and in the absence of costly TaqMan probes. More importantly, this method exhibited higher specificity compared with conventional TaqMan. Data from the present study indicated that the modified stem-loop RT-qPCR method was able to differentiate between the let-7 family members that differ by a single nucleotide, which thereby provides a method to investigate the specific function of each family member and the potential cooperation between these members, as well as the role let-7 family members in various pathological conditions, particularly in the process of tumour generation and progression. In addition, this method is able to discriminate mature miRNAs from their longer progenitors (16). These characteristics and merits may facilitate the use of this modified stem-loop RT-based qPCR into clinical applications and may be further expanded to detect and distinguish between miRNA congeners differing by as little as a single nucleotide.
Acknowledgements
The present study was supported in part by The Special Fund for the Development of Shenzhen Strategic Emerging Industries (grant no. CXZZ20130517100617030).
References
- 1.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 2.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced Accumulation of Specific MicroRNAs in Colorectal Neoplasia11Note: Susan M. O'Connor and Nicholas G. van Holst Pellekaan contributed equally to this work. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 9.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 10.Rodini CO, Suzuki DE, Saba-Silva N, Cappellano A, de Souza JE, Cavalheiro S, Toledo SR, Okamoto OK. Expression analysis of stem cell-related genes reveal OCT4 as a predictor of poor clinical outcome in medulloblastoma. J Neurooncol. 2012;106:71–79. doi: 10.1007/s11060-011-0647-9. [DOI] [PubMed] [Google Scholar]
- 11.Sakurai M, Miki Y, Masuda M, Hata S, Shibahara Y, Hirakawa H, Suzuki T, Sasano H. LIN28: A regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. J Ster Biochem Mol Biol. 2011;131:101–106. doi: 10.1016/j.jsbmb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling; Proc Natl Acad Sci USA; 2010; pp. 1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 MicroRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 14.Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F, Yuan J, Chen Z, Yang A, Wang H. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One. 2010;5:e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. P Natl Acad Sci USA. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 18.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 19.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 20.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 21.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family; Proc Natl Acad Sci USA; 2008; pp. 3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu L, Schwartz P, Scarampi L, Rutherford T, Canuto EM, Yu H, Katsaros D. MicroRNA let-7a: a potential marker for selection of paclitaxel in ovarian cancer management. Gynecol Oncol. 2011;122:366–371. doi: 10.1016/j.ygyno.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 23.Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, et al. microRNA-34c is a novel target to treat dementias. Embo J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]