Abstract
Polymorphisms in the cytochrome P (CYP) 450 family may cause adverse drug responses in individuals. Cytochrome P450 2C19 (CYP2C19) is a member of the CYP family, where the presence of the 681 G>A, 636 G>A and 806 C>T polymorphisms result in the CYP2C19*2, CYP2C19*3 and CYP2C19*17 alleles, respectively. In the current study, the frequency of the CYP2C19*2, CYP2C19*3 and CYP2C19*17 alleles in an Iranian population cohort of different ethnicities were examined and then compared with previously published frequencies within other populations. Allelic and genotypic frequencies of the CYP2C19 alleles (*2, *3 and *17) were detected using polymerase chain reaction (PCR)-restriction fragment length polymorphism analysis, PCR-single-strand conformation polymorphism analysis and DNA sequencing from blood samples of 1,229 unrelated healthy individuals from different ethnicities within the Iranian population. The CYP2C19 allele frequencies among the Iranian population were 21.4, 1.7, and 27.1% for the CYP2C19*2, CYP2C19*3 and CYP2C19*17 alleles, respectively. The frequency of the homozygous A/A variant of the CYP2C19*2 allele was significantly high and low in the Lur (P<0.001) and Caspian (P<0.001) ethnicities, respectively. However, the frequency of the homozygous A/A variant of the CYP2C19*3 allele was not detected in the Iranian cohort in the current study. The frequency of the heterozygous G/A variant of the CYP2C19*3 allele had the significantly highest and lowest frequency in the Fars (P<0.001) and Lur (P<0.001) groups, respectively. The allele frequency of the homozygous T/T variant of the CYP2C19*17 allele was significantly high in the Caspian (P<0.001) and low in the Kurd (P<0.05) groups. The frequency of the CYP2C19 alleles involved in drug metabolism, may improve the clinical understanding of the ethnic differences in drug responses, resulting in the advancement of the personalized medicine among the different ethnicities within the Iranian population.
Keywords: CYP2C19*3 allele, CYP2C19, Iranian populations, Fars, Turk, Caspian, Lur, Kurd
Introduction
The cytochrome P450 enzymes have an important role in the metabolism of various therapeutic agents, in which different enzymatic activities lead to variability in drug responses among individuals (1). Cytochrome P450 2C19 (CYP2C19) is a highly polymorphic isoenzyme of the cytochrome P450 superfamily, affecting the metabolism of an extensive range of therapeutic drugs (2). The CYP2C19 gene, which includes nine exons and eight introns, is located at the 10q24.1–10q24.3 locus of chromosome 10, where coding sequences is 1,473 bp and resulting in a protein of 490 amino acid residues (3,4). Approximately 25 genetic variants in the exonic region of the CYP2C19 have been identified (4). CYP2C19 is involved in metabolizing several important therapeutic drugs, including omeprazole, lansoprazole, proguanil, propranolol, imipramine, mephenytoin, chloroguanide, hexabarbitone, diazepam and certain antidepressants (1,5). Common variants of the CYP2C19 gene are associated with impaired drug metabolism. CYP2C19*2 and CYP2C19*3 were identified in individuals who exhibited a reduced capability for metabolizing the probe drugs, and variant CYP2C19*17 is associated with ultra-rapid metabolism of CYP2C19 substrates (3).
The principal detrimental allele, CYP2C19*2, results from a guanine (G) to adenine (A) transition at position 681 in exon 5 (rs4244285), producing an aberrant splice site and it represents the most frequent CYP2C19 defect in all populations (6). CYP2C19*2 and CYP2C19*3 are the most common alleles, encoding enzymes with decreased activity (7). CYP2C19*3 (636G>A) is considered the most important allele, in which a point mutation in exon 4 results in a premature stop codon, and therefore nonfunctional protein (1,4).
The prevalence of the CYP2C19 poor metabolizer (PM) phenotype is 2-5% among Caucasians and Africans, and ~15% in Asians (8), while CYP2C19*3 is considered to be an Asian mutation (1). CYP2C19*2 and *3 alleles have been proposed to explain <50%, to >90%, of the PM phenotype (2).
The CYP2C19*17 allele was previously reported to be associated with high CYP2C19 activity, and identified at 18-28% in European populations, in 17-18% of Africans and in 0.3–4% of Asian populations (9). CYP2C19*17 is a −806 C>T single nucleotide polymorphism that causes specific nuclear protein binding to the 5′-flanking region. This binding results in increased gene transcription and high enzyme activities (10).
In this study, the frequency of the CYP2C19*2, *3, and *17 was examined among an Iranian cohort of different ethnicities.
Materials and methods
Specimen collection and ethical approval
A total of 1,229 blood specimen of unrelated healthy donors were obtained from Iranians through the Special Medical Centre (SMC; Tehran, Iran) including: 180 Fars, 110 Turk (Azari), 73 Caspian (Mazani, Gilaki), 80 Lure and 95 Kurd individuals to examine the CYP2C19*2 allele; 120 Fars, 82 Turk (Azari), 75 Caspian (Mazani, Gilaki), 73 Lure and 70 Kurd individuals to examine the CYP2C19*3 allele; and 156 Fars, 56 Turk (Azari), 32 Caspian (Mazani, Gilaki), 13 Lure and 14 Kurd individuals to examine the CYP2C19*17 allele. Blood samples (2 ml) with ethylenediaminetetraacetic acid were collected from participants. Informed consent to participate in genetic and molecular analyses, and consent to publish results were obtained from individuals. The Medical Ethics Committee of the SMC specifically approved this study (approval no. AA/27/2008). The exclusion criteria to select individuals in this study were any background of familial and sporadic cancer, metabolic, nuclear and mitochondrial DNA-associated disorders.
Genomic DNA extraction and primer sequences
Genomic DNA from blood samples was extracted using the MBST salting-out kit (CinnaGen, Tehran, Iran). The oligonucleotide forward and reverse primers used for the amplification of CYP2C19*2, *3, and *17 alleles were from previous published studies as follows: CYP2C19*2, forward 5′-AATTACAACCAGAGCTTGGC-3′ and reverse 5′-TATCACTTTCCATAAAAGCAAG-3′ (11); CYP2C19*3, forward 5′-AACATCAGGATTGTAAGCAC-3′ and reverse 5′-TCAGGGCTTGGTCAATATAG-3′ (11); and CYP2C19*17, forward 5′-GCCCTTAGCACCAAATTCTC-3′ and reverse 5′-ATTTAACCCCCTAAAAAAACACG-3′ primers (10).
Restriction fragment length polymorphism (RFLP)
Genotyping analysis of the CYP2C19*2, *3 and *17 alleles was performed using polymerase chain reaction-RFLP (PCR-RFLP). The PCR amplification was performed using 60 ng genomic DNA, 0.3 U Taq DNA polymerase (CinnaGen), 5 pmol each primers, 10X PCR buffer, 1.5 mM MgCl2 and 0.5 mM dNTP. The reaction mixture was initially denatured at 95°C for 3 min, followed by 35 cycles of 95°C for 1 min, 54°C for 1 min, and 72°C for 2 min for CYP2C19*2; 35 cycles of 94°C for 50 sec, 54.2°C for 50 sec, and 72°C for 50 sec for CYP2C19*3; 35 cycles of 94°C for 1 min, 56.3°C for 1 min, and 72°C for 1 min for CYP2C19*17; and all had and a final extension at 72°C for 10 min.
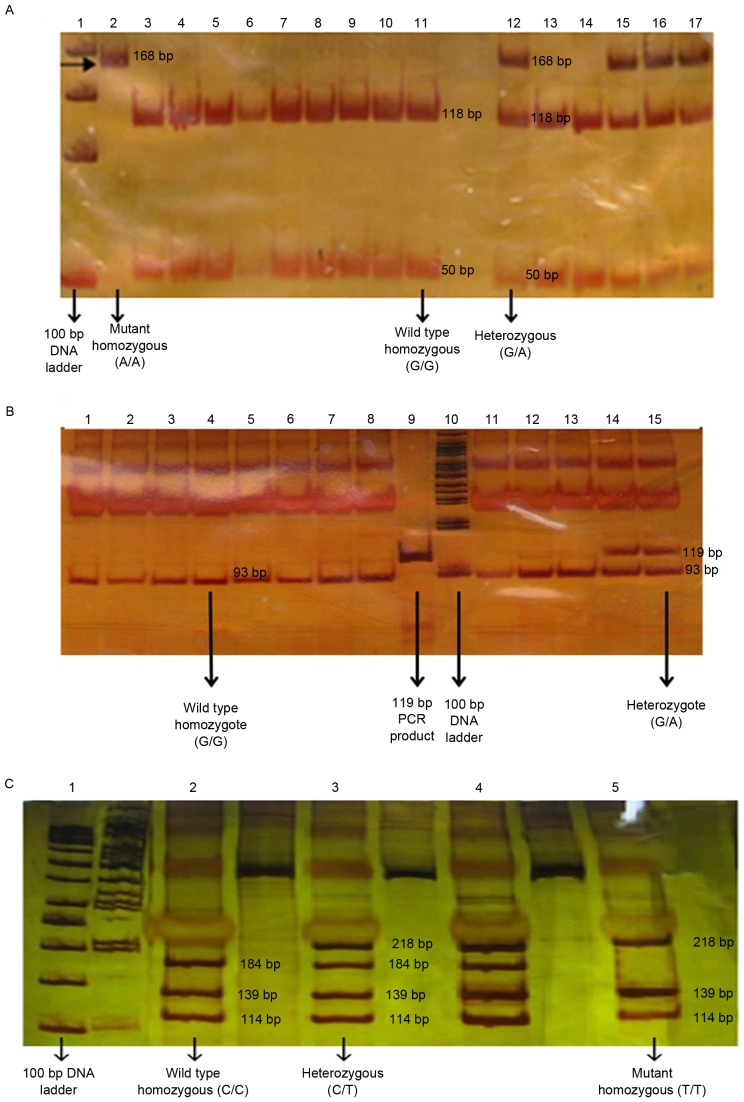
The 168-bp, 119-bp and 473-bp amplified fragments for *2, *3 and *17 alleles, respectively, were run on a 1.5% agarose gel and then stained using silver nitrate (CinnaGen) prior to restriction digestion. Restriction endonucleases, including SmaI, BamHI or LweI, were then added to PCR products in a 31 µl reaction volume consisting of 10 µl PCR products, 2 µl 10X SmaI or BamHI or LweI buffer (Thermo Fisher Scientific, Inc.), 1 µl SmaI or BamHI or LweI enzyme (Thermo Fisher Scientific, Inc.) and 18 µl double distilled water, and were then incubated at 37°C for 16 hr. The digested PCR products were separated by 8% polyacrylamide gel electrophoresis (PAGE) at 180 V for 110 min. The DNA bands were then visualized using silver nitrate staining. The PCR products with length of 168-bp, 119-bp, and 473-bp were subsequently digested with restriction endonuclease SmaI, BamHI, and LweI, respectively.
Single-strand conformation polymorphism (SSCP)
The PCR-amplified DNA fragments were denatured prior to loading on a polyacrylamide gel. Briefly, 10 µl PCR mixture was mixed with 7 µl denaturation buffer (990 µl of 100% formamide, 10 µl of 1 M sodium hydroxide, and a few granules of bromophenol blue). Samples were then incubated at 45°C for 30 min, and 12 µl SSCP color (CinnaGen) was added to sample after 23 min. The samples were analyzed using 6% PAGE at 80 V for 16 h for *2 and *3, and 150 V for 16 h for *17 and then stained by silver nitrate. The homozygous (as mutated or wild type) and heterozygous variants of the CYP2C19*2, *3, and *17 were identified as several bands.
Sequencing analysis
The PCR products were sequenced with forward and reversed primers on an automated ABI 3100 sequencing machine (Applied Biosystems, Kavosh Fanavaran Kawsar Company, Iran). All DNA fragments were then sequenced and analyzed using the Finch TV program (Geospiza, Inc., Seattle, WA, USA) in order to confirm any nucleotide variations (Fig. 1).
Figure 1.
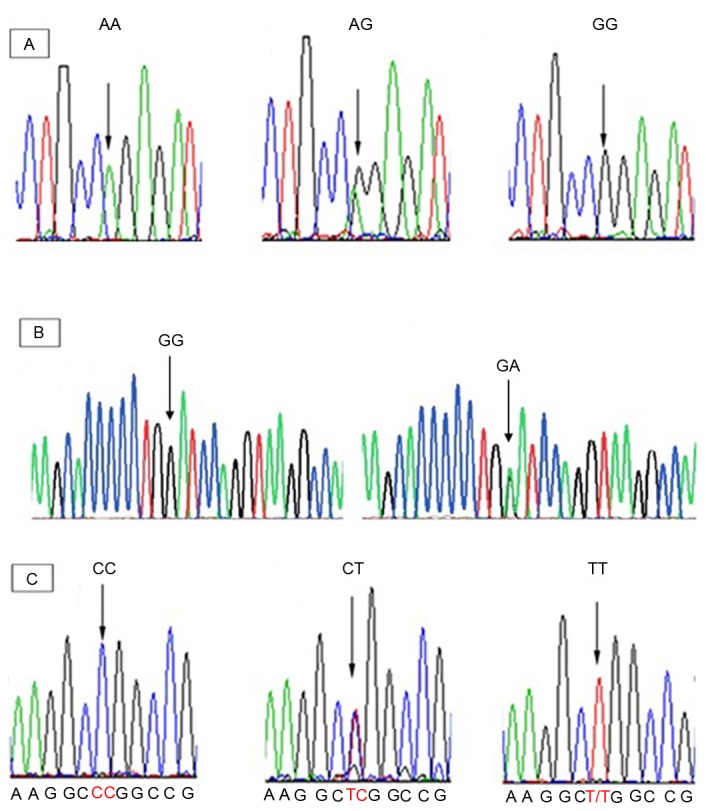
DNA sequencing chromatograms of CYP2C19*2, *3 and *17. (A) Homozygous mutant CYP2C19*2 (A/A), heterozygous mutant CYP2C19*2 (G/A) and wild-type CYP2C19 (G/G). (B) Wild-type CYP2C19 (G/G), and heterozygous mutant CYP2C19*3 (G/A). (C) Wild-type CYP2C19 (C/C), heterozygous mutant CYP2C19*17 (C/T) and homozygous mutant CYP2C19*17 (T/T). CYP2C19, cytochrome P450 2C19.
Statistical analysis
The statistical analyses were conducted using SPSS (version 22; IBM Corp., Armonk, NY, USA) software to perform χ2 analysis of the association between the frequency of alleles among the different ethnicities within the Iranian population and confidence interval test (95%) was used to calculate the frequency of alleles. P<0.05 was considered to indicate a statistically significant difference.
Results
Allelic and genotypic frequency distributions of CYP2C19*2, *3 and *17 were analyzed using blood samples of 1,229 unrelated healthy individuals from the Iranian population with different ethnicities. The homozygous (mutated and wild type) and heterozygous variants of the CYP2C19*2, *3 and *17 were analyzed using the PCR-RFLP and PCR-SSCP (Figs. 2 and 3).
Figure 2.
PCR-RFLP analysis. (A) The PCR products were disgusted with SmaI, producing 50-bp and 118-bp fragments for wild type allele, and one fragment of 168-bp for the allele carrying the CYP2C19*2681 G>A substitution. (B) The PCR product is disgusted with BamHI, producing 26 and 93-bp fragments for the wild type allele, and one fragment of 119-bp for the allele carrying the CYP2C19*3636 G>A substitution. (C) The PCR product was disgusted with LweI, producing 114, 184, 34 and 139-bp fragments for the wild type allele, and three fragments in size 218, 139 and 114-bp for the allele carrying the CYP2C19*17-806 C>T substitution. DNA fragments with size of >50 bp were not visible. PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; CYP2C19, cytochrome P450 2C19.
Figure 3.
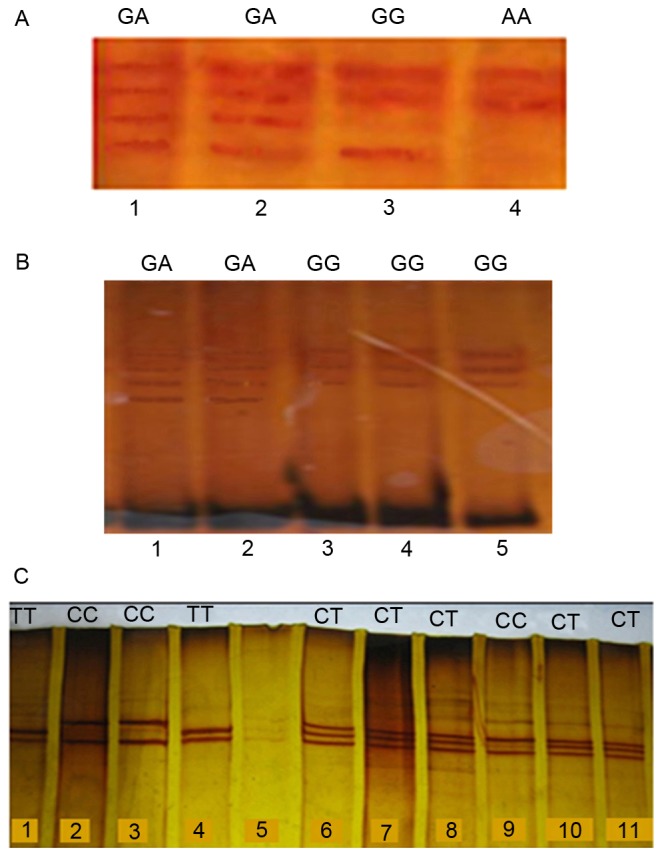
Polymerase chain reaction-single-strand conformation polymorphism analysis of the (A) CYP2C19*2 (B) CYP2C19*3 and (C) CYP2C19*17). Two, three and four bands in each lane represent the homozygous mutant, homozygous wild-type, and heterozygous alleles of the CYP2C19*2 and *3, and *17. CYP2C19, cytochrome P450 2C19.
RFLP and SSCP for CYP2C19*2
The frequency of the extensive metabolizer (EM; G/G), intermediate metabolizer (IM; G/A) and PM (A/A) of CYP2C19*2 were 63.6, 30.1 and 6.3%, respectively (Table I), in which the frequency of the homozygous A/A variants of the CYP2C19*2 allele was significantly highest in Lure individuals (P<0.001), while Caspian individuals had the lowest frequency (P<0.001) in comparison with rest of Iranian ethnicities. The frequency of the heterozygous G/A variant of CYP2C19*2 was significantly highest in Lure individuals (P<0.001) and Caspians had the lowest frequency (P<0.001) in comparison with rest of Iranian ethnicities. Furthermore, the frequency of the wild-type G/G variant of CYP2C19*2 was significantly high in Caspians (P<0.001) and low in Lures (P<0.001) in comparison with other ethnicities.
Table I.
Statistical analysis of the CYP2C19 (*2, *3 and *17) allelic and genotypic frequencies (%) among different ethnic groups in Iran.
| A, CYP2C19*2 | ||||||
|---|---|---|---|---|---|---|
| Genotype frequency (%) | ||||||
| Ethnicity | CYP2C19*2 allele frequency (%) | G/G | G/A | A/A | χ2 | P-value |
| Fars | 15.3 | 72.8 (66.1–79.4) | 23.9 (17.2–30.6) | 3.3 (1.1–6.1) | 137.4 | <0.001a |
| Turk | 25.0 | 58.2 (49.1–67.3) | 33.6 (24.5–42.7) | 8.2 (3.6–13.6) | 41.2 | <0.001a |
| Caspian | 9.6 | 83.6 (74–91.8) | 13.7 (5.5–21.9) | 2.7 (0.0 −6.8) | 84.1 | <0.001a |
| Lure | 35.0 | 41.3 (31.3–52.5) | 47.5 (37.5–58.8) | 11.3 (5.0 −18.8) | 18.0 | <0.001a |
| Kurd | 26.3 | 55.8 (46.3–66.3) | 35.8 (25.3–45.3) | 8.4 (3.2–14.7) | 32.2 | <0.001a |
| Total population | 21.4 | 63.6 (59.9–67.7) | 30.1 (26–33.8) | 6.3 (4.5–8.6) | 269.0 | <0.001a |
| B, CYP2C19*3 | ||||||
| Genotype frequency (%) | ||||||
| Ethnicity | CYP2C19*3 allele frequency (%) | G/G | G/A | A/A | χ2 | P-value |
| Fars | 2.9 | 94.2 (89.2–98.3) | 5.8 (1.7–10.8) | 0.0 | 93.6 | <0.001a |
| Turk | 1.8 | 96.3 (91.5–100.0) | 3.6 (0.0–8.5) | 0.0 | 70.4 | <0.001a |
| Caspian | 1.3 | 97.3 (93.3–100.0) | 2.6 (0.0–6.7) | 0.0 | 67.2 | <0.001a |
| Lure | 0.6 | 98.6 (95.9–100.0) | 1.3 (0.0–4.1) | 0.0 | 69.0 | <0.001a |
| Kurd | 1.4 | 97.1 (92.9–100.0) | 2.8 (0.0–7.1) | 0.0 | 62.2 | <0.001a |
| Total population | 1.7 | 96.4 (94.5–98.1) | 3.5 (1.9–5.5) | 0.0 | 362.1 | <0.001a |
| C, CYP2C19*17 | ||||||
| Genotype frequency | ||||||
| Ethnicity | CYP2C19*17 allele frequency (%) | C/C | C/T | T/T | χ2 | P-value |
| Fars | 28.2 | 51.9 (44.2–60.3) | 39.7 (32.1–47.4) | 8.3 (3.8–12.8) | 47.3 | <0.001a |
| Turk | 26.9 | 53.8 (42.9–67.9) | 38.4 (25–50) | 7.6 (1.8–14.3) | 19.9 | <0.001a |
| Caspian | 17.1 | 75.0 (59.4–87.5) | 15.6 (3.1–28.1) | 9.3 (0.0–21.9) | 25.1 | <0.001a |
| Lure | 46.1 | 15.3 (0.0–38.5) | 76.9 (53.8–100.0) | 7.6 (0.0–23.1) | 11.2 | 0.004a |
| Kurd | 21.4 | 64.2 (35.7–85.7) | 28.5 (7.1–50) | 7.1 (0.0–21.4) | 7.0 | 0.030a |
| Total population | 27.1 | 54.2 (48.3- 60.1) | 37.6 (31.7–43.5) | 8.1 (5.2–11.4) | 88.7 | <0.001a |
Different genotype frequencies of each allele were statistically analyzed for each ethnicity using χ2 test, and the genotype frequency of each ethnicity was then compared with the total population (last row).
Statistically significant. Values shown in brackets represent 95% confidence interval. CYP2C19, cytochrome P450 2C19.
RFLP and SSCP for CYP2C19*3
The prevalence of the A/A homozygous variant (PM) of the CYP2C19*3 was undetectable among different ethnicities, indicating the absence of this genotype within the Iranian population, while homozygous G/G (EM) and heterozygous G/A (IM) frequencies were 96.4 and 3.57%, respectively (Table I). The frequency of the G/A variant (IM) of the CYP2C19*3 allele was significantly high among the Fars population (χ2=93.633; P<0.001), while Lures had the low frequency (P<0.001) in comparison with other ethnicities. However, the frequency of the heterozygous CYP2C19*3 G/A variant in Caspians was similar to that of Kurds. Furthermore, the frequency of the wild-type G/G variant (EM) of the CYP2C19*3 was significantly highest in Lures (χ2=69.055; P<0.001), and lowest in Fars (P<0.001) in comparison with other ethnicities.
RFLP and SSCP for CYP2C19*17
The frequency of the C/C (EM), C/T (IM) and T/T [ultra-rapid metabolizer (UM)] variants of CYP2C19*17 were 54.24, 37.64 and 8.12%, respectively, in the total population (Table I). The frequency of the homozygous T/T variant (UM) of the CYP2C19*17 allele was significantly highest among the Caspians (P<0.001), while Kurds had the lowest frequency (P<0.001) in comparison with rest of Iranian ethnicities; the Lures and Turks demonstrated a similar percentage, which was close to that of Kurds. Additionally, the frequency of the heterozygous C/T variant (IM) of the CYP2C19*17 allele among was the highest the Lures (P=0.004), but Caspians had the lowest frequency in comparison with other Iranian ethnicities (P<0.001; Table I; Fig. 4).
Figure 4.
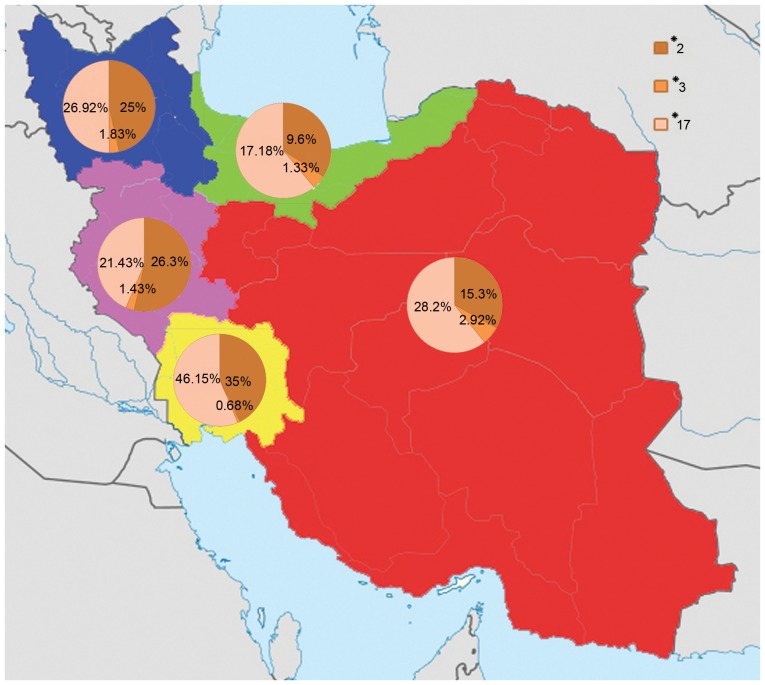
The CYP2C19*2, *3 and *17 alleles distribution among different Iranian ethnicities including Fars, Turk, Caspian, Lure and Kurd populations. The highest frequency of the CYP2C19*2 allele was found in Lure individuals (35%) and the lowest frequency was found in Caspian individuals (9.6%). The highest frequency of the CYP2C19*3 allele was found in Fars individuals (2.92%) and the lowest frequency was found in Lure individuals (0.68%). The highest frequency of the CYP2C19*17 allele was found in Lure individuals (46.15%) and the lowest frequency was found in Caspian individuals (17.18%). The red, blue, green, yellow and pink colors indicate the Fars, Turk, Caspian, Lure and Kurd regions. CYP2C19, cytochrome P450 2C19.
Discussion
The present study provides comprehensive data regarding distribution of the allelic and genotypic frequency of the CYP2C19 among Iranian population of different ethnicities. The variant alleles of the CYP2C19 family including CYP2C19*2, CYP2C19*3 and CYP2C19*17 are associated with variation in adverse drug reactions among populations according to race and ethnic backgrounds. Many of these polymorphic genes encode inactive enzymes that may cause adverse drug reactions among individuals because of their poor metabolic activities (12). CYP2C19 is the main factor for the metabolism of drugs, including omeprazole, lansoprazole, imipramine, propranolol, mephenytoin, chloroguanide, hexabarbitone, diazepam, proguanil and certain antidepressants (1,5). Polymorphisms in CYP2C19 may produce non-functional alleles and therefore no enzyme activity to metabolize these drugs correctly. The most important alleles, CYP2C19*2, CYP2C19*3 and CYP2C19*17, carry polymorphisms at 681 G>A in exon 5, 636 G>A in exon 4 and −806 C>T in the 5′ flanking region, respectively (3,13,14). CYP2C19*2 results in a splicing defect, CYP2C19*3 in a premature stop codon and CYP2C19*17 in increased gene transcription (13,14).
CYP2C19*2 is the most common allele among the Asian population and its prevalence is varies in different region of Asia. The CYP2C19*2 allele was observed with a frequency of 21.4% in the Iranian population, which is higher than the Swedish (14.4%) (15), German (15%) (16), Ethiopian (13.6%) (17) and Zimbabwean (13.1%) (18) populations, while not as high as reported in other populations, including Japanese (23%) (19) and Chinese-Taiwanese (32%) (19). The population with most frequent detection of the CYP2C19*2 allele in the previous reports that were examined was Filipino (39%; Table II). The prevalence of the CYP2C19*2 increases steeply from Western Asia and Iran to India, reaching its maximum (>75%) in Melanesian populations (20).
Table II.
Comparison of allele frequencies of CYP2C19*2 and CYP2C19*3 reported from different populations.
| Author, year | Population | NA | *2 frequency (%) | *3 frequency (%) | Method | (Refs.) |
|---|---|---|---|---|---|---|
| Dehbozorgi et al, 2017 | Iranian | 1916 | 21.4 | 1.7 | PCR-RFLP, PCR-SSCP and sequencing | Present study |
| Jurima-Romet et al, 1996 | Canadian (Inuit) | 304 | 11 | 0 | PCR-RFLP | (21) |
| Bathum et al, 1998 | Danish | 478 | 16.1 | 0 | Oligonucleotide ligation assay | (22) |
| Kurzawski et al, 2006 | Polish | 250 | 11.6 | ND | (29) | |
| Rudberg et al, 2008 | Norwegian | 664 | 18.1 | 0.6 | PCR-RFLP | (28) |
| Yamada et al, 1998 | Swedish | 166 | 14.4 | 0.7 | PCR-RFLP | (15) |
| Brockmöller et al, 1995 | German | 280 | 15 | 0 | PCR-RFLP | (16) |
| Ruas et al, 1997 | Portuguese | 306 | 13 | 0 | PCR-RFLP | (30) |
| Hoskins et al, 1998 | Australian | 198 | 14.6 | 0 | PCR-RFLP | (31) |
| Goldstein et al, 1997 | Saudi Arabia | 194 | 15 | 0 | PCR-RFLP | (19) |
| Goldstein et al, 1997 | Japanese | 106 | 23 | 10.4 | PCR-RFLP | (19) |
| Herrlin et al, 1998 | Korean | 206 | 20.9 | 11.6 | PCR-SSCP | (23) |
| Goldstein et al, 1997 | Chinese-Taiwanese | 236 | 32 | 5.5 | PCR-RFLP | (19) |
| Goldstein et al, 1997 | Filipino | 104 | 39 | 7.7 | PCR-RFLP | (19) |
| Lamba et al, 2000 | North Indian | 242 | 30 | 0 | PCR-RFLP | (32) |
| Hamdy et al, 2002 | Egyptians | 494 | 11 | 0.2 | AFNCRAS | (1) |
| Herrlin et al, 1998 | Bantu-Tanzanian | 502 | 17.9 | 0.6 | PCR-SSCP | (23) |
| Persson et al, 1996 | Ethiopian | 228 | 13.6 | 1.8 | PCR-RFLP | (17) |
| Dandara et al, 2001 | Venda | 304 | 21.7 | 0 | PCR-RFLP | (18) |
| Dandara et al, 2001 | Zimbabwean | 336 | 13.1 | 0 | PCR-RFLP | (18) |
| Goldstein et al, 1997 | African-Americans | 216 | 25 | 0 | PCR-RFLP | (19) |
CYP2C19, cytochrome P450 2C19; NA, number of alleles analyzed; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SSCP, single-strand conformation polymorphism; AFNCRAS, allele-specific fluorogenic 5′ nuclease chain reaction assay-sequencing; ND, not detected.
CYP2C19 is a clinically important enzyme in metabolism of different drugs, including antiplatelet drug, clopidogrel. Among individuals treated with clopidogrel, carriers of CYP2C19*2, CYP2C19 reduced-function alleles, exhibited markedly lower levels of the active metabolite of the drug, decreased platelet inhibition, and a higher rate of subsequent cardiovascular events, compared with non-carriers. However, the CYP2C19*17 allele is reported significantly related with an increased response to clopidogrel and high risk of bleeding. Thus, identification and functional analysis of CYP2C19 genetic polymorphisms are important for improving the understanding of safer drug therapy (2).
Furthermore, the CYP2C19*3 allele was observed with a prevalence of 1.7% among the Iranian population in the present study, which is higher than the previously reported frequency in the Canadian (0%) (21) and Danish (0%) (22) populations, but not as high as reported in other populations, including Japanese (10.4%) (19) and Korean (11.6%) (23), and was demonstrated to be of an approximately similar frequency to the Ethiopian population (1.8%; Table II) (17).
The frequency of CYP2C19*3 as an Asian mutation (1) increases from the west to the east of Asia (Table II; Fig. 5). According to previous studies, the highest frequency of the CYP2C19*3 allele was reported in the Indonesian populations of South-East Asia (37%) (24), followed by the Iruna population from New Guinea (34%; Fig. 5) (24). These results suggest that the prediction of the CYP2C19*3 allele is necessary in drug research and therapy, and the effects of the CYP2C19*3 allele on drug metabolism should be investigated, because side effects of drugs are comparatively common among these countries (25,26). In fact, individuals with CYP2C19*3 (A/A) genotype have no ability to metabolize drugs completely, consequently resulting in incomplete and poor metabolism of the drugs, drug accumulation in the blood and drug toxicity.
Figure 5.
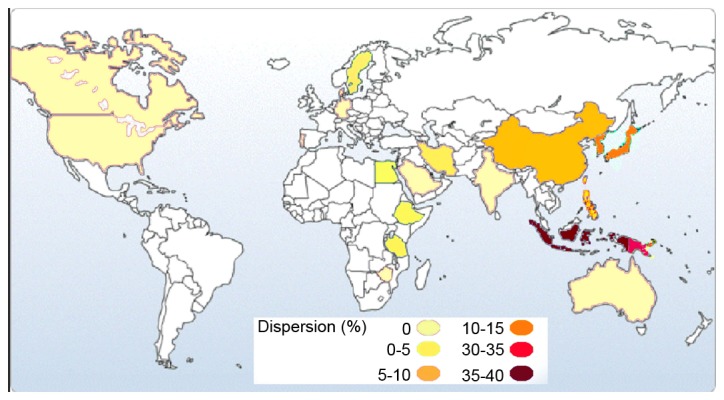
Frequency of the CYP2C19*3 allele in different countries around the world. The highest incidence of the CYP2C19*3 allele has been reported in Indonesians (37%). The second highest CYP2C19*3 frequency has been reported in the Iruna population from New Guinea (34%). It seems that the allelic frequency of CYP2C19*3 increases from the west to the east of Asia. CYP2C19, cytochrome P450 2C19.
In the current study, the CYP2C19*17 allele was identified with a prevalence of 27.1% among the Iranian population, which was higher than most countries included in Table III, including the Danish (20.1%) (27) and Norwegian (22%) (28) population, and was similar in frequency to the Polish population (27.2%) (29).
Table III.
Comparison of allele frequencies of CYP2C19*17 reported from different populations.
| Author, year | Population | NA | *17 frequency (%) | Method | (Refs.) |
|---|---|---|---|---|---|
| Dehbozorgi et al, 2017 | Iranian | 542 | 27.1 | PCR-RFLP, PCR-SSCP and sequencing | Present study |
| Pedersen et al, 2010 | Danish | 552 | 20.1 | Quantitative PCR | (27) |
| Kurzawski et al, 2006 | Polish | 250 | 27.2 | PCR-RFLP | (29) |
| Rudberg et al, 2008 | Norwegian | 664 | 22 | PCR-RFLP | (28) |
| Ramsjö et al, 2010 | Swedish | 370 | 20 | Quantitative PCR | (33) |
| Sugimoto et al, 2008 | Japanese | 530 | 1.3 | PCR-RFLP | (14) |
| Kim et al, 2010 | Korean | 542 | 1.5 | Multiplex pyrosequencing | (34) |
| Chen et al, 2008 | Chinese-Taiwanese | 800 | 1.2 | Direct sequencing | (35) |
| Egyptians | 494 | ND | |||
| Sim et al, 2006 | Ethiopian | 380 | 17.9 | Sequencing | (10) |
| Kearns et al, 2010 | African-Americans | 228 | 21 | PCR-RFLP | (36) |
CYP2C19, cytochrome P450 2C19; NA, number of alleles; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SSCP, single-strand conformation polymorphism; ND, not detected.
In summary, the determination of the allelic and genotypic frequencies of the CYP2C19 gene among different ethnicities may provide data to be used to personalize treatments in order to improve the efficiency of the therapeutic outcomes and decrease the appearance of adverse effects, and therefore, facilitate the advancement of personalized medicine. Genotyping of the CYP2C19*2, *3 and *17 alleles among different ethnicities within the Iranian population, including Fars, Turks, Caspians, Lures and Kurds is an important for avoiding the side effect of drugs and drug interactions, and may lead to improved survival and decreased drug modality risk.
Acknowledgements
This work was supported by Dr M. Houshmand (National Institute of Genetic Engineering and Biotechnology, Tehran, Iran), to whom we give our grateful appreciation.
References
- 1.Hamdy SI, Hiratsuka M, Narahara K, El-Enany M, Moursi N, Ahmed MS, Mizugaki M. Allele and genotype frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. Br J Clin Pharmacol. 2002;53:596–603. doi: 10.1046/j.1365-2125.2002.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, An N, Wang H, Gao Y, Liu D, Bian T, Zhu J, Chen C. Evaluation of the effects of 20 nonsynonymous single nucleotide polymorphisms of CYP2C19 on S-mephenytoin 4′-hydroxylation and omeprazole 5′-hydroxylation. Drug Metab Dispos. 2011;39:830–837. doi: 10.1124/dmd.110.037549. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhry A, Kochhar R, Kohli K. Genetic polymorphism of CYP2C19 & therapeutic response to proton pump inhibitors. Indian J Med Res. 2008;127:521–530. [PubMed] [Google Scholar]
- 4.Yin T, Miyata T. Pharmacogenomics of clopidogrel: Evidence and perspectives. Thromb Res. 2011;128:307–316. doi: 10.1016/j.thromres.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Blaisdell J, Mohrenweiser H, Jackson J, Ferguson S, Coulter S, Chanas B, Xi T, Ghanayem B, Goldstein JA. Identification and functional characterization of new potentially defective alleles of human CYP2C19. Pharmacogenetics. 2002;12:703–711. doi: 10.1097/00008571-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Buzoianu AD, Trifa AP, Popp RA, Militaru MS, Militaru CF, Bocşan CI, Farcaş MF, Pop IV. Screening for CYP2C19*2, *3 and *4 gene variants in a Romanian population study group. Farmacia. 2010;58:806–818. [Google Scholar]
- 7.Beitelshees AL, Horenstein RB, Vesely MR, Mehra MR, Shuldiner AR. Pharmacogenetics and clopidogrel response in patients undergoing percutaneous coronary interventions. Clin Pharmacol Ther. 2011;89:455–459. doi: 10.1038/clpt.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA, Roden DM, Klein TE, Shuldiner AR, Clinical Pharmacogenetics Implementation Consortium Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90:328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gawrońska-Szklarz B, Adamiak-Giera U, Wyska E, Kurzawski M, Gornik W, Kaldonska M, Drozdzik M. CYP2C19 polymorphism affects single-dose pharmacokinetics of oral pantoprazole in healthy volunteers. Eur J Clin Pharmacol. 2012;68:1267–1274. doi: 10.1007/s00228-012-1252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–113. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Ikebuchi J, Yamada M, Ogura Y, Yamamoto Y, Nishimura A, Nishi K, Yamada K, Irizawa Y. Individual difference in drug metabolism and disposition: Toxicological significance of genotypes and phenotypes of S-mephenytoin 4′-hydroxylase (CYP2C19) Int Congress Series. 2003;1239:589–592. doi: 10.1016/S0531-5131(02)00389-8. [DOI] [Google Scholar]
- 12.Shastry BS. Pharmacogenetics and the concept of individualized medicine. Pharmacogenomics J. 2005;6:16–21. doi: 10.1038/sj.tpj.6500338. [DOI] [PubMed] [Google Scholar]
- 13.Nakamoto K, Kidd JR, Jenison RD, Klaassen CD, Wan YJ, Kidd KK, Zhong XB. Genotyping and haplotyping of CYP2C19 functional alleles on thin-film biosensor chips. Pharmacogenet Genomics. 2007;17:103–114. doi: 10.1097/FPC.0b013e32801152c2. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto K, Uno T, Yamazaki H, Tateishi T. Limited frequency of the CYP2C19*17 allele and its minor role in a Japanese population. Br J Clin Pharmacol. 2008;65:437–439. doi: 10.1111/j.1365-2125.2007.03057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada H, Dahl ML, Lannfelt L, Viitanen M, Winblad B, Sjöqvist F. CYP2D6 and CYP2C19 genotypes in an elderly Swedish population. Eur J Clin Pharmacol. 1998;54:479–481. doi: 10.1007/s002280050497. [DOI] [PubMed] [Google Scholar]
- 16.Brockmöller JJ, Rost K, Gross D, Schenkel A, Roots I. Phenotyping of CYP2C19 with enantiospecific HPLC-quantification of R- and S-mephenytoin and comparison with the intron4/exon5 G->A-splice site mutation. Pharmacogenetics. 1995;5:80–88. doi: 10.1097/00008571-199504000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Persson I, Aklillu E, Rodrigues F, Bertilsson L, Ingelman-Sundberg M. S-mephenytoin hydroxylation phenotype and CYP2C19 genotype among Ethiopians. Pharmacogenetics. 1996;6:521–526. doi: 10.1097/00008571-199612000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Dandara C, Masimirembwa CM, Magimba A, Sayi J, Kaaya S, Sommers DK, Snyman JR, Hasler JA. Genetic polymorphism of CYP2D6 and CYP2C19 in east-and southern African populations including psychiatric patients. Eur J Clin Pharmacol. 2001;57:11–17. doi: 10.1007/s002280100282. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein JA, Ishizaki T, Chiba K, de Morais SM, Bell D, Krahn PM, Evans DA. Frequencies of the defective CYP2C19 alleles responsible for the mephenytoin poor metabolizer phenotype in various Oriental, Caucasian, Saudi Arabian and American black populations. Pharmacogenetics. 1997;7:59–64. doi: 10.1097/00008571-199702000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics. 2009;19:170–179. doi: 10.1097/FPC.0b013e32831ebb30. [DOI] [PubMed] [Google Scholar]
- 21.Jurima-Romet M, Goldstein JA, LeBelle M, Aubin RA, Foster BC, Walop W, Rode A. CYP2C19 genotyping and associated mephenytoin hydroxylation polymorphism in a Canadian Inuit population. Pharmacogenetics. 1996;6:329–339. doi: 10.1097/00008571-199608000-00006. [DOI] [PubMed] [Google Scholar]
- 22.Bathum L, Andersen-Ranberg K, Boldsen J, Brøsen K, Jeune B. Genotypes for the cytochrome P450 enzymes CYP2D6 and CYP2C19 in human longevitY. Role of CYP2D6 and CYP2C19 in longevity. Eur J Clin Pharmacol. 1998;54:427–430. doi: 10.1007/s002280050487. [DOI] [PubMed] [Google Scholar]
- 23.Herrlin K, Massele AY, Jande M, Alm C, Tybring G, Abdi YA, Wennerholm A, Johansson I, Dahl ML, Bertilsson L, Gustafsson LL. Bantu Tanzanians have a decreased capacity to metabolize omeprazole and mephenytoin in relation to their CYP2C19 genotype. Clin Pharmacol Ther. 1998;64:391–401. doi: 10.1016/S0009-9236(98)90070-4. [DOI] [PubMed] [Google Scholar]
- 24.Hsu HL, Woad KJ, Woodfield DG, Helsby NA. A high incidence of polymorphic CYP2C19 variants in archival blood samples from Papua New Guinea. Hum Genomics. 2008;3:17–23. doi: 10.1186/1479-7364-3-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blendon RJ, Schoen C, DesRoches C, Osborn R, Zapert K. Common concerns amid diverse systems: Health care experiences in five countries. Health Aff (Millwood) 2003;22:106–121. doi: 10.1377/hlthaff.22.3.106. [DOI] [PubMed] [Google Scholar]
- 26.Lindley CM, Tully MP, Paramsothy V, Tallis RC. Inappropriate medication is a major cause of adverse drug reactions in elderly patients. Age and Ageing. 1992;21:294–300. doi: 10.1093/ageing/21.4.294. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen RS, Brasch-Andersen C, Sim SC, Bergmann TK, Halling J, Petersen MS, Weihe P, Edvardsen H, Kristensen VN, Brøsen K, Ingelman-Sundberg M. Linkage disequilibrium between the CYP2C19*17 allele and wildtype CYP2C8 and CYP2C9 alleles: Identification of CYP2C haplotypes in healthy Nordic populations. Eur J Clin Pharmacol. 2010;66:1199–1205. doi: 10.1007/s00228-010-0864-8. [DOI] [PubMed] [Google Scholar]
- 28.Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther. 2008;83:322–327. doi: 10.1038/sj.clpt.6100291. [DOI] [PubMed] [Google Scholar]
- 29.Kurzawski M, Gawrońska-Szklarz B, Wrześniewska J, Siuda A, Starzyńska T, Droździk M. Effect of CYP2C19*17 gene variant on Helicobacter pylori eradication in peptic ulcer patients. Eur J Clin Pharmacol. 2006;62:877–880. doi: 10.1007/s00228-006-0183-2. [DOI] [PubMed] [Google Scholar]
- 30.Ruas JL, Lechner MC. Allele frequency of CYP2C19 in a Portuguese population. Pharmacogenetics. 1997;7:333–335. doi: 10.1097/00008571-199708000-00009. [DOI] [PubMed] [Google Scholar]
- 31.Hoskins JM, Shenfield GM, Gross AS. Relationship between proguanil metabolic ratio and CYP2C19 genotype in a Caucasian population. Br J Clin Pharmacol. 1998;46:499–504. doi: 10.1046/j.1365-2125.1998.00807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamba JK, Dhiman RK, Kohli KK. CYP2C19 genetic mutations in North Indians. Clin Pharmacol Ther. 2000;68:328–335. doi: 10.1067/mcp.2000.109365. [DOI] [PubMed] [Google Scholar]
- 33.Ramsjö M, Aklillu E, Bohman L, Ingelman-Sundberg M, Roh HK, Bertilsson L. CYP2C19 activity comparison between Swedes and Koreans: Effect of genotype, sex, oral contraceptive use, and smoking. Eur J Clin Pharmacol. 2010;66:871–877. doi: 10.1007/s00228-010-0835-0. [DOI] [PubMed] [Google Scholar]
- 34.Kim KA, Song WK, Kim KR, Park JY. Assessment of CYP2C19 genetic polymorphisms in a Korean population using a simultaneous multiplex pyrosequencing method to simultaneously detect the CYP2C19*2, CYP2C19*3, and CYP2C19*17 alleles. J Clin Pharm Ther. 2010;35:697–703. doi: 10.1111/j.1365-2710.2009.01069.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Qin S, Xie J, Tang J, Yang L, Shen W, Zhao X, Du J, He G, Feng G. Genetic polymorphism analysis of CYP2C19 in Chinese Han populations from different geographic areas of mainland China. Pharmacogenomics. 2008;9:691–702. doi: 10.2217/14622416.9.6.691. [DOI] [PubMed] [Google Scholar]
- 36.Kearns GL, Leeder JS, Gaedigk A. Impact of the CYP2C19*17 allele on the pharmacokinetics of omeprazole and pantoprazole in children: Evidence for a differential effect. Drug Metab Dispos. 2010;38:894–897. doi: 10.1124/dmd.109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]