Abstract
Sevoflurane is a commonly used inhalation anesthetic. Sevoflurane-induced neuroapoptosis and cognitive impairments in animals are widely reported, however, the underlying molecular mechanisms remain largely unknown. The results of the present study demonstrated that sevoflurane anesthesia induced spatial memory impairments in rats, as determined by the Morris water maze test. Mechanistically, the current study demonstrated that sevoflurane administration significantly enhanced the expression of microRNA (miR)-188-3p. Furthermore, inhibition of miR-188-3p using lentiviral miR-188-3p inhibitors attenuated sevoflurane-induced cognitive impairments in rats. The present study also demonstrated that miR-188-3p targeted MDM2 proto-oncogene (MDM2) and negatively regulated the expression of MDM2, as determined by luciferase assays, reverse transcription-quantitative polymerase chain reaction and western blot analysis. Furthermore, decreased abundance of MDM2 following transfection with miR-188-3p mimics was associated with increased stability of p53 protein. Suppression of p53 activity using the specific p53 inhibitor pifithrin-α alleviated sevoflurane-induced neuroapoptosis. These results indicate that the miR-188-3p-MDM2-p53 axis may have a critical role in sevoflurane-induced cognitive dysfunction. Therefore, miR-188-3p may be a potential target for the treatment of sevoflurane-induced cognitive impairment.
Keywords: sevoflurane, microRNA-188-3p, cognitive impairment, MDM2 proto-oncogene, p53
Introduction
Isoflurane and sevoflurane are commonly used anesthetics in clinical surgery (1). However, increasing evidence from epidemiological studies indicates that children <4 years that are exposed to anesthetics may be at an increased risk of developing cognitive disabilities and learning difficulties (2–4). It has been reported that sevoflurane treatment induces neuroapoptosis and degeneration in neonatal rats, which consequently leads to pathological alternations in the hippocampus (5,6). The safety of anesthetics in surgical procedures has received a large amount of attention, however, the underlying molecular mechanism by which sevoflurane induces neurodegeneration remains largely unknown.
Accumulating evidence has demonstrated that cognitive impairments induced by surgical anesthesia are associated with alterations in the expression of a variety of genes involved in brain development (6,7). In recent years, dysregulation of microRNAs (miRNAs/miRs) has been implicated in anesthesia-associated cognitive dysfunction (8–10). MiRNAs are small, noncoding RNAs that negatively regulate gene expression through inhibiting the translation of transcripts or inducing the degradation of target mRNA (11–15). It has been well-documented that aberrant expression of miRNAs is associated with the development of human cancers, and miRNAs therefore serve as potential biomarkers for cancer diagnosis and treatment (13,16–23). Additionally, an increasing number of miRNAs, including miR-665, miR-572 and miR-34a, have been demonstrated to be associated with anesthetic-induced hippocampal apoptosis (24–26). As miRNAs have important roles in the regulation of cell death and differentiation, the identification of novel miRNAs that participate in the neurodegeneration caused by anesthetics exposure, and understanding the functional molecular mechanisms of these miRNAs, is critical.
A recent study by Zhou et al (6) screened the differentially expressed miRNAs following sevoflurane exposure. Among these, the expression of miRNA-34c was significantly downregulated in the hippocampus of developing rat brains following sevoflurane treatment. miR-34c is reported to be a direct downstream target of p53 and to regulate the expression of Bcl-2 and Bcl-2-associated X in response to the sevoflurane administration (25). In addition to the downregulated miRNAs, miR-188-3p has been identified as one of the most significantly increased miRNAs following sevoflurane treatment; however, the functional molecular mechanism of miR-188-3p in sevoflurane anesthesia-induced remains largely unknown.
In the present study, the expression level of miR-188-3p in the hippocampus of rats following exposure to sevoflurane was detected. To determine the functional mechanism of miR-188-3p, the downstream targets of miR-188-3p were predicted by databases. The results of the present study demonstrated that the expression of miR-188-3p was significantly increased following sevoflurane exposure, and that miR-188-3p may be implicated in sevoflurane-induced apoptosis in the rat brain through negative regulation of the MDM2 proto-oncogene (MDM2)-p53 pathway.
Materials and methods
Cell lines and animals
SH-SY5Y neuroblastoma cells, which are subcloned from the SK-N-SH neuroblastoma cell line, were obtained from Leibniz Institut DSMV-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). The cells were cultured in a mixture of Dulbecco's modified Eagle's medium (DMEM) and F12 (1:1) supplemented with 10% fetal bovine serum (all from Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were cultured at 37°C in a humidified atmosphere of 5% CO2. Animal experiments were performed with Sprague-Dawley rats (n=120; male; 7-weeks-old; weight, 240±10 g) that were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The animals were housed under a 12 h light/dark cycle with a room temperature (21±1°C) and humidity (60±5%). Rats were allowed ad libitum access to water and food. The experiments were approved by the Animal Care and Ethical Committee of Cangzhou Central Hospital (Cangzhou, China) and was performed in accordance with the Guide for the Care and Use of Laboratory Animals (27).
Sevoflurane exposure and animal transduction
The rats were randomly divided into two groups: A control group (n=8) receiving regular air inhalation for 6 h and a sevoflurane group (n=8) receiving 2.5% sevoflurane (Shanghai Hengrui Pharmaceutical Co., Ltd., Shanghai, China) at 600 µg/kg/min in 100% O2 at the identical conditions for 6 h in a plastic container. The levels of sevoflurane and oxygen were detected by the gas monitor. The lentivirus expressing miR-188-3p or miR-188-3p inhibitor was purchased from the Guangzhou RiboBio Co., Ltd. (Guangzhou, China). For transduction of animals with miR-188-3p mimics, miR-188-3p inhibitor or respective controls, 10 µl lentivirus-miR-188-3p mimics/inhibitor or control miRNA (20 µM) was given intracerebroventricularly using a Hamilton microsurgically gauged syringe into the left lateral cerebral ventricles with a pre-drilled skull micro-hole (10×10 µm). Following transfection for 120 h, the following experiments were performed.
Morris water maze task
The learning and memory ability of rats exposed to sevoflurane and regular air was assessed by the Morris water maze task, as previously described (28). Briefly, rats in each group were trained to swim for 5 days in a circular water pool (200 cm in diameter, 60 cm in depth). A hidden circular platform of ~10 cm in diameter and 2 cm below the water surface was submerged in the pool filled with warm water. Rats were forced to swim and find the platform within 60 sec. When rats failed to locate the hidden platform within the time limit, animals were gently guided. The time taken to locate the platform, the number of platform crossings and the swimming speed of each rat on their first attempt on day 5, which was recorded with the ANY-maze Video Tracking System version 5.1 (Stoelting Co., Wood Dale, IL, USA), was used to evaluate the learning ability of the rats.
Cell transfection
miR-188-3p mimics (5′-CUCCCACAUGCAGGGUUUGCA) and negative control (NC) miRNA (5′-UUUGUACUACACAAAAGUACUG) (Guangzhou RiboBio Co., Ltd.) was dissolved with RNase-free water. SH-SY5Y cells were seeded in 6-well plates at a density of 1×105/well overnight at 37°C. miRNAs were transfected using the NanoFectin Transfection Reagent (Shanghai ExCell Biology, Inc., Shanghai, China), with a final concentration of 100 nM. The mixture of transfection reagent and miRNAs was incubated at room temperature for 25 min prior to addition to the cell culture. Following transfection for 96 h, the cells were harvested and the expression level of miR-188-3p was detected by RT-qPCR.
miR-188-3p target prediction
TargetScan (www.targetscan.org, release 7.1) and miRNA.org (http://www.microrna.org, August 2010 release) were used for scanning the candidate targets of miR-188-3p. Basic information of miR-188-3p was submitted online and the potential targets of miR-188-3p were presented.
Luciferase assay
The wild-type (WT) and mutant 3′-untranslated region (3′-UTR) of MDM2, containing the predicted binding site of miR-188-3p, were constructed into the pMIR-REPORT firefly luciferase vector (Ambion; Thermo Fisher Scientific, Inc.). For the luciferase experiments, cells were seeded into the 96-well plate at the density of 1,000 cells/well. Cells were then co-transfected with 100 nM miR-188-3p or NC miRNA in the presence of 50 µg luciferase vector containing either WT or mutant forms of the 3′-UTR of MDM2 using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). Following transfection for 36 h, the luciferase activity was determined and normalized with the activity of Renilla luciferase using a Luciferase Reporter assay kit (BioVision, Inc., Milpitas, CA, USA). The experiment was performed in triplicate.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
The rats were sacrificed 6 h after the last sevoflurane exposure by decapitation. The hippocampal tissues were dissected and stored in the liquid nitrogen. For the RT-qPCR assay, the tissues were ground into powder. Total RNA and miRNA extraction from the hippocampal tissues was performed with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and an miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany), respectively. The RNA quality and quantity were detected using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). RT was performed at 42°C for 15 min using a stem-loop RT primer (miRcute Plus miRNA First-Strand cDNA Synthesis kit, KR211-01; Tiangen Biotech Co., Ltd., Beijing, China) that hybridized to miR-188-3p. qPCR was performed using the SsoFast™ EvaGreen® Supermix kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and the iQ5 Real-Time PCR System (Bio-Rad Laboratories, Inc.). GAPDH and U6 were used as the internal control for mRNA and miRNA respectively and quantification used the 2−∆∆Cq method (29). The reaction conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min. The following primer sequences were used: miR-188-3p, 5′-ATTATTGGCTCCCACATGCAGGG-3′ (forward) and 5′-ATCCAGTGCAGGGTCCGAGG-3′ (reverse); MDM2, 5′-TACAGGGACGCCATCGAATC-3′ (forward) and 5′-TGAAGTGCATTTCCAATAGTCAGC-3′ (reverse); p53, 5′-TACTCCCCTGCCTCAACAAG-3′ (forward) and 5′-CGCTATCTGAGCAGCGCTCAT3′ (reverse); U6, 5′-GCTTCGGCAGCACATATACTAA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); and GAPDH, 5′-TGTGTCCGTCGTGGATCTGA-3′ (forward) and 5′-CCTGCTTCACCACCTTCTTGA-3′ (reverse). The experiment was repeated three times.
Terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling (TUNEL) assay
Hippocampal tissues from rats exposed to the sevoflurane treatment for 6 h were fixed with 4% paraformaldehyde at room temperature for 10 min. Tissues were dehydrated in a grade series of ethanol and embedded in paraffin. Sections (3 µm) were obtained by cutting the tissues with the microtome (JungRM 2035; Leica Microsystems GmbH, Wetzlar, Germany). The paraffin-embedded tissues were dewaxed by incubating the slides at 55°C for 30 min, and then washed in a series of ethanol (100, 95, 80, 75 and 50%) for 2 min, respectively. The slides were incubated with 20 µg/ml proteinase K for 30 min at room temperature and then rinsed with PBS for 10 min at room temperature. TUNEL reaction mixture (50 µl, TUNEL kit; Roche Diagnostics GmbH, Mannheim, Germany) was added to the tissues and incubated at room temperature for 2 h. Subsequently, tissues were washed twice with pre-cooled PBS followed by staining with 25 µg/ml propidium iodide (PI) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min at room temperature. Then 10 fields of view were observed from each sample with an Olympus FLUOVIEW FV10i confocal microscope (Olympus Corporation, Tokyo, Japan).
Western blot analysis
The hippocampal tissues of rats exposed to sevoflurane treatment for 6 h were homogenized in NP-40 lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing 1% phenylmethylsulfonyl fluoride. The samples were centrifuged at 13,000 × g for 15 min at 4°C and the supernatant was collected. The protein concentration was determined by a BCA protein assay (Bio-Rad Laboratories, Inc.). Protein samples (50 mg/lane) were separated by 15% SDS-PAGE. Subsequently, proteins were transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk at room temperature for 1 h and subsequently incubated with the indicated primary antibodies at room temperature for 2 h, followed by incubation with secondary antibodies at room temperature for 1 h. Protein bands were detected using the Pierce ECL Plus Western Blotting Substrate system (Pierce; Thermo Fisher Scientific, Inc.) and visualized with ImageMaster 2D Platinum 7.0 software (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). The antibodies used in the current study were as follows: Rabbit anti-GAPDH (1:2,000, cat. no. ab70699), rabbit anti-MDM2 (1:2,000, cat. no. ab137413), mouse anti-p53 (DO-1, 1:500, cat. no. ab1101) (all from Abcam, Cambridge, UK), goat anti-rabbit IgG (H+L)-HRP conjugated secondary antibody (cat. no. 170–6515, 1:5,000), and goat anti-mouse IgG (H+L)-HRP conjugated secondary antibody (cat. no. 170–6516, 1:5,000) (both from Bio-Rad Laboratories, Inc.).
Exposure of SH-SY5Y cells to sevoflurane and cell apoptosis
SH-SY5Y cells (2×105) were incubated with or without 10 µM pifithrin-α (04–0040; Yeasen Biotechnology, Co., Ltd., Shanghai, China) for 12 h at 37°C. Then the cells were treated with 5 µM sevoflurane for 30 min at 37°C and the cell apoptosis was determined using an Alexa Fluor 488 Annexin V/Dead Cell Apoptosis kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Briefly, cells were harvested and washed with pre-cooled PBS. Cells were centrifuged at 1,000 × g for 5 min at 4°C and resuspended in annexin-binding buffer. Subsequently, Annexin V-fluorescein isothiocyanate (5 µl) was added to ~1×105 cells and incubated at room temperature for 15 min, followed by the addition of 1 µl PI (100 µg/ml). Cell apoptosis wad detected with a BD FACSCalibur™ flow cytometer and analyzed with the CFlow Plus Software, version l.0.172.9 (BD Biosciences, Franklin Lakes, NJ, USA).
Statistical analysis
Data are presented as the mean ± standard deviation of three independent experiments. Statistical analysis was performed by Student's t-test or one-way analysis of variance followed by Tukey's post hoc comparison test using SPSS 19.0 software (IBM Corp., Armonk, NY, USA). The plots were generated using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Sevoflurane anesthesia induces spatial memory impairments in rats
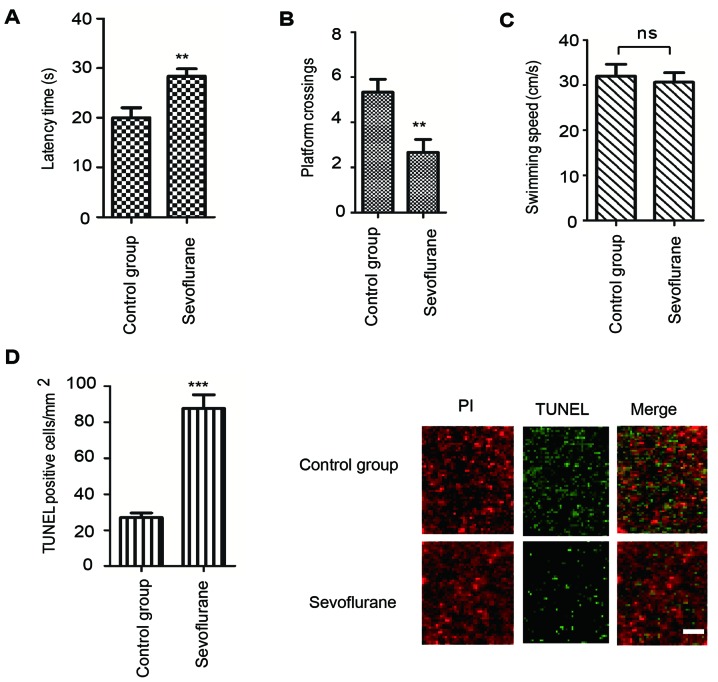
The Morris water maze test is considered to be a highly sensitive method of determining cognitive function to examine hippocampus-dependent learning and memory (30). To determine the effect of sevoflurane exposure on the memory ability of rats, the animals in each group were trained to locate the hidden platform. As demonstrated in Fig. 1A, the latency time taken to locate the hidden platform was significantly higher for rats treated with sevoflurane compared with the control group. Consistently, a decreased number of platform crossings in the sevoflurane group was observed in comparison with the control group (Fig. 1B). However, no significant differences in swimming speed were observed between the two groups of rats (Fig. 1C). These results demonstrated that sevoflurane exposure led to cognitive impairment in rats.
Figure 1.
Sevoflurane anesthesia induced spatial memory impairment in rats. (A) Latency time to locate the hidden platform following sevoflurane anesthesia. (B) Number of platform crossings in the probe trial tests. (C) Swimming speed of rats when locating the hidden platform. (D) Neuroapoptosis rate of the hippocampal tissues with or without sevoflurane treatment was determined by TUNEL analysis. Scale bar, 30 µm. Data are presented as the mean ± standard deviation, n=8. ***P<0.001 vs. control group. TUNEL, terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling; ns, not significant.
To determine whether sevoflurane-induced cognitive dysfunction in rats was associated with neuroapoptosis, apoptosis was investigated by TUNEL staining of the hippocampal tissues following sevoflurane exposure. The results demonstrated that, compared with the control group, sevoflurane administration for 6 h significantly increased the proportion of TUNEL-positive cells in the hippocampus of rats (Fig. 1D). These results indicated that sevoflurane exposure induced neuroapoptosis and subsequent cognitive impairment in rats.
Sevoflurane anesthesia upregulates the expression of miR-188-3p
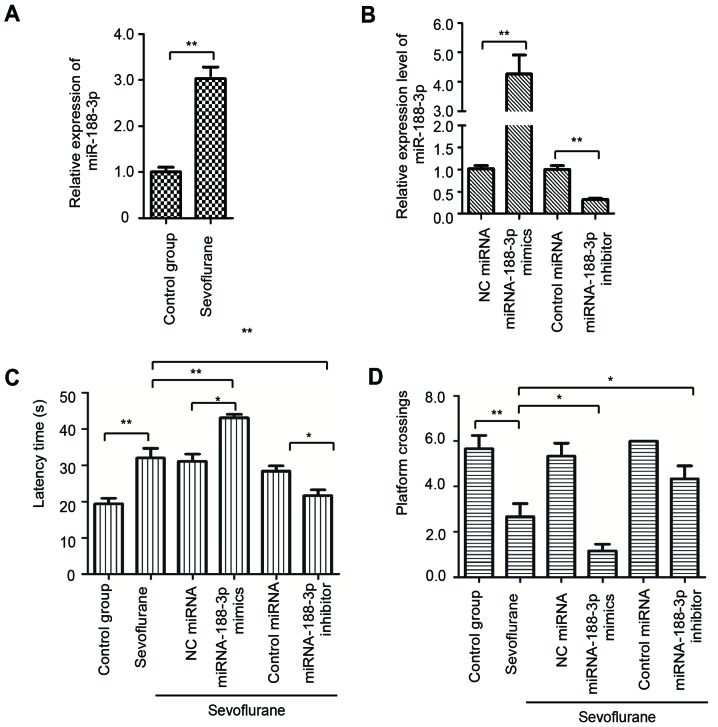
As the present study reported sevoflurane-induced cognitive dysfunction in rats, the potential underlying mechanisms of this effect were subsequently investigated. It has been well-documented that miRNAs have important roles in the regulation of gene expression in response to sevoflurane treatment (6,31–35), and bioinformatics analyses identified miR-188-3p as one of the most significantly upregulated miRNAs following sevoflurane administration (6). To confirm this observation, the expression level of miR-188-3p in the hippocampal tissue of rats with or without sevoflurane treatment was determined by RT-qPCR. As demonstrated in Fig. 2A, the expression of miR-188-3p was significantly increased in the hippocampus of rats following sevoflurane exposure, compared with the control group.
Figure 2.
Sevoflurane exposure upregulated the expression of miR-188-3p. (A) miR-188-3p expression level in the hippocampal tissues of rats with or without sevoflurane treatment was detected by RT-qPCR. (B) Successful transduction of rats with miR-188-3p mimics or miR-188-3p inhibitor was determined by RT-qPCR. Following transduction of rats with miR-188-3p mimics or miR-188-3p inhibitor and exposure to sevoflurane, the (C) latency time and (D) number of platform crossings when locating the hidden platform were recorded; n=8/group. *P<0.05 and **P<0.01 as indicated. miR/miRNA, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control.
To validate the association between upregulated miR-188-3p expression in the hippocampus and sevoflurane-induced cognitive impairments, rats were transfected with lentiviruses containing miR-188-3p mimics or inhibitor. The expression level of miR-188-3p following transduction with miR-188-3p mimics, miR-188-3p inhibitor and the respective controls was confirmed by RT-qPCR (Fig. 2B). The Morris water maze test was performed to determine the learning and memory ability of rats harboring overexpression or depletion of miR-188-3p following sevoflurane treatment. The data indicated that, following exposure to sevoflurane, rats with overexpression of miR-188-3p exhibited an increased latency time compared with the rats transduced with NC miRNA and the sevoflurane-only group (Fig. 2C). By contrast, the rats treated with miR-188-3p inhibitor exhibited a lower latency time compared with the control miRNA group and the sevoflurane-only group (Fig. 2C). Similarly, overexpression of miR-188-3p in rats treated with sevoflurane decreased the number of platform crossings, while depletion of miR-188-3p significantly increased the number of platform crossings, compared with the sevoflurane-only group (Fig. 2D). These results demonstrated the critical involvement of miR-188-3p in sevoflurane-induced cognitive dysfunction in rats.
MDM2 is a direct target of miR-188-3p
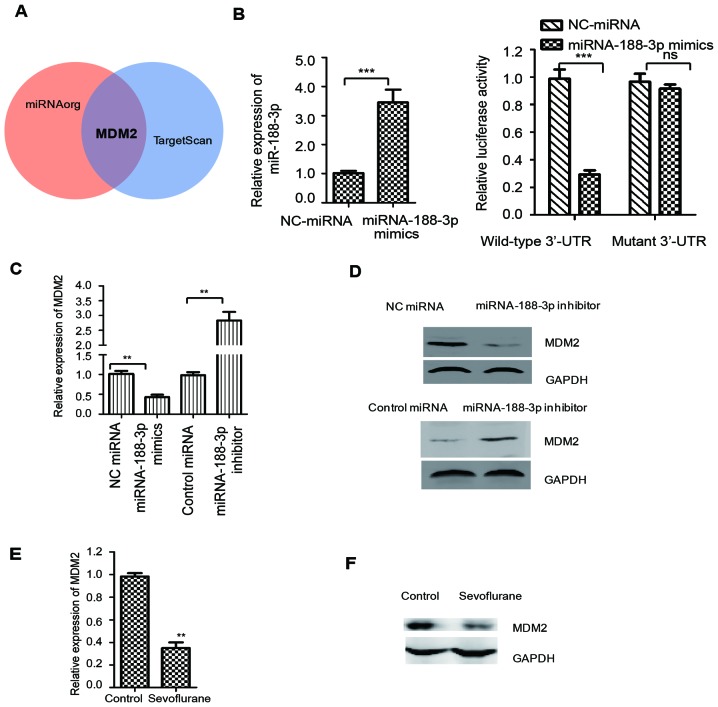
To further investigate the underlying molecular mechanism by which miR-188-3p may regulate sevoflurane-induced cognitive dysfunction, the downstream targets of miR-188-3p were predicted using TargetScan and miRNA databases. Among the putative candidates, MDM2 was predicted as one of the downstream targets of miR-188-3p (Fig. 3A). To confirm this result, a luciferase assay was performed to investigate the binding between miR-188-3p and the 3′-UTR of MDM2. As indicated in Fig. 3B, decreased luciferase activity was observed in SH-SY5Y neuroblastoma cells that were co-transfected with miR-188-3p mimics and the WT 3′-UTR of MDM2, compared with those co-transfected with NC miRNA and the WT 3′UTR (Fig. 3B). No significant difference was observed between cells co-transfected with the mutant 3′-UTR of MDM2 and miR-188-3p mimics or NC miRNA (Fig. 3B). These results confirmed that miR-188-3p specifically bound to the 3′-UTR of MDM2 mRNA.
Figure 3.
MDM2 is a downstream target of miR-188-3p. (A) Candidate targets of miR-188-3p were predicted using miRNA and TargetScan databases. MDM2 was selected as a putative target of miR-188-3p. (B) SH-SY5Y neuroblastoma cells were co-transfected with a luciferase vector containing a wild-type or mutant version of the MDM2 3′UTR and NC miRNA or miR-188-3p mimics, and the luciferase activity was measured. The expression level of miR-188-3p was detected by RT-qPCR as shown in left panel. (C) mRNA expression of MDM2 was detected in the hippocampal tissues of rats that were treated with miR-188-3p mimics or miR-188-3p inhibitor after sevoflurane administration, n=8/group. (D) Protein expression of MDM2 in the hippocampal tissues of rats transduced with miR-188-3p mimics, miR-188-3p inhibitor and the respective controls was determined by western blot analysis. GAPDH was used as the loading control, n=8/group. (E) mRNA and (F) protein levels of MDM2 in the hippocampal tissues of rats with or without sevoflurane treatment were detected by reverse transcription-quantitative polymerase chain reaction and western blot analysis, respectively, n=8/group. **P<0.01 and ***P<0.001 as indicated. MDM2, MDM2 proto-oncogene; miR/miRNA, microRNA; UTR, untranslated region; NC, negative control; ns, not significant.
To confirm this conclusion, the expression of MDM2 in the hippocampal tissues of rats treated with miR-188-3p mimics or inhibitor, and the respective controls, was determined by RT-qPCR and western blotting. As demonstrated in Fig. 3C, compared with the NC miRNA group, the mRNA abundance of MDM2 was significantly decreased in rats harboring miR-188-3p mimics. Consistent with this result, the mRNA level of MDM2 was increased in rats that were treated with miR-188-3p inhibitor, compared with the control miRNA group (Fig. 3C). Western blot analysis was performed to determine the protein levels of MDM2 in hippocampal tissues from rats treated with miR-188-3p mimics or inhibitor. The results demonstrated that overexpression of miR-188-3p suppressed the expression of MDM2, while downregulation of miR-188-3p increased the expression of MDM2, compared with their relative control groups (Fig. 3D). These results indicated that miR-188-3p may target MDM2 and negatively regulate its expression. To further confirm the involvement of MDM2 in sevoflurane-induced neuroapoptosis, the expression level of MDM2 was detected following treatment of rats with sevoflurane. As demonstrated in Fig. 3E and F, the mRNA and protein expression of MDM2 was markedly decreased following sevoflurane treatment, compared with the control group. These results indicate that sevoflurane treatment may negatively regulate the expression of MDM2.
miR-188-3p regulates the MDM2-p53 signaling pathway
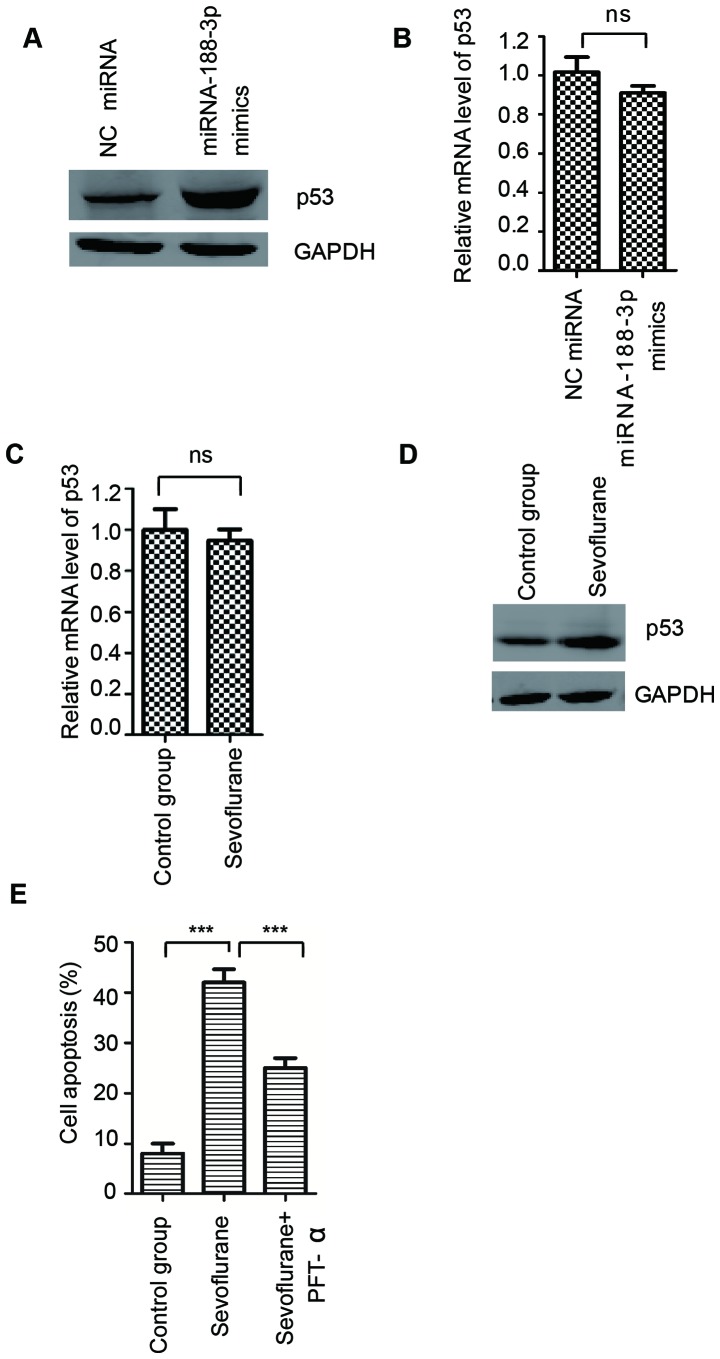
It has been well-documented that MDM2 is an E3 ubiquitin ligase that mediates the polyubiquitylation and degradation of p53 (36). To determine whether the negative regulation of MDM2 by miR-188-3p affects the expression of p53, the protein and mRNA expression of p53 was determined in hippocampal tissues from rats transduced with miR-188-3p mimics or NC miRNA. Western blotting results demonstrated that overexpression of miR-188-3p enhanced the protein expression of p53, compared with the NC miRNA group (Fig. 4A). However, the mRNA expression level of p53 was not significantly altered following overexpression of miR-188-3p (Fig. 4B). To investigate the effect of sevoflurane on the expression of p53, the protein and mRNA expression of p53 in the hippocampal tissues of rats with or without sevoflurane treatment was also determined. The results demonstrated that the protein expression, but not the mRNA expression, of p53 was increased following sevoflurane treatment, compared with the control group (Fig. 4C and D). These results indicated that the activation of p53 may be increased following sevoflurane exposure.
Figure 4.
miR-188-3p overexpression stabilized p53 and contributed to the neuroapoptosis induced by sevoflurane. (A) Protein and (B) mRNA levels of p53 were detected by western blot analysis and RT-qPCR, respectively, in the hippocampal tissues of rats that were treated with miR-188-3p mimics or NC miRNA, n=8/group. (C) mRNA and (D) protein levels of p53 in the hippocampal tissues of rats with or without sevoflurane treatment were detected by RT-qPCR and western blot analysis, respectively, n=8/group. (E) SH-SY5Y neuroblastoma cells were exposed to sevoflurane with or without the specific inhibitor of p53, PFT-α, and the cell apoptosis rate was determined by flow cytometry. ***P<0.001 as indicated. miR/miRNA, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control; PFT-α, pifithrin-α; ns, not significant.
Enhanced expression of p53 is associated with cell apoptosis. To determine whether the increased stability of p53 induced by miR-188-3p contributed to sevoflurane-induced neuroapoptosis, SH-SY5Y neuroblastoma cells exposed to sevoflurane were treated with pifithrin-α, the specific inhibitor of p53, and the rate of cell apoptosis rate was measured by flow cytometry. The results demonstrated that sevoflurane treatment induced cell apoptosis compared with the control group, while inhibition of p53 suppressed sevoflurane-induced cell apoptosis (Fig. 4E). These results indicated that sevoflurane enhanced the expression of miR-188-3p, which negatively regulated the MDM2-p53 pathway and the stabilization of p53 and may partially contribute to the cell apoptosis induced by sevoflurane.
Discussion
Anesthesia-induced cognitive impairment is considered to be the most common type of postoperative cognitive impairment. Previous studies have demonstrated that extended exposure to anesthetics resulted in excessive apoptosis of neurons and decreased the self-renewal capacity of neural stem cells, which consequently led to learning and memory deficits in animals (37–41). miRNA dysregulation has been previously associated with neurodegeneration induced by anesthesia (6,15,24). The present study demonstrated that miR-188-3p was upregulated in the hippocampus of rats following sevoflurane administration. Furthermore, the results indicated that overexpression of miR-188-3p may promote cognitive dysfunction by targeting MDM2 and increasing the stability of p53, as inhibition of the activity of p53 suppressed sevoflurane-induced neuroapoptosis.
Numerous miRNAs have been reported to be associated with anesthesia-induced cognitive dysfunction. Among these, miR-665 antagonized sevoflurane anesthesia-induced neurodegeneration in rats via the phosphatidylinositol 3-kinase/Akt signaling pathway by targeting insulin-like growth factor 2 (24). In addition, miR-34c was reported to be involved in sevoflurane-induced neural apoptosis through the mitochondrial pathway in the hippocampus of developing rat brains (6) and miR-572 was demonstrated to improve early postoperative cognitive dysfunction by downregulating the expression of neural cell adhesion molecule 1 (25). The present study investigated the function of miR-188-3p in sevoflurane-induced cognitive impairment, as this miRNA was reported to be one of the most significantly upregulated miRNAs following sevoflurane exposure in a previous study (6). The results of the present study demonstrated that increased expression of miR-188-3p was observed in the hippocampal tissues of rats that received sevoflurane treatment compared with control rats. It has been reported that the expression of miR-34c was decreased in response to sevoflurane treatment, and bioinformatics analyses indicated that miR-34c was a direct downstream target of p53 (6). The upstream regulators that mediate the overexpression of miR-188-3p in sevoflurane exposure require further investigation.
Using TargetScan and miRNA databases, MDM2 was predicted and confirmed as a downstream target of miR-188-3p. MDM2 is a well-established oncogene and an E3 ubiquitin-protein ligase that is responsible for the ubiquitylation of p53 (42,43). Increased expression of MDM2 was reported to promote the degradation of p53 and attenuate p53-dependent cell cycle arrest and cell apoptosis (43). Additionally, a p53-independent role of MDM2, via the Notch1 signaling pathway, was also reported to be associated with apoptosis inhibition and cell proliferation (44). In the present study, miR-188-3p overexpression reduced the expression of MDM2, which consequently upregulated the protein expression of p53. Furthermore, a specific inhibitor of p53 significantly reversed the cell apoptosis induced by sevoflurane. These results indicated that regulation of the MDM2-p53 axis by miR-188-3p may have a critical role in sevoflurane-induced neurodegeneration.
In conclusion, the results of the present study demonstrated that sevoflurane treatment enhances the expression of miR-188-3p, which may subsequently regulate the MDM2-p53 pathway and contribute to cell apoptosis following sevoflurane anesthesia. Therefore, small interfering RNA targeting miR-188-3p may have therapeutic potential and alleviate anesthetic-induced neuronal toxicity.
References
- 1.Sinner B, Becke K, Engelhard K. General anaesthetics and the developing brain: An overview. Anaesthesia. 2014;69:1009–1022. doi: 10.1111/anae.12637. [DOI] [PubMed] [Google Scholar]
- 2.DiMaggio C, Sun LS, Li G. Early childhood exposure to anesthesia and risk of developmental and behavioral disorders in a sibling birth cohort. Anesth Analg. 2011;113:1143–1151. doi: 10.1213/ANE.0b013e3182147f42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ing C, DiMaggio C, Whitehouse A, Hegarty MK, Brady J, von Ungern-Sternberg BS, Davidson A, Wood AJ, Li G, Sun LS. Long-term differences in language and cognitive function after childhood exposure to anesthesia. Pediatrics. 2012;130:e476–e485. doi: 10.1542/peds.2011-3822. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Li DY, Zhao SM, Zheng ZJ, Hu J, Li ZZ, Xiong SB. Rutin attenuates isoflurane-induced neuroapoptosis via modulating JNK and p38 MAPK pathways in the hippocampi of neonatal rats. Exp Ther Med. 2017;13:2056–2064. doi: 10.3892/etm.2017.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng X, Liu JJ, Zhou X, Song FH, Yang XY, Chen XS, Huang WQ, Zhou LH, Ye JH. Single sevoflurane exposure decreases neuronal nitric oxide synthase levels in the hippocampus of developing rats. Br J Anaesth. 2012;109:225–233. doi: 10.1093/bja/aes121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Xian D, Xia J, Tang Y, Li W, Chen X, Zhou Z, Lu D, Feng X. MicroRNA-34c is regulated by p53 and is involved in sevoflurane-induced apoptosis in the developing rat brain potentially via the mitochondrial pathway. Mol Med Rep. 2017;15:2204–2212. doi: 10.3892/mmr.2017.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Z, Lu XF, Shao C, Zhang C, Yang J, Ma T, Zhang LC, Cao JL. The effects of sevoflurane anesthesia on rat hippocampus: A genomic expression analysis. Brain Res. 2011;1381:124–133. doi: 10.1016/j.brainres.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Goto G, Hori Y, Ishikawa M, Tanaka S, Sakamoto A. Changes in the gene expression levels of microRNAs in the rat hippocampus by sevoflurane and propofol anesthesia. Mol Med Rep. 2014;9:1715–1722. doi: 10.3892/mmr.2014.2038. [DOI] [PubMed] [Google Scholar]
- 9.Luo T, Yin S, Shi R, Xu C, Wang Y, Cai J, Yue Y, Wu A. miRNA expression profile and involvement of Let-7d-APP in aged rats with isoflurane-induced learning and memory impairment. PLoS One. 2015;10:e0119336. doi: 10.1371/journal.pone.0119336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry. 2014;19:410–416. doi: 10.1038/mp.2013.196. [DOI] [PubMed] [Google Scholar]
- 11.Fu L, Jin L, Yan L, Shi J, Wang H, Zhou B, Wu X. Comprehensive review of genetic association studies and meta-analysis on miRNA polymorphisms and rheumatoid arthritis and systemic lupus erythematosus susceptibility. Hum Immunol. 2016;77:1–6. doi: 10.1016/j.humimm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Ma H, Wu Y, Yang H, Liu J, Dan H, Zeng X, Zhou Y, Jiang L, Chen Q. MicroRNAs in oral lichen planus and potential miRNA-mRNA pathogenesis with essential cytokines: A review. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122:164–173. doi: 10.1016/j.oooo.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Mizuguchi Y, Takizawa T, Yoshida H, Uchida E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol Res. 2016;46:391–406. doi: 10.1111/hepr.12606. [DOI] [PubMed] [Google Scholar]
- 14.Organista-Nava J, Gomez-Gomez Y, Illades-Aguiar B, Leyva-Vazquez MA. Regulation of the miRNA expression by TEL/AML1, BCR/ABL, MLL/AF4 and TCF3/PBX1 oncoproteins in acute lymphoblastic leukemia (Review) Oncol Rep. 2016;36:1226–1232. doi: 10.3892/or.2016.4948. [DOI] [PubMed] [Google Scholar]
- 15.Wen MM. Getting miRNA therapeutics into the target cells for neurodegenerative diseases: A mini-review. Front Mol Neurosci. 2016;9:129. doi: 10.3389/fnmol.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang QX, Zhu YQ, Zhang H, Xiao J. Altered MiRNA expression in gastric cancer: A systematic review and meta-analysis. Cell Physiol Biochem. 2015;35:933–944. doi: 10.1159/000369750. [DOI] [PubMed] [Google Scholar]
- 17.Ganju A, Khan S, Hafeez BB, Behrman SW, Yallapu MM, Chauhan SC, Jaggi M. miRNA nanotherapeutics for cancer. Drug Discov Today. 2017;22:424–432. doi: 10.1016/j.drudis.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Zhang YH, Deng Q, Li Y, Huang T, Zhou S, Cai YD. Cancer-Related triplets of mRNA-lncRNA-miRNA revealed by integrative network in uterine corpus endometrial carcinoma. Biomed Res Int. 2017;2017:3859582. doi: 10.1155/2017/3859582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nalluri JJ, Barh D, Azevedo V, Ghosh P. miRsig: A consensus-based network inference methodology to identify pan-cancer miRNA-miRNA interaction signatures. Sci Rep. 2017;7:39684. doi: 10.1038/srep39684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lodewijk L, Prins AM, Kist JW, Valk GD, Kranenburg O, Rinkes IH, Vriens MR. The value of miRNA in diagnosing thyroid cancer: A systematic review. Cancer Biomark. 2012;11:229–238. doi: 10.3233/CBM-2012-0273. [DOI] [PubMed] [Google Scholar]
- 21.Srivastava K, Srivastava A. Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PLoS One. 2012;7:e50966. doi: 10.1371/journal.pone.0050966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harquail J, Benzina S, Robichaud GA. MicroRNAs and breast cancer malignancy: An overview of miRNA-regulated cancer processes leading to metastasis. Cancer Biomark. 2012;11:269–280. doi: 10.3233/CBM-120291. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Lv S, Mi Y, Wang L, Wang G. Neuroprotective effect of miR-665 against sevoflurane anesthesia-induced cognitive dysfunction in rats through PI3K/Akt signaling pathway by targeting insulin-like growth factor 2. Am J Transl Res. 2017;9:1344–1356. [PMC free article] [PubMed] [Google Scholar]
- 24.Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert S, Jung K. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013;15:695–705. doi: 10.1016/j.jmoldx.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Lv S, Mi Y, Wang L, Wang G. Neuroprotective effect of miR-665 against sevoflurane anesthesia-induced cognitive dysfunction in rats through PI3K/Akt signaling pathway by targeting insulin-like growth factor 2. Am J Transl Res. 2017;9:1344–1356. [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X, Liu S, Li J, Fan X, Chen Y, Bi X, Liu S, Deng X. MicroRNA-572 improves early post-operative cognitive dysfunction by down-regulating neural cell adhesion molecule 1. PLoS One. 2015;10:e0118511. doi: 10.1371/journal.pone.0118511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang XL, Du BX, Chen J, Liu L, Shao WB, Song J. MicroRNA-34a negatively regulates anesthesia-induced hippocampal apoptosis and memory impairment through FGFR1. Int J Clin Exp Pathol. 2014;7:6760–6767. [PMC free article] [PubMed] [Google Scholar]
- 28.Committee for the Update of the Guide for the Care and Use of Laboratory Animals, Institute for Laboratory Animal Research, Division on Earth and Life Studies, National Research Council of the National Academies, corp-author. Guide for the Care and Use of Laboratory Animals. 8th. The National Academies Press; Washington, DC: 2011. [Google Scholar]
- 29.Vorhees CV, Williams MT. Morris water maze: Procedures for assessing spatial and related forms of learning and memory. Nat Protoc. 2006;1:848–858. doi: 10.1038/nprot.2006.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Hu X, Guan W, Luan L, Li B, Tang Q, Fan H. Isoflurane anesthesia promotes cognitive impairment by inducing expression of beta-amyloid protein-related factors in the hippocampus of aged rats. PLoS One. 2017;12:e0175654. doi: 10.1371/journal.pone.0175654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Liu X, Wang R, Luo H, Qin G, Wang LU, Ye Z, Guo Q, Wang E. Circulating microRNAs indicate cardioprotection by sevoflurane inhalation in patients undergoing off-pump coronary artery bypass surgery. Exp Ther Med. 2016;11:2270–2276. doi: 10.3892/etm.2016.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Li G, Li B, Chen Q, Lv D, Liu J, Ma J, Sun N, Yang L, Fei X, Song Q. Sevoflurane represses the self-renewal ability by regulating miR-7a, 7b/Klf4 signalling pathway in mouse embryonic stem cells. Cell Prolif. 2016;49:609–617. doi: 10.1111/cpr.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, Zhang Z, Wang Y, Chen C, Xu X, Yu H, Peng M. Altered hippocampal microRNA expression profiles in neonatal rats caused by sevoflurane anesthesia: MicroRNA profiling and bioinformatics target analysis. Exp Ther Med. 2016;12:1299–1310. doi: 10.3892/etm.2016.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi W, Li D, Guo Y, Zhang Y, Huang B, Li X. Sevoflurane inhibits the migration and invasion of glioma cells by upregulating microRNA-637. Int J Mol Med. 2016;38:1857–1863. doi: 10.3892/ijmm.2016.2797. [DOI] [PubMed] [Google Scholar]
- 36.Jiang J, Chen Z, Yang Y, Yan J, Jiang H. Sevoflurane downregulates IGF1 via microRNA98. Mol Med Rep. 2017;15:1863–1868. doi: 10.3892/mmr.2017.6219. [DOI] [PubMed] [Google Scholar]
- 37.Jin F, Wang Y, Wang X, Wu Y, Wang X, Liu Q, Zhu Y, Liu E, Fan J, Wang Y. Bre Enhances osteoblastic differentiation by promoting the Mdm2-mediated degradation of p53. Stem Cells. 2017;35:1760–1772. doi: 10.1002/stem.2620. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Lv J, Li X, Meng Q, Yang Q, Ma W, Li Y, Ke ZJ. Sevoflurane decreases self-renewal capacity and causes c-Jun N-terminal kinase-mediated damage of rat fetal neural stem cells. Sci Rep. 2017;7:46304. doi: 10.1038/srep46304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodama M, Satoh Y, Otsubo Y, Araki Y, Yonamine R, Masui K, Kazama T. Neonatal desflurane exposure induces more robust neuroapoptosis than do isoflurane and sevoflurane and impairs working memory. Anesthesiology. 2011;115:979–991. doi: 10.1097/ALN.0b013e318234228b. [DOI] [PubMed] [Google Scholar]
- 40.Tagawa T, Sakuraba S, Kimura K, Mizoguchi A. Sevoflurane in combination with propofol, not thiopental, induces a more robust neuroapoptosis than sevoflurane alone in the neonatal mouse brain. J Anesth. 2014;28:815–820. doi: 10.1007/s00540-014-1822-x. [DOI] [PubMed] [Google Scholar]
- 41.Yang ZJ, Wang YW, Li CL, Ma LQ, Zhao X. Pre-treatment with a Xingnaojing preparation ameliorates sevoflurane-induced neuroapoptosis in the infant rat striatum. Mol Med Rep. 2015;11:1615–1622. doi: 10.3892/mmr.2014.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, Xia J, Chen Y, Zhang J. Sevoflurane-Induced endoplasmic reticulum stress contributes to neuroapoptosis and BACE-1 expression in the developing brain: The role of eIF2α. Neurotox Res. 2017;31:218–229. doi: 10.1007/s12640-016-9671-z. [DOI] [PubMed] [Google Scholar]
- 43.Bond GL, Hu W, Levine AJ. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- 44.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 45.Pettersson S, Sczaniecka M, McLaren L, Russell F, Gladstone K, Hupp T, Wallace M. Non-degradative ubiquitination of the Notch1 receptor by the E3 ligase MDM2 activates the Notch signalling pathway. Biochem J. 2013;450:523–536. doi: 10.1042/BJ20121249. [DOI] [PubMed] [Google Scholar]