Abstract
Coronary artery disease (CAD) is a major cause of mortality in India, more importantly the young Indians. Combinatorial and integrative approaches to evaluate pathways and genes to gain an improved understanding and potential biomarkers for risk assessment are required. Therefore, 608 genes from the CADgene database version 2.0, classified into 12 functional classes representing the atherosclerotic disease process, were analyzed. Homology analysis of the unique list of gene ontologies (GO) from each functional class gave 8 GO terms represented in 11 and 10 functional classes. Using disease ontology analysis 80 genes belonging to 8 GO terms, using FunDO suggested that 29 of them were identified to be associated with CAD. Extended network analysis of these genes using STRING version 9.1 gave 328 nodes and 4,525 interactions of which the top 5% had a node degree of ≥75 associated with pathways including the ErbB signaling pathway with epidermal growth factor receptor (EGFR) gene as the central hub. Evaluation of EFGR protein levels in age and gender-matched 342 CAD patients vs. 342 control subjects demonstrated significant differences [controls=149.76±2.47 pg/ml and CAD patients stratified into stable angina (SA)=161.65±3.40 pg/ml and myocardial infarction (MI)=171.51±4.26 pg/ml]. Logistic regression analysis suggested that increased EGFR levels exhibit 3-fold higher risk of CAD [odds ratio (OR) 3.51, 95% confidence interval [CI] 1.96–6.28, P≤0.001], upon adjustment for hypertension, diabetes and smoking. A unit increase in EGFR levels increased the risk by 2-fold for SA (OR 2.58, 95% CI 1.25–5.33, P=0.01) and 3.8-fold for MI (OR 3.82, 95% CI 1.94–7.52, P≤0.001) following adjustment. Thus, the use of ontology mapping and network analysis in an integrative manner aids in the prioritization of biomarkers of complex disease.
Keywords: coronary artery disease, gene ontology, disease ontology, protein-protein interaction, network topology, hubs
Introduction
Coronary artery disease (CAD) is a complex multifactorial condition and is considered to be the leading cause of morbidity and mortality worldwide (1). CAD is particularly prevalent in India, where the prevalence of CAD has increased 4-fold in the last 40 years (2). Various clinical risk factors, including hypertension, diabetes, metabolic disorder, lifestyle, dyslipidemia and heredity are accepted as contributing to this rising threat to public health in India (3). Several genome-wide associations and global gene expression studies have been performed to improve the understanding of the underlying causal factors for CAD (4–6). The resulting data from these studies have confirmed the association of hundreds of genes. However, biological validation of all the genes is impractical and identifying individual disease-specific genes from these complex data sets is difficult. However, the use of informatic approaches may aid the prioritization of molecules that may have biological and functional relevance to CAD.
Therefore, network-based approaches have been extensively used for candidate gene prioritization, prioritizing drug targets and identification of regulatory genes (7–10). Network approaches use protein-protein interaction (PPI) to identify the inter-dependencies between biological functions and this aids in the identification of molecules associated with the phenotype. Previous studies used novel network-based approaches to identify key genes involved in cardiovascular diseases (9,11–13). These studies have used genome-wide association studies or gene expression profiles along with network analysis to comprehensively explain the cardiovascular disease process. A study conducted by Makinen et al (9) and a recent study by Zhao et al (10) used an integrative approach to identify novel biological processes and their key regulatory genes in CAD. Alexander et al (14) used network centrality measures, such as node degree, shortest path, and clustering coefficient to predict the candidate biomarker. The Approach for Candidate Gene Retrieval method by Yilmaz et al (15) was developed to unravel the gene-disease associations and for selection of candidate genes. In this approach, a similarity value is assigned to each gene based on the number of gene ontology (GO) terms shared between diseases. Thus, prompted by all these studies, the present study integrated GO term and network based approach for candidate gene prioritization followed by validation of the key protein on a case-control cohort.
In the current study of biomarker prioritization, the following steps were undertaken. As an initial step, the CAD-associated genes were selected from a CADgene database used for identifying common GO terms across the CAD functional categories. The genes that were part of common GO and disease ontology (DO) formed the seed genes for network construction. The network was based on the analysis of topological parameters to understand the backbone network and the hub molecules. Finally, pathway enrichment analysis and validation in laboratory settings on samples were performed. The findings of the present study revealed that the ErbB signalling pathway and EGF receptor (EGFR) had a central role. The measurement of the protein level of EGFR in biological samples also confirmed its increased level in CAD subjects compared with controls. This confirmed that the combined approach of ontology and network analysis may be effectively used for the identification of key molecules in complex diseases such as CAD.
Materials and methods
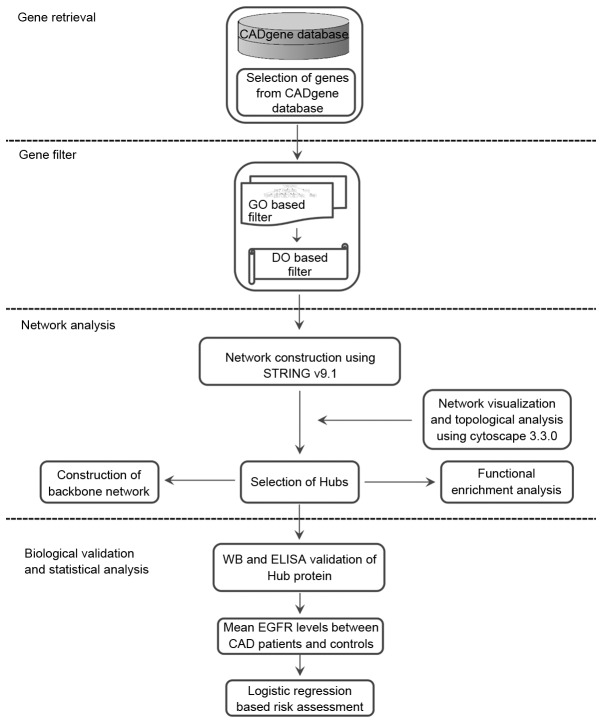
The present study delineated the approach that may be adopted for the identification of key molecules involved in disease conditions based on the ontological filtration method. The workflow diagram presented in Fig. 1 depicts the steps taken for the identification of key genes.
Figure 1.
An overview of the biomarker prioritization method. The method of biomarker prioritization involves the following: Step 1, selection of CAD related genes from CADgene database; Step 2, gene filtration based on GO and DO; Step 3, the gene set obtained was used for network construction, analysis, identification of hub and pathway enrichment analysis; and, Step 4, biological validation using WB, ELISA and statistical assessment of the biomarker. GO, gene ontology; DO, disease ontology; WB, western blot analysis; CAD, coronary artery disease; EGFR, epidermal growth factor receptor.
Selection of CAD-associated genes
The CADgene database was used to retrieve the list of genes that are associated with CAD. The database contained 604 candidate gene lists categorized based on the 12 functional categories, including endothelial integrity, gender difference, homocysteine metabolism, immune and inflammation, lipid lipoproteins metabolism, metalloproteinase and ECM, oxidation-reduction state, renin-angiotensin system, glucose metabolism, thrombosis, vascular smooth muscle cell abnormalities and an uncategorized group (consisting of genes which are not part of above mentioned categories) associated with the development of atherosclerotic processes (16). Constructing a disease-specific network and using its topological parameters to rank the hub genes is one of the widely used approaches in biomarker prioritization studies. The present study adopted a different approach, of selecting genes based on common GO terms. This method may improve the current understanding the most important molecular and biological processes that, upon dysregulation, affect the atherosclerotic function. Thus, gene filtration based on GO terms was used in the present study.
Gene filtration
GO filter
The GO terms of all the 604 genes belonging to 12 functional categories were extracted from UniProtKB (http://www.uniprot.org). The common GO IDs across all 12 categories were identified. The gene ontologies that were present in a minimum of 10 common functional categories were used as a cut-off. The genes belonging to these ontologies were identified and subjected to next level of filtration subsequent to selecting the common genes across all the ontologies.
DO filter
The overlapping genes obtained following GO filtration were subjected to DO enrichment analysis using FunDO (http://fundo.nubic.northwestern.edu/) (17). The FunDO enriches each gene to its corresponding disease phenotype and statistical significance of the enrichment was calculated and corrected by Bonferroni test. The genes enriched for coronary artery disease, hypertension, diabetes mellitus, and obesity were used as a seed set for network construction and further analysis.
Construction and analysis of network
Network generation
The 29 seed genes obtained following DO filtration were used for extended network construction. The PPI network was constructed using STRING version 9.1 (https://string-db.org/) (18) with a confidence score of 0.7. STRING is a functional PPI database, which uses data from heterogeneous sources, such as the neighborhood, gene fusions, co-occurrence, co-expression, experiments, databases and literature mining. The PPIs obtained were used for the construction and analysis of the network using Cytoscape version 3.0.2. The network was analyzed using ‘Network analyzer’ and ‘CentiscaPe’ plugins available in Cytoscape (19,20). The highly connecting nodes were identified as hub genes.
Topology-based gene prioritization
The extended PPI network for the seed genes was constructed using STRING version 9.1 and visualized using Cytoscape version 3.0.2. The topological parameters such as node degree (ND), betweenness centrality (BC) and eigenvector measures were used for prioritization and identification of the hubs. The nodes in the PPI network represented genes or proteins and the edges represented the type of interactions, which revealed the functional association between two nodes. ND denotes the number of direct interacting partners in the network. BC denotes essentiality of the node in a given network and it is quantified using the number of shortest paths passing through it. Eigenvector ranks the node based on the importance of the interacting neighbors considering the quantity and quality of the connections.
Pathway enrichment analysis
In order to understand the biological importance of the hubs, pathway analysis was performed using ClueGO version 1.8 (21). ClueGO aids in the clustering and visualization of functionally connected genes and presents as a chart. The right-sided hypergeometric test was used for the enrichment with a Benjamini-Hochberg correction and a κ score of 0.3 to link the terms.
Biomarker evaluation
The key gene, namely EGFR, which was identified based on this approach was biologically validated using western blot analysis and ELISA in a case-control cohort.
Selection of study population
The study subjects included 342 CAD patients and 342 age and gender-matched controls were selected from the ongoing Indian Atherosclerosis Research Study (IARS) (22). Patients were recruited between October 2005 and February 2012. CAD patients at age ≤45 years at onset for males and ≤50 years for females were selected. The presence of disease condition was evaluated based on angiogram and subjects with >70% stenosis in any one of the coronary artery or >50% in two or more arteries or subjects undergoing coronary artery bypass graft surgery were selected for the study. The controls subjects were healthy volunteers without clinical signs of CAD as confirmed by normal electrocardiogram pattern. None of the participants enrolled in the study had inflammatory disorders, concomitant infections or any other major illness, such as cancer, liver or renal disease. Written informed consent was obtained and ethical approval was granted by the Ethics Committee of the Thrombosis Research Institute (Bengaluru, India).
Clinical assessment
A total of 5 ml of venous blood samples were collected after 12 to 14 h of fasting in the morning h with minimum stasis, using evacuated collection tubes (Vacuette®; Greiner Bio-One International GmbH, Vienna, Austria). To separate the plasma, the collected blood samples were centrifuged at 2,000 × g for 20 min at 25°C. A total of 500 µl aliquots of plasma samples were stored at −80°C. The level of total cholesterol was investigated using a Total Cholesterol assay kit (cat. no. CH200; Randox Laboratories Ltd., Antrim, UK) and the level of triglycerides was investigated using a Triglycerides assay kit (cat. no. TR210; Randox Laboratories Ltd.), and both total cholesterol and triglyceride levels were then determined using a Cobas-Fara II Clinical Chemistry Auto analyzer (Roche Diagnostics GmbH, Mannheim, Germany). For the estimation of high density lipoprotein-cholesterol (HDL-c), the non-HDL fractions were precipitated with a mixture of 2.4 mmol/l phosphotungstic acid and 39 mmol/l magnesium chloride (Bayer Diagnostics India Ltd., Baroda, India) prior to estimation. Low density lipoprotein (LDL) concentration was calculated using Friedewald's formula as previously described (23). The inter-assay coefficient of variation for commercial controls and normal human pool serum/plasma was 4.9–7.0% for total cholesterol, 6.1–7.7% for triglycerides, and 7.1–12.2% for HDL-c.
Estimation of EGFR expression levels in plasma samples
The hub protein EGFR was validated following three independent western blot analyses, where patients with CAD (n=20) and an equal number of age and gender matched controls were included. Total plasma protein levels were estimated using Bradford reagent (cat. no. 500–0205; Bio-Rad Laboratories, Inc., Hercules, CA, USA). An equal amount of 50 µg of protein mixed with gel loading buffer (cat. no. ER07; Genei Laboratories, Pvt., Ltd., Bengaluru, Karnataka, India) and then heat denatured at 95°C for 5 min prior to being loaded on to 10% SDS PAGE and subsequently transferred onto a PVDF membrane. Membranes were blocked via incubation for 1 h at 4°C with 5% non-fat milk solution with gentle agitation. The membranes were then incubated with EGFR primary antibodies (cat. no. 352901; 1:1,000; BioLegend, Inc., San Diego, CA, USA) overnight at 4°C. Following this, the membranes were then incubated with horseradish peroxidase conjugated secondary antibodies for 1 h at room temperature (cat. no. sc-2005; 1:10,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) following several washes using 1X Tris-buffered saline-Tween-20. The same blot was reprobed for loading control GAPDH (cat. no. sc-48167; Santa Cruz Biotechnology, Inc.). Images were scanned using ChemiDoc XR Plus (Bio-Rad Laboratories, Inc.). The densitometric analysis of bands was performed by using ImageJ software version 1.49 (National Institutes of Health, Bethesda, MD, USA) and a ratio for EGFR/GAPDH was calculated. The plasma level of EGFR was quantified in 342 CAD patients and 342 control samples using an ELISA kit (cat. no. SEK10001; Sino Biological Inc., Beijing, China).
Statistical analysis
Statistical analysis was performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used for plotting the data. Results are presented as frequencies for categorical variables and mean ± standard error for all continuous variables. P<0.05 was considered to indicate a statistically significant difference. Probability-probability plot and Shapiro-Wilks test were used to evaluate the distributions of continuous variables. Independent Student's t-test, univariate analysis and one-way analysis of variance followed by Tukey's post hoc analysis were performed on continuous variables. Categorical variables were analyzed by Pearson's χ2 and Fisher's Exact test. The Pearson's partial correlation analysis was performed to investigate the correlation between EGFR and risk factors of CAD. The association of biomarkers with CAD was assessed using logistic regression analysis.
Results
Selection of CAD-associated genes
The GO enrichment for 604 genes from the CADgene database yielded 4,547 GO (biological process and functions). The GO similarity search between all 12 functional categories led to the identification of 1,516 corresponding GO terms.
Identification of conserved molecular functions across different functional classes associated with CAD
The GO homology of 604 genes (4,547 terms of biological processes and functions) across 12 different functional categories associated with disease yielded 1,516 terms shared between ≥2 classes. Further identification of highly conserved GO terms resulted in 8 GO terms of which GO:0008270 was associated with 11 functional disease categories. The following GO IDs: GO:0046872, GO:0045944, GO:0043066, GO:0008284, GO:0008285, GO:0042803 and GO:0006805 were associated with 10 functional disease categories (Table I). The search for associated genes in these 8 GO terms yielded 268 genes, 80 of which were overlapping across the 8 GO terms.
Table I.
Gene ontology-based similarities between 12 CAD functional classes.
| Categories | GO ID | GO term | No. of categories |
|---|---|---|---|
| Endothelial integrity + gender difference + glucose metabolism + homocysteine metabolism + immune and inflammation + lipid lipoproteins metabolism + metalloproteinase and ECM + uncategorized group + Oxidation-reduction state + renin-angiotensin system + thrombosis | GO:0008270 | Zinc ion binding | 11 |
| Endothelial integrity + glucose metabolism + homocysteine metabolism + immune and inflammation + lipid lipoproteins metabolism + metalloproteinase and ECM + uncategorized group + oxidation-reduction state + thrombosis + VSMCA | GO:0046872 | Metal ion binding | 10 |
| Endothelial integrity + gender difference + glucose metabolism + immune and inflammation + lipid lipoproteins metabolism + metalloproteinase and ECM + uncategorized group + oxidation-reduction state + renin-angiotensin system + VSMCA | GO:0045944 | Positive regulation of transcription from RNA polymerase II | 10 |
| Endothelial integrity + homocysteine metabolism + immune and inflammation + lipid lipoproteins metabolism + metalloproteinase and ECM + uncategorized group + oxidation-reduction state + renin-angiotensin system + thrombosis + VSMCA | GO:0043066 | Negative regulation of apoptotic process | 10 |
| Endothelial integrity + gender difference + glucose metabolism + immune and inflammation + lipid lipoproteins metabolism + metalloproteinase and ECM + uncategorized group + oxidation-reduction state + renin-angiotensin system + thrombosis | GO:0008284 | Positive regulation of cell proliferation | 10 |
| Endothelial integrity + glucose metabolism + immune and inflammation + lipid lipoproteins metabolism + metalloproteinase and ECM + uncategorized group + oxidation-reduction state + renin-angiotensin system + thrombosis + VSMCA | GO:0008285 | Negative regulation of cell proliferation | 10 |
| Endothelial integrity + gender difference + homocysteine metabolism + immune and inflammation + lipid lipoproteins metabolism + uncategorized group + oxidation-reduction state + renin-angiotensin system + thrombosis + VSMCA | GO:0042803 | Protein homodimerization activity | 10 |
| Endothelial integrity + gender difference + homocysteine metabolism + immune and inflammation + lipid lipoproteins metabolism + metalloproteinase and ECM + uncategorized group + oxidation-reduction state + renin-angiotensin system + thrombosis | GO:0006805 | Xenobiotic metabolic process | 10 |
The first column contains the CAD disease functional classes; second and third column represents the common GO ID/Term. The fourth column represents the number of common functional categories for the GO ID. CAD, coronary artery disease; ECM, extracellular matrix; VSMCA, vascular smooth muscle cell activation.
Molecular enrichment of disease-associated genes
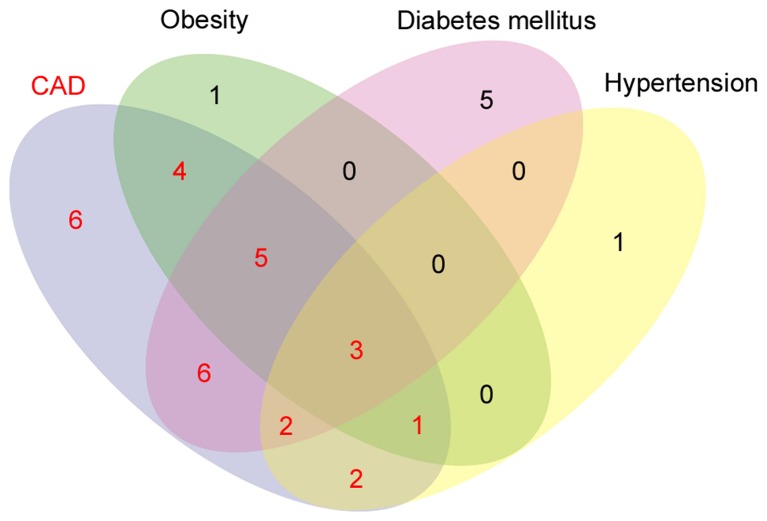
DO filtration using FunDO analysis revealed that 80 genes were significantly enriched for 153 different diseases, with 29 genes being significantly enriched (P≤0.05) for CAD and were considered as seeds for the network construction and analysis. The DO analysis identified 29 genes associated with CAD, 21 genes associated with diabetes mellitus, 14 genes associated with obesity and 9 genes associated with hypertension. The Venn analysis (Fig. 2) revealed 23 genes were shared between these 4 disease phenotypes (CAD + obesity=4; CAD + diabetes=6; CAD + hypertension=2; CAD + obesity + diabetes=5; CAD + obesity + hypertension=1; CAD + hypertension + diabetes=2; CAD + obesity + hypertension + diabetes=3) and 6 genes, including GATA binding protein 2, cystathionine-β-synthase, insulin like growth factor 1 (IGF1) receptor, alcohol dehydrogenase 1B (class I), β polypeptide, S100 calcium binding protein B and prostaglandin-endoperoxide synthase 2 were specific for CAD, 5 genes, such as purinergic receptor P2X 7, ATP binding cassette subfamily G member 1, glutathione S-transferase pi 1, nicotinamide phosphoribosyltransferase and heat shock protein family A (Hsp70) member 1A were specific for diabetes mellitus, interleukin-6 (IL-6) was specific for obesity and gene GCLC was specific for hypertension.
Figure 2.
Venn analysis of genes associated with CAD, obesity, diabetes mellitus and hypertension. CAD shared 23 genes with obesity, diabetes mellitus and hypertension (CAD + obesity=4; CAD + diabetes=6; CAD + hypertension=2; CAD + obesity + diabetes=5; CAD + obesity + hypertension=1; CAD + hypertension + diabetes=2; CAD + obesity + hypertension + diabetes=3) and 6 genes were CAD-specific. The seed gene set selected for the analysis has been highlighted in red. CAD, coronary artery disease.
Network analysis for hub nodes identification
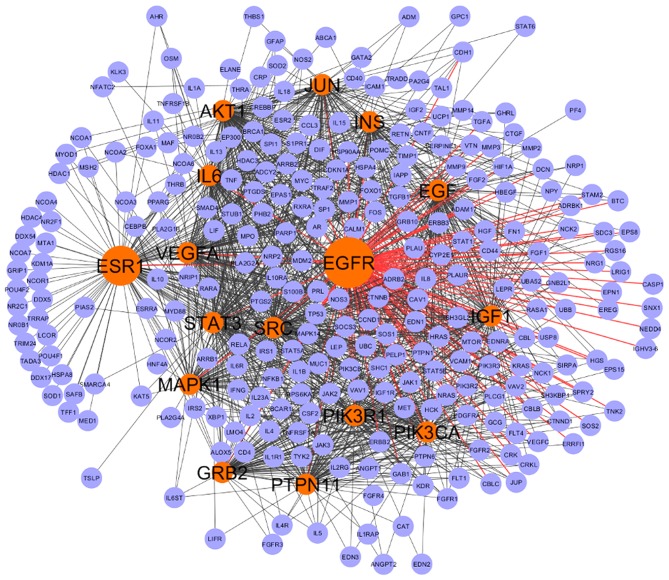
The extended network constructed with 29 seed genes had 328 nodes and 4,525 interactions, of which 5% of nodes (16 genes; Fig. 3 and Table II) with degree ≥75, BC=1,194.82 and eigenvector=0.124 were considered to be the backbone of the network. The EGFR node was the top scoring based on the centrality measures and formed the central hub of the network. Thus, the CAD network was observed to be EGFR centered with 16 key genes forming the backbone of the network.
Figure 3.
CAD backbone network. The backbone network involved 16 hub genes interacting with 294 genes. The hub genes are colored in red and rest of the interactors are in violet. EGFR appears to have the largest centrality measures. CAD, coronary artery disease; EGFR, epidermal growth factor receptor.
Table II.
Key hub genes selected using topological parameters like BC, degree, and Eigen vector.
| Gene | Node degree | BC | Eigenvector |
|---|---|---|---|
| EGFR | 115 | 5399.54 | 0.1574 |
| ESR1 | 114 | 9262.62 | 0.1403 |
| SRC | 111 | 4376.24 | 0.1564 |
| AKT1 | 110 | 4054.04 | 0.1615 |
| STAT3 | 103 | 3077.36 | 0.1585 |
| IL6 | 103 | 2919.17 | 0.1480 |
| EGF | 96 | 2784.97 | 0.1405 |
| VEGFA | 95 | 2948.25 | 0.1407 |
| IGF1 | 85 | 1561.65 | 0.1385 |
| JUN | 84 | 1996.45 | 0.1276 |
| PIK3CA | 83 | 1194.82 | 0.1343 |
| INS | 82 | 2582.59 | 0.1274 |
| MAPK1 | 82 | 1891.13 | 0.1316 |
| GRB2 | 82 | 1494.86 | 0.1235 |
| PIK3R1 | 80 | 1238.81 | 0.1261 |
| PTPN11 | 75 | 1476.50 | 0.1239 |
BC, node degree, and eigenvector were the topological parameters considered for selection of candidate genes. A cut-off of node degree ≥75, BC ≥1194.82 and eigenvector ≥0.124 were considered as key genes forming backbone of the network. EGFR was the super-hub with the highest centrality. BC, betweenness centrality; EGFR, epidermal growth factor receptor; ESR1, estrogen receptor 1; SRC, Src protein–tyrosine kinase; AKT1, Akt serine-threonine kinase; STAT3, signal transducer and activator of transcription 3; IL, interleukin; EGF, epidermal growth factor; VEGFA, vascular endothelial growth factor A; IGF1, insulin-like growth factor 1; JUN, Jun proto-oncogene, AP-1 transcription factor subunit; PIK3CA, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform; INS, insulin; MAPK1, mitogen-activated protein kinases 1; GRB2, growth factor receptor-bound protein 2; PIK3R1, phosphatidylinositol 3-kinase regulatory subunit α; PTPN11, protein tyrosine phosphatase, non-receptor type 11.
Pathway enrichment analysis
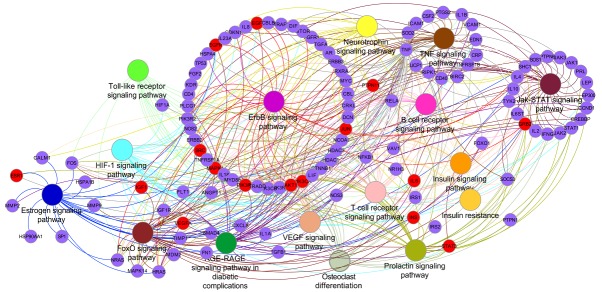
To gain an improved understanding of the pathways that may be affected by the top 105 genes having node degree >25, BC >123.09 and eigenvector >0.045, pathway enrichment analysis was performed. It was observed that 16 pathways were significantly enriched. The top 10 pathways were as follows: ErbB signaling pathway, advanced glycation endproducts/receptor for advanced glycation endproducts signaling pathway in diabetic complications, hypoxia-inducible factor 1 signaling pathway, Janus kinase/signal transducers and activators of transcription signaling (STAT) pathway, forkhead box protein O1 signaling pathway, prolactin signaling pathway, T cell receptor signaling pathway, estrogen signaling pathway, tumor necrosis factor signaling pathway and neurotrophin signaling pathway (Fig. 4).
Figure 4.
Gene and pathway interaction networks obtained after functional enrichment analysis using ClueGO, a Cytoscape plug-in. The red nodes are the hub genes; remaining genes are presented in violet color. The same color code was used for pathway nodes and their corresponding edges to highlight the cross talk between the pathways. All the represented pathways here are highly significant following a Benjamini-Hochberg correction. Pathway enrichment analysis and visualization was performed in Cytoscape version 3.0.2.
Clinical summary of study subjects
Table III shows the complete details of clinical profiles and matching age distribution, with a higher frequency of males (70.2%) than females (29.8%). Hypertension, diabetes mellitus and current smoking were significantly higher in the patient population compared with the control. The frequency of metabolic syndrome was higher in CAD patients (44.7%) in comparison to controls (26.4%). Total cholesterol and LDL levels were significantly lower in CAD patients, which may be attributed to higher statin usage (68.5%) in the group.
Table III.
Baseline characteristics of the study population used for EGFR validation experiment.
| Characteristics | Control (n=342) | CAD patients (n=342) | P-value |
|---|---|---|---|
| Age, (years) | 41.51±0.26 | 41.51±0.26 | 1 |
| Males, n (%) | 240 (70.2) | 240 (70.2) | 1 |
| Females, n (%) | 102 (29.8) | 102 (29.8) | |
| BMI (kg\m2) | 26.07±0.26 | 25.84±0.22 | 0.481 |
| Waist Circumference (cm) | 91.68±0.65 | 87.94±0.55 | <0.001 |
| SBP (mmHg) | 122.24±0.89 | 117.76±0.82 | <0.001 |
| DBP (mmHg) | 81.11±0.58 | 77.75±0.47 | <0.001 |
| FBS (mg/dl) | 94.31±1.92 | 114.70±3.41 | <0.001 |
| Age at onset (years) | 40.88±0.297 | ||
| Stable angina, n (%) | 182 (53.2) | ||
| Myocardial infarction, n (%) | 160 (46.8) | ||
| Number of diseased vessels, n (%) | |||
| 0 | 36 (10.5) | ||
| 1 | 93 (27.2) | ||
| 2 | 88 (25.7) | ||
| 3 | 125 (36.5) | ||
| Current Smoking, n (%) | 62 (18.3) | 148 (43.3) | <0.001 |
| Alcohol habit, n (%) | 68 (20.1) | 89 (26) | 0.065 |
| Hypertension, n (%) | 38 (11.3) | 127 (37.1) | <0.001 |
| Diabetes, n (%) | 48 (14.2) | 118 (34.7) | <0.001 |
| Total cholesterol (mg/dl) | 171.58±2.03 | 152.36±2.99 | <0.001 |
| Triglyceride (mg/dl) | 154.95±5.42 | 170.71±4.69 | 0.03 |
| HDL-c (mg/dl) | 40.11±0.498 | 34.76±0.596 | <0.001 |
| LDL-c (mg/dl) | 101.28±1.65 | 83.88±2.72 | <0.001 |
| Metabolic syndrome, n (%) | 90 (26.4) | 153 (44.7) | <0.001 |
| Statins, n (%) | – | 226 (68.5) | – |
| EGFR (pg/ml) | 154.11±2.68 | 161.93±2.54 | 0.007 |
Continuous variables are presented as mean ± standard error and categorical variable as frequencies. CAD, coronary artery disease; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; FBS, fasting blood sugar; HDL-c, high density lipid cholesterol; LDL-c, low density lipid cholesterol; EGFR, epidermal growth factor receptor.
EGFR validation
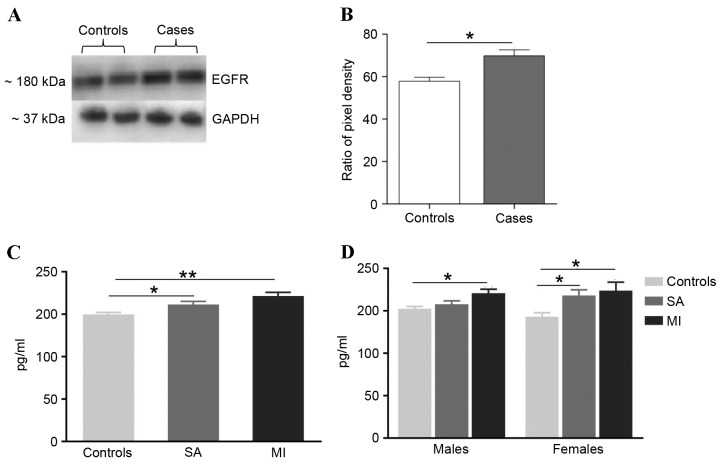
The hub protein was validated using western blot analysis (Fig. 5A and B). The EGFR level was significantly (P≤0.05) upregulated in CAD patients compared with controls. EGFR levels were measured in 342 CAD patients and 342 controls using an ELISA, which identified a significant increase of EGFR levels in CAD patients (Fig. 5C and D). Comparison of EGFR levels between controls (149.76±2.47 pg/ml) and CAD patients stratified into stable angina (SA; 161.65±3.40 pg/ml) and myocardial infarction (MI; 171.51±4.26 pg/ml) demonstrated significant increase in the patient groups (Fig. 5C). Gender-based analysis demonstrated that the EGFR levels remained higher in male CAD patients (SA: 152.39±2.93 vs. 157.99±3.78 pg/ml and MI: 152.39±2.93 vs. 170.92±4.70 pg/ml) and female CAD patients (SA: 143.16±4.54 vs. 168.21±6.64 pg/ml and MI: 143.16±4.54 vs. 173.92±9.99 pg/ml). However, the difference between control and SA in males was not statistically significant (Fig. 5D).
Figure 5.
EGFR levels confirmed by western blot analysis and ELISA in CAD patients and control subjects. (A) Western blotting images of plasma EGFR and loading control GAPDH were assayed in CAD patients and controls. (B) Quantification of relative intensity levels of EGFR in CAD patients and controls adjusted for loading control. ImageJ software was used. (C) ELISA-based comparison of mean EGFR levels between the CAD patients and control groups. A significantly higher level of EGFR was observed in the MI and SA groups in comparison with controls. (D) EGFR levels were only significantly different between male MI groups in comparison with male control group. The EGFR levels were significantly different between female MI and SA groups when compared with controls. *P≤0.05, **P≤0.001. EGFR, epidermal growth factor receptor; CAD, coronary artery disease; MI, myocardial infarction; SA, stable angina.
Correlation of EGFR levels with risk factors
The correlation of EGFR levels with lipids, body mass index, waist circumference and fasting blood sugar was performed using correlation analysis. EGFR did not show any correlation with any of the above-mentioned risk factors (data not shown) prior to and following adjustment for the conventional risk factors.
Association of EGFR with CAD
Logistic regression analysis was adopted to estimate the association of EGFR with CAD. Higher EGFR levels demonstrated a 3-fold higher risk of CAD [OR 3.51 (95% CI 2.09–5.87); P≤0.001], which remained unaffected following adjustment for the conventional risk factors [hypertension, diabetes and current smoking; OR 3.51 (95% CI 1.96–6.28); P≤0.001]. Stratified analysis demonstrated that a unit increase in EGFR levels increased the risk 2-fold in SA patients [OR 2.73 (95% CI 1.47–5.07); P≤0.001] and risk in MI subjects increases four times [OR 4.25 (95% CI 2.28–7.89); P≤0.001] prior to adjustment. Upon adjustment, the ratio remained same for SA [OR 2.58 (95% CI 1.25–5.33); P=0.01] cases and demonstrated a decrease in risk in subjects with MI [OR 3.82 (95% CI 1.94–7.52); P≤0.001]. Details are presented in Table IV.
Table IV.
Association of risk between high EGFR levels and CAD based on OR.
| A, CAD | |||||||
|---|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | ||||||
| Model | OR | 95% CI | P-value | Model | OR | 95% CI | P-value |
| EGFR | 3.51 | 2.09–5.87 | ≤0.001 | EGFR | 3.51 | 1.96–6.28 | ≤0.001 |
| B, Stable angina | |||||||
| Unadjusted | Adjusted | ||||||
| Model | OR | 95% CI | P-value | Model | OR | 95% CI | P-value |
| EGFR | 2.73 | 1.47–5.07 | ≤0.001 | EGFR | 2.58 | 1.25–5.33 | 0.01 |
| C, Myocardial infarction | |||||||
| Unadjusted | Adjusted | ||||||
| Model | OR | 95% CI | P-value | Model | OR | 95% CI | P-value |
| EGFR | 4.25 | 2.28–7.89 | ≤0.001 | EGFR | 3.82 | 1.94–7.52 | ≤0.001 |
Association of EGFR levels with CAD, stable angina, and myocardial infarction. The logistic regression models were adjusted for hypertension, diabetes mellitus and current smoking status. EGFR, epidermal growth factor receptor; CAD, coronary artery disease; OR, odds ratio; CI, confidence intervals.
Discussion
Network-based approaches for the identification of biomarkers for different diseases have been previously investigated (24,25). These approaches used the genes associated with the specific condition to construct the network and selected the hub molecules that may potentially be associated with the disease condition. The current study proposed a two step approach to filter the CAD genes based on functional similarities of genes using GO and DO to identify specific genes associated with CAD. The information thus obtained was then translated to prioritize genes and identify possible underlying molecular pathways. CAD genes that were associated with 12 functional categories associated with the development of atherosclerotic process were selected from the CADgene database (16). The initial analytical step involved the identification of 8 common ontologies (Table I) across these 12 functional groups. The 8 common ontologies identified interactions between the functional categories involved in disease development (Table I).
The top zinc ion binding (GO:0008270) ontology was enriched across 11 functional categories, which is in agreement with a previous study that demonstrated that zinc ion binding has a crucial role in signal transduction in its protein bound form or the unbound form (26). The zinc ion (Zn+2) is demonstrated to maintain endothelial integrity by regulating nuclear factor-κB (NF-κB), which expresses focal adhesion molecules on endothelial cells, a characteristic feature of CAD initiation (27–29). A previous study also suggested that deficiency in Zn+2 may lead to the abnormal production of superoxide dismutase and result in impaired removal of reactive oxygen species (ROS) from the system, leading to oxidative stress (30). However, if present in excess Zn+2 increases the concentration of phosphorylated EGFR, which is followed by NF-κB-mediated endothelial dysfunction (31,32). These previous findings emphasize the importance of Zn+2 binding function in CAD.
The other top ontologies that were observed to be shared between the CAD functional categories were ‘dysregulation in cell proliferation’ [positive (GO:0008284) and negative regulation (GO:0008285) of cell proliferation] and ‘negative regulation of the apoptotic process’ (GO:0043066). Disruptions in these processes have been demonstrated to have an important role in manifestation of atherosclerosis. Previous studies have demonstrated that excessive cell proliferation leads to the local accumulation of cells (hypercellularity) in the intima of the arterial wall and macrophage apoptosis is an important feature of atherosclerotic plaque development (33,34). The ontology ‘protein homodimerization activity’ (GO: 0042803) is a characteristic feature of vascular smooth muscle cell (VSMC) death receptor while exposed to ROS-generated oxLDL (35,36).
CAD is a multifactorial condition and based on the current evidence, CAD is associated with at least 3 major risk factors: Hypertension, diabetes mellitus and obesity. It is expected that genes common between these risk factors and CAD are major contributors to disease development. Therefore, in the second step of the present study, genes commonly enriched between CAD and its risk factors were selected as the seed set (29 genes; Fig. 2). The extended main network was constructed using 29 seed genes, which had 4,525 interactions between 328 nodes. The top 5% nodes, with node degree ≥75, BC ≥1,194.82 and eigenvector ≥0.124 were considered as key genes, forming the backbone of the network (Fig. 3). Topological measures of PPI networks are widely used for understanding the disease process and the identification of key disease genes. Our previous study used topological measures in identification of key inflammatory genes and regulators of inflammation in CAD (7). Similarly, in a previous study by Sarajlic et al (11) topological measures were used to identify key drug targets and driver genes in cardiovascular disease. In an integrative bioinformatics analysis Zhang et al (37) applied topological analysis on significantly differentially regulated genes. Furthermore, Zhang et al (37) used BC and stress centrality to determine the importance of genes and modules were used for the functional validation. The centrality-lethality rule suggests that the node with the highest number of connections in the network is more functionally essential and may have lethal effects upon elimination (38,39). The network was constructed and analyzed considering these topological parameters in the present study, which demonstrated EGFR to be the central hub along with 15 genes [estrogen receptor 1 (ESR1), Src protein-tyrosine kinase (Src), Akt serine-threonine kinase (AKT1), STAT3, interleukin (IL)6, epidermal growth factor (EGF), vascular endothelial growth factor A (VEGFA), IGF1, Jun proto-oncogene, AP-1 transcription factor subunit (JUN), phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (PIK3CA), insulin (INS), mitogen-activated protein kinase 1 (MAPK1), growth factor receptor-bound protein 2 (GRB2), phosphatidylinositol 3-kinase regulatory subunit α (PIK3R1) and protein tyrosine phosphatase, non-receptor type 11 (PTPN11); Table II]. This backbone network of EGFR and 15 genes had 4,346 interactions with 294 nodes out of 328 nodes, indicating that 96% of the interactions were formed by these genes (Fig. 3).
The central role of EGFR in atherosclerosis has been supported by previous studies (40,41). Activated EGFR is involved in regulating multiple cellular aspects, such as cell proliferation, migration and apoptosis (31,32,35,36,42,43). Lamb et al (44) demonstrated the presence of EGFR on peripheral monocytes and its co-localization in the atherosclerotic lesion, causing chemotaxis of monocytes and monocytes-derived macrophage proliferation in high cholesterol diet rabbits. It has also been previously reported to be expressed on smooth muscle cells (SMC) localized in intima and media of aorta in subjects with advanced disease (45). In a retrospective case-control study, EGFR (R497K) polymorphism has been demonstrated to be associated with acute coronary syndrome (46).
ESR1 is a key member of nuclear receptor family and expressed in wide variety of cells including endothelial cells, VSMCs and macrophages (47). ESR1 was demonstrated to have an atheroprotective role by stimulating endothelial nitric oxide (NO) production and re-endothelialization (48). However, the non-functional ESR1 has been demonstrated to be associated with premature CAD (49). Src is a tyrosine protein kinase that acts downstream of the EGFR signaling. It is considered to be a focal adhesion molecule and has been demonstrated to regulate vasoconstriction (50). Higher serum levels of Src have been detected in CAD subjects (51). AKT serine/threonine kinase 1 (Akt1) acts downstream to the Src and its active form are highly abundant in heart tissue, endothelial cells, platelets and SMC (52–54). Triple knockout mice for apolipoprotein E−/−, scavenger receptor class B type I−/− and Src−/− demonstrated reduced ROS generation and lipid oxidation compared with Src+/+ suggesting a pro-atherogenic role for the Akt1 gene (52).
MAPK1 belongs to the Ser-Thr kinases protein family and has been previously reported in different features of cardiac modeling and regulation of inflammation, cell proliferation and differentiation (55,56). The high activity of Erk1/2 has been observed in T-lymphocytes from CAD patients, including ST-elevation myocardial infarction (STEMI), Non-STEMI and unstable angina (57). A previous study demonstrated the EGFR-mediated cross-talk between MAPK and Akt1 signaling, which combined had an important role in abnormal vascular remodeling (58).
IGF1 is a survival factor hormone peptide, which interacts with IGF1R and insulin receptor. IGF1 maintains endothelial integrity by stimulating NO expression; however, upon endothelial dysfunction, it stimulates SMC proliferation (59–61). Although, SMC proliferation provides stability to damaged endothelium, uncontrolled proliferation leads to vascular complications. Higher levels of IGF1 have been identified in patients with heart failure on ACE inhibitors with ejection fraction <40%. An association of increased levels of IGF1 with cardiovascular disease risk was identified previously (62).
PTPN11 is a cytoplasmic protein tyrosine phosphatase expressed throughout multiple tissue types. PTPN11 enhances the cell proliferation under the influence of EGF, platelet-derived growth factor (PDGF) and INS. INS facilitates binding of PTPN11 with insulin receptor substrate 1 (IRS1) and works as an adaptor for the insulin receptor and IRS1 (63). PDGF stimulates binding of PTPN11 with GRB2 and may subsequently enhance SMC proliferation with a decrease in endothelial cell migration and proliferation (63–65) that may lead to the acceleration of atherosclerosis.
PIK3CA and PIK3R1 were identified to be novel genes as to the best of our knowledge, no previous studies have identified their association with atherosclerosis. However, PIK3R1 has been demonstrated to have a role in insulin resistance in high-fat diet-obese mice (66), which has been identified to be an underlying process involved in atherosclerosis. The other molecules (IL-6, VEGFA, JUN and STAT3) in the network were primarily associated with inflammatory processes. These genes were also observed to be the hub genes based on our previous inflammatory driven network study (7).
In order to investigate the class of pathways being regulated, the pathway mapping of the top 105 genes with node degree >25, BC >123.09 and eigenvector >0.045 was performed. A significant enrichment of 16 pathways was observed (Fig. 4). Previous studies have demonstrated the involvement of these pathways in the atherosclerotic process (67–76). The pathway enrichment network revealed the interaction with key 16 genes that are colored as red nodes, 8 out of the 16 key genes were involved in the ErbB signaling pathway. This revealed the importance of EGFR and its regulating pathway in CAD conditions. Finally, the biological validation of EGFR was performed using a protein estimation assay. EGFR protein expression levels were significantly higher in CAD cases when compared with the control, specifically in terms of patients with MI complications. To understand the possible contributors to the EGFR expression levels, correlation analysis was performed.
In conclusion, the present study combined multiple methods in a stepwise manner to unravel the complexity of CAD and identify the key coronary artery disease genes. Ontology-based gene filtration was used in the process, which is a fundamental strength of the present study as constructing the network from an unfiltered gene set may lead to false positive reports. However, the limitation of the present study is that it requires a prior knowledge of the disease process. This issue could be addressed by utilizing disease ontological information in gene filtration. The topological parameters revealed that 16 hub genes formed the backbone of the network, with EGFR as a central molecule. The roles of EGFR and the remaining 15 hub genes were validated based on evidence in the literature and the biological validation of EGFR expression levels supported its role in CAD patients.
Acknowledgements
The authors would like to thank the Bharati foundation, Thrombosis Research Institute (TRI), London, foundation Bey, Switzerland, Tata Social Welfare Trust, India (grant no. TSWT/IG/SNB/JP/Sdm) for providing financial assistance. The authors are grateful to the late Professor Vijay V. Kakkar OBE, (Thrombosis Research Institute, London, UK; and Thrombosis Research Institute, Bangalore, India) for his guidance and suggestions, and would like to thank Dr Jayashree Shanker (Thrombosis Research Institute, Bangalore, India) and Dr Lakshmi Mundkur (Thrombosis Research Institute, Bangalore, India) for their valuable suggestions and comments that improved the manuscript. The authors also thank all the staff of TRI and participants of IARS from Narayana Hrudayalaya, Bangalore and the Asian Heart Centre (Mumbai, India) for their contribution.
References
- 1.Achari V, Thakur AK. Association of major modifiable risk factors among patients with coronary artery disease-a retrospective analysis. J Assoc Physicians India. 2004;52:103–108. [PubMed] [Google Scholar]
- 2.Krishnan MN. Coronary heart disease and risk factors in India- on the brink of an epidemic? Indian Heart J. 2012;64:364–367. doi: 10.1016/j.ihj.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enas EA, Singh V, Munjal YP, Bhandari S, Yadave RD, Manchanda SC. Reducing the burden of coronary artery disease in India: Challenges and opportunities. Indian Heart J. 2008;60:161–175. [PubMed] [Google Scholar]
- 4.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta NN. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Circ Cardiovasc Genet. 2011;4:327–329. doi: 10.1161/CIRCGENETICS.111.960989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arvind P, Jayashree S, Jambunathan S, Nair J, Kakkar VV. Understanding gene expression in coronary artery disease through global profiling, network analysis and independent validation of key candidate genes. J Genet. 2015;94:601–610. doi: 10.1007/s12041-015-0548-3. [DOI] [PubMed] [Google Scholar]
- 7.Nair J, Ghatge M, Kakkar VV, Shanker J. Network analysis of inflammatory genes and their transcriptional regulators in coronary artery disease. PLoS One. 2014;9:e94328. doi: 10.1371/journal.pone.0094328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofree M, Shen JP, Carter H, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makinen VP, Civelek M, Meng Q, Zhang B, Zhu J, Levian C, Huan T, Segrè AV, Ghosh S, Vivar J, et al. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10:e1004502. doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Chen J, Freudenberg JM, Meng Q, Rajpal DK, Yang X. Network-based identification and prioritization of key regulators of coronary artery disease loci. Arterioscler Thromb Vasc Biol. 2016;36:928–941. doi: 10.1161/ATVBAHA.115.306725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarajlic A, Janjić V, Stojković N, Radak D, Przulj N. Network topology reveals key cardiovascular disease genes. PLoS One. 2013;8:e71537. doi: 10.1371/journal.pone.0071537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camargo A, Azuaje F. Linking gene expression and functional network data in human heart failure. PLoS One. 2007;2:e1347. doi: 10.1371/journal.pone.0001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diez D, Wheelock AM, Goto S, Haeggström JZ, Paulsson-Berne G, Hansson GK, Hedin U, Gabrielsen A, Wheelock CE. The use of network analyses for elucidating mechanisms in cardiovascular disease. Mol Biosyst. 2010;6:289–304. doi: 10.1039/B912078E. [DOI] [PubMed] [Google Scholar]
- 14.Alexander MR, Moehle CW, Johnson JL, Yang Z, Lee JK, Jackson CL, Owens GK. Genetic inactivation of IL-1 signaling enhances atherosclerotic plaque instability and reduces outward vessel remodeling in advanced atherosclerosis in mice. J Clin Invest. 2012;122:70–79. doi: 10.1172/JCI62827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz S, Jonveaux P, Bicep C, Pierron L, Smail-Tabbone M, Devignes MD. Gene-disease relationship discovery based on model-driven data integration and database view definition. Bioinformatics. 2009;25:230–236. doi: 10.1093/bioinformatics/btn612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Liu W, Liao Y, Cheng L, Liu Q, Ren X, Shi L, Tu X, Wang QK, Guo AY. CADgene: A comprehensive database for coronary artery disease genes. Nucleic Acids Res. 2011;39(Database Issue):D991–D996. doi: 10.1093/nar/gkq1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du P, Feng G, Flatow J, Song J, Holko M, Kibbe WA, Lin SM. From disease ontology to disease-ontology lite: Statistical methods to adapt a general-purpose ontology for the test of gene-ontology associations. Bioinformatics. 2009;25:i63–i68. doi: 10.1093/bioinformatics/btp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database Issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 20.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanjilal S, Rao VS, Mukherjee M, Natesha BK, Renuka KS, Sibi K, Iyengar SS, Kakkar VV. Application of cardiovascular disease risk prediction models and the relevance of novel biomarkers to risk stratification in Asian Indians. Vasc Health Risk Manag. 2008;4:199–211. doi: 10.2147/VHRM.S2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 24.Li J, Lin X, Teng Y, Qi S, Xiao D, Zhang J, Kang Y. A comprehensive evaluation of disease phenotype networks for gene prioritization. PLoS One. 2016;11:e0159457. doi: 10.1371/journal.pone.0159457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Sahni N, Yi S. Comparative analysis of protein interactome networks prioritizes candidate genes with cancer signatures. Oncotarget. 2016;7:78841–78849. doi: 10.18632/oncotarget.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little PJ, Bhattacharya R, Moreyra AE, Korichneva IL. Zinc and cardiovascular disease. Nutrition. 2010;26:1050–1057. doi: 10.1016/j.nut.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Meerarani P, Reiterer G, Toborek M, Hennig B. Zinc modulates PPARgamma signaling and activation of porcine endothelial cells. J Nutr. 2003;133:3058–3064. doi: 10.1093/jn/133.10.3058. [DOI] [PubMed] [Google Scholar]
- 28.Litwin M, Clark K, Noack L, Furze J, Berndt M, Albelda S, Vadas M, Gamble J. Novel cytokine-independent induction of endothelial adhesion molecules regulated by platelet/endothelial cell adhesion molecule (CD31) J Cell Biol. 1997;139:219–228. doi: 10.1083/jcb.139.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross R. Atherosclerosis-an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 30.Wu CY, Steffen J, Eide DJ. Cytosolic superoxide dismutase (SOD1) is critical for tolerating the oxidative stress of zinc deficiency in yeast. PLoS One. 2009;4:e7061. doi: 10.1371/journal.pone.0007061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu W, Bromberg PA, Samet JM. Zinc ions as effectors of environmental oxidative lung injury. Free Radic Biol Med. 2013;65:57–69. doi: 10.1016/j.freeradbiomed.2013.05.048. [DOI] [PubMed] [Google Scholar]
- 32.Wu W, Jaspers I, Zhang W, Graves LM, Samet JM. Role of Ras in metal-induced EGF receptor signaling and NF-kappaB activation in human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1040–L1048. doi: 10.1152/ajplung.00390.2001. [DOI] [PubMed] [Google Scholar]
- 33.Orekhov AN, Andreeva ER, Mikhailova IA, Gordon D. Cell proliferation in normal and atherosclerotic human aorta: Proliferative splash in lipid-rich lesions. Atherosclerosis. 1998;139:41–48. doi: 10.1016/S0021-9150(98)00044-6. [DOI] [PubMed] [Google Scholar]
- 34.Tabas I. Macrophage apoptosis in atherosclerosis: Consequences on plaque progression and the role of endoplasmic reticulum stress. Antioxid Redox Signal. 2009;11:2333–2339. doi: 10.1089/ars.2009.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy NJ, Bennett MR. The regulation of vascular smooth muscle cell apoptosis. Cardiovasc Res. 2000;45:747–755. doi: 10.1016/S0008-6363(99)00275-8. [DOI] [PubMed] [Google Scholar]
- 36.Mallat Z, Tedgui A. Apoptosis in the vasculature: Mechanisms and functional importance. Br J Pharmacol. 2000;130:947–962. doi: 10.1038/sj.bjp.0703407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Li X, Tai J, Li W, Chen L. Predicting candidate genes based on combined network topological features: A case study in coronary artery disease. PLoS One. 2012;7:e39542. doi: 10.1371/journal.pone.0039542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 39.Barábasi AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 40.Makki N, Thiel KW, Miller FJ., Jr The epidermal growth factor receptor and its ligands in cardiovascular disease. Int J Mol Sci. 2013;14:20597–20613. doi: 10.3390/ijms141020597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreier B, Gekle M, Grossmann C. Role of epidermal growth factor receptor in vascular structure and function. Curr Opin Nephrol Hypertens. 2014;23:113–121. doi: 10.1097/01.mnh.0000441152.62943.29. [DOI] [PubMed] [Google Scholar]
- 42.Samarakoon R, Higgins PJ. Integration of non-SMAD and SMAD signaling in TGF-beta1-induced plasminogen activator inhibitor type-1 gene expression in vascular smooth muscle cells. Thromb Haemost. 2008;100:976–983. [PMC free article] [PubMed] [Google Scholar]
- 43.Holbro T, Hynes NE. ErbB receptors: Directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 44.Lamb DJ, Modjtahedi H, Plant NJ, Ferns GA. EGF mediates monocyte chemotaxis and macrophage proliferation and EGF receptor is expressed in atherosclerotic plaques. Atherosclerosis. 2004;176:21–26. doi: 10.1016/j.atherosclerosis.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Tamura R, Miyagawa J, Nishida M, Kihara S, Sasada R, Igarashi K, Nakata A, Yamamori K, Kameda-Takemura K, Yamashita S, Matsuzawa Y. Immunohistochemical localization of Betacellulin, a member of epidermal growth factor family, in atherosclerotic plaques of human aorta. Atherosclerosis. 2001;155:413–423. doi: 10.1016/S0021-9150(00)00576-1. [DOI] [PubMed] [Google Scholar]
- 46.Gao L, Zhou B, Zhang L, Wei YS, Wang YY, Liang WB, Lv ML, Pan XM, Chen YC, Rao L. R497K polymorphism in epidermal growth factor receptor gene is associated with the risk of acute coronary syndrome. BMC Med Genet. 2008;9:74. doi: 10.1186/1471-2350-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maha R, Imen K, Rania A, Najla K, Nouha AB, Leila A, Ahmed R. Association analysis of polymorphisms in EGFR, HER2, ESR1 and THRA genes with coronary artery diseases. Egyptian J Med Human Genet. 2016;18:241–248. [Google Scholar]
- 48.Arnal JF, Fontaine C, Billon-Galés A, Favre J, Laurell H, Lenfant F, Gourdy P. Estrogen receptors and endothelium. Arterioscler Thromb Vasc Biol. 2010;30:1506–1512. doi: 10.1161/ATVBAHA.109.191221. [DOI] [PubMed] [Google Scholar]
- 49.Sudhir K, Chou TM, Chatterjee K, Smith EP, Williams TC, Kane JP, Malloy MJ, Korach KS, Rubanyi GM. Premature coronary artery disease associated with a disruptive mutation in the estrogen receptor gene in a man. Circulation. 1997;96:3774–3777. doi: 10.1161/01.CIR.96.10.3774. [DOI] [PubMed] [Google Scholar]
- 50.Min J, Reznichenko M, Poythress RH, Gallant CM, Vetterkind S, Li Y, Morgan KG. Src modulates contractile vascular smooth muscle function via regulation of focal adhesions. J Cell Physiol. 2012;227:3585–3592. doi: 10.1002/jcp.24062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.V A, Nayar PG, Murugesan RSS, Krishnan J, Ahmed SS. A systems biology and proteomics-based approach identifies SRC and VEGFA as biomarkers in risk factor mediated coronary heart disease. Mol Biosyst. 2016;12:2594–2604. doi: 10.1039/C6MB00264A. [DOI] [PubMed] [Google Scholar]
- 52.Kerr BA, Ma L, West XZ, Ding L, Malinin NL, Weber ME, Tischenko M, Goc A, Somanath PR, Penn MS, et al. Interference with akt signaling protects against myocardial infarction and death by limiting the consequences of oxidative stress. Sci Signal. 2013;6:ra67. doi: 10.1126/scisignal.2003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernández-Hernando C, Ackah E, Yu J, Suárez Y, Murata T, Iwakiri Y, Prendergast J, Miao RQ, Birnbaum MJ, Sessa WC. Loss of Akt1 leads to severe atherosclerosis and occlusive coronary artery disease. Cell Metab. 2007;6:446–457. doi: 10.1016/j.cmet.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J, Somanath PR, Razorenova O, Chen WS, Hay N, Bornstein P, Byzova TV. Akt1 regulates pathological angiogenesis, vascular maturation and permeability in vivo. Nat Med. 2005;11:1188–1196. doi: 10.1038/nm1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu R, van Berlo JH, York AJ, Vagnozzi RJ, Maillet M, Molkentin JD. DUSP8 regulates cardiac ventricular remodeling by altering ERK1/2 signaling. Circ Res. 2016;119:249–260. doi: 10.1161/CIRCRESAHA.115.308238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao J, Li L, Peng L. MAPK1 up-regulates the expression of MALAT1 to promote the proliferation of cardiomyocytes through PI3K/AKT signaling pathway. Int J Clin Exp Pathol. 2015;8:15947–15953. [PMC free article] [PubMed] [Google Scholar]
- 57.Indolfi C, Gasparri C, Vicinanza C, De Serio D, Boncompagni D, Mongiardo A, Spaccarotella C, Agosti V, Torella D, Curcio A. Mitogen-activated protein kinases activation in T lymphocytes of patients with acute coronary syndromes. Basic Res Cardiol. 2011;106:667–679. doi: 10.1007/s00395-011-0172-1. [DOI] [PubMed] [Google Scholar]
- 58.Eguchi S, Iwasaki H, Ueno H, Frank GD, Motley ED, Eguchi K, Marumo F, Hirata Y, Inagami T. Intracellular signaling of angiotensin II-induced p70 S6 kinase phosphorylation at Ser(411) in vascular smooth muscle cells. Possible requirement of epidermal growth factor receptor, Ras, extracellular signal-regulated kinase, and Akt. J Biol Chem. 1999;274:36843–36851. doi: 10.1074/jbc.274.52.36843. [DOI] [PubMed] [Google Scholar]
- 59.Bayes-Genis A, Conover CA, Schwartz RS. The insulin-like growth factor axis: A review of atherosclerosis and restenosis. Circ Res. 2000;86:125–130. doi: 10.1161/01.RES.86.2.125. [DOI] [PubMed] [Google Scholar]
- 60.Tsukahara H, Gordienko DV, Tonshoff B, Gelato MC, Goligorsky MS. Direct demonstration of insulin-like growth factor-I-induced nitric oxide production by endothelial cells. Kidney Int. 1994;45:598–604. doi: 10.1038/ki.1994.78. [DOI] [PubMed] [Google Scholar]
- 61.Muniyappa R, Walsh MF, Rangi JS, Zayas RM, Standley R, Ram JL, Sowers JR. Insulin like growth factor 1 increases vascular smooth muscle nitric oxide production. Life Sci. 1997;61:925–931. doi: 10.1016/S0024-3205(97)00594-8. [DOI] [PubMed] [Google Scholar]
- 62.Chisalita SI, Dahlström U, Arnqvist HJ, Alehagen U. Increased IGF1 levels in relation to heart failure and cardiovascular mortality in an elderly population: Impact of ACE inhibitors. Eur J Endocrinol. 2011;165:891–898. doi: 10.1530/EJE-11-0584. [DOI] [PubMed] [Google Scholar]
- 63.Wong L, Johnson GR. Epidermal growth factor induces coupling of protein-tyrosine phosphatase 1D to GRB2 via the COOH-terminal SH3 domain of GRB2. J Biol Chem. 1996;271:20981–20984. doi: 10.1074/jbc.271.35.20981. [DOI] [PubMed] [Google Scholar]
- 64.Seki N, Hashimoto N, Suzuki Y, Mori S, Amano K, Saito Y. Role of SRC homology 2-containing tyrosine phosphatase 2 on proliferation of rat smooth muscle cells. Arterioscler Thromb Vasc Biol. 2002;22:1081–1085. doi: 10.1161/01.ATV.0000022878.37277.EC. [DOI] [PubMed] [Google Scholar]
- 65.Bell L, Madri JA. Effect of platelet factors on migration of cultured bovine aortic endothelial and smooth muscle cells. Circ Res. 1989;65:1057–1065. doi: 10.1161/01.RES.65.4.1057. [DOI] [PubMed] [Google Scholar]
- 66.McCurdy CE, Schenk S, Holliday MJ, Philp A, Houck JA, Patsouris D, MacLean PS, Majka SM, Klemm DJ, Friedman JE. Attenuated Pik3r1 expression prevents insulin resistance and adipose tissue macrophage accumulation in diet-induced obese mice. Diabetes. 2012;61:2495–2505. doi: 10.2337/db11-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramasamy R, Yan SF, Schmidt AM. Receptor for AGE (RAGE): Signaling mechanisms in the pathogenesis of diabetes and its complications. Ann N Y Acad Sci. 2011;1243:88–102. doi: 10.1111/j.1749-6632.2011.06320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maziere C, Mazière JC. Activation of transcription factors and gene expression by oxidized low-density lipoprotein. Free Radic Biol Med. 2009;46:127–137. doi: 10.1016/j.freeradbiomed.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 69.Grote K, Luchtefeld M, Schieffer B. JANUS under stress-role of JAK/STAT signaling pathway in vascular diseases. Vascul Pharmacol. 2005;43:357–363. doi: 10.1016/j.vph.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 70.Feinberg MW. OutFOXing myeloid cells in atherosclerosis with FoxOs. Circ Res. 2013;112:978–982. doi: 10.1161/CIRCRESAHA.113.301159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reuwer AQ, Twickler MT, Hutten BA, Molema FW, Wareham NJ, Dallinga-Thie GM, Bogorad RL, Goffin V, Smink-Bol M, Kastelein JJ, et al. Prolactin levels and the risk of future coronary artery disease in apparently healthy men and women. Circ Cardiovasc Genet. 2009;2:389–395. doi: 10.1161/CIRCGENETICS.109.853572. [DOI] [PubMed] [Google Scholar]
- 72.Matsumoto T, Sasaki N, Yamashita T, Emoto T, Kasahara K, Mizoguchi T, Hayashi T, Yodoi K, Kitano N, Saito T, et al. Overexpression of Cytotoxic T-Lymphocyte-Associated Antigen-4 prevents atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 2016;36:1141–1151. doi: 10.1161/ATVBAHA.115.306848. [DOI] [PubMed] [Google Scholar]
- 73.Dreux AC, Lamb DJ, Modjtahedi H, Ferns GA. The epidermal growth factor receptors and their family of ligands: Their putative role in atherogenesis. Atherosclerosis. 2006;186:38–53. doi: 10.1016/j.atherosclerosis.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 74.Murphy E. Estrogen signaling and cardiovascular disease. Circ Res. 2011;109:687–696. doi: 10.1161/CIRCRESAHA.110.236687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hopkins PN. Molecular biology of atherosclerosis. Physiol Rev. 2013;93:1317–1542. doi: 10.1152/physrev.00004.2012. [DOI] [PubMed] [Google Scholar]
- 76.Caporali A, Emanueli C. Cardiovascular actions of neurotrophins. Physiol Rev. 2009;89:279–308. doi: 10.1152/physrev.00007.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]