Abstract
Background
Restrictive cardiomyopathy (RCM) is a rare cardiomyopathy characterized by impaired diastolic ventricular function resulting in a poor clinical prognosis. Rarely, heritable forms of RCM have been reported and mutations underlying RCM have been identified in genes that govern the contractile function of the cardiomyocytes.
Methods and Results
We evaluated 8 family members across four generations by history, physical examination, electrocardiography and echocardiography. Affected individuals presented with a pleitropic syndrome of progressive RCM, atrioventricular septal defects, and a high prevalence of atrial fibrillation. Exome sequencing of 5 affected members identified a single novel missense variant in a highly conserved residue of FLNC (p.V2297M). FLNC encodes Filamin C, a protein that acts as both a scaffold for the assembly and organization of the central contractile unit of striated muscle, and also as a mechanosensitive signaling molecule during cell migration and shear stress. Immunohistochemical analysis of Filamin C localization in cardiac tissue from an affected family member revealed a diminished localization at the z-disk, whereas traditional localization at the intercalated disk was preserved. Stem-cell derived cardiomyocytes mutated to carry the effect allele had diminished contractile activity when compared to controls.
Conclusion
We have identified a novel variant in FLNC as pathogenic variant for familial RCM, a finding that further expands upon the genetic basis of this rare and morbid cardiomyopathy.
Keywords: restrictive cardiomyopathy, atrial fibrillation, mutation, stem cell
Subject codes: Genetics, Contractile Function, Cardiomyopathy
Introduction
Restrictive cardiomyopathy (RCM) is a rare cardiomyopathy characterized by preserved systolic function with impaired diastolic filling due to abnormal ventricular relaxation.1 Affected patients typically present with heart failure, palpitations, syncope, or thromboembolic events. Echocardiography usually reveals a normal or modestly reduced ejection fraction, biatrial enlargement, and a restrictive profile with Doppler imaging. RCM may result from inflammation, infiltrative diseases such as sarcoidosis and amyloidosis, or storage diseases including hemochromatosis or Fabry’s disease that result in secondary RCM. In addition, a significant genetic contribution to RCM has been reported. Since the initial discovery of a familial form of RCM in 1998,2 several variants have been reported in cardiac contractile proteins.3–9 Additional evidence exists for variation in genes associated with amyloidosis, but whether presentation of RCM in these cases is primary or secondary remains unclear.
Here, we report a four generation family that was clinically diagnosed with a variety of cardiac pathologies that include atrioventricular septal defects, a bicuspid aortic valve, RCM, and a high prevalence of atrial fibrillation.
Methods
Clinical and Pathological Evaluation
Written informed consent was obtained from all family members prior to clinical evaluation. The study was approved by the Institutional Review Board at Massachusetts General Hospital.
Family members were characterized using a standardized questionnaire, physical examination, 12-lead ECG and echocardiography. Individuals were considered affected if they exhibited evidence of RCM, atrial fibrillation, or atrioventricular septal defects.
Individual II-5 died during an attempted cardiac transplant and their heart was preserved for pathological analyses. Hematoxylin and eosin, Masson’s trichrome, and Congo red histological stains of paraffin embedded samples, as well as transmission electron microscopy of formaldehyde-fixed, resin-embedded tissue sections, were performed according to standard techniques.
DNA Isolation, Whole Exome Sequencing, and Bioinformatic Analyses
Venous blood samples were collected in acid citrate dextrose vacutainers from all enrolled family members and isolated using the Gentra Puregene Blood Kit (Qiagen, Germany). Whole exome sequencing was performed on five affected family members (II-2, III-2, IV-3, II-5 and II-7). Individual II-2 was sequenced using third generation Agilent SureSelect Target Enrichment assay followed by pair-end sequenced on the Illumina HiSeq 2000 platform at Perkin Elmer (Waltham, MA, USA). After the enrollment of additional family members, whole exome sequencing was performed on individuals III-2, IV-3, II-5, and II-7 using the fourth generation Agilent SureSelect assay (Agilent Technologies Inc, USA) and pair-end sequencing on the Illumina HiSeq 2500 platform at the Beijing Genomics Institute (Shenzhen, China).
The BWA software package was used to map the sequenced reads to the human genome (NCBI Build 37, hg19).10 The MarkDuplicates function of the Picard software package was used identify duplicate reads. The GATK software package (The Broad Institute, Inc., Cambridge, MA) was used to spot-check alignments and to recalibrate base qualities, this software was also used to call single nucleotide polymorphisms (SNVs) through its UnifiedGenotyper tool.11 These variants were then filtered according to the following analyses: transmission of variants with disease in all family members, cardiac gene expression profiles, interspecies conservation, predicted effect on protein function, and potential involvement in cardiac-related functional pathways (Supplemental Table 1, Supplemental Table 2). Initially, SNVs with a quality score <20 or a strand bias >60 were excluded. After filtering for common variants with a MAF >0.1% (dbSNP, 1000 Genomes Project, Exome Variant Server, ExAC) and exclusion of synonymous variants, only variants with transmission in all 5 sequenced family members were considered as potential candidate genes. PolyPhen-2,12 SIFT,13 and MutationTaster14 were used to predict the effects of mutation on protein function and Phylop was utilized to examine vertebrate conservation.15 The protein expression, expressed in reads per kilobase of transcript per million reads mapped (RPKM) was collected from the GTEx Portal.16 Finally, transmission was confirmed by Sanger sequencing of amplicons derived from genomic DNA isolated from the peripheral blood of all family members.
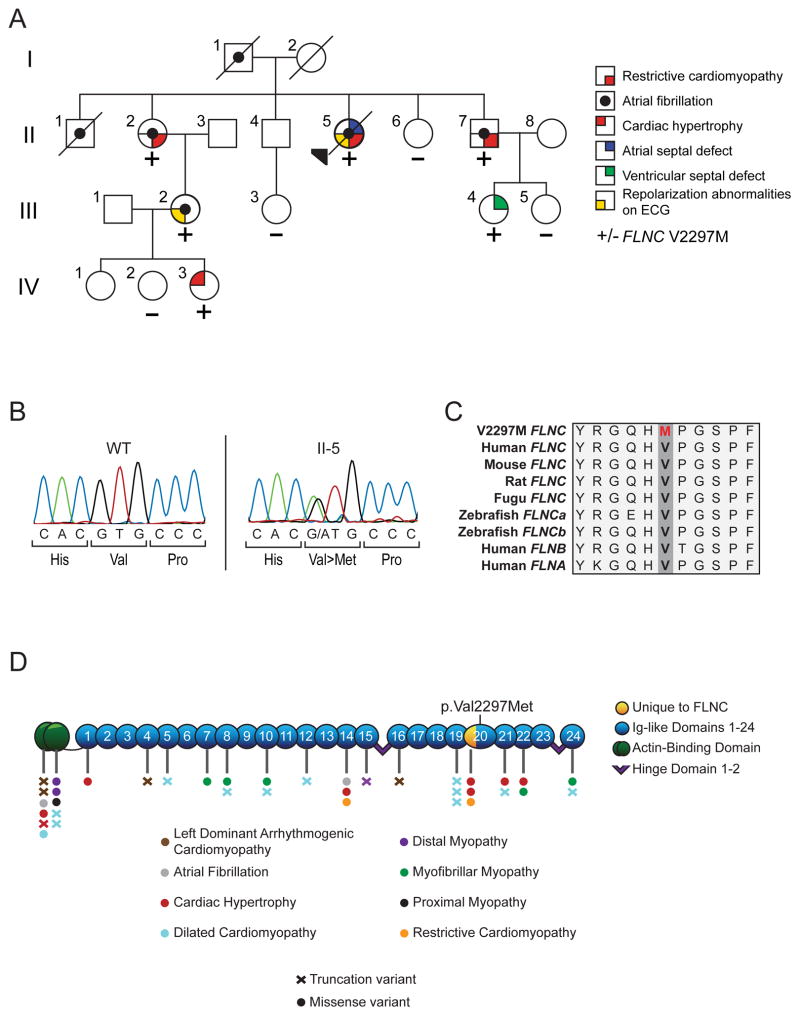
The conservation across species of FLNC flanking the identified p.V2297M mutation (Figure 1C) was accomplished using the ClustalW2 alignment tool. References sequences are as follows: Human FLNC-NP_001449.3, Mouse FLNC-NP_001074654.1, Rat FLNC-NP_001178791.1, Fugu FLNC-XP_011616004.1, Zebrafish FLNCa-XP_009296453.1, Zebrafish FLNCb-XP_009298482.1, Human FLNB-NP_001157789.1, and Human FLNA-NP_001447.2.
Figure 1. Transmission of the FLNC p.V2297M variant with cardiac phenotypes.
A: Pedigree detailing the spectrum of myocardial phenotypes observed in the family. +/− denotes carrier versus wildtype genotype at amino acid 2297. B: Chromatogram obtained from Sanger sequencing reaction of amplicon obtained from the genomic DNA of II-5. C: Amino acid sequence alignment of FLNC in vertebrate species. Red denotes the p.V2297M variant observed. D: Depiction of FLNC functional domains overlain with known variants and associated phenotypes.
Site Directed Mutagenesis
A full length FLNC clone harboring a C-term myc-DDK tag in vector pCMV6 (RC212462) was purchased from Origene (USA). Site directed mutagenesis to produce the p.V2297M mutation was accomplished using the Gibson Assembly system from New England Biolabs (USA). The primers used in these reactions are located in Supplemental Table 3. Sanger sequencing was used to confirm successful final assembly and to screen for unintended mutations.
Immunohistochemistry
Formalin-fixed explanted heart tissue sections from Individual II-5 were de-embedded and rehydrated, then subjected to a 30 minute steam chamber incubation in a solution of 10mM sodium citrate supplemented with 0.05% Tween20 for antigen retrieval. Samples were first labeled with primary antibodies against FLNC (HPA006135, Sigma-Aldrich, USA), MYH2 (sc-53096, Santa Cruz Biotechnology, USA), or α-actinin (ab9465, Abcam, USA) and then labeled with a fluorescent secondary antibody (anti-rabbit Alexa Fluor® 488 (A11008), anti-mouse Alexa Fluor® 546 (A11003) Life TechnologiesUSA). Samples were then mounted using VECTASHIELD hard-set mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (H-1200, Vector Laboratories, CA, USA) and imaged on a Zeiss 510 LSM using a 63X Zeiss Plan-APOCHROMAT 1.4NA oil immersion lens.
C2C12 cells were cultured in a 37°C incubator with 5% CO2 on sterilized glass coverslips coated with poly-L-lysine (P4707, Sigma-Aldrich, USA) according to conditions described by ATCC. At 75% confluency, cells were transfected with the wild-type FLNC or p.V2297M FLNC vectors using the Lipofectamine LTX kit (Life Technologies, CA, USA) according the manufacturer’s instructions. At 24 hours post-transfection cells were fixed using 3.5% paraformaldehyde and stained first with an anti-DDK antibody (TA50011, Origene, MD, USA), then a fluorescent secondary antibody (A11029, Life Technologies, USA). Cells were counterstained for F-actin using 0.02uM rhodamine-labeled phalloidin (Sigma-Aldrich, USA), then mounted and imaged as described above.
Contractile activity measurements in stem cell-derived cardiomyocytes
Human embryonic stem cells (hESCs) (WA07 (H7); WiCell, USA) were maintained and differentiated into a cardiomyocyte lineage by standard protocols known to produce ventricular cardiomyocytes with an immature phenotype.17,18 To introduce a single base pair mutation at Chr7: 128494628 (G>A) in hESCs, we designed a single guide RNA (sgRNA) targeting the vicinity of Chr7: 128494628 using the online CRISPR Design tool (http://crispr.mit.edu/) and cloned the sgRNA sequence into plasmid PX458 (Addgene, USA; 48138),19 which carries the nuclease spCas9 fused with an EGFP reporter. A single-stranded oligodeoxynucleotide (ssODN) was synthesized and used as a repair template for homology-directed repair. To initiate genome editing, approximately 8 × 106 hESCs were electroporated with the PX458-sgRNA construct (13 μg) and the ssODN (0.5 nmol). 48 hours after electroporation, hESCs positive for EGFP signal were sorted with a BD FACSAria II cell sorter and allowed for expansion into single cell-derived colonies. PCR amplification of the region surrounding Chr7: 128494628 followed by Sanger sequencing was performed to identify colonies with the expected mutation (G>A). Off target regions were identified using the CRISPR Design tool and are listed in Supplemental Table 4. The highest scoring coding off-target was present in FLNA on the X chromosome, which was Sanger sequenced in all clones with no observed mutation (Supplemental Figure 1A). The sequences of the sgRNA and ssODN are as follows:
sgRNA - TACCGTGGCCAGCACGTGCC
ssODN - CGCCTACAGCGTGCGCTTTGTGCCCCAGGAAATGGGGCCCCATACGGTCGCTGTCAAGTACCGTGGCCAGCACATGCCCGGCAGCCCCTTTCAGTTCACTGTGGGGCCGCTGGGTGAAGGTGGTGCCCACAAGGTGCGGGCCGGAGGCACAGGGCT
All phenotypic measurements were performed at 12 days post differentiation in three independent clones. Contractile measurements were conducted as previously described.20 Briefly, differentiated myocytes were plated at single cell density on a flexible substrate. Cells were field paced at 1hz using a C-Pace EP stimulator (Ion Optix Inc, USA). Images were acquired at approximately 90 fps on a Nikon A1R confocal microscope under DIC conditions. Pairwise similarity comparison was performed to identify frames in which maximal perturbation (contraction) was observed. Identified frames were used to measure the long axis of the myocyte during maximal contraction and relaxation.
Gene expression and protein analysis
Quantitative real-time PCR was performed using a BioRad CFX384 using the TaqMan™ Gene Expression Master Mix (ThermoFisher, USA). Taqman probes were purchased from ThermoFisher for FLNC (cat#: 4331182), and housekeeping genes GAPDH (cat#:4331182) and TBP (cat#:4331182). Relative expression was calculated using the ΔΔCt method.
Sub-cellular differential detergent fractionation was performed using protocols from Cold Spring Harbor,21 then quantified using the Pierce BCA Protein Assay Kit (ThermoFisher, USA). Total protein was harvested using Laemmli buffer, then sonicated using a Bioruptor Plus (Diagenode, USA). SDS-PAGE was performed using BioRad TGX gels, and transferred to nitrocellulose membrane using a Trans-Blot Turbo Transfer System (BioRad, USA). Equivalent loading was analyzed by staining with PonceauS (Sigma-Aldrich, USA) before probing with anti-FLNC (Sigma-Aldrich, USA, 1:2500) and anti-Rabbit-HRP (1:10,000) antibodies. Measurement of chemiluminescence was performed using the Clarity ECL Western Blotting Substrate (BioRad, USA) and imaged on an Amersham Imager 600 (GE Healthcare, USA).
Statistical analyses
Statistical analyses were conducted using Graphpad Prism 7 (GraphPad Software, Inc., USA). All datasets were tested for normality using D’Agostio & Pearson normality tests. Those with normal distributions were subjected to Student’s t-test, whereas those with non-normal distributions were subjected to non-parametric analyses using Mann-Whitney tests. P-values of less than 0.05 were considered significant.
Results
Clinical identification of familial RCM
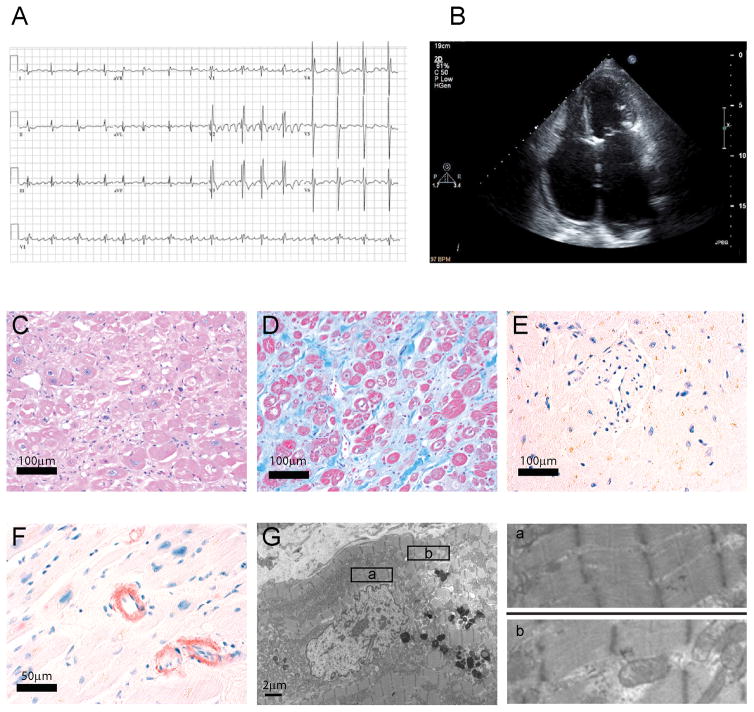
The proband in the family (II-5) presented with a severe clinical course of progressive heart failure that culminated in a cardiac transplant (Figure 1A, Figure 2). The explanted heart showed a moderate interstitial fibrosis and myocyte hypertrophy. Pathologic assessment also showed a mild amyloid deposition in only the atria (Figure 2C–G), findings consistent with previous studies on exclusively atrial amyloid deposition in patients with persistent atrial fibrillation.22 Since there was no evidence for storage disease or deposition disorder, the pathological findings were consistent with a primary RCM.
Figure 2. Clinical and pathological findings for patient II-5.
A. Electrocardiogram is notable for coarse atrial fibrillation, ventricular, hypertrophy and incomplete right bundle branch block pattern. B. Echocardiogram notable for massively enlarged atria with left atrial dimensions of 46×51×82mm. Histologically the right atrial myocardium demonstrated myocyte hypertrophy with enlarged nuclei on hematoxylin and eosin stain (C) and extensive fibrosis (blue) on trichrome stain (D). On Congo red stains, there was no amyloid in the left ventricle (E) but focal amyloid in the right atrium (F) indicating isolated atrial amyloid. By transmission electron microscopy of the left ventricle myocardium (G and enlargements in panels a, and b) there was no evidence of a storage disorder or appreciable alteration in sarcomeric architecture.
The patient’s sister (II-2) presented aged 51 with a pulmonary embolism and atrial fibrillation. Since then, she has had recurrent and highly symptomatic atrial fibrillation despite treatment with multiple antiarrhythmic medications including flecainide, propafenone, dofetilide and sotalol. She has also had more than 30 cardioversions, three pulmonary vein ablation procedures, and ultimately implantation of a permanent pacemaker. Over the ensuing years, she has also developed progressive symptoms of congestive heart failure so coronary disease was excluded by coronary angiography. Serial echocardiography revealed no cardiac hypertrophy or LV dilatation but progressive atrial dilation and a restrictive filling pattern using tissue Doppler imaging. At age 58, the patient suffered from two embolic strokes within 3 days prompting surgical amputation of the left atrial appendage. Pathological analysis revealed moderate to focally severe interstitial/replacement fibrosis and myocyte hypertrophy without evidence of storage disease or significant deposition disorder. In aggregate, her presentation was consistent with a diagnosis of RCM.
The daughter of patient II-2 was diagnosed with congestive heart failure and atrial fibrillation (III-2). Recently, an echocardiography of this patient’s 14-year old daughter (IV-3) revealed focal hypertrophy of the papillary muscles and a highly trabeculated ventricle. The patient’s father (I-1) and the oldest brother (II-1) were also reported to have had heart failure and a cardiomyopathy. They died at ages 42 and 25 of a stroke and sudden cardiac death, respectively; however, other electrocardiographic and echocardiographic data is unavailable on these two family members. Finally, another brother (II-7) was also diagnosed with RCM and atrial fibrillation. For purposes of genetic variant segregation with disease, we employed a tiered approach, where first only individuals with diagnosed RCM were considered affected, followed by a secondary hypothesis that the causative variant underlies a cardiac spectrum disorder which includes RCM. For this second hypothesis, all family members with documented cardiac abnormalities (II-2, II-5, II-7, III-2, IV-3) were considered affected for purposes of genetic analyses. Patient II-6 is 54 years of age, had no history of cardiac disease, a normal cardiac evaluation and was therefore considered unaffected. None of the affected family members exhibited an overt skeletal muscle myopathy. All other family members were considered to be of unknown status. Clinical characteristics for all individuals are listed in Table 1.
Table 1.
Clinical Characteristics of AF-523 Family Members
| ECG | Echocardiography | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | ID | Sex | Age | PR | QRS | QT | Other | LVEF | LA | LVID | RCM | FLNC mutation | Comment |
| I-1 | 523.01 | M | 58 | − | − | − | AF | − | − | − | NA | Stroke at age 42 | |
| I-2 | 523.02 | F | 72 | − | − | − | − | − | − | NA | No known cardiac disease | ||
| II-1 | 523.1 | M | 25 | − | − | − | AF | − | − | − | NA | SCD at age 25 | |
| II-2 | 329 | F | 51 | − | 83 | 402 | AF | 70 | 46 | 38 | + | + | Sick sinus syndrome, pacemaker, stroke |
| II-4 | 523.3 | M | 60 | − | − | − | − | − | − | NA | |||
| II-5 | 523.4 | F | 51 | − | 98 | 455 | AF, VT | 67 | 49 | 41 | + | + | Pacemaker, ASD, stroke |
| II-6 | 523.5 | F | 54 | − | − | − | − | − | − | − | |||
| II-7 | 523.6 | M | 44 | 238 | 104 | 410 | AF | 60 | 41 | 38 | + | + | Pulmonary hypertension, ICD |
| III-2 | 523.21 | F | 37 | 214 | 86 | 498 | AF, T wave inversions | 52 | 38 | 47 | ? | + | CHF at age |
| III-3 | 523.31 | − | − | − | − | − | − | − | |||||
| III-4 | 523.61 | F | 17 | 114 | 80 | 459 | 71 | 33 | 43 | + | VSD | ||
| III-5 | 523.62 | F | 15 | 144 | 76 | 457 | 66 | 34 | 48 | − | |||
| IV-1 | 523.211 | F | 20 | − | − | − | − | − | − | − | |||
| IV-2 | 523.212 | F | 19 | 118 | 66 | 427 | 62 | 28 | 39 | − | |||
| IV-3 | 523.213 | F | 14 | 146 | 78 | 412 | T wave inversions | 61 | − | 47 | + | Papillary muscle hypertrophy | |
AF, atrial fibrillation; VT, ventricular tachycardia; LVEF, left ventricular ejection fraction; LA, left atrial diameter; CM, cardiomyopathy; ICD, implanted cardioverter-defibrillator; CHF, congestive heart failure
ECG and echocardiography parameters were shown from the day of study enrollment
Identification of a FLNC mutation as a causative variant for familial RCM
To identify a mutation associated with RCM in this family, genomic DNA was isolated from peripheral blood samples of 5 family members with RCM and/or other cardiovascular defects and subjected to whole exome sequencing. We included the latter in order to address the hypothesis that the causative mutation might underlie a cardiac spectrum disorder. Exomes were sequenced in excess of 100X coverage, except individual II-2, which had average coverage of 48x due to the use of older enrichment and sequencing technologies. For individual II-2, 70.09% of the targeted regions reached 20X coverage, a number that was improved to an average of 98.17% in the remaining samples (Supplemental Table 5). An average of 20,566 variants were identified in each individual, of which ~9855 were missense, nonsense, stopgain or splice site variants (Supplemental Table 6). Of these variants, 34 were shared between the three individuals diagnosed with RCM, while only 11 were shared amongst the five sequenced family members. Within the three RCM affected individuals (II-2, II-5 and II-7), no variation in known cardiomyopathy genes, excluding FLNC p.V2297M, was uncovered (Supplemental Table 2).
Sanger sequencing was then performed for the 5 novel variants in all individuals in order to identify which transmitted with cardiac abnormalties. Of the three which transmit with cardiovascular abnormalities, only variants in PLOD3 and FLNC were predicted to be damaging by Polyphen-2. After examination of cardiac expression and vertebrate conservation, we identified a single candidate variant for further analysis in FLNC (c.6889 G>A, Figure 1B). FLNC p.V2297M is highly conserved with a PhyloP conservation score of 5.775 (Figure 1C) and cardiac expression of 264.1 RPKM in the left ventricle, both the highest values of all candidate variants. It was predicted to be probably damaging according to PolyPhen-2, and it was the only variant shared among affected family members that was not present in the ExAC or EVS databases. We next wished to address the possibility that independent variants or modifiers might underlie the congenital manifestations in this family. However, no pathogenic variants in genes known to underlie congenital cardiac disorders were uncovered in individual II-4.
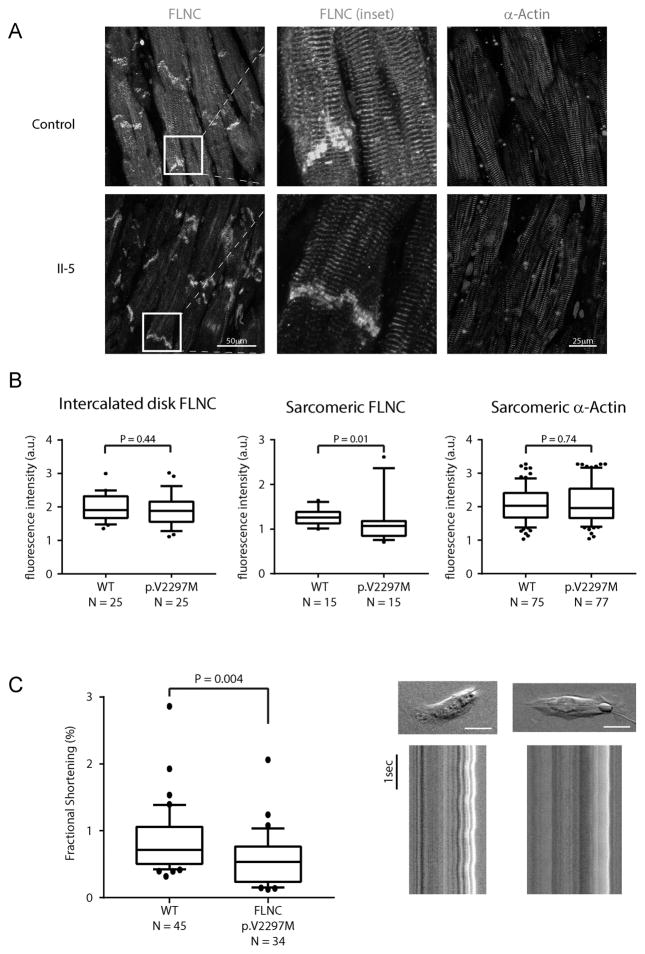
Formalin-fixed, paraffin-embedded ventricular samples from individual II-5 and a RCM-free control were sectioned and labeled for FLNC and the sarcomeric markers α-sarcomeric actin (Figure 3A). In the control samples, FLNC displayed a typical subcellular distribution, namely a high level of association with the intercalated disks and with the z-disk region within individual sarcomeres. In contrast, although FLNC continues to be localized to intercalated disk in the samples from individual II-5, there is a markedly diminished association of FLNC with the sarcomeric architecture (Figure 3A, B). To investigate this finding in vitro, a full length clone of p.V2297M was generated by site directed mutagenesis of the wild type FLNC clone RC212462. To analyze localization patterns, we transfected C2C12 myoblasts with overexpression constructs and at 24 hours post transfection we performed fluorescence immunolabeling by detecting the FLAG epitope and F-actin (Supplemental Figure 2). Although undifferentiated C2C12 myoblasts do not form sarcomeres, both wild-type and mutant FLNC localized to the stress fiber architecture equivalently, the major thin filament cytoskeletal component in non-myocytes. Given this colocalization, it appears that p.V2297M retains its actin binding ability conferred by the N-terminal actin binding domain, suggesting an alternative mechanism for the perturbed localization in the sarcomeres of mature myocardium.
Figure 3. Cellular consequences of FLNC p.V2297M variant.
A: Localization of FLNC (green) and α-MHC (red) in ventricular free wall tissue obtained from a control sample and in the failing heart of II-5. Images were obtained from independent tissue sections B: Quantification of fluorescence intensity for intercalated disk resident FLNC, sarcomeric FLNC, and sarcomeric α-actin. Images from all samples were obtained with equivalent acquisition conditions. N represents number of independent intercalated disks or myofibrils examined. C: Left panel: Comparison of fractional shortening measured from singularized stem cell derived cardiomyocytes. N represents the combined number of cells analyzed in three independent experiments. Error bars represent the SEM. Right panel: Representative images of isolated cardiomyocytes. Red line represents the major axis for fractional shortening measurement. Scale bar represents 20μm. Lower images represent line scans of the major axes for wild-type and mutant cardiomyocytes displayed over time.
We then utilized CRISPR-Cas9 combined with homology driven repair to generate bi-allelic insertions of p.V2297M in human embryonic stem cells (Supplemental Figure 1A). We differentiated these cells toward a ventricular cardiomyocyte lineage (MYL3 expression, ~2070 RPKM) and assayed the contractile function of field-paced myocytes plated on a pliable substrate. FLNC- p.V2297M myocytes exhibited significantly reduced contractility as measured by fractional shortening (p=0.004) suggesting an impairment of the contractile apparatus due to this mutation (Figure 3C). Expression of FLNC was equivalent in both wild-type and p.V2297M cardiomyocytes, with no differences in protein expression level or subcellular fraction (Supplemental Figure 2B,C).
Discussion
In the present study, we have identified a novel mutation in FLNC, p.V2297M, as a potential cause of RCM in a family. This variant is absent from control datasets and cosegregates with disease in a family with strong evidence for a single genetic etiology. Data from a stem cell derived cardiomyocyte model and immunohistochemical studies in patient samples provide supportive data regarding the damaging effect of p.V2297M on cardiomyocyte function. As mutations in FLNC are known to cause cardiomyopathies, and computational evidence supports a deleterious effect, p.V2297M should be classified as a pathogenic variant according to the guidelines of the American College of Medical Genetics and Genomics.23
Filamin C in the context of other known causes of RCM
We have identified a novel cause of RCM due to a mutation in Filamin C. Previous identification of RCM-causing mutations have centered on those in the contractile apparatus of the cardiac muscle. These include the myosin heavy chain 7 motor subunit, the contractile anchor cardiac alpha actin, the regulatory subunits troponin I and troponin T, and the scaffolding protein titin.
FLNC encodes Filamin C, one of three filamin family members found in humans. While similar to previously identified RCM candidate proteins as a component of the sarcomeric architecture, the function of Filamin C is distinct, as it acts as a scaffolding and signaling protein instead of directly governing contractile force of the myocyte. All three family members of filamin have a highly conserved N-terminal actin binding domain followed by a series of Ig repeats and C-terminal domains that enable association with integrins and isoform-specific membrane receptors. Expression of the 291kD FLNC protein, in contrast to the ubiquitous expression of FLNB and endothelial enrichment of FLNA, is most prevalent in the striated muscle tissues. In that context, FLNC dimers play a critical role for the linkage of the sarcomeric architecture to the mechanosensory system mediated by integrin based cell-cell or cell-extracellular matrix attachments.24 In addition to this role, FLNC also localizes to the intercalated disk, the structure found at the border of adjacent myocytes, where it is thought to aid in cell migration and cytoskeletal remodeling. To perform these tasks, Filamin C has been demonstrated to bind with a multitude of other sarcomeric and intercalated disk components. V2297 is located in domain 20, part of the carboxy-terminal structure of FLNC which has been defined as the interaction site for Xin25, KY26, myotilin27, and multiple cadherins.28,29 Although the disruptions in the amino-terminal actin binding domains that lead to the skeletal muscle-centric myofibrillar and distal myopathy are mechanistically understood, the list of interactors in the carboxy-terminal region is likely incomplete. While identification of the interactome is technically challenging given the density of protein complexes in the sarcomere and the detergent-insoluble nature of many protein components, full identification of FLNC protein-protein interactions, and how known variation affects these interactions, should be the focus of future study. Until then, the biological role for the carboxy-terminal interactions, as well as the functional consequences of their disruption, remains largely unclear.
Role of Filamin C in the heart
Mutations in FLNC were first reported as a cause for myofibrillar familial myopathies30,31 and distal myopathies,32,33 which are both defects in the structure or function of skeletal muscle tissue. None of these reports indicated any observed cardiac phenotype. More recently, the impact of FLNC variation on the heart has been recognized, including reports on dilated cardiomyopathy,34–36 hypertrophic cardiomyopathy,37,38 atrial fibrillation,37,39 and restrictive cardiomyopathy.9 The phenotypic spectrum associated with FLNC variation is intriguing, suggesting that variants act through divergent and tissue specific manner and/or that variants are modified by other genetic factors present in the given family. Although sequencing results suggested some possible genetic modifiers in the present study, including a variant in TRPV4,40 a gene which increases in expression with atrial fibrillation, a concrete link between modifiers and phenotype would require a larger sample size or greatly expanded functional modeling. In the present study, we support the role for FLNC variation in RCM, while adding evidence that FLNC variation should be considered in manifestations of congenital heart defects. Although most reported variants in FLNC, as well as those reported for FLNA and FLNB, are in distinct domains of the peptide than p.V2297M, many previously reported mutations result in a mislocalization or aggregation of the filamin protein.39,41 A comprehensive listing of known FLNC variation in skeletal and cardiac myopathies is displayed in Figure 1D and listed in Supplemental Table 7. Animal models of filamin deficiency also exhibit both skeletal and cardiac phenotypes.42–44
To address this possibility in the present report, we examined the subcellular distribution of FLNC in ventricular tissue obtained from the failing heart of individual II-5 following transplant. As evidenced in patient samples harboring the p.V2297M FLNC mutation, the Filamin C protein displayed diminished sarcomeric localization when compared to controls. This mislocalization does not appear to be due to aggregation of FLNC, as has been reported for HCM, or altered actin binding capability, since localization to F-actin rich stress fibers was observed in our exogenous overexpression system. Instead, it is likely that altered interactions between mutant FLNC with additional sarcomeric components may underlie the onset of RCM. Uncovering these altered interactions may be critical to understanding the altered contractile states found both clinically and in the stem-cell derived cardiomyocytes. Alternatively, the recognition and rapid degradation of p.V2297M FLNC could lead to a haploinsufficiency-mediated phenotype similar to those observed in DCM-mediating truncation mutations. Ideally, primary tissue samples would provide conclusive information regarding transcript and protein levels, but tissue samples appropriate for these measurements are not available. Examination of expression in stem cell derived cardiomyocyte systems revealed equivalent total protein levels, suggesting that loss-of-function and not haploinsufficiency at the production level may underlie the pathogenic nature of this variant. There is a definitive reduction in fractional shortening in the ES-CM model which can be the result of reduced contractile activity, or impaired sarcomeric relaxation. Given the inhibited diastolic filling characteristic of RCM, the second of these is the presumed mechanism in the present study. In sum, given the altered localization of FLNC in patient samples and perturbed contractile function of p.V2297M cardiomyocytes, we would classify this as a loss-of-function variant of unclear molecular mechanism. The further definition of the mechanism underlying the p.V2297M-mediated myopathy will likely prove critical to translating these findings into future clinical interventions for RCM or other FLNC-related myopathies.
Clinical utility of FLNC mutations in RCM
The diagnosis of primary RCM is still a challenge since there is an overlap of clinical and pathological findings between other cardiac diseases that makes it difficult to specifically differentiate RCM from other cardiomyopathies, pericarditis, or storage diseases.1 Given that there are a series of known causes for RCM, (5, 26) the latest HRS/EHRA consensus statement from 2011 recommends consideration of genetic testing for patients in whom a cardiologist has established a clinical index of suspicion for RCM based on examination of the patient’s clinical history, family history, and electrographic/echocardiographic phenotype (Class IIb).45 We believe that our findings provide an additional gene for inclusion on future RCM genetic testing panels.
Conclusion
We have identified a mutation in a striated muscle specific actin-binding protein, Filamin C, as the cause of pleiotropic syndrome of familial RCM, atrial fibrillation, and congenital heart defects. Our findings further expand upon the filaminopathies and expand on the role for Filamin C in cardiac development and function.
Supplementary Material
Clinical Perspective.
Restrictive cardiomyopathy (RCM) is a rare and severe cardiomyopathy associated with a poor long-term prognosis. Although not traditionally considered to be an inherited disorder, recent studies have identified a series of genetic causes for RCM. In the present work, we utilized whole exome sequencing to identify a pathogenic mutation in a family with a spectrum of cardiovascular disorders including RCM, atrial fibrillation and congenital heart defects. The missense variant resides in the FLNC gene, which encodes a critical component of the contractile apparatus of the cardiomyocyte. Functional modeling of the FLNC mutation in stem cell-derived cardiomyocytes revealed impaired contractility, a finding which supports the role of this variant in RCM. The present study supports the role for expanded genetic testing of FLNC in RCM and provides additional insights into the pathogenesis of this rare cardiomyopathy.
Acknowledgments
Funding sources
Dr. Tucker was supported by an institutional training grant from the National Institutes of Health (T32HL007208) and by support from the George L. Nardi, MD Memorial Research Fund. Dr. Clauss was supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme (PIOF-GA-2012-328352) and by the German Centre for Cardiovascular Research (DZHK). This work was supported by grants from the National Institutes of Health to Dr. Ellinor (1RO1HL092577, R01HL128914, K24HL105780). Dr. Ellinor is also supported by an Established Investigator Award from the American Heart Association (13EIA14220013) and by the Fondation Leducq (14CVD01). Dr. Lubitz is supported by grants from the NIH (K23HL114724) and by a Doris Duke Charitable Foundation Clinical Scientist Development Award (2014105).
Footnotes
Conflict of Interest Disclosures
Dr. Ellinor is a principal investigator on a grant from Bayer Healthcare to study the genetic architecture of atrial fibrillation.
References
- 1.Mogensen J, Arbustini E. Restrictive cardiomyopathy. Curr Opin Cardiol. 2009;24:214–20. doi: 10.1097/hco.0b013e32832a1d2e. [DOI] [PubMed] [Google Scholar]
- 2.Goldfarb LG, Park KY, Cervenáková L, Gorokhova S, Lee HS, Vasconcelos O, et al. Missense mutations in desmin associated with familial cardiac and skeletal myopathy. Nat Genet. 1998;19:402–3. doi: 10.1038/1300. [DOI] [PubMed] [Google Scholar]
- 3.Kaski JP, Syrris P, Burch M, Tomé-Esteban M-T, Fenton M, Christiansen M, et al. Idiopathic restrictive cardiomyopathy in children is caused by mutations in cardiac sarcomere protein genes. Heart. 2008;94:1478–84. doi: 10.1136/hrt.2007.134684. [DOI] [PubMed] [Google Scholar]
- 4.Hoedemaekers YM, Caliskan K, Majoor-Krakauer D, van de Laar I, Michels M, Witsenburg M, et al. Cardiac beta-myosin heavy chain defects in two families with non-compaction cardiomyopathy: linking non-compaction to hypertrophic, restrictive, and dilated cardiomyopathies. Eur Heart J. 2007;28:2732–7. doi: 10.1093/eurheartj/ehm429. [DOI] [PubMed] [Google Scholar]
- 5.Mogensen J, Kubo T, Duque M, Uribe W, Shaw A, Murphy R, et al. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111:209–16. doi: 10.1172/JCI16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menon SC, Michels VV, Pellikka PA, Ballew JD, Karst ML, Herron KJ, et al. Cardiac troponin T mutation in familial cardiomyopathy with variable remodeling and restrictive physiology. Clin Genet. 2008;74:445–54. doi: 10.1111/j.1399-0004.2008.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peled Y, Gramlich M, Yoskovitz G, Feinberg MS, Afek A, Polak-Charcon S, et al. Titin mutation in familial restrictive cardiomyopathy. Int J Cardiol. 2014;171:24–30. doi: 10.1016/j.ijcard.2013.11.037. [DOI] [PubMed] [Google Scholar]
- 8.Huby A-C, Mendsaikhan U, Takagi K, Martherus R, Wansapura J, Gong N, et al. Disturbance in Z-disk mechanosensitive proteins induced by a persistent mutant myopalladin causes familial restrictive cardiomyopathy. J Am Coll Cardiol. 2014;64:2765–76. doi: 10.1016/j.jacc.2014.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodehl A, Ferrier RA, Hamilton SJ, Greenway SC, Brundler MA, Yu W, et al. Mutations in FLNC are Associated with Familial Restrictive Cardiomyopathy. Hum Mutat. 2016;37:269–279. doi: 10.1002/humu.22942. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 15.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye J, Tucker NR, Weng L-C, Clauss S, Lubitz SA, Ellinor PT. A Functional Variant Associated with Atrial Fibrillation Regulates PITX2c Expression through TFAP2a. Am J Hum Genet. 2016;99:1281–1291. doi: 10.1016/j.ajhg.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, et al. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell. 2016;39:480–490. doi: 10.1016/j.devcel.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kijlstra JD, Hu D, Mittal N, Kausel E, van der Meer P, Garakani A, et al. Integrated Analysis of Contractile Kinetics, Force Generation, and Electrical Activity in Single Human Stem Cell-Derived Cardiomyocytes. Stem Cell Reports. 2015;5:1226–1238. doi: 10.1016/j.stemcr.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsby M, Makowski G. Differential detergent fractionation of eukaryotic cells. Cold Spring Harb Protoc. 2011;2011 doi: 10.1101/pdb.prot5592. prot5592. [DOI] [PubMed] [Google Scholar]
- 22.Röcken C, Peters B, Juenemann G, Saeger W, Klein HU, Huth C, et al. Atrial amyloidosis: an arrhythmogenic substrate for persistent atrial fibrillation. Circulation. 2002;106:2091–7. doi: 10.1161/01.cir.0000034511.06350.df. [DOI] [PubMed] [Google Scholar]
- 23.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou A-X, Hartwig JH, Akyürek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113–23. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 25.van der Ven PFM, Ehler E, Vakeel P, Eulitz S, Schenk JA, Milting H, et al. Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin c and Mena/VASP. Exp Cell Res. 2006;312:2154–67. doi: 10.1016/j.yexcr.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Beatham J, Romero R, Townsend SKM, Hacker T, van der Ven PFM, Blanco G. Filamin C interacts with the muscular dystrophy KY protein and is abnormally distributed in mouse KY deficient muscle fibres. Hum Mol Genet. 2004;13:2863–74. doi: 10.1093/hmg/ddh308. [DOI] [PubMed] [Google Scholar]
- 27.van der Ven PF, Wiesner S, Salmikangas P, Auerbach D, Himmel M, Kempa S, et al. Indications for a novel muscular dystrophy pathway. gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J Cell Biol. 2000;151:235–48. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson TG, Chan YM, Hack AA, Brosius M, Rajala M, Lidov HG, et al. Filamin 2 (FLN2): A muscle-specific sarcoglycan interacting protein. J Cell Biol. 2000;148:115–26. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guyon JR, Kudryashova E, Potts A, Dalkilic I, Brosius MA, Thompson TG, et al. Calpain 3 cleaves filamin C and regulates its ability to interact with gamma- and delta-sarcoglycans. Muscle Nerve. 2003;28:472–83. doi: 10.1002/mus.10465. [DOI] [PubMed] [Google Scholar]
- 30.Shatunov A, Olivé M, Odgerel Z, Stadelmann-Nessler C, Irlbacher K, van Landeghem F, et al. In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy. Eur J Hum Genet. 2009;17:656–63. doi: 10.1038/ejhg.2008.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vorgerd M, van der Ven PFMM, Bruchertseifer V, Löwe T, Kley RA, Schröder R, et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams DR, Reardon K, Roberts L, Dennet X, Duff R, Laing NG, et al. A new dominant distal myopathy affecting posterior leg and anterior upper limb muscles. Neurology. 2005;64:1245–54. doi: 10.1212/01.WNL.0000156524.95261.B9. [DOI] [PubMed] [Google Scholar]
- 33.Duff RM, Tay V, Hackman P, Ravenscroft G, McLean C, Kennedy P, et al. Mutations in the N-terminal actin-binding domain of filamin C cause a distal myopathy. Am J Hum Genet. 2011;88:729–40. doi: 10.1016/j.ajhg.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinstein E, Gutierrez-Fernandez A, Tzur S, Bormans C, Marcu S, Tayeb-Fligelman E, et al. Congenital dilated cardiomyopathy caused by biallelic mutations in Filamin C. Eur J Hum Genet. 2016;24:1792–1796. doi: 10.1038/ejhg.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Begay RL, Tharp CA, Martin A, Graw SL, Sinagra G, Miani D, et al. FLNC Gene Splice Mutations Cause Dilated Cardiomyopathy. JACC Basic to Transl Sci. 2016;1:344–359. doi: 10.1016/j.jacbts.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortiz-Genga MFMFMF, Cuenca SS, Dal Ferro M, Zorio E, Salgado-Aranda R, Climent V, et al. Truncating FLNC Mutations Are Associated With High-Risk Dilated and Arrhythmogenic Cardiomyopathies. J Am Coll Cardiol. 2016;68:2440–2451. doi: 10.1016/j.jacc.2016.09.927. [DOI] [PubMed] [Google Scholar]
- 37.Gómez J, Lorca R, Reguero JR, Morís C, Martín M, Tranche S, et al. Screening of the Filamin C Gene in a Large Cohort of Hypertrophic Cardiomyopathy Patients. Circ Cardiovasc Genet. 2017;10:e001584. doi: 10.1161/CIRCGENETICS.116.001584. [DOI] [PubMed] [Google Scholar]
- 38.Valdés-Mas R, Gutiérrez-Fernández A, Gómez J, Coto E, Astudillo A, Puente DA, et al. Mutations in filamin C cause a new form of familial hypertrophic cardiomyopathy. Nat Commun. 2014;5:5326. doi: 10.1038/ncomms6326. [DOI] [PubMed] [Google Scholar]
- 39.Valdés-Mas R, Gutiérrez-Fernández A, Gómez J, Coto E, Astudillo A, Puente DA, et al. Mutations in filamin C cause a new form of familial hypertrophic cardiomyopathy. Nat Commun. 2014;5:5326. doi: 10.1038/ncomms6326. [DOI] [PubMed] [Google Scholar]
- 40.Düzen IV, Yavuz F, Vuruskan E, Saracoglu E, Poyraz F, Göksülük H, et al. Leukocyte TRP channel gene expressions in patients with non-valvular atrial fibrillation. Sci Rep. 2017;7:9272. doi: 10.1038/s41598-017-10039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Löwe T, Kley RA, van der Ven PFM, Himmel M, Huebner A, Vorgerd M, et al. The pathomechanism of filaminopathy: altered biochemical properties explain the cellular phenotype of a protein aggregation myopathy. Hum Mol Genet. 2007;16:1351–8. doi: 10.1093/hmg/ddm085. [DOI] [PubMed] [Google Scholar]
- 42.Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006;26:6522–34. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, et al. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci U S A. 2006;103:19836–41. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–67. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- 45.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1308–39. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.