Abstract
To adapt to changing environments, bacteria have evolved numerous pathways that activate stress response genes. In Gram-positive bacteria, the stressosome, a cytoplasmic complex, relays external cues and activates the sigma B regulon. The stressosome is structurally well-characterized in Bacillus, but how it senses stress remains elusive. Here, we report a genome-wide N-terminomic approach in Listeria that strikingly led to the discovery of 19 internal translation initiation sites and 6 miniproteins, among which one, Prli42, is conserved in Firmicutes. Prli42 is membrane-anchored and interacts with orthologues of Bacillus stressosome components. We reconstituted the Listeria stressosome in vitro and visualized its supramolecular structure by electron microscopy. Analysis of a series of Prli42 mutants demonstrated that Prli42 is important for sigma B activation, bacterial growth following oxidative stress and for survival in macrophages. Taken together, our N-terminonic approach unveiled Prli42 as a long-sought link between stress and the stressosome.
Listeria monocytogenes is a human pathogen that has emerged as a model organism in infection biology1. The genome of L. monocytogenes strain EGD-e was sequenced in 2001 and predicted to encode 2846 open reading frames (ORFs)2. Tiling array-based transcriptomic analyses then explored bacterial transcription3,4, and RNA-sequencing was used to generate genome-wide transcription start (TSS) and termination site (TTS) maps5,6, making this model bacterium ideally suited for further exploration of the regulation of gene expression, at the level of translation. Ribosome profiling in Escherichia coli and Bacillus subtilis recently revealed a wealth of information about translational regulation, including translational pausing at Shine–Dalgarno (SD)-like sequences7. While pause sites can be differentiated from internal translation initiation sites (TISs) in mammalian cells using pulse-chase experiments8, it has been challenging to do so with precision in bacteria9,10. In contrast, N-terminal proteomics-based approaches can unambiguously detect bacterial TISs on a genome-wide scale. N-terminal COFRADIC (combined fractional diagonal chromatography) is one such method that isolates N-terminal peptides via two sequential chromatographic separations11–14. Interestingly, in prokaryotes, N-terminal peptides are formylated and this modification can be exploited to identify TISs (ref. 15).
Here, we report the first global TIS map of L. monocytogenes using an N-terminomics-based proteogenomic approach that combines COFRADIC and treatment with a peptide deformylase (PDF) inhibitor, actinonin16 (Fig. 1a). This genome-wide map yielded 62% genome coverage of predicted TIS and led to unexpected discoveries, such as previously unidentified TIS including 19 internal TIS and 6 miniproteins. We characterized one of these miniproteins, Prli42, in detail, as a tail-anchored membrane protein that relays oxidative stress signals to the stressosome to activate the general stress-sensing pathway, the sigma B regulon.
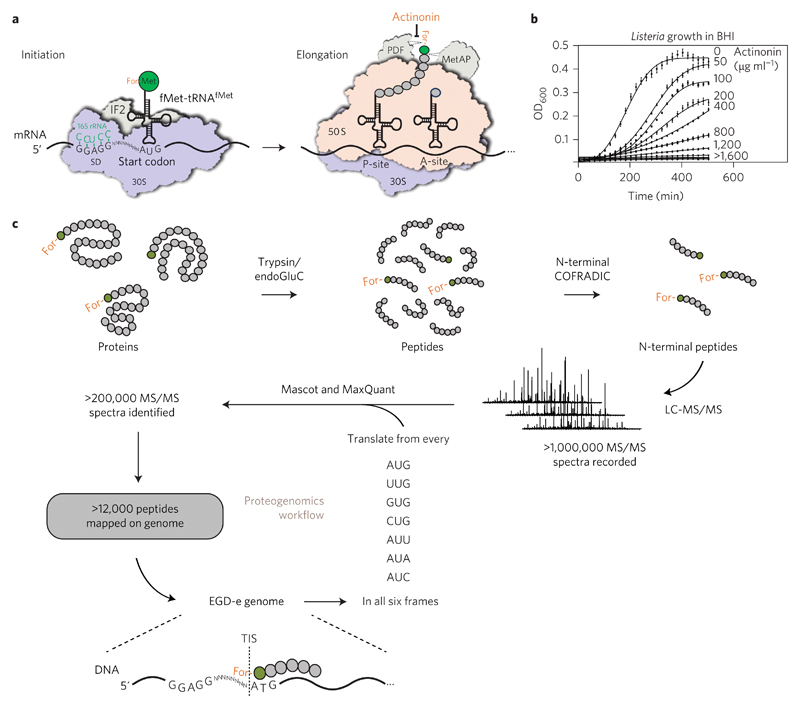
Figure 1. A proteogenomic approach to map translation initiation sites (TISs).
a, Schematic representation of bacterial protein translation. b, Dose-dependent Listeria growth inhibition by actinonin. Values are expressed as mean ± s.e.m. of three replicates from one representative experiment. c, Schematic representation of our proteogenomic approach. L. monocytogenes EGD-e was grown in BHI medium under different conditions and actinonin (50 μg ml−1) was added for 90 min before collecting the bacteria. Extracted proteins were digested with trypsin or endoGluC and (formylated) N-terminal peptides were isolated by COFRADIC before LC-MS/MS analysis. Over 1,000,000 MS/MS spectra were recorded and searched against a database generated by six-frame translation of the EGD-e genome sequence from every possible start codon. Two different search algorithms—Mascot and MaxQuant—were used to identify more than 12,000 peptides that were mapped on the Listeria genome, leading to the identification of TISs at a single-codon resolution.
Results
A proteogenomic approach to map TISs in Listeria
To map the translational landscape of L. monocytogenes strain EGD-e, we used N-terminal COFRADIC followed by mapping of the identified peptide sequences onto the Listeria genome to locate the corresponding TISs (Fig. 1). To this end, we first created a protein sequence database by translating the L. monocytogenes EGD-e genome in all six reading frames. This allowed us to work independently of the original genome annotation, a requirement for the identification of novel ORFs. We grew Listeria in exponential phase at 20 °C and 37 °C and in stationary phase at 37 °C. We added the PDF inhibitor actinonin to all three conditions at sub-inhibitory concentrations to increase TIS identification (Fig. 1b), and compared identified TISs with an untreated control (37 °C, exponential phase). Equal quantities of bacteria from the three growth conditions were mixed, proteins were extracted and, after blocking free NH2, digested into peptides with either trypsin or endoproteinase GluC (endoGluC). Due to their different cleavage specificities (trypsin after K/R or endoGluC after E/D), these enzymes generate complementary N-terminal peptides and thus increased coverage of our N-terminome (Supplementary Fig. 1a). We then isolated formylated peptides using COFRADIC and performed liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. We identified the spectra using two different algorithms (Mascot, from Matrix Science, and MaxQuant17) (Fig. 1c). Based on published data3,18, we predicted that more than 90% of all previously annotated Listeria proteins should be expressed in the growth conditions used, while 231 proteins would not be expressed (Supplementary Table 1). We estimated that 314 of the 2,846 predicted Listeria proteins would not be detected due to peptides that are too short or too long for mass-spectrometry identification (Supplementary Fig. 1a and Supplementary Table 1). Furthermore, protein secretion leads to removal of the N-terminal signal sequence, thereby preventing the detection of N-terminal peptides for 309 predicted secreted proteins (Supplementary Table 1 and ref. 19). We therefore estimated that 2,109 out of 2,846 annotated Listeria TISs (74%) could theoretically be detected by our approach.
Over 1,000,000 MS/MS spectra were recorded in over 200 LC-MS/MS runs, leading to the identification of 12,000 unique peptides that were mapped onto the genome (Fig. 1c). Actinonin treatment resulted in a sevenfold increase (20.5%) in peptides starting with a formyl-Meti, compared to control (2.8% formyl-Meti peptides). We identified 1,401 TISs, which we represented visually on a publically available whole-genome browser (http://nterm.listeriomics.pasteur.fr) that integrates our proteogenomic data with all previous transcriptomic data from tiling arrays and RNA-Seq3,5,6 (Fig. 2a).
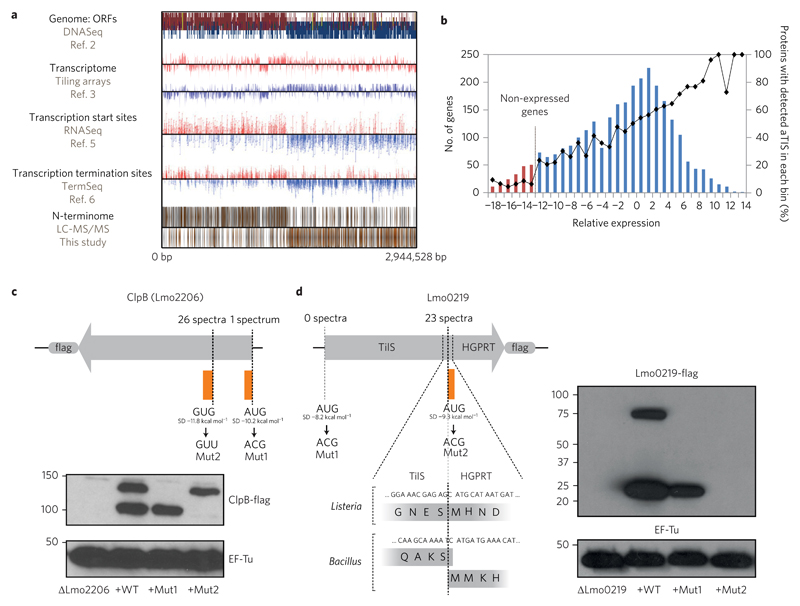
Figure 2. An integrated map of Listeria TISs and deviations from the current genome annotation.
a, Mapped N-terminal peptides are visually represented on a publically available browser (http://nterm.listeriomics.pasteur.fr). b, Correlation of the N-terminomics data with previous transcriptomics data. The histogram shows summed expression values for each gene measured by tiling arrays under the growth conditions that we analysed. For each bin the percentage of proteins with detected aTIS was calculated and plotted over the histogram. c, Validation of identified internal TISs (iTISs) in ClpB through complementation of L. monocytogenes LO28 strain deficient for ClpB with C-terminally flag-tagged substitution variants of the protein (in either start codon) by immunoblot. d, Validation of identified iTISs in Lmo0219 by ectopic expression of C-terminally flag-tagged substitution variants of Lmo0219 (in either start codon) by immunoblot. The elongation factor EF-Tu was used as loading control in both immunoblots. Immunoblots in c and d are representative of two independent experiments.
Divergence from previously annotated TISs
The majority of detected N-terminal peptides correspond to previously annotated TISs (aTISs)2. Our data validated predicted start codons for 1,322 L. monocytogenes ORFs, which are evenly spread across the genome and not enriched for particular start codons or Shine–Dalgarno (SD) sequences (Supplementary Fig. 1b,c and Supplementary Table 2). Furthermore, TIS abundance correlated well with gene transcription data, and we identified primarily cytoplasmic and membrane proteins (Fig. 2b, Supplementary Fig. 1d and Supplementary Table 1)19. We identified 72 genes with potential leaderless transcripts (TSS located fewer than ten nucleotides upstream from the observed TIS5, Supplementary Table 3), 13 of which lacked a second TIS. Leaderless transcripts can mediate a rapid stress response20 and, interestingly, two of the genes we found with putative leaderless transcripts play a role in sigma B signalling (for example, regulators of sigma B RsbU and RsbV). Our approach also identified small deviations from and several errors in previous annotation (detailed in Supplementary Fig. 2 and Supplementary Tables 4–6).
Detection of internal TISs (iTISs)
We identified 19 N-terminal peptides that fell within annotated ORFs revealing in-frame translation of shorter protein forms (Supplementary Table 7)5. We identified iTISs in aspartokinase II (Lmo1235)21 and the heat shock protein ClpB (LmO2206)22, proteins previously known to have two forms in other bacterial species. In E. coli, the long and short isoforms of ClpB cooperate for efficient protein disaggregation23. To confirm the iTIS in Listeria ClpB, we complemented a ΔclpB strain with a ClpB-flag. Immunoblotting validated the presence of two isoforms, one long and one short. We then mutated the annotated AUG or the internal GUG start codons of ClpB. In these mutants, only the short- or long-form protein is produced, respectively, demonstrating that internal translation of ClpB occurs in Listeria (Fig. 2c).
Another iTIS occurs in the bifunctional protein Lmo0219, which seems to be a Listeria-specific gene fusion. The N-terminal portion encodes a putative tRNAIle lysidine synthetase (TilS), while the C-terminal portion may function as a hypoxanthine-guanine phosphoribosyltransferase (HGPRT)24. In Bacillus, these two enzymes form a complex and are produced from distinct genes with overlapping stop and start codons (Fig. 2d). In Listeria, however, the ORFs have fused in-frame with an iTIS to produce HGPRT. Interestingly, when we mutated the internal start codon, we could not isolate clones that expressed the full-length protein. Thus, loss of HGPRT could destabilize the TilS-HGPRT fusion protein. More experiments are required to determine if this is the case (Fig. 2d). Taken together, our data suggest that alternative translation initiation events may be more common in bacteria than previously accepted.
Discovery of Listeria miniproteins
The initial annotation of the Listeria genome excluded ORFs of fewer than 40 amino acids2. This strategy reduced the number of incorrectly annotated proteins, but probably excluded several important small protein-coding genes. Here, we identified six miniproteins that were previously annotated either as small RNAs (rli24, rli41 and rli42) or 5′ untranslated regions (5′ UTRs of lmo0669 and lmo1064) (Supplementary Table 8). Their predicted three-dimensional structure and topology were determined (Supplementary Fig. 3). Two of the small ORFs were predicted to be encoded by the small RNA rli41, which probably functions as a small bicistronic mRNA (ref. 3). Prli42 is encoded by the previously annotated small RNA rli42 and is described below. Prli24, another miniprotein, is encoded by rli24, a small RNA upstream of a hypothetical RNA ligase gene (lmo0257). We identified a 35-amino-acid miniprotein in the 5′ UTR of Listeria corA (lmo1064). Interestingly, ribosome profiling revealed a similar ORF in the 5′ UTR of corA in E. coli25. Finally, we detected a small ORF in the 5′ UTR of a hypothetical oxidoreductase (Lmo0669).
Miniprotein Prli42 is conserved and interacts with Listeria orthologues of components from the stressosome
Prli42 is a 31-amino-acid protein, and computational analysis using the TOPCONS software26 suggests that it is a tail-anchored membrane protein (Supplementary Fig. 3). When we compiled a database of all potential small bacterial ORFs between 20 and 60 amino acids and performed a blastP search on L. monocytogenes EGD-e Prli42, we identified orthologous sequences of Prli42 in many other Firmicutes, including Bacilli and Staphylococci species (Fig. 3a and Supplementary Table 9). To characterize Prli42 we first sought interaction partners of the flag-tagged protein by co-immunoprecipitation followed by LC-MS/MS analysis. Interestingly, we identified RsbR (Lmo0889), RsbS (Lmo0890) and three RsbR-like proteins (Lmo0799, Lmo0161 and Lmo1642) as interaction partners of Prli42 (Fig. 3b and Supplementary Table 10) in two independent analyses. We identified other Prli42 partners as well, namely t-RNA synthetases and hypothetical proteins (Fig. 3b and Supplementary Table 10). RsbR (regulator of sigma B protein) and RsbS are components of the stressosome, a supramolecular complex involved in bacterial stress signalling and activation of the general stress factor, sigma B, in Bacillus. Interestingly, nearly three-quarters of species expressing Prli42 also displayed RsbR, RsbS and RsbT homologues (Supplementary Table 9).
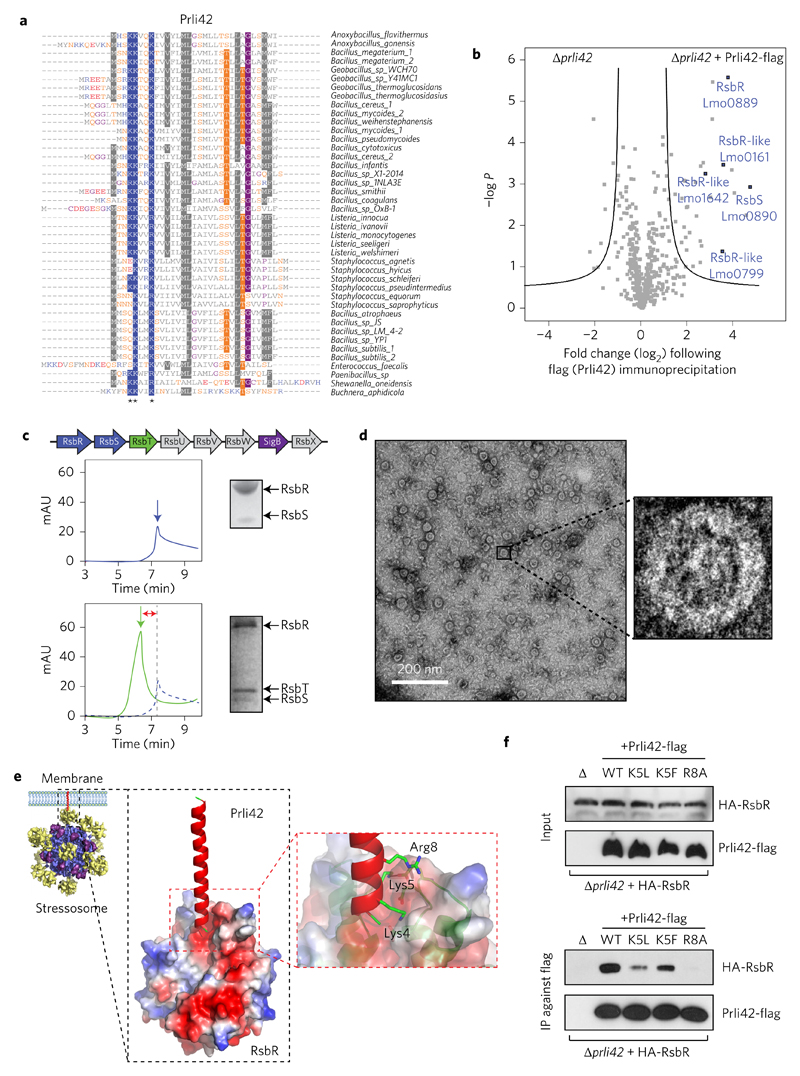
Figure 3. Miniprotein Prli42 is highly conserved and interacts with Listeria orthologues of the stressosome.
a, Multiple sequence alignment of orthologous sequences of Prli42. Amino acids are highlighted when conserved in more than 55% of the sequence. Stars indicate the conserved basic residues (Lys4, Lys5 and Arg8). Colour code for amino acids is as follows: grey, hydrophobic; blue, positively charged; red, negatively charged; orange, neutral; purple, glycine. b, Identification of Prli42 protein interaction partners. The volcano plot shows the intensity fold change (in log2) in the Prli42-expressing strain compared to the control deletion strain on the x axis. The pull-down was performed in triplicate and a t-test was performed to calculate −log P values for each protein, indicated on the y axis. Black lines indicate the boundary of significance as set by Perseus software (FDR = 0.05 and S0 = 1) c, Assembly of the stressosome. The panels show elution from a Superdex-200 sizing column. The RsbR:RsbS complex has an apparent mass of 1.5 MDa (top). The RsbR:RsbS: RsbT complex has an apparent mass of 1.8 MDa (bottom). mAU, milli arbitrary units. d, Electron micrographs of the RsbR:RsbS:RsbT complex, revealing a fourfold symmetry in the central cavity of the supramolecular assembly. e, Model of the Prli42–RsbR interaction from docking of Prli42 to the homodimeric N-terminal domain of Listeria RsbR, obtained through homology modelling from the known N-terminal RsbR structure of B. subtilis (PDB ID: 2BNL). f, WT, K5L, K5F and R8A variants of Prli42-flag were expressed in the Prli42 deletion strain together with HA-RsbR, flag pull-down followed by immunoblotting against the HA-tag. The stressosome was assembled at least three different times, and the elution profiles shown are representative. The experiment in c was subsequently imaged by EM. Immunoblots in f are representative of three independent experiments.
Listeria RsbR, RsbS and RsbT form a supramolecular complex in vitro
The formation of the stressosome in Listeria has not yet been reported. The cryo-electron microscopy (cryo-EM) structure of the Bacillus stressosome complex has been solved and consists of a core complex composed of the C-terminal STAS (sulfate transporter and anti-sigma factor antagonist) domains of RsbR and RsbS with N-terminal RsbR homodimeric protrusions that are proposed to act as sensors27. The core domain sequesters the kinase RsbT, and releases it upon detection of environmental stresses. This initiates a signalling cascade that culminates in sigma B-mediated induction of factors that help the bacteria to counteract and resist stress28. To determine if a similar complex is formed in Listeria, we individually purified Listeria RsbR, RsbS and RsbT (Lmo0891). We then reassembled the structure by first combining RsbR and RsbS, similar to the purification of the Bacillus stressosome (Fig. 3c)27. RsbR and RsbS formed a core stressosome complex of a higher molecular weight than each individual protein. The addition of RsbT (Supplementary Fig. 4) led to a larger complex (Fig. 3c), which we then visualized using EM (Fig. 3d). When imaged, the supramolecular complex was reminiscent of the Bacillus stressosome, revealing that the stressosome assembles in Listeria.
Prli42 interacts with RsbR via its conserved N-terminal Lys5 and Arg8
To gain deeper insight into the molecular interaction between Prli42 and RsbR, their association was simulated through computational docking. We took advantage of the crystal structure of the homodimeric N-terminal domain of Bacillus RsbR to build a structural model for Listeria RsbR (ref. 29; Supplementary Fig. 4d,e). We made the assumption that the N-terminal domain would interact with Prli42 because the N-terminal dimers of RsbR lie on the accessible exterior of the stressosome and are thought to be involved in signal transduction27. We then simulated docking of the predicted structure of Prli42 with that of Listeria RsbR. The optimal model showed an interaction of the basic N-terminal tail of Prli42, including conserved residues Lys4, Lys5 and Arg8, with an acidic patch formed between two RsbR subunits, on the periphery of the stressosome (Fig. 3e).
To test this interaction model, we constructed Listeria strains that expressed Prli42-flag, in which Lys5 and Arg8 residues were replaced by an alanine residue. Because K5A reduces Prli42 stability, we mutated Lys5 to leucine or phenylalanine (Supplementary Fig. 4f) and tested wild type (WT), K5L, K5F and R8A for their interaction with RsbR. We were able to validate our proteomics results by immunoprecipitation and immunoblot, showing that WT Prli42 can interact with RsbR while the Prli42–RsbR interaction was reduced in K5L and K5F mutants and completely abrogated by the R8A mutation (Fig. 3f). In addition, mass-spectrometry following immunoprecipitation of Prli42 R8A-flag demonstrated that RsbR and its paralogues no longer co-immunoprecipitated with Prli42 (Supplementary Fig. 4g and Supplementary Table 10). Together, these data establish the crucial role of the Lys5 and Arg8 residues of Prli42 in binding RsbR, in support of our interaction model.
Prli42 can anchor RsbR to the bacterial membrane
To test whether Prli42 was membrane-anchored, we performed cellular fractionation. This experiment indicated that Prli42 localized strictly to the membrane fraction (Fig. 4a). We then visualized RsbR and estimated that RsbR is 60% membrane-associated and 40% cytoplasmic (Fig. 4a and Supplementary Fig. 5b). Because Prli42 is membrane-localized and the interaction with RsbR probably occurs at the interface of the membrane and the cytoplasm, we hypothesized that Prli42 was involved in anchoring RsbR to the bacterial membrane. We fractionated WT, Δprli42 and Δprli42+flag-Prli42 and observed that, upon deletion of Prli42, ~40% of RsbR shifts from the bacterial membrane fraction to the bacterial cytoplasm (Fig. 4b,c and Supplementary Fig. 5b). This shift is nearly but not completely restored by complementation with Prli42 (45% RsbR membrane) (Fig. 4b,c and Supplementary Fig. 5b). Taken together, our data show that Prli42 is needed, directly or indirectly, for close association of RsbR with the membrane.
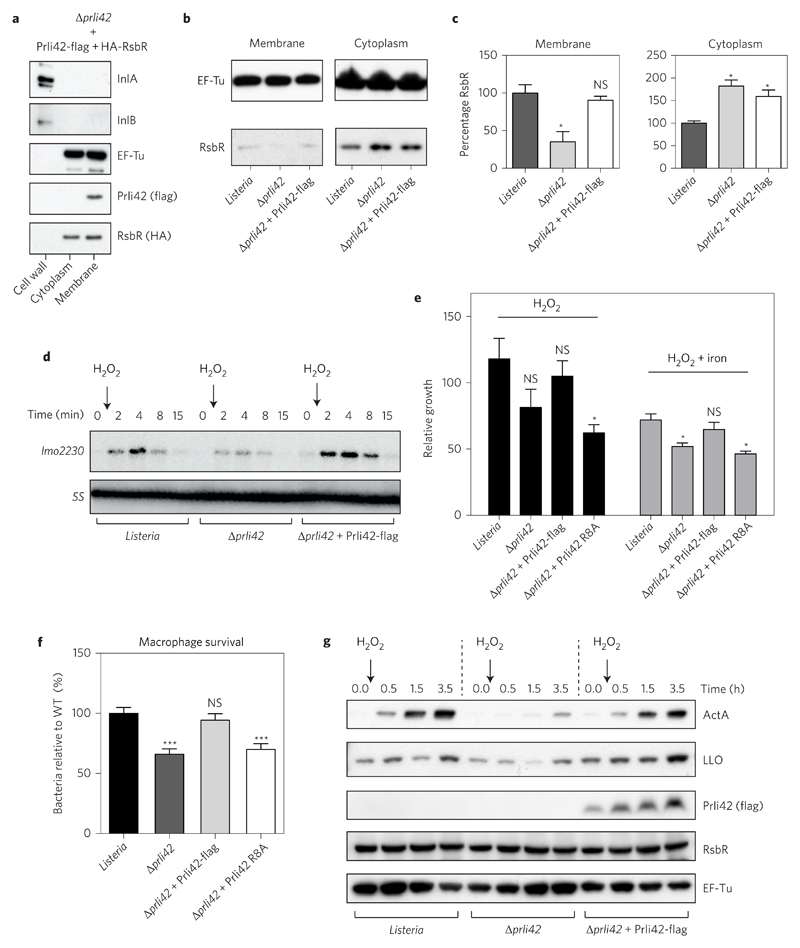
Figure 4. Membrane miniprotein Prli42 partially tethers RsbR to the bacterial membrane and is essential for the sigma B signalling following oxidative stress and survival in macrophages.
a, Prli42-flag and HA-RsbR were expressed in Δprli42 bacteria. Cellular fractions were analysed by immunoblot. InlA, InlB and EF-Tu were used as controls for fractionation. b, Localization of endogenous RsbR was analysed by immunoblot after cellular fractionation. EF-Tu was used as a loading control. c, Quantification of RsbR levels in membrane or cytosolic fractions relative to EF-Tu and normalized to levels in WT bacteria using ImageJ. Values are expressed as mean ± s.e.m. of three independent experiments. d, 0.05% H2O2 was added to exponential cultures and sigma B pathway activation was assessed by northern blot using the lmo2230 gene and 5S as a loading control. e, Bacteria were grown in BHI medium and 0.05% H2O2 ± iron (2.5 μg ml–1 FeC6H5O7) was added to cultures in exponential growth. The number of living bacteria was monitored by counting c.f.u. For each time point and for each strain, the number of c.f.u. is shown relative to the number of c.f.u. at time zero. Results are expressed as mean ± s.e.m. of four independent experiments. f, BMDMs were infected and cells were lysed at 24 h post-infection, and the released bacteria were counted. WT c.f.u. average was normalized to 100 to cross-compare experiments between macrophages from different mice. Results are expressed as mean ± s.e.m. of five independent experiments. g, Expression of ActA and listeriolysin O (LLO) following H2O2 treatment (as in d) was analysed by immunoblot and is representative of three experiments. Lysates were from total bacterial pellet. Flag, RsbR and EF-Tu were used as controls. Immunoblots and northern blot shown in a, b, d and g are representative of three independent experiments. For all graphs, significance was determined by ordinary one-way analysis of variance followed by Dunnett’s multiple comparison test, and significance is shown relative to WT in each condition (***P < 0.001; *P < 0.05; NS, not significant).
Prli42 is essential for the sigma B-mediated stress response to oxidative stress
Because the stressosome mediates the stress response in Bacillus and previous work from our laboratory has shown that Prli42 transcription is upregulated during bacterial growth in blood3, we hypothesized that Prli42 could affect sigma B signalling in Listeria. We thus monitored sigma B signalling following hydrogen peroxide treatment during bacterial growth, conditions similar to the oxidative stress encountered by bacteria during growth in blood. Following hydrogen peroxide treatment, Δprli42 induced far less expression of lmo2230, a sigma B-dependent gene, than WT or complemented bacteria (Fig. 4d and Supplementary Fig. 5d). To determine whether Prli42 also had a functional effect on bacterial growth following oxidative stress, we exposed bacteria to hydrogen peroxide and hydrogen peroxide combined with iron to generate reactive oxygen species (ROS) via the Fenton reaction, and assessed their survival. Listeria Δprli42 was significantly more sensitive to oxidative stress than the WT and complemented strains (Fig. 4e). Interestingly, a mutant of Prli42 R8A, for which the interaction between Prli42 and RsbR is impaired, is also more sensitive to oxidative stress than WT (Fig. 4e), revealing the functional relevance of the Prli42–RsbR complex to the stress response. Futhermore, mutations in Prli42 do not affect bacterial growth following several other stresses, suggesting that Prli42 is particularly important for oxidative stress (Supplementary Fig. 5a,c). Taken together, the Prli42–RsbR interaction partitions a subset of RsbR to the membrane, and absence of this interaction sensitizes bacteria to oxidative stress.
Prli42 is essential for survival in macrophages
Because Prli42 is transcriptionally upregulated during bacterial growth in blood and Prli42 interacts with components of the stressosome, we investigated whether the Listeria stressosome is involved in virulence. Survival in bone-marrow-derived macrophages (BMDMs) constitutes an oxidative stress as macrophages can produce ROS to target intracellular bacteria. We thus monitored the survival of bacteria in BMDMs. The survival of the Listeria Δprli42 strain was significantly attenuated in macrophages relative to WT Listeria. This phenotype was complemented when Prli42 expression was restored and the Prli42-R8A mutant phenocopied Prli42 deletion (Fig. 4f).
Sigma B controls virulence factor expression in Listeria in concert with the master regulator of virulence, PrfA (ref. 30). We therefore assayed virulence factor expression following oxidative stress to assess whether Prli42 affects virulence factor production. Following oxidative stress, WT bacteria displayed an increased level of both ActA and listeriolysin O, two essential virulence factors (Fig. 4g). In contrast, Δprli42 bacteria expressed lower levels of virulence factors, a phenotype that was rescued by complementation. In summary, the Listeria stressosome plays an important role in pathogenesis, and Prli42 mediates this effect through increased expression of virulence factors in response to oxidative stress.
Discussion
N-terminal proteogenomics: alternative bacterial TIS and miniproteins
Using N-terminal proteogenomics we mapped the translational landscape of L. monocytogenes and obtained an overall coverage of 62%, which is similar to the number of ORFs detected in ribosome profiling of E. coli and B. subtilis7,25,31. Our data revealed additional start codons in proximity to aTISs, suggesting that ribosomes initiate translation with a certain flexibility. Although slight differences in TIS may not result in functionally different protein isoforms, protein stability could be affected according to the bacterial N-end rule32. We also detected 19 TISs located within ORFs. Only a handful of such alternative in-frame TISs have been described in bacteria, for example, ClpB (ref. 22) and aspartokinase II (ref. 21). Interestingly, short- and long-protein isoforms generated by internal translation initiation are often part of the same protein complex. In Salmonella SsaQ, a component of the Salmonella type III secretion system (T3SS) is translated in two isoforms; the short form stabilizes the long form and the complex is required for efficient type III secretion33. This is also the case for the TilS-HGPRT fusion protein24, suggesting that use of internal TISs might be a common mechanism in bacteria to regulate protein complex formation. Here, we identified six miniproteins in Listeria. In other bacteria, a few miniproteins have already been discovered, often serendipitously34. Many miniproteins act at the bacterial membrane and modulate the activity of larger membrane proteins or complexes. Several alter the transport of small molecules; for example, during the oxidative stress response in E. coli, a miniprotein (MntS) blocks a manganese exporter (MntP) during manganese limitation35. Another miniprotein in E. coli, MgrB, has been implicated in stress signalling, but to our knowledge, a stress-signalling miniprotein has yet to be identified in Gram-positive bacteria36,37.
Discovery of Prli42, a protein involved in the bacterial stress response
We have shown that Prli42 is a membrane miniprotein that tethers a component of the stressosome, RsbR, to the membrane. The EM structure of the Bacillus stressosome demonstrates that the N-terminal domain of RsbR forms homodimeric protrusions that are proposed to sense stress, but how this occurs mechanistically has remained elusive. Here, we have shown that Prli42 binding to RsbR requires conserved basic residues in its N-terminal cytoplasmic tail. We have also shown that Prli42 itself is involved in the stress response, and that its role is to facilitate the transmission of stress signals through the bacterial membrane. The current hypothesis in the field is that the RsbR N-terminal protrusions are thought to act as sensors and are accessible for binding to small molecules or other proteins to relay stress signals to the stressosome27. As it is unclear how the stressosome might detect externally applied stresses from its cytoplasmic location, we hypothesize that Prli42 could itself act as a channel or interact with a membrane channel, thereby positioning RsbR to sense a signal. We also propose that Prli42 could be the first member of a class of long-sought functional membrane miniproteins, each specialized to respond to different types of stress or small molecules. Additionally, there are five RsbR homologues in Listeria, among which one (Lmo0799) has been characterized in detail as a blue-light sensor and is thought to sense ROS and activate sigma B (ref. 38). Deletion of this sensor has specific downstream effects on stress signalling, suggesting that different paralogues of RsbR could play both unique and complementary roles in response to various stresses. An important future direction will be to determine the molecular composition of individual stressosomes to understand how these isoforms work together within the bacterial cell.
We have shown that Prli42 and the Listeria stressosome are important for virulence factor expression and consequently survival in macrophages. These data are in line with previous reports showing that sigma B regulates virulence factor expression3,30,39,40. A recent paper has characterized a putative stressosome in the Gram-negative bacterium Vibrio brasiliensis41. The RsbR of this stressosome binds haem and effectively acts as an oxygen sensor. In Bacillus, the haem-binding domain of RsbR is occluded and RsbR proteins cluster into two phylogenetically separate clades in which Vibrio species are distant from Listeria and Bacillus species. Prli42 is conserved in Bacillus and Listeria, but is absent in species related to V. brasiliensis (Supplementary Table 11). Because Prli42 levels increase during infection in the blood, a hypoxic condition, and the mutant is less able to survive oxidative stress, it is tempting to speculate that Listeria and Bacillus RsbR would require Prli42 to bind haem, perhaps after inducing a conformational change, whereas V. brasiliensis RsbR can bind haem by itself. Gram-positive and Gram-negative stressosomes thus could have distinct strategies for stress sensing. Future structure–function comparisons of all three stressosomes will undoubtedly shed light on these hypotheses. Taken together, our study reveals a link between a previously uncharacterized tail-anchored membrane miniprotein, the Gram-positive bacterial stress response, and the role of the stressosome in virulence in an important human pathogen.
Methods
Bacterial strains, plasmids, primers and growth conditions
For standard experiments, L. monocytogenes was grown overnight in brain heart infusion (BHI) medium (Difco) at 37 °C while shaking at 200 r.p.m. Overnight cultures were collected or diluted 1:50 in fresh BHI and grown at 37 °C until exponential phase (optical density at 600 nm (OD600) of 1.0). When required, 7 μg ml−1 chloramphenicol or 5 μg ml−1 erythromycin was added to the culture medium.
For the control COFRADIC analysis, overnight cultures were diluted 1:75 into 50 ml BHI and grown at 37 °C until exponential phase (OD600 = 1.2). For protein extraction, 6 × 1010 bacteria were pelleted by centrifugation for 20 min at 3,400g at 4 °C, washed with 50 ml of cold PBS, and stored at −80 °C. For the main COFRADIC analysis, similarly diluted cultures were grown until exponential phase at 20 °C (OD600 = 0.3) or 37 °C (OD600 = 1.0) or early stationary phase at 37 °C (OD600 = 3.0), after which actinonin (Sigma) was added to all cultures to a concentration of 50 μg ml−1. Following further incubation for 90 min, bacteria were collected by centrifugation, and aliquots with 2.5 × 1010 bacteria were stored at −80 °C.
To generate Listeria mutants, standard techniques for DNA manipulation were used. A Prli42 deletion mutant was constructed using the pMAD shuttle plasmid42 as described previously43, and confirmed by DNA sequencing. Sequencing also indicated an additional point mutation in lmo1026 (C196→T). For the construction of pAD-based plasmids, fragments obtained by PCR with EGD-e genomic DNA or synthetic DNA (Gblocks from Integrated DNA Technologies) as template were cloned into the XmaI/SalI sites of the pAD vector44 derived previously from the pPL2 vector45. To obtain start codon mutants of CplB or LmO219, site-directed mutagenesis was performed by inverse PCR using pAD-CplB WT-FLAG or pAD-LmO219 WT-FLAG as DNA template. Plasmids were verified by sequencing with primers pPL2-Rv and pPL2 Fw and were transformed into L. monocytogenes by electroporation. Integration in the chromosome was verified by PCR using primers NC16 and PL95 (ref. 46). To obtain pP1-HA-RsbR, DNA of the HA-tagged rsbR was synthetized (Gblock from Integrated DNA Technologies), then amplified by PCR and cloned into the SalI/SmaI sites of pP1. Strains and primers used in this study are listed in Supplementary Table 12.
Protein extraction and isolation of N-terminal peptides by COFRADIC
Bacterial pellets were re-dissolved in 1 ml lysis buffer containing 50 mM sodium phosphate pH 7.4, 100 mM NaCl, 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate and complete protease inhibitor (Roche Diagnostics, 1 tablet per 100 ml) and lysed by sonication (10 cycles of 10 s, 20% amplitude). For the actinonin COFRADIC analysis, pellets containing equal amounts of bacteria from all three growth conditions were mixed and lysed together. Extracts were cleared by centrifugation for 20 min at 16,000g at 4 °C, and the supernatant (soluble fraction) contained 2.6 and 4.2 mg of total protein for the preliminary and main analysis, respectively, as measured by the Biorad protein assay. The pellet, with primarily insoluble membrane and cell wall components, was also retained, and during the downstream processing steps, proteins in this fraction were treated similarly to the proteins in the soluble fraction. Only desalting steps (buffer swaps) were conducted differently: proteins in the soluble fraction were desalted on disposable NAP-10 columns (GE Healthcare), and proteins in the insoluble fraction were desalted by a combination of centrifugation and re-dissolution steps.
The next steps of the N-terminal COFRADIC procedure were essentially performed as described before12. Briefly, guanidinium hydrochloride (Gu.HCl) was added dry to the soluble fraction to a final concentration of 4 M while the insoluble fraction was re-dissolved in 1 ml of lysis buffer. Proteins were reduced and alkylated by incubation with 15 mM tris(2-carboxyethyl)phosphine (TCEP) and 30 mM iodoacetamide for 20 min at 37 °C. After desalting in 50 mM sodium phosphate buffer at pH 8 with or without 2 M Gu.HCl (for the soluble and insoluble fraction, respectively), free amino groups were acetylated by incubation with 20 mM of an N-hydroxysuccinimide (NHS) ester of trideutero-acetate (provided by K. Gevaert and B. Ruttens, VIB/Ghent University) for 1 h at 30 °C, followed by addition of 10 mM glycine to quench residual NHS esters and 120 mM hydroxylamine to reverse potential O-acetylation and incubation for 20 min at room temperature. The protein samples were desalted in 20 mM ammonium bicarbonate, boiled for 5 min and put on ice for 5 min. During the control COFRADIC analysis, proteins in both fractions were digested overnight with sequencing-grade trypsin (Promega) (enzyme:substrate of 1:100 (wt/wt) for the soluble fraction; 20 μg for the insoluble fraction) at 37 °C. For the actinonin analysis, half of both fractions was digested with trypsin, while the other half was digested with similar amounts of sequencing-grade Glu-C (Promega).
The resulting peptide mixtures were incubated with 1,250 mU Q-cyclase (Qiagen,) to drive the formation of N-terminal pyroglutamate to completion and with 625 mU activated pGAPase (Qiagen) to proteolytically remove this residue. N-terminal peptides in the trypsin-digested samples were then pre-enriched by strong cation exchange (SCX) chromatography on disposable SCX cartridges (Agilent Technologies) in a buffer containing 50% acetonitrile in 10 mM sodium phosphate, pH 3.0. N-terminal peptides, which were not retained on the SCX resin under these conditions, were collected in the run-through fraction, concentrated by vacuum drying, and re-suspended in 10 mM ammonium acetate pH 5.5 in 2% acetonitrile. Because Glu-C generated peptides do not end on a positively charged arginine residue, N-terminal peptides cannot be pre-enriched by SCX under these conditions and therefore the SCX step was omitted for the Glu-C digested samples. Methionine residues were then oxidized by incubating the peptides with 0.5% H2O2 for 30 min at 30 °C, followed by immediate injection of the samples on a capillary reversed-phase–high-performance liquid chromatography (RP-HPLC) column (Zorbax 300SB-C18, 2.1 mm internal diameter, 150 mm length, Agilent Technologies) for the first COFRADIC separation. Peptides were separated by a linear gradient of acetonitrile (from 2 to 70% in 100 min) and peptides that eluted between 20 and 92 min were collected in 18 primary COFRADIC fractions of 4 min each. Each primary fraction was incubated four times with 15 nmol 2,4,6-trinitrobenzenesulfonic acid (TNBS), and each fraction was re-separated by RP-HPLC under the same conditions as during the primary separation. N-terminal peptides, which cannot be modified by TNBS, eluted from the column during the same time interval and were collected in 16 fractions of 0.5 min. Internal and C-terminal peptides, which were modified by TNBS, underwent a hydrophobic shift and were not collected. Secondary fractions with 12 min difference in retention time were pooled to a total of 33 samples for LC-MS/MS per analysis.
LC-MS/MS and data analysis
Secondary fractions were dried completely in a vacuum concentrator and re-dissolved in 50 μl solvent A (0.1% formic acid in water/acetonitrile (98:2, vol/vol)) of which 2–4 μl was used for LC-MS/MS analysis on an Ultimate 3000 HPLC system (Dionex) in line connected to an LTQ Orbitrap Velos mass spectrometer (Thermo Electron). Trapping was performed at 10 μl min−1 for 4 min in solvent A on a PepMapTM C18 column (0.3 mm inner diameter (ID) × 5 mm (Dionex)) and following back-flushing from the trapping column, the sample was loaded on a reverse-phase column (made in-house, 75 μm ID ×150 mm, 3 μm beads C18 Reprosil-HD, Dr Maisch). Peptides were eluted by a linear increase from 2 to 55% solvent B (0.08% formic acid in water/acetonitrile (2:8, vol/vol)) over 30 min at a constant flow rate of 300 nl min−1. The mass spectrometer was operated in data-dependent mode as described previously47, automatically switching between MS and MS/MS acquisition for the ten most abundant ion peaks per MS spectrum.
From the MS/MS data in each LC run, Mascot generic files (mgf) were created using the Mascot Distiller software (version 2.5.0.0, Matrix Science) and generated peak lists were searched with Mascot using the Mascot Daemon interface (version 2.4.0, Matrix Science) against two databases with L. monocytogenes EGD-e protein sequences generated in house. The first database contained 4,414 sequences with a high potential to be translated. This database was created starting from 2,846 L. monocytogenes EGD-e (ORFs predicted in 2001 upon sequencing of the Listeria genome2,18 (downloaded from http://www.ncbi.nlm.nih.gov/, RefSeq ID NC_003210). For 588 of these ORFs, the annotated sequence was replaced by a potential longer sequence generated by in-frame back-translation from the annotated translation initiation codon to the first transcription start sites of the mRNA reported in ref. 5. Also, 157 potential ORFs present in 5′ UTR regions of annotated genes were added this way. Finally, a total of 676 and 735 potential ORFs were added by three-frame translation of small RNA (sRNA) and antisense RNA (asRNA) molecules, respectively (as described in ref. 18) from any of the known start codons (ATG, GTG, TTG, CTG, ATT, ATA, ATC). Note that, for all potential ORFs, a minimal length of six amino acids was required to be included. The second database contained 589,911 sequences with a minimum length of six amino acids, generated by six-frame translation of the EGD-e genome from any of the known start codons (ATG, GTG, TTG, CTG, ATT, ATA, ATC). Spectra from trypsin-digested samples were searched in both databases in combination with Arg-C/P and semi-Arg-C/P enzyme settings (with one missed cleavage allowed), leading to a total of four different searches for these samples. In all searches, oxidation of methionine (+15.994915 Da), trideuteron-acetylation of lysine (+45.029395 Da) and carbamidomethylation of cysteine (+57.021464 Da) were set as fixed modifications, while variable modifications included acetylation (+42.010565 Da), trideuteron-acetylation (+45.029395 Da) and formylation (+27.994915 Da) of N termini and pyroglutamate formation of N-terminal glutamine residues (−17.026549 Da). Mass tolerance of the precursor ions was set to 10 ppm, and mass tolerance of the fragment ions was set to 0.5 Da. The peptide charge was set to 1+, 2+ or 3+, and the C13 setting of Mascot was set to 1. Spectra from Glu-C generated samples were searched in the same two databases, but in combination with Glu-C, semi-Glu-C and ‘none’ as enzyme settings (with one missed cleavage allowed), leading to a total of six different searches. Other search parameters were identical to the trypsin searches except for the additional variable modification of N-termini by trinitrobenzene (+210.986535 Da). Only peptides that were ranked first and scored above the threshold score set at 99% confidence were withheld. The estimated false discovery rate was calculated as described previously48 by searching decoy databases (reversed versions of both databases made with DBToolkit49) and was found to be a maximum of 0.68% for the trypsin searches and 2.93% for the Glu-C searches. For processing of all MS data, the ms_lims software platform was used50. Potential false positive peptide identifications were selected and automatically removed by the Peptizer software application51 if more than one of the following four rules were fulfilled: the MASCOT homology threshold was higher than the MASCOT identity threshold or the peptide ion score, multiple confident hits were identified, b-ion coverage was lower than 10%, and y-ion coverage was lower than 20%. Furthermore, peptides shorter than six amino acids were removed in any case. In this way, 6,583 (3%) of the identified MS/MS spectra were not used for further analysis. The overall efficiency of the COFRADIC procedure is reflected by the fraction of Peptizer validated peptides with blocked alpha-amines. This fraction ranged from 53% for the endoGluC digested samples to 85% for the trypsin-digested samples. The higher enrichment in the latter samples is the result of the SCX pre-enrichment step that was performed for the tryptic peptides, described above. The effect of actinonin treatment was evaluated by comparing the average fraction of For-Met starting peptides in the (trypsin-digested) treated samples (20.5%) versus the control (2.8%). The number of recorded, identified and removed spectra as well as the number of N-terminally blocked and N-terminally formylated peptides are indicated for each data set in Supplementary Table 13. Identified peptides from the control and actinonin analyses were combined, and 13,582 N-terminally formylated, acetylated or trideutero-acetylated peptides were mapped on the L. monocytogenes EGD-e genome, using an in-house script. A total of 121 N-terminally modified peptides were mapped onto multiple loci on the EGD-e genome, while the large majority were mapped onto a single locus. Of these, mapping of 9,895 non-methionine starting peptides was straightforward, while for 557 (out of 3,685) peptides starting with methionine, successful mapping was only achieved when non-AUG start codons were considered. The MS data of mapped peptides have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository52 with the data set identifier PXD000890 and DOI 10.6019/PXD000890, and mapped peptides are visualized in our home-made genome browser on http://nterm.listeriomics.pasteur.fr. For the detection of previously annotated TISs, the identification of a methionine-starting peptide or non-methionine-starting peptide that mapped exactly to the corresponding start codon or downstream codon, respectively, was considered sufficient, independent of the N-terminal modification status of the peptide. For any other TIS reported here for the first time, detection was strictly based on the identification of a Meti-starting peptide for non-AUG start codons and a formyl-Meti-starting peptide for AUG start codons. Furthermore, identification of these peptides by a second search algorithm was required to increase the confidence of our proteogenomic workflow. Therefore, in parallel to the above Mascot-based workflow, all searches were repeated using the MaxQuant software (version 1.5.3.8), with default search settings including a false discovery rate set at 1% on both the peptide and protein level with a mass tolerance for precursor and fragment ions of 4.5 and 20 ppm, respectively, during the main search. Fixed and variable modifications were identical to the above Mascot parameters, and searches in both databases were combined with identical enzyme settings, leading to two different searches for the trypsin-digested samples and three different searches for the Glu-C-treated samples. Novel TISs were only withheld if the corresponding N-terminal peptide was also identified by MaxQuant as indicated by its presence in one of the ‘evidence’ output files.
Shine–Dalgarno profiling
For each mapped peptide, the first codon as well as the genome sequence 20 bp upstream were extracted and used to calculate the hybridization free energy against the consensus anti-Shine–Dalgarno sequence ‘AUCACCUCCUUU’53 using the UNAFold software (v3.8. with default parameters, 37 °C)54.
RAST re-annotation of the Listeria genome
Annotation of the L. monocytogenes EGD-e genome sequence was performed using the RAST annotation software55. A total of 2,903 protein-coding ORFs were detected, that is, 57 more than the 2,846 ORFs that were annotated by the Listeria consortium in 2001 (ref. 2); 2,591 ORFs were found to have the same start and stop positions in both annotations, 244 ORFs had a different start position in each annotation, and three ORFs had a different stop position in the RAST annotation because they were described as pseudogenes in the original genome annotation. Twenty-five ORFs were unique to the latter annotation, whereas 66 ORFs were only found in the RAST annotation. Thus, in addition to the original genome annotation2, RAST annotation provided us with a list of predicted TISs to compare to our N-terminome analysis. It also helped us to reduce from 737 to 376 the number of ORFs described as hypothetical or unknown proteins. The RAST annotation data are included in Supplementary Table 9 and RAST-predicted ORFs are shown in the genome browser when they deviate from the original genome annotation (http://nterm.listeriomics.pasteur.fr).
Gene expression analysis
L. monocytogenes EGD-e gene expression data from ref. 3 (three data sets from ArrayExpress E-MEXP-2138: BHI exponential phase at 30 °C, BHI exponential phase at 37 °C and BHI stationary phase at 37 °C) and ref. 18 (one data set from ArrayExpress E-MTAB-1676: BHI exponential phase at 37 °C) were analysed together. Normalized expression values from both ‘BHI exponential phase at 37 °C’ data sets were averaged and summed with the values from the other two data sets to mimic mixing of the samples in our N-terminome analysis (Supplementary Table 1). The histogram of the summed expression values is shown in Fig. 2b and for each bin the per cent of proteins with detected aTISs was calculated and plotted over the histogram. A clear correlation between the transcriptomics and N-terminomics data was observed, except for the lowest expressed genes (up to bin −13) for which the per cent remained lower than 10%. Therefore, when the cumulative per cent of detected aTISs was calculated for each protein, the first 230 genes for which this value remained lower than 10% were considered as non-expressed (Supplementary Table 1).
Phylogenetic analysis of Prli42
To detect potential orthologues of Prli42 in other bacterial genomes, all complete bacterial genomes were downloaded from the RefSeq database (5,000 genomes, version from February 2016)56. For each genome, a list of possible ORFs was generated by six-frame translation from any of the known start codons (ATG, GTG, TTG, CTG, ATT, ATA, ATC) to the first stop codon. Lists of potential small ORFs with a size between 20 and 60 amino acids were then extracted. From these lists, BlastP databases with small ORFs for each genome were created. Prli42 was searched against our in-house databases using BlastP with default parameters57. Each blast hit with an e-value below 0.1 and similarities above 25% was retained. The list of potential Prli42 orthologues was manually curated for each species before using MAFFT for multiple sequence alignment. The search for RsbR, RsbS and RsbT homologues was carried out using the lists of annotated ORFs for all 5,000 genomes provided on the RefSeq database. From these lists, we created BlastP databases and performed a BlastP search with default parameters and no additional cutoffs.
Prli42-flag co-immunoprecipitation and LC-MS/MS analysis
Exponential cultures (20 ml) of L. monocytogenes EGD-e Δprli42, L. monocytogenes EGD-e Δprli42 Prli42-flag and L. monocytogenes EGD-e Δprli42 Prli42-R8A-flag were washed once with 50 ml PBS and re-dissolved in 13.5 ml lysis buffer (50 mM Tris HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.5 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF)). Nutanolysin (410 μg) (Sigma) was added to each sample and samples were incubated for 1 h at 37 °C to digest the cell wall. Triton X-100 was added to both samples to a concentration of 1% as well as 1 ml of a protease inhibitor stock solution (1 complete protease inhibitor tablet (Roche) in 10 ml of water). Samples were sonicated by four pulses of 15 s at an amplitude of 20% and centrifuged at 4 °C for 15 min at 16,000g. The supernatant of both samples (containing 2 mg of total protein) was recovered and 125 μl of settled M2 anti-flag beads (Sigma, washed three times with 1 ml of lysis buffer) were added to both lysates. Each lysate was split into three aliquots and immunoprecipitation was performed in triplicate overnight on a rotating wheel at 4 °C. Beads were collected by centrifugation at 4 °C for 2 min at 2,000g, washed once with 1 ml lysis buffer and three times with 1 ml digestion buffer (20 mM Tris HCl pH 8.0, 2 mM CaCl2). Beads were resuspended in 100 μl digestion buffer and incubated with 1 μg sequencing-grade trypsin (Promega) at 37 °C to release bound proteins. After 4 h of incubation, beads were removed and another 1 μg of trypsin was added to the supernatant for further overnight protein digestion. Peptides in every replicate were purified on Omix C18 tips (Agilent), dried and re-dissolved in 15 μl 0.1% formic acid in water/acetonitrile (98:2, vol/vol), of which 6 μl was injected for LC-MS/MS analysis using the same instrument and settings as described above, except that peptides were eluted from the LC column by a linear increase from 2 to 55% solvent B over 120 min and that a ‘top 15’ method was used. Data analysis was performed with MaxQuant (version 1.5.3.8)17 with default search settings including a false discovery rate set at 1% on both the peptide and protein level. Spectra were searched against a database of 2,846 proteins from L. monocytogenes strain EGD-e (RefSeq ID NC_003210, downloaded from http://www.ncbi.nlm.nih.gov/). The mass tolerance for precursor and fragment ions were set to 4.5 and 20 ppm, respectively, during the main search. Enzyme specificity was set as C terminal to arginine and lysine, also allowing cleavage at proline bonds with a maximum of two missed cleavages. Oxidation of methionine residues was set as variable modification. Matching between runs was enabled with an alignment time window of 20 min and a match time window of 1 min. Only proteins with at least one unique or razor peptide were retained, leading to the identification of 771 Listeria proteins. Proteins were quantified by the MaxLFQ algorithm integrated in the MaxQuant software58. A minimum ratio count of two unique or razor peptides was required for quantification. Further data analysis was performed with the Perseus software (version 1.5.2.6) after loading the protein groups file from MaxQuant. Proteins only identified by site, reverse database hits and contaminants were removed, and replicates of both samples were grouped. Proteins with fewer than three valid values in at least one group were removed, and missing values were imputed from a normal distribution around the detection limit. Then, t-tests were performed (false discovery rate, FDR = 0.05, and exchangeability factor, S0 = 1) for pairwise comparison and to generate the volcano plots depicted in Fig. 3b and Supplementary Fig. 4g (Supplementary Table 10).
Prli42-flag co-immunoprecipitation and western blot against HA-RsbR
Exponential cultures (15 ml) of L. monocytogenes EGD-e Δprli42 and EGD-e Δprli42 Prli42-flag WT, K5L, K5F or R8A, all co-expressing HA-RsbR, were washed once with 15 ml PBS. One-third of the bacteria in each culture were lysed in 150 μl 2× diluted Tricine sample buffer (Biorad) supplemented with 20 mM dithiothreitol (DTT), of which 30 μl (corresponding to 10% of the pull-down input) was used for SDS–PAGE and immunoblotting of the input fraction. Two-thirds of the bacteria in each culture were re-dissolved in 10 ml lysis buffer (50 mM Tris HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.5 mM AEBSF). Mutanolysin (300 μg) (Sigma) was added to each sample and samples were incubated for 1 h at 37 °C to digest the cell wall. Triton X-100 was added to both samples to a concentration of 1%, as well as 1 ml of a protease inhibitor stock solution (one complete protease inhibitor tablet (Roche) in 10 ml of water). Samples were sonicated by three pulses of 15 s at an amplitude of 20% and centrifuged at 4 °C for 15 min at 16,000g. The supernatant of both samples was recovered, and 35 μl of settled M2 anti-flag beads (Sigma, washed three times with 1 ml lysis buffer) was added to each lysate. Immunoprecipitation was performed overnight on a rotating wheel at 4 °C. Beads were collected by centrifugation at 4 °C for 3 min at 3,500g, washed twice with 1 ml lysis buffer, and proteins were eluted in 70 μl 2× diluted Tricine sample buffer (Biorad) and incubated for 5 min at room temperature. Beads were removed, DTT was added to a concentration of 20 mM, and samples were used for analysis by SDS–PAGE and immunoblotting against Prli42-flag and HA-RsbR using mouse anti-flag (M2, Sigma) and rabbit anti-HA (Y11, Santa Cruz Biotechnology) as primary antibodies, respectively.
Cellular fractionation
Listeria proteins were fractionated into three compartments (cell wall, membrane and cytoplasm) as described previously59. A 2 ml volume of an exponential culture was pelleted, and the supernatant (SN) was precipitated on ice with 16% trichloracetic acid. Bacteria were washed once in 2 ml PBS and once in 2 ml TMS (Tris, Mg, sucrose) buffer (10 mM Tris HCl pH 6.9, 10 mM MgCl2 and 0.5 M sucrose) and incubated in TMS buffer (TMS buffer containing 60 μg mutanolysin (Sigma) and 2 mM AEBSF) for 1 h at 37 °C to digest the cell wall. Protoplasts were pelleted by centrifugation for 5 min at 15,000g, and the supernatant containing cell wall proteins (CN) was TCA precipitated. Protoplasts were lysed by four freeze–thaw cycles in 200 μl of 100 mM Tris pH 7.5, 100 mM NaCl and 10 mM MgCl2 and the membrane (MB) and cytoplasmic (CY) fractions were separated by centrifugation at 4 °C for 15 min at 16,000g. The membrane fraction was subsequently resuspended in 200 μl radioimmunoprecipitation assay (RIPA) buffer. Equal percentages of each fraction were analysed by SDS–PAGE and western blotting.
SDS–PAGE and western blotting
Bacteria from 2 ml exponential (for ClpB) or overnight (for Lmo0219) cultures were collected by centrifugation and lysed in 150 μl or 200 μl 2× diluted Tricine sample buffer (Biorad) supplemented with 20 mM DTT, respectively, by sonication (two cycles of 15 s, 20% amplitude). Samples were boiled for 5 min, centrifuged for 5 min at 16,000g and 20 μl (for ClpB) or 30 μl (for Lmo0219) was loaded onto a 4–15% Mini-PROTEAN TGX Stain-Free gel (Biorad). Proteins were separated in Tris/Tricine/SDS buffer (Biorad) at 120 V and transferred onto a polyvinylidene fluoride (PVDF) membrane using the Pierce G2 Fast Blotter (Thermo) with standard settings. Samples after cellular fractionation or co-immunoprecipitation were analysed similarly, except that for the detection of Prli42, for which 16.5% Mini-PROTEAN Tris-Tricine gels (Biorad) were used. Protein detection was performed using standard protocols for membrane blocking and antibody incubation, and proteins were revealed using Pierce ECL 2 Western Blotting Substrate (Fisher Scientific). The following primary antibodies were used: mouse monoclonal anti-flag (M2, Sigma), mouse monoclonal anti-HA (6E2, Cell Signaling Technology) and rabbit polyclonal anti-HA (Y11, Santa Cruz Biotechnology); home-made mouse monoclonal antibody raised against Internalin A L7.7 (ref. 60), rabbit polyclonal antibody raised against EF-Tu (ref. 61) and rabbit polyclonal antibody raised against Internalin B R25 or RsbR. Anti-mouse and anti-rabbit HRP-conjugated antibodies (AbCys) were used as secondary antibodies.
Three-dimensional modelling of Listeria miniproteins
Three-dimensional (3D) modelling of the identified Listeria miniproteins (Supplementary Table 8) involved a template search, selection and alignment, followed by model building and model evaluation using PHYRE Intensive62, MODELLER63 and LOMETS64 programs. Models were cross-checked using the ccp4 (ref. 65) suite MOLREP66 to thread peptide sequences onto selected templates and produce a final model. MOLPROBITY was used to analyse the geometry and stereochemical properties of all 3D models. PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger) and COOT67 were used for the visualization and analysis of all protein structures. All final structural figures were generated with PyMOL.
Structural modelling of Listeria RsbR and docking with Prli42
The 3D model of L. monocytogenes EGD-e RsbR was constructed from the X-ray structure of the N-terminal domain of B. subtilis RsbR using residues 1–136 (PDB ID: 2BNL) as a template. The final model of Listeria RsbR was generated by MOLREP. Initial docking of Prli42 and RsbR was accomplished using AutoDock Vina68 and GRAMM-X69 to predict potential binding modes. The final model of RsbR-Prli42 was aligned to the structure of B. subtilis RsbR in the cryo-EM reconstruction of the stressosome at medium resolution (EMD-1558, EMD-1552 and EMD-1556) to assist in fine-tuning of the docking model. The electrostatic surface prediction of RsbR and Prli42 was calculated using the Adaptive Poisson–Boltzman Software program70 to demonstrate surface complementarity between RsbR and Prli42. This was used as a guide to adjust the final model for chemical accuracy. Iterative adjustment of the final docking model was made in COOT.
Protein purification
GST-RsbR, RsbS and RsbT proteins were expressed in BL21 (DE3) Gold competent cells (Novagen) grown at 37 °C. All contructs were induced with 0.6 mM IPTG at an OD of 0.7–0.8 and collected after 12 h of induction at 16 °C. Cells were resuspended in 20 mM Tris pH 8.5, 150 mM NaCl, 1 mM DTT and lysed by sonication at 4 °C. Soluble fractions were recovered from cell debris by ultracentrifugation at 40,000g. After glutathione affinity chromatography and thrombin cleavage, proteins were purified to homogeneity by size exclusion chromatography (Superdex-200, GE) in 20 mM Tris pH 8.5, 1 mM DTT. After gel filtration columns, proteins were immediately used for crystallization. Alternatively, proteins were flash-frozen in liquid nitrogen and stored at −80 °C.
Proteins complex
The purified RsbR and RsbS were combined in a 1:2 ratio, that is, 2.5 mg of RsbR and 5 mg of RsbS, and incubated at 4 °C for 2 h before being subjected to a gel filtration (Superdex-200, GE) column pre-equilibrated in 20 mM Tris pH 8.5, 1 mM DTT. Only those fractions containing RsbR-RsbS proteins in the void volume fractions were pooled and concentrated by centrifugation using a 30 kDa molecular weight cutoff centrifugal filter (Vivaspin). Purified RsbR:RsbS complex was combined with an excess of RsbT and incubated at 4 °C for 2 h followed by gel filtration using a Superdex S200 10/300 column (GE Healthcare) pre-equilibrated in 20 mM Tris HCl pH 8.5, 150 mM NaCl, 1 mM adenosine diphosphate, 1 mM DTT. Only those fractions containing RsbR-RsbS-RsbT proteins in the void volume were pooled and concentrated by centrifugation using a 30 kDa molecular weight cutoff centrifugal filter (Vivaspin).
Electron microscopy
Visualization of the stressosome suprastructure was achieved by negative-stain EM. Briefly, 10 μl of a 0.01 mg ml−1 RsbR-RsbT-RsbS sample were absorbed on glow discharged-treated carbon-coated copper grids and air-dried for 1 min. After absorption, excess samples were blotted and washed away with three drops of water followed by negative staining with 2% uranyl acetate, pH 4.5. Samples were imaged using an FEI Tecan T12 Biotwin electron microscope operating at 100 keV at the Ultrapole of the Institut Pasteur.
H2O2 and FeC6H5O7 treatment
Overnight cultures were diluted 1:100 into 25 ml BHI and grown at 37 °C until exponential phase (OD600 = 1), after which 0.05% H2O2 (Sigma-Aldrich) or 2.5 μg ml−1 ferric citrate (FeC6H5O7, Sigma) were added to all cultures. At various time intervals, aliquots were collected, diluted and plated onto BHI plates to determine the number of colony-forming units (c.f.u.).
Bacterial growth following stresses
Overnight cultures were diluted 1:25 into BHI +0.05% H2O2 or 4% NaCl or 200 μM CoSO4 or 400 μM CuSO4 or 0.025% Triton and 2% ethanol. OD600 values was measured using a TECAN sunrise microplate reader.
Preparation of total RNA
The bacteria were grown in BHI at 37 °C under constant agitation until OD600 = 1. H2O2 was added at a concentration of 0.05%, and bacteria were collected at different time points, pelleted and frozen at −80 °C. RNAs were extracted according to the FastRNA Pro protocol (Qbiogene), with slight modifications: three phenol/chloroform extractions in the presence of 300 μl Tris HCl pH 7.5, 100 mM were performed rather than using the Fast Pro Solution and chloroform extraction. Fast prep was used twice for 45 s at speed 6.0, and sodium acetate 0.3 M was added for RNA precipitation.
qRT–PCR
For each sample, 10 μg of RNA was treated with DNaseI (Turbo DNA-free kit, Ambion). RNAs (500 ng) were reverse-transcribed with Quantiscript Reverse Transcriptase (QuantiTect Reverse Transcription kit, Qiagen). Quantitative reverse-transcription–PCR (qRT–PCR) reactions were carried out and quantified with SYBR Green master mix on a C1000 Touch CFX384 machine (Biorad). Expression levels of lmo2230 were normalized to the L. monocytogenes rpoB gene, and the fold change was calculated using the delta-delta CT method. All samples were evaluated in triplicate and in at least three independent experiments.
Northern blot
After separation on agarose gels 1.5% containing 20 mM guanidine thiocyanate, 5 μg of total RNA was transferred onto Nytran N membranes (GE Healthcare) and crosslinked with ultraviolet at 245 nm. For detection of lmo2230, γP32-ATP labelled lmo2230-REV oligodeoxyribonucleotides was used in Ultrahyb-Oligo Hybridization buffer (Ambion). For detection of the 5S, α-ATP labelled RNA probe was transcribed from PCR product generated using T7-anti-5S and anti-5S-rev oligonucleotides and used in ULTRAhyb Ultrasensitive Hybridization Buffer (Ambion). The experiment was reproduced three times.
Bacterial infection of macrophages
Primary BMDMs were obtained from female C57BL/6J WT mice. BMDMs were grown in Roswell Park Memorial Institute medium (Gibco) supplemented with 10% fetal calf serum (FCS, BioSera), 2 mM glutamine (Gibco), 1 mM sodium pyruvate (Gibco), 10 mM HEPES (Sigma-Aldrich), 50 μM β-mercaptoethanol, 100 U ml−1 penicillin/streptomycin (Gibco) and homemade L929 cell-conditioned medium 20% (vol/vol). After 3 days of culture, fresh complete medium containing L929-cell conditioned medium was added to the growing macrophages. On day 7, cells were washed and seeded in complete medium without antibiotic and incubated for 24 h before bacterial challenge. For infection of BMDMs, bacteria were grown in BHI overnight then regrown to exponential phase and added to the cells at a multiplicity of infection (MOI) of one, centrifuged at 900g for 1 min and incubated at 37 °C for 1 h. To follow a synchronized population of bacteria, extracellular bacteria were killed through incubation of the host cells with Dulbecco’s modified eagle medium containing 20 μg ml−1 gentamicin (Sigma-Aldrich) for the duration of infection. For enumeration of intracellular bacteria, infected cells were lysed with 0.1% Triton X-100 for 5 min and dilution series were plated onto BHI agar.
Animal ethics statement
This study was carried out in strict accordance with French national and European laws and conformed to the Council Directive on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes (86/609/Eec). Experiments that relied on laboratory animals were performed in strict accordance with the Institut Pasteur’s regulations for animal care and use protocol, which was approved by the Animal Experiment Committee of the Institut Pasteur (approval no. 03–49).
Supplementary Material
Supplementary information is available for this paper.
Acknowledgements
This work was supported by grants to P.C. (European Research Council (ERC) Advanced Grant BacCellEpi (670823), ANR BACNET (BACNET 10-BINF-02-01), ANR Investissement d’Avenir Programme (10-LABX-62-IBEID), Human Frontier Science Program (HFSP; RGP001/2013), ERANET Infect-ERA PROANTILIS (ANR-13-IFEC-0004-02) and the Fondation le Roch les Mousquetaires) and by a grant to M.G.P from the Spanish Ministry of Economy and Competitiveness (BIO2014-55238-R). The authors thank E. Gouin and L. Maranghi for essential technical support, C. O’Byrne and J. Johansson for discussions, C. Thireau for technical support, the Pasteur Ultrapole and C. Rapisarda for help with the EM. The authors thank T. Msadek for providing the L. monocytogenes LO28 ΔclpB strain and the Pasteur Proteomics platform, in particular M. Matondo-Bouzanda and T. Chaze. F.I. received financial support from a Pasteur-Roux Fellowship. L.R. was supported by an HFSP long-term fellowship. A.H.W. was supported by an EMBO long-term fellowship (ALTF 732-2010) and an Institut Carnot–Pasteur Maladies Infectieuses fellowship. P.C. is a Senior International Research Scholar of the Howard Hughes Medical Institute.
Footnotes
Data availability. The mass spectrometry data for the mapped peptides have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with data set identifier PXD000890 and DOI 10.6019/PXD000890, and mapped peptides are visualized in our homemade genome browser at http://nterm.listeriomics.pasteur.fr. All other data that support the findings of this study are available from the corresponding author upon request.
Author contributions
P.C. initiated, conceived and supervised the project. F.I. initiated the project and performed the proteomics analysis and validation of the proteomics work and docking model. N.R. identified the oxidative stress phenotype, constructed nearly all the bacterial strains and performed the analysis of sigma B signalling. L.R. performed the macrophage experiments, the fractionation experiments and the virulence experiments. C.B. made the proteogenomics pipeline and is responsible for the bioinformatic analysis of the paper. M.D. performed the northern blots of Sigma B signalling. J.M. constructed the initial bacterial strains for validation. F.G.d.P. and M.G.P. contributed essential reagents. A.H.W. reconstituted the stressosome and imaged it using EM, and performed the docking model and all of the structural biology. L.R. and P.C. wrote the paper, with editing help and discussions from N.R., M.D., F.I. and A.H.W.
Reprints and permissions information is available at www.nature.com/reprints.
Competing interests
The authors declare no competing financial interests.
References
- 1.Cossart P. Illuminating the landscape of host–pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glaser P, et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 3.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 4.Archambaud C, et al. Impact of lactobacilli on orally acquired listeriosis. Proc Natl Acad Sci USA. 2012;109:16684–16689. doi: 10.1073/pnas.1212809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wurtzel O, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dar D, et al. Term-Seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science. 2016;352:aad9822. doi: 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 2016;14:686–694. doi: 10.1016/j.celrep.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015;11:13–21. doi: 10.1016/j.celrep.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gevaert K, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 12.Staes A, et al. Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat Protoc. 2011;6:1130–1141. doi: 10.1038/nprot.2011.355. [DOI] [PubMed] [Google Scholar]
- 13.Bland C, Hartmann EM, Christie-Oleza JA, Fernandez B, Armengaud J. N-terminal-oriented proteogenomics of the marine bacterium Roseobacter denitrificans Och114 using N-succinimidyloxycarbonylmethyl)tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP) labeling and diagonal chromatography. Mol Cell Proteomics. 2014;13:1369–1381. doi: 10.1074/mcp.O113.032854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakahigashi K, et al. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 2016;23:193–201. doi: 10.1093/dnares/dsw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bienvenut WV, Giglione C, Meinnel T. Proteome-wide analysis of the amino terminal status of Escherichia coli proteins at the steady-state and upon deformylation inhibition. Proteomics. 2015;15:2503–2518. doi: 10.1002/pmic.201500027. [DOI] [PubMed] [Google Scholar]
- 16.Chen DZ, et al. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry. 2000;39:1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 18.Becavin C, et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio. 2014;5:e00969–14. doi: 10.1128/mBio.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renier S, Micheau P, Talon R, Hebraud M, Desvaux M. Subcellular localization of extracytoplasmic proteins in monoderm bacteria: rational secretomics-based strategy for genomic and proteomic analyses. PLoS ONE. 2012;7:e42982. doi: 10.1371/journal.pone.0042982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malys N, McCarthy JE. Translation initiation: variations in the mechanism can be anticipated. Cell Mol Life Sci. 2011;68:991–1003. doi: 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen NY, Paulus H. Mechanism of expression of the overlapping genes of Bacillus subtilis aspartokinase II. J Biol Chem. 1988;263:9526–9532. [PubMed] [Google Scholar]
- 22.Park SK, et al. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 23.Nagy M, et al. Synergistic cooperation between two ClpB isoforms in aggregate reactivation. J Mol Biol. 2010;396:697–707. doi: 10.1016/j.jmb.2009.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin TH, Hu YN, Shaw GC. Two enzymes, TilS and HprT, can form a complex to function as a transcriptional activator for the cell division protease gene ftsH in Bacillus subtilis. J Biochem. 2014;155:5–16. doi: 10.1093/jb/mvt081. [DOI] [PubMed] [Google Scholar]
- 25.Oh E, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsirigos KD, Peters C, Shu N, Kall L, Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marles-Wright J, et al. Molecular architecture of the ‘stressosome,’ a signal integration and transduction hub. Science. 2008;322:92–96. doi: 10.1126/science.1159572. [DOI] [PubMed] [Google Scholar]
- 28.Marles-Wright J, Lewis RJ. The stressosome: molecular architecture of a signalling hub. Biochem Soc Trans. 2010;38:928–933. doi: 10.1042/BST0380928. [DOI] [PubMed] [Google Scholar]
- 29.Murray JW, Delumeau O, Lewis RJ. Structure of a nonheme globin in environmental stress signaling. Proc Natl Acad Sci USA. 2005;102:17320–17325. doi: 10.1073/pnas.0506599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milohanic E, et al. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- 31.Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dougan DA, Truscott KN, Zeth K. The bacterial N-end rule pathway: expect the unexpected. Mol Microbiol. 2010;76:545–558. doi: 10.1111/j.1365-2958.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- 33.Yu XJ, Liu M, Matthews S, Holden DW. Tandem translation generates a chaperone for the Salmonella type III secretion system protein SsaQ. J Biol Chem. 2011;286:36098–36107. doi: 10.1074/jbc.M111.278663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu Rev Biochem. 2014;83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin JE, Waters LS, Storz G, Imlay JA. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet. 2015;11:e1004977. doi: 10.1371/journal.pgen.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lippa AM, Goulian M. Perturbation of the oxidizing environment of the periplasm stimulates the PhoQ/PhoP system in Escherichia coli. J Bacteriol. 2012;194:1457–1463. doi: 10.1128/JB.06055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiensuu T, Andersson C, Ryden P, Johansson J. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol Microbiol. 2013;87:909–924. doi: 10.1111/mmi.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, et al. Rsbv of Listeria monocytogenes contributes to regulation of environmental stress and virulence. Arch Microbiol. 2013;195:113–120. doi: 10.1007/s00203-012-0855-5. [DOI] [PubMed] [Google Scholar]
- 40.Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J Bacteriol. 2003;185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia X, Wang JB, Rivera S, Duong D, Weinert EE. An O2-sensing stressosome from a Gram-negative bacterium. Nat Commun. 2016;7:12381. doi: 10.1038/ncomms12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellin JR, et al. A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proc Natl Acad Sci USA. 2013;110:13132–13137. doi: 10.1073/pnas.1304795110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balestrino D, et al. Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. Appl Environ Microbiol. 2010;76:3625–3636. doi: 10.1128/AEM.02612-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hastings JW, Morin JG. Calcium-triggered light emission in Renilla. A unitary biochemical scheme for coelenterate bioluminescence. Biochem Biophys Res Commun. 1969;37:493–498. doi: 10.1016/0006-291x(69)90942-5. [DOI] [PubMed] [Google Scholar]
- 47.Eskandarian HA, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 48.Kall L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 49.Martens L, Vandekerckhove J, Gevaert K. DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics. 2005;21:3584–3585. doi: 10.1093/bioinformatics/bti588. [DOI] [PubMed] [Google Scholar]
- 50.Helsens K, et al. Ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics. 2010;10:1261–1264. doi: 10.1002/pmic.200900409. [DOI] [PubMed] [Google Scholar]
- 51.Helsens K, Timmerman E, Vandekerckhove J, Gevaert K, Martens L. Peptizer, a tool for assessing false positive peptide identifications and manually validating selected results. Mol Cell Proteomics. 2008;7:2364–2372. doi: 10.1074/mcp.M800082-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Vizcaino JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma J, Campbell A, Karlin S. Correlations between Shine–Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol. 2002;184:5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 55.Aziz RK, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. Refseq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42:D553–D559. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jonquieres R, Bierne H, Fiedler F, Gounon P, Cossart P. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of Gram-positive bacteria. Mol Microbiol. 1999;34:902–914. doi: 10.1046/j.1365-2958.1999.01652.x. [DOI] [PubMed] [Google Scholar]
- 60.Mengaud J, et al. Antibodies to the leucine-rich repeat region of internalin block entry of Listeria monocytogenes into cells expressing E-cadherin. Infect Immun. 1996;64:5430–5433. doi: 10.1128/iai.64.12.5430-5433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Archambaud C, Gouin E, Pizarro-Cerda J, Cossart P, Dussurget O. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol Microbiol. 2005;56:383–396. doi: 10.1111/j.1365-2958.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- 62.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 63.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 64.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 66.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 67.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 68.Trott O, Olson AJ. Autodock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.