Abstract
New Guinea shows human occupation since ~50 thousand years ago (kya), independent adoption of plant cultivation ~10 kya, and great cultural and linguistic diversity today. We performed genome-wide SNP genotyping on 381 individuals from 85 language groups in Papua New Guinea (PNG) and find a sharp divide originating 10-20 kya between lowland and highland groups, and a lack of non-New Guinean admixture in the latter. All highlanders share ancestry within the last 10 kya, with major population growth in the same period, suggesting population structure was reshaped following the Neolithic lifestyle transition. However, genetic differentiation between groups in PNG is much stronger than in comparable regions in Eurasia, demonstrating that such a transition does not necessarily limit the genetic and linguistic diversity of human societies.
The island of New Guinea contains some of the earliest archaeological evidence for modern humans outside of Africa, dating back to approximately 50 kya (1). Starting ~10 kya, systematic plant cultivation was developed in its central mountain range (2), approximately coinciding with similar, independent developments in the Near East, East Asia and the Americas. Today, the country of Papua New Guinea (PNG) occupies the eastern half of the island and northern Island Melanesia, and is the most linguistically diverse country with approximately 850 languages (3). About half belong to the Trans-New Guinea phylum, spoken across all of the highlands and large parts of the lowlands and hypothesized to have spread alongside plant cultivation (4).
The Sahul continent appears to have been isolated from the rest of the world at least until the last few thousand years (5, 6), so its prehistory likely represents an independent instance of human genetic and cultural evolution over ~50 ky. Genetic studies are increasingly indicating that agriculture, languages and culture in Eurasia and Africa have primarily spread through the movement of people (7–10), and it is of great interest to understand if the shift from a hunter-gatherer to a sedentary, cultivation-based lifestyle in New Guinea – which we here refer to as a Neolithic transition – followed similar patterns.
We genotyped 381 individuals from 85 language groups across PNG at 1.7 million genome-wide markers (Figs. 1A, S1, tables S1, S2, (11)), and analysed 39 previously generated high-coverage whole-genome sequences (6, 12), including the PNG samples from the HGDP-CEPH panel (which we find consists of one highland and one lowland subset (fig. S2, table S3, (11))).
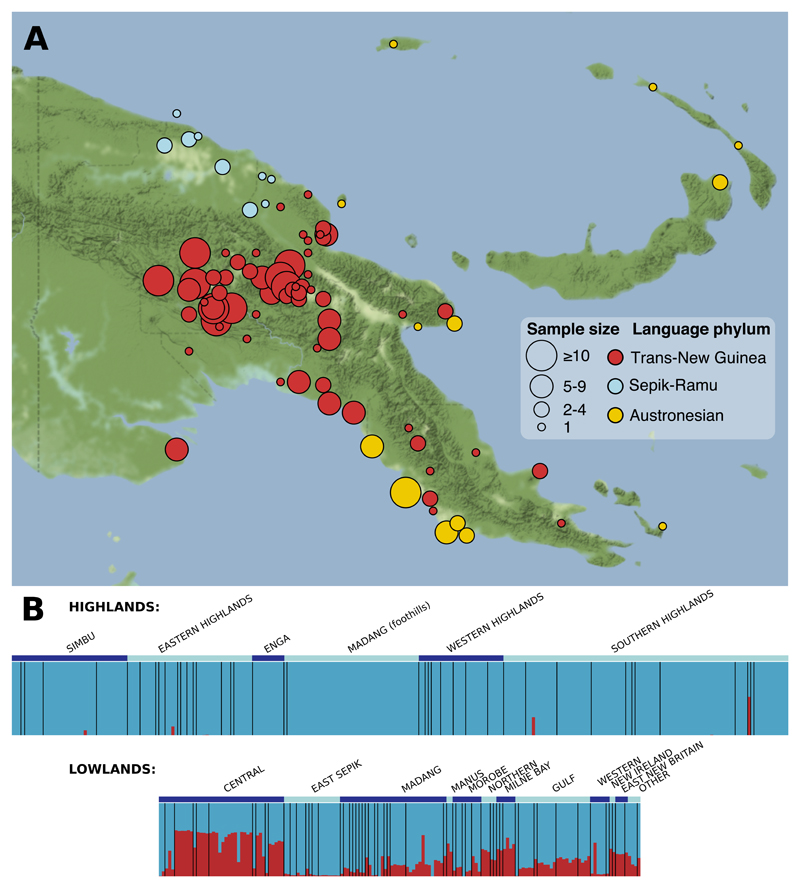
Fig. 1. PNG samples.
(A) Each language group is represented by a circle, the area of which indicates the number of genotyped individuals and the colour the top-level language phylum. 39 individuals are not included as the specific language is unknown or the two parents are from different language groups. Also see fig. S1. (B) Papuan (blue) and Southeast Asian (red) ancestry proportions as estimated by ADMIXTURE (K=2 with 504 East Asian individuals from the 1000 Genomes Project, also see fig. S3); individuals are grouped by province and then language group (separated by black bars). Ancestry proportions correlate strongly (r = 0.988) with those estimated using f4-ratios (11).
We first examined the impact of external gene flow to PNG, particularly that derived from Holocene migrations from Southeast Asia (13). Highlanders show no excess shared ancestry with Asians relative to Aboriginal Australians (D-statistics (14), Z > 2 (11), and ADMIXTURE (15)), except for four individuals who likely reflect recent admixture via the lowlands (Fig. 1B). We also find no mitochondrial or Y chromosomes of recent non-Sahul origin in any highlander (figs. S4, S5). The lowlands, however, harbour widespread Southeast Asian ancestry, with substantially higher levels in Austronesian speakers than in non-Austronesian speakers (mean of 38.7% vs 11.6%, p = 1.4×10−13, Wilcoxon rank sum test). The lowest levels (mean of 4.3%) are found in northern groups speaking Sepik-Ramu phylum languages. Our results thus demonstrate a variable Southeast Asian genetic impact on different parts of PNG, and independence of highlander ancestry from non-Sahul sources.
Papuans diverged genetically from Aboriginal Australians long before rising sea levels separated New Guinea and Australia ~8 kya, and different groups across Australia display a uniform relationship to Papuans (6). When accounting for Southeast Asian admixture using admixture graphs and D-statistics (14), we similarly find that all genotyped Papuan individuals share a uniform relationship to Aboriginal Australians (fig. S6), revealing a lack of genetic continuity across Sahul.
The strongest genetic separation within PNG appears to be that between the mainland and the Bismarck archipelago islands (New Britain and New Ireland) (Figs. 2B, S7), consistent with previous studies (16). Highlanders fall into three clusters: one western, one eastern and one corresponding to a small set of Angan language groups from the south-eastern highlands (Figs. 2A, S8), the last showing evidence of genetic isolation (fig. S9).
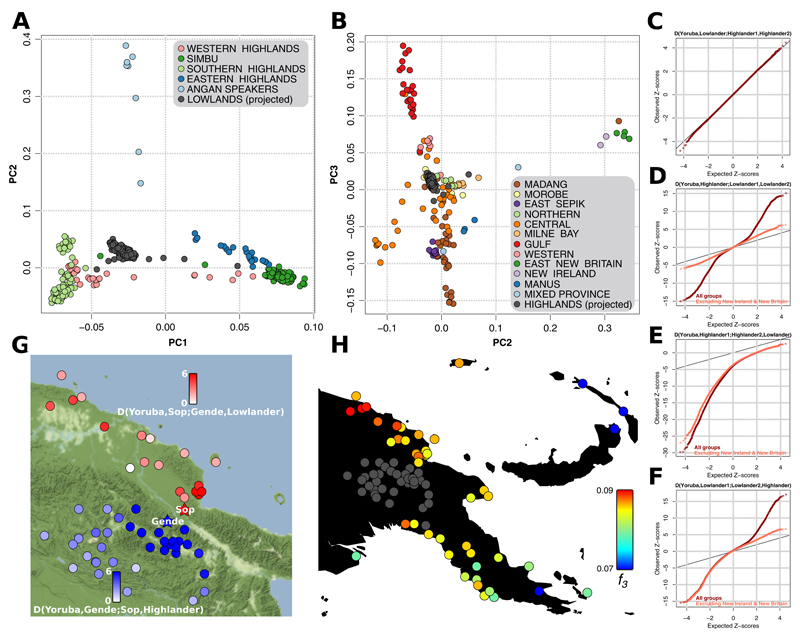
Fig. 2. PNG Population structure.
(A) When projected onto principal components constructed with only highlander genotypes, all lowlanders (excepting a few outliers) group uniformly. Also see fig. S10. (B) When projected onto principal components constructed with only lowlander genotypes, all highlanders (excepting a few outliers) group uniformly. Also see fig. S11. (C-F) Quantile-quantile plots comparing Z-scores from D-statistics relating highlanders and lowlanders to those expected under a normal distribution (11). (C) Lowlanders are equally similar to different highlander groups. (D) Highlanders have stronger affinity to some lowlander groups than to others. (E) Highlanders are more similar to each other than to lowlanders. (F) Lowlanders are not always more similar to each other than to highlanders. (G) Z-scores (capped at 6) of two different D-statistics, the first measuring if the highland Gende speakers are more similar to the lowland Sop speakers, living just 40 km away, or to other highlanders (blue meaning more highlander similarity), and the second if Sop are more similar to Gende or to other lowlanders (red meaning more lowlander similarity). (H) Genetic affinity of highlanders (treated as a single group, in grey) to different lowland groups measured by the outgroup f3 statistic f3(Highlanders,X;Aboriginal Australian) (red meaning higher affinity). C-H were calculated after masking lowlander genomes for Southeast Asian ancestry.
To compare highlanders and lowlanders, we masked lowlander genomes for Southeast Asian ancestry tracts, achieving a misclassification rate of <0.5% (11). We find no differences in the affinity of lowlanders to different highlander groups using PCA and D-statistics (Figs. 2A,C, S10, (11)). Thus, all highlanders, regardless of geographic location, seem to form a clade relative to lowlanders. In line with this, there is not a single D-statistic (at Z > 3) in which a highlander group is more similar to any lowlander group than to any other highlander group (Fig. 2E, 2G).
In contrast, highlanders as a group are not equally similar to all lowlanders (Fig. 2D), displaying slightly higher affinity to groups from the Sepik river region (Fig. 2H). This is surprising linguistically, as the local Sepik-Ramu languages are unrelated to the Trans-New Guinea languages of the highlands. There is, however, archaeological evidence for Holocene cultural contact between the two regions (17). While highlanders are all similar amongst themselves, the same is not true of lowlanders (Fig. 2F) – both southern and northern lowlanders are more similar to highlanders than they are to each other.
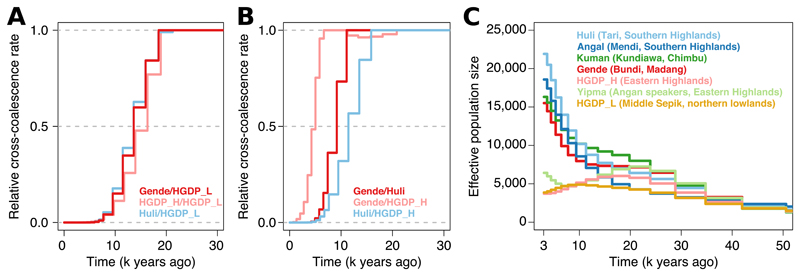
To investigate when present-day groups in PNG separated, we applied MSMC (18) to whole-genome sequences for six highland groups and one Sepik lowlands group. We used 10x Genomics linked-read whole-genome sequencing (19) to physically phase eight genomes (table S4), and also analysed perfectly phased male X chromosomes (11). The results suggest that highlanders and Sepik lowlanders separated 10-20 kya. All splits within the highlands seem to have occurred within the last ~10 kya (Figs. 3A, B, S12). A Y-chromosomal phylogeny similarly revealed shared ancestry across groups within these timescales (fig. S13). We also find evidence of major increase in effective population sizes in most highlander groups in the last 10 ky (Figs. 3c, S14), using SMC++ (20) and MSMC. Sepik lowlanders do not share this increase, consistent with anthropological records of lower lowland population densities, likely linked to widespread malaria (21).
Fig. 3. Time depth of population separation and growth in PNG.
(A) Cross-coalescence curves between highlanders and a northern lowlands Middle Sepik group suggests a split time between 10 and 20 kya. (B) Cross-coalescence curves between highland groups suggest split times within the last ~10 ky (Huli representing the western cluster, Gende and HGDP_H the eastern, figs. S8, S10). These were inferred using MSMC on genomes physically phased using linked-read sequencing. Also see fig. S12. (C) Effective population size histories inferred using SMC++ on five genomes per group. Also see fig. S14.
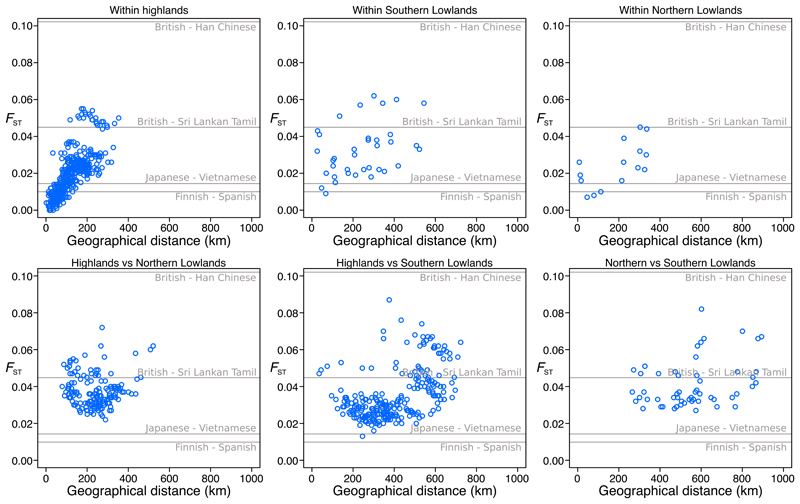
Genetic differentiation is much stronger in PNG than in regions of similar size in Eurasia, where FST values between major populations within Europe or East Asia are generally 1% or less (Fig. 4). Within the highlands, a sampled area about the size of Denmark, FST between eastern and western groups are 2-3%, and values between the Angan-speaking and other groups reach 4-5% (as high as between European and South Asian populations). Within each of the eastern and western highland clusters, values are below 2% but many are above 1%. Levels of FST in the lowlands are also high, suggesting that cultural-linguistic factors, rather than terrain, drive the differentiation. Between the highland, northern lowland and southern lowland regions, differentiation is even higher. Structure is stronger for the Y chromosome than for the mitochondrial genome, suggesting lower male effective population sizes and/or more female movement between groups (fig. S15).
Fig. 4. Genetic differentiation in PNG.
Geographical distance between groups plotted against FST, after masking lowlander genomes for Southeast Asian ancestry. Grey lines indicate FST between selected 1000 Genomes Project populations.
Our results confirm the independent evolution of Sahul for most of the last 50 ky, and the independence of New Guinea from Australia for much of this time. Present-day mainland population structure, marked by a very sharp highland-lowland division, does not date back to the initial peopling of Sahul, but instead appears to have formed within the last 20 ky. Highland structure formed subsequently, mostly within the last 10 ky, which is within the general timescale of the spread of cultivation (2) and the Trans-New Guinea languages (4). We thus propose that an expansion of cultivating groups across the highlands could explain our observations, including the uniform relationship of highlanders to lowlanders and the recent increase in population sizes. Our data also suggest higher diversity in the western than the eastern highlands (figs. S9,S14), consistent with a hypothesized origin of cultivation in the former (22). Our results thus suggest that, as in many other parts of the world, the spread of cultivation in PNG was associated with an expansion of peoples and a reshaping of population structure.
The strong genetic differentiation within PNG, however, sets it apart from other parts of the world that also underwent Neolithic lifestyle transitions. Ancient DNA studies in Europe and the Near East have documented a gradual but dramatic decrease in differentiation, showing that the genetic homogeneity of present-day west Eurasia emerged in the last few thousand years (10, 23). FST values in PNG fall between those of hunter-gatherers and present-day populations of west Eurasia, suggesting that a transition to cultivation alone does not necessarily lead to genetic homogenization.
A key difference might be that PNG had no Bronze Age, which in west Eurasia was driven by an expansion of herders and led to massive population replacement, admixture and cultural and linguistic change (7, 8), or Iron Age like that linked to the expansion of Bantu-speaking farmers in Africa (24). Such cultural events have resulted in rapid Y chromosome lineage expansions due to increased male reproductive variance (25), but we consistently find no evidence for this in PNG (fig. S13). Thus, in PNG, we may be seeing the genetic, linguistic and cultural diversity that sedentary human societies can achieve in the absence of massive technology-driven expansions.
Supplementary Material
One Sentence Summary.
A Neolithic transition reshaped population structure in Papua New Guinea, but in comparison to other areas of the world, present-day genetic differentiation remains high.
Acknowledgments
We thank all sample donors who contributed to this study, T. Parks, A. V. Hill, J. B. Clegg, D. Higgs, D. J. Weatherall, O. Bunari, A. Spencer, J. Barker, R. Spark and P. Sill for assistance in sample collection and discussion, Jonathan Friedlaender for background information on the HGDP-CEPH samples and the Wellcome Trust Sanger Institute genotyping and sequencing facilities for generating data, especially Michael Quail, David Jackson and Steven Leonard for generating 10x Genomics data. A.B., Y.X., M.S.S. and C.T.-S. were supported by the Wellcome Trust (098051). A.J.M. was supported by a Wellcome Trust Clinical Research Training grant (106289/Z/14/Z). S.J.O., A.J.M., and K.A. were supported by a Wellcome Trust Core Award (090532/Z/09/Z) and K.A. was supported by a European Research Council Advanced Grant to Adrian VS Hill (294557). The array genotypes are available for population history studies (EGA accession EGAS00001001587). The 10x Genomics sequencing data are available as two sets, the HGDP-CEPH samples with no restrictions (ENA accession ERP015796) and the others for population history studies (EGA accession EGAS00001001853).
References
- 1.O'Connell JF, Allen J. The process, biotic impact, and global implications of the human colonization of Sahul about 47,000 years ago. J Archaeol Sci. 2015;56:73–84. [Google Scholar]
- 2.Denham T, Barton H. The emergence of agriculture in New Guinea: a model of continuity from pre-existing foraging practices. In: Kennett DJ, Winterhalder B, editors. Behavioral Ecology and the Transition to Agriculture. 2006. pp. 237–264. [Google Scholar]
- 3.Lewis MP, Simons GF, Fennig CD. Ethnologue: Languages of the World. Nineteenth edition. 2016. [Google Scholar]
- 4.Pawley A. The chequered career of the Trans New Guinea hypothesis: recent research and its implications. In: Pawley A, Attenborough R, Golson J, Hide R, editors. Papuan Pasts: cultural, linguistic and biological histories of Papuan-speaking peoples. 2005. pp. 67–107. [Google Scholar]
- 5.Bergström A, Nagle N, Chen Y, McCarthy S, Pollard MO, Ayub Q, Wilcox S, Wilcox L, van Oorschot RA, McAllister P, Williams L, et al. Deep Roots for Aboriginal Australian Y Chromosomes. Curr Biol. 2016;26:809–813. doi: 10.1016/j.cub.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malaspinas AS, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergström A, Athanasiadis G, Cheng JY, Crawford JE, Heupink TH, et al. A genomic history of Aboriginal Australia. Nature. 2016;538:207–214. doi: 10.1038/nature18299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, Fu Q, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allentoft ME, Sikora M, Sjogren KG, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlstrom T, Vinner L, Malaspinas AS, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 9.de Filippo C, Bostoen K, Stoneking M, Pakendorf B. Bringing together linguistic and genetic evidence to test the Bantu expansion. Proc Biol Sci. 2012;279:3256–3263. doi: 10.1098/rspb.2012.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, Connell S, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supplementary materials at the Science website.
- 12.Raghavan M, Steinrucken M, Harris K, Schiffels S, Rasmussen S, DeGiorgio M, Albrechtsen A, Valdiosera C, Avila-Arcos MC, Malaspinas AS, Eriksson A, et al. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science. 2015;349:aab3884. doi: 10.1126/science.aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duggan AT, Stoneking M. Recent developments in the genetic history of East Asia and Oceania. Curr Opin Genet Dev. 2014;29:9–14. doi: 10.1016/j.gde.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedlaender JS, Friedlaender FR, Reed FA, Kidd KK, Kidd JR, Chambers GK, Lea RA, Loo JH, Koki G, Hodgson JA, Merriwether DA, et al. The genetic structure of Pacific Islanders. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swadling P, Wiessner P, Tumu A. Prehistoric stone artefacts from Enga and the implication of links between the highlands, lowlands and islands for early agriculture in Papua New Guinea. Le Journal de la Société des Océanistes. 2008:271–292. [Google Scholar]
- 18.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, Kyriazopoulou-Panagiotopoulou S, Masquelier DA, Merrill L, Terry JM, Mudivarti PA, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terhorst J, Kamm JA, Song YS. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 2017;49:303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley ID. Population-Change and Distribution in Papua-New-Guinea - an Epidemiological Approach. J Hum Evol. 1983;12:125–132. [Google Scholar]
- 22.Feil DK. The Evolution of Highland Papua New Guinea Societies. Cambridge University Press; 1987. [Google Scholar]
- 23.Skoglund P, Malmstrom H, Omrak A, Raghavan M, Valdiosera C, Gunther T, Hall P, Tambets K, Parik J, Sjogren KG, Apel J, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 24.Huffman TN. Archaeology and Ethnohistory of the African Iron-Age. Annu Rev Anthropol. 1982;11:133–150. [Google Scholar]
- 25.Poznik GD, Xue Y, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, Ayub Q, McCarthy SA, Narechania A, Kashin S, Chen Y, et al. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat Genet. 2016;48:593–599. doi: 10.1038/ng.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourke RM, Harwood T, editors. Food and agriculture in Papua New Guinea. ANU E Press, The Australian National University; 2009. [Google Scholar]
- 27.Glottolog. Nuclear Trans-New Guinea family. 2017 http://glottolog.org/resource/languoid/id/nucl1709.
- 28.Vines AP. An epidemiological sample survey of the Highlands, Mainland, and Islands Regions of the Territory of Papua and New Guinea. DM Thesis, University of Sydney, Australia; 1967. [DOI] [PubMed] [Google Scholar]
- 29.Ward RG. Distribution and density of population. In: Ward RG, Lea DAM, Ploeg M, editors. An Atlas of Papua New Guinea. 1970. pp. 8–11. [Google Scholar]
- 30.Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR, Serjeantson SW, Bana-Koiri J, Bhatia K, Alpers MP, Boyce AJ, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321:744–750. doi: 10.1038/321744a0. [DOI] [PubMed] [Google Scholar]
- 31.Oppenheimer SJ, Higgs DR, Weatherall DJ, Barker J, Spark RA. Alpha thalassaemia in Papua New Guinea. Lancet. 1984;1:424–426. doi: 10.1016/s0140-6736(84)91754-9. [DOI] [PubMed] [Google Scholar]
- 32.Oppenheimer SJ, Gibson FD, Macfarlane SB, Moody JB, Hendrickse RG. Iron Supplementation and Malaria. Lancet. 1984;1:389–390. doi: 10.1016/s0140-6736(84)90434-3. [DOI] [PubMed] [Google Scholar]
- 33.Oppenheimer SJ, Hill AV, Gibson FD, Macfarlane SB, Moody JB, Pringle J. The interaction of alpha thalassaemia with malaria. Trans R Soc Trop Med Hyg. 1987;81:322–326. doi: 10.1016/0035-9203(87)90253-7. [DOI] [PubMed] [Google Scholar]
- 34.Hill AV, Bowden DK, Trent RJ, Higgs DR, Oppenheimer SJ, Thein SL, Mickleson KN, Weatherall DJ, Clegg JB. Melanesians and Polynesians share a unique alpha-thalassemia mutation. Am J Hum Genet. 1985;37:571–580. [PMC free article] [PubMed] [Google Scholar]
- 35.Hill AV, Bowden DK, Flint J, Whitehouse DB, Hopkinson DA, Oppenheimer SJ, Serjeantson SW, Clegg JB. A population genetic survey of the haptoglobin polymorphism in Melanesians by DNA analysis. Am J Hum Genet. 1986;38:382–389. [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden DK, Hill AV, Higgs DR, Oppenheimer SJ, Weatherall DJ, Clegg JB. Different hematologic phenotypes are associated with the leftward (-alpha 4.2) and rightward (-alpha 3.7) alpha+-thalassemia deletions. J Clin Invest. 1987;79:39–43. doi: 10.1172/JCI112804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv: 1207.3907. 2012 [Google Scholar]
- 40.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 41.Weissensteiner H, Pacher D, Kloss-Brandstatter A, Forer L, Specht G, Bandelt HJ, Kronenberg F, Salas A, Schonherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reich D, Patterson N, Kircher M, Delfin F, Nandineni MR, Pugach I, Ko AMS, Ko YC, Jinam TA, Phipps ME, Saitou N, et al. Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania. American Journal of Human Genetics. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loh PR, Danecek P, Palamara PF, Fuchsberger C, A R Y, K F H, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PL, Aximu-Petri A, Prufer K, de Filippo C, Meyer M, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skoglund P, Posth C, Sirak K, Spriggs M, Valentin F, Bedford S, Clark GR, Reepmeyer C, Petchey F, Fernandes D, Fu Q, et al. Genomic insights into the peopling of the Southwest Pacific. Nature. 2016;538:510–513. doi: 10.1038/nature19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, Skoglund P, et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song S, Sliwerska E, Emery S, Kidd JM. Modeling Human Population Separation History Using Physically Phased Genomes. Genetics. 2017;205:385–395. doi: 10.1534/genetics.116.192963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood JW. The Genetic Demography of the Gainj of Papua-New-Guinea. 2. Determinants of Effective Population-Size. Am Nat. 1987;129:165–187. [Google Scholar]
- 55.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 56.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 57.Kayser M, Brauer S, Weiss G, Schiefenhovel W, Underhill P, Shen P, Oefner P, Tommaseo-Ponzetta M, Stoneking M. Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am J Hum Genet. 2003;72:281–302. doi: 10.1086/346065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.