Abstract
Recent evidence indicates that human cancer cells reactivate the expression of latent human endogenous retroviral (HERV) proteins. However, the extent to which cancer patients mount de novo immune responses against expressed HERV elements is unclear. In this study, we determined the extent of HERV-K env expression in human breast cancer (BC) and whether both humoral and cell-mediated immunity against HERV-K can be found in BC patients. We found HERV-K env protein expression in 88% of BC (n = 119) but not in normal breast (n = 76) tissues. ELISA screening assays detected significant titers of anti–HERV-K env IgG in a large proportion of BC patients. T-cell responses against HERV-K were also detected in peripheral blood mononuclear cells (PBMC) from BC patients stimulated with autologous dendritic cells pulsed with HERV-K env SU antigens. These responses included induction of T-cell proliferation (P = 0.0043), IFN-γ production measured by enzyme-linked immunospot (P < 0.0001), and multiplex cytokine secretion (P = 0.0033). Multiplex cytokine analysis found a T-helper 1 cytokine response, including interleukin (IL)-2 (P = 0.0109), IL-6 (P = 0.0396), IL-8 (P = 0.0169), and IP-10 (P = 0.0045) secretion during in vitro stimulation of BC PBMC with HERV-K antigen. We also found HERV-K–specific CTLs that were capable of lysing target cells expressing HERV-K env protein in BC patients but not in normal female controls without cancer. These findings suggest that retroviral gene products are capable of acting as tumor-associated antigens activating both T-cell and B-cell responses in BC patients.
Introduction
Breast cancer (BC) is the most common cancer in women. The chance of developing invasive BC at some time in a woman’s life is about one in eight, and there are over 2 million BC survivors in the United States. It was estimated that in 2007, more than 178,000 new cases of invasive BC would be diagnosed among women, and about 41,000 women would die from the disease, according to the American Cancer Society. Although early detection and improved treatment have reduced the mortality rates of BC, it is still the second leading cause of cancer death in American women.
Clinical trials for a variety of malignant diseases have shown that T-cell therapy may be effective and even curative for some patients. Many tumor-associated antigens (TAA) have been identified, such as melanoma antigens gp100 and MART-1 (1, 2), and their ability to induce antitumor T-cell immunity has been shown in clinical studies. Antigens being targeted in T-cell responses against BC, such as HER2/neu, p53, carcinoembryonic antigen (CEA), and telomerase, are overexpressed self-antigens. One of the main hurdles encountered is the need to overcome self-tolerance mechanisms that limit the immune response against these types of tumor antigens. In contrast, viral antigens may be better suited to trigger stronger antitumor T-cell responses due to their foreign nature. Although only a few human cancers are associated with a viral etiology, recent evidence indicates that endogenous retroviruses silenced under normal conditions throughout our lifetime are induced in cancer cells and may be a new source of viral-like tumor antigens.
Human endogenous retroviruses (HERV) comprise up to 8.3% of the human genome, as determined by the Human Genome Sequencing Project (3). The most biologically active HERVs are members of the HERV-K superfamily, which are transcriptionally active in several human cancer tissues (4, 5), and in tumor cell lines, most notably the human BC cell line T47D (6, 7). We recently showed the expression of full-length HERV-K transcripts in human BC (8, 9).
Although the HERV-K envelope (env) transcripts are expressed in BC cells, no studies have reported the expression of HERV-K env protein in actual specimens of breast ductal epithelial tumors in humans. In addition, it is not known whether HERV-K is immunogenic in human BC. At present, only one report has documented the presence of HERV-K–specific CTL responses in human cancer (10). However, whether HERV-K itself primes an antitumor T-cell response has not been directly addressed. In this study, we show that HERV-K env proteins are expressed in invasive ductal breast carcinomas using immunohistochemistry. We also show that BC patients mount cellular and humoral immune responses against HERV-K env, characterized by the presence of elevated levels of antigen-specific T-cell proliferation, cytokine production, HERV-K–specific CTL activity, and presence of anti-HERV-IgG in patient sera.
Materials and Methods
HERV-K expression in BC cell lines and tissues
Human breast tissues and peripheral blood mononuclear cells (PBMC) from BC patients (Table 1), patients with other cancers, or healthy female controls were obtained from the University of Alabama at Birmingham and The M. D. Anderson Cancer Center according to approved Institutional Review Board protocols. HLA typing for HLA-A0201+ (HLA-A2.1) was performed by flow cytometry using anti-human HLA-A2-PE antibody (BD PharMingen). Multiple breast tissue arrays (n = 182) that included normal breast tissues (n = 56), hyperplastic breast tissues (n = 7), and BC tissues (n = 119) were obtained from Cybrdi, Inc.8 Individual slides prepared from 34 BC tissues (diagnosed by a pathologist) and 20 normal breast tissues collected from women undergoing reduction mammoplasty (without cancer, as diagnosed by a pathologist) were obtained from The University of Texas Health Science Center at Houston. Human BC cell lines were obtained from the American Type Culture Collection.
Table 1.
Patient characteristics
| Patient sample | Age/sex | Diagnosis | ER, PR, HER2/neu, nuclear grade, LN status |
Date of diagnosis (month/year) |
|---|---|---|---|---|
| BC1 | 64/F | IDC + SCC | N, N, N, 3, N | Jul-06 |
| BC2 | 66/F | IDC | P, P, N, 2, N | Sep-06 |
| BC3 | 49/F | IDC | P, P, N, 2, P | Aug-06 |
| BC4 | 43/F | DCIS | P, P, nd, 3, N | Aug-06 |
| BC5 | 55/F | DCIS + IDC + ILC | P, P, N, 2, P | Oct-06 |
| BC6 | 63/F | IDC + DCIS | P, P, N, 3, P | Mar-07 |
| BC7 | 42/F | DCIS + IDC | P, P, N, 2, nd | Mar-07 |
| BC8 | 51/F | IMuC | P, N, nd, N, P | Apr-07 |
| BC9 | 34/F | MSCT | N/A | Apr-07 |
| BC10 | 51/F | IMC, Mixed D + L | P, P, N, 1, N | Mar-07 |
| BC11 | 54/F | IDC | N, N, P, 3, P | May-07 |
| BC12 | 54/F | IDC | P, N, P, 2, P | Apr-07 |
| BC13 | 52/F | IDC | P, P, N, 3, P | May-07 |
| BC14 | 47/F | IDC | P, N, N, 3, P | May-07 |
| BC15 | 63/F | IDC | P, P, N, 3, N | Mar-07 |
| BC16 | 41/F | IDC | N, N, N, 3, N | June, 07 |
| BC17 | 56/F | IDC | P, P, N, 2, N | Aug. 07 |
| BC18 | 56/F | IDC | P, N, N, nd, P | Nov. 07 |
| BC19 | 76/F | IDC | P, N, nd, 3, N | Nov. 07 |
| BC20 | 42/F | DCIS + IDC | P, P, nd, 2, N | Nov. 07 |
| BC21 | 58/F | IDC | P, P, N, 3, P | Nov. 07 |
| BC50 | 51/F | IDC | nd | Dec-92 |
| BC51 | 59/F | IDC | nd | Apr-04 |
| BC52 | 40/F | DCIS + ILC | nd | Jun-04 |
| BC53 | 54/F | DCIS + IDC | nd | Mar-03 |
| BC54 | 41/F | ILC | nd | Sep-04 |
| BC55* | 41/F | IDC | Negative | Apr-04 |
| BC56 | 49/F | IDC | Negative | Jan-00 |
| BC57 | 55/F | DCIS | Negative | Mar-04 |
| BC58 | 32/F | IDC | Negative | Dec-03 |
| BC59 | 55/F | IDC + colon C | Positive | 1990 + 2004 |
| BC60 | 38/F | IDC | Positive | 1993 + 2000 |
| BC61 | 49/F | DCIS | Negative | 1997 |
| BC262 | 42/F | DCIS | Negative | Nov-04 |
| BC63 | 79/F | IDC | Negative | 1994 |
| BC9824 | 52/F | IDC + recurrent | Positive | 1996 + 1998 |
Abbreviations: LN, lymph node; N, negative; P, positive; N/A, not available; nd, not determined; SCC, squamous cell carcinoma; ILC, invasive lobular carcinoma; IMuC, invasive mucinous carcinoma; MSCT, malignant spindle cell tumor; IMC, invasive mammary carcinoma; Mixed D + L, mixed ductal and lobular; colon C, colon cancer.
Lymph node status only was determined for samples BC55 through BC9824.
Synthesis of HERV-K env fusion proteins, antibodies, and cRNA
HERV-K env surface fusion proteins were expressed from the pGEX-6p1 plasmid (K10G17) in BL-21 (DE3) or pQE30 plasmid (K10Q18) in M15 Escherichia coli at 18°C overnight with 1 mmol/L isopropyl-l-thio-B-d-galactopyranoside. Bacterial pellets were harvested, disrupted by lysozyme treatment followed by sonication, clarified by 0.2 µm filtration, and affinity purified with glutathione-Sepharose FF using an ÄKTA fast protein liquid chromatography (FPLC; GE Healthcare; ref. 11). The purified K10Q18 fusion proteins were used to immunize 6- to 8-week-old BALB/c mice. Hybridoma cells were derived from the splenocytes of immunized mice by fusion with a myeloma cell line using standard techniques as described previously (11, 12). Several monoclonal antibodies (mAb; clones 6H5, 4D1, 4E11, 6E11, and 4E6) against HERV-K env SU protein were generated in our laboratory. ELISA and immunoblot screening revealed that mAb 6H5 had the highest specificity and sensitivity toward HERV-K. 6H5 hybridoma ascites was collected from pristane-primed mice, precipitated twice with saturated ammonium sulfate, clarified by 0.2 µm filtration, and affinity purified with protein G-Sepharose HP by FPLC. The eluate fractions containing 6H5 mAb were dialyzed against PBS, concentrated, and filter sterilized before testing. HERV-K cRNA was produced by in vitro transcription (8). Fusion proteins and cRNA were used for production of in vitro stimulated (IVS) cells.
Isolation of anti–HERV-K env IgG from BC patient sera
HERV-K-glutathione S-transferase (GST) fusion protein was coupled to CNBr-activated Sepharose 4B as per the manufacturer’s recommendations (GE Healthcare) and packed onto a gravity-feed chromatography column. HERV-K–positive human BC patient serum was precipitated twice with saturated ammonium sulfate, clarified by 0.2 µm filtration, and affinity purified with this HERV-K-SU-GST resin. The eluate fraction containing patient anti–HERV-K antibody was used to detect 6H5 immunoprecipitates of HERV-K from both MCF-7 and MDA-MB-231 cell lines.
Isolation of HERV-K env from BC patient sera
HERV-K env protein was immunoprecipitated from HERV-K–positive human patient serum using 6H5 as the capture antibody. Patient serum (250 µL) was precleared with 30 µL of protein G-Sepharose FF beads. The cleared supernatant was incubated overnight at 4°C with a 1:1,000 dilution of 6H5 and then incubated at room temperature for 1 h with 30 µL of protein G-Sepharose FF beads to bind the HERV-K env-6H5 complex. The antigen-antibody-bead complex was washed with alternating radioimmunoprecipitation assay and high-salt buffers, boiled in 20 µL of Laemmli sample buffer for 10 min, loaded onto a SDS-PAGE gel for electrophoresis, transferred onto nitrocellulose, and blotted with 6H5 to detect the captured HERV-K env antigen.
Immunohistochemical analysis, ELISA, and Western blot assays
Immunohistochemical analysis was performed on tissue microarray slides using an LV-1 Autostainer universal staining system (Dako; ref. 11). ELISA, Western blot, and immunoprecipitation assays have been described previously (11, 12).
Preparation and fluorescence-activated cell sorting analysis of dendritic cells
Dendritic cells (DC) were generated from adherent or CD14-positive PBMC isolated by magnetic cell sorting with CD14 MicroBeads (Miltenyi Biotec) followed by incubation for 6 d with granulocyte macrophage colony-stimulating factor (GM-CSF) and interleukin (IL)-4 (1,000 units/mL). The immature DCs were harvested and transfected with K-SU protein or cRNA followed by treatment with tumor necrosis factor-α (TNF-α) to induce DC maturation. Immature or mature DCs, with or without prior pulsing with HERV-K env protein, were phenotyped on day 7 using CD86-PE/CD209-PerCP-Cy5.5/CD83-APC (BD Biosciences) and analyzed on a BD FACSCalibur system. DCs stained with only secondary antibody served as negative controls.
IVS of T cells and T-cell proliferation assay
Autologous PBMCs (1 × 106/mL) were added to transfected DC at a DC to PBMC ratio of 1:30 on day 0. DCs were transfected using the N-[1-(2,3-dioleoyloxyl)propyl]-N,N,N-trimethylammoniummethyl sulfate (DOTAP) liposomal transfection reagent. DCs were incubated with a protein/DOTAP mixture at a ratio of 100 µg protein (or cRNA) to 100 AL DOTAP for 4 h at 37°C. Cultures were incubated in the presence of IL-2 (10 units/mL) for 7 d to generate 1-week IVS cells. T-cell proliferation was evaluated in the PBMC or IVS cells by restimulation for 72 h with DC pulsed with no added protein, K-SU protein (or cRNA) or E6 control protein (or cRNA), at a DC to PBMC or IVS ratio of 1:30. After 72 h of incubation, the supernatants were collected and tested for cytokine secretion by bead array assay (see details in section of cytokine bead array), and the cells were pulsed with 1 µCi of [3H]thymidine and incubated for another 18 h at 37°C. Cells were then harvested onto filter papers, transferred to scintillation vials with scintillation fluid, and run on a beta counter.
Enzyme-linked immunospot and CTL assays
An IFN-γ or granzyme B (GrB) enzyme-linked immunospot (ELISPOT) assay to measure cytokine-secreting cells in response to antigen was performed using a commercial kit (Biosource International). ELISPOT plates were coated with 10 µg/mL of purified anti-human IFN-γ or GrB capture antibody and incubated for 24 h at 4°C. Plates were then blocked for 2 h and PBMC or IVS cells were plated at 1 × 105 per well with DCs pulsed with the different proteins or cRNA plated at 5 × 103 per well. Plates were incubated for 24 h at 37°C. Plates were washed and incubated with the detection antibodies for 2 h at 25°C. Plates were washed again and incubated with streptavidin horseradish peroxidase for 2 h at 25°C. Plates were washed again and developed by adding AEC substrate solution and incubating for 10 to 60 min at 25°C. Plates were washed thoroughly with water and allowed to dry, and spots were counted using an ELISPOT reader (C.T.L. Technologies). CTL assays were performed in round-bottomed 96-well plates using a standard 4-h 51Cr-release assay (13) using 5 × 104 target cells. Anti-HLA antibody (U.S. Biological) was added to some wells to block HLA-A. Supernatants (100 µL) from each well were removed and counted in a gamma counter.
Multiplex cytokine bead array analysis
PBMC or IVS cells were stimulated with DC pulsed with no added protein, K-SU protein (or cRNA) or control E6 protein (or cRNA), as was done for T-cell proliferation assays. The supernatants were collected after 72 h of incubation and tested for IFN-γ, IL-2, IL-4, GM-CSF, IL-6, IL-8, TNF-α, and IP-10 using a multiplex cytokine bead array analysis kit (BD Biosciences). The samples were analyzed on a BD FACSArray bioanalyzer.
Statistical analysis
Analyses were done using GraphPad Prism version 4. Statistical significance between groups was determined by a χ2 test, a McNemar’s test, a Fisher’s exact test, or a Student’s t test. A P value of <0.05 is considered to be statistically significant.
Results
The expression of HERV-K env protein in BC patients
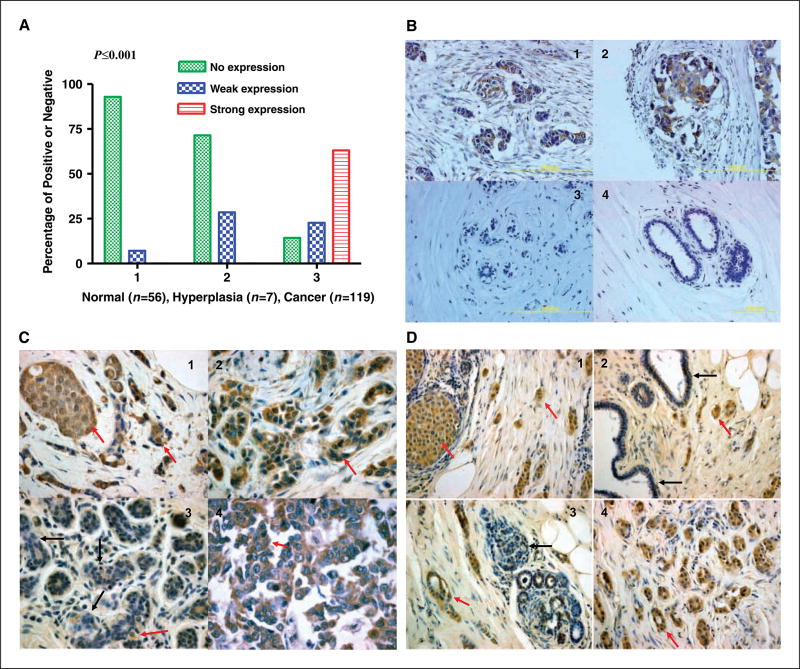
HERV-K recombinant fusion protein, K10Q18, was used to immunize mice to generate antibodies, as described previously (8, 11). A hybridoma clone (6H5) had greater sensitivity against K-SU protein than against HERV-E env surface protein, another HERV family member (cloned into pQE30 vector; data not shown). After we confirmed the specificity and sensitivity of mAb 6H5, immunohistochemistry was used to detect the expression of HERV-K env protein in BC cell lines and tissues. Under identical staining conditions, we stained BC and normal breast tissue specimens (multiple tissue arrays) for HERV-K env protein expression by immunohistochemical analysis using 6H5 to determine the specificity of expression in cancer cells. Expression of HERV-K env protein was found in 102 of 119 (85.7%) invasive ductal carcinoma (IDC) samples, whereas only weak expression was found in 4 of 56 (7.14%) normal breast tissues and in 2 of 7 (28.57%) hyperplasia breast tissues, obtained from multiple BC tissue microarray slides under identical staining conditions (Fig. 1A). Representative photomicrographs of stained samples showing significantly higher HERV-K expression in cancer tissues over benign and normal breast tissues (n = 182; P < 0.001, χ2 test) are shown in Fig. 1B. Figure 1B (panels 1 and 2) shows moderate to strong staining for HERV-K env protein in the invasive tumor cases in comparison with normal breast tissue samples (panel 3 and 4) that were negative. We next determined the association between HERV-K env protein expression in BC tissues (n = 34) obtained from patients with IDC, and associated age, race, tumor size, estrogen receptor (ER), progesterone receptor (PR), HER2/neu status, nuclear grade, and metastatic lymph node involvement data. More than 91% (31 of 34 cases) of biopsies containing >70% tumor cells had intermediate or strong expression of HERV-K env protein. Examples of positively stained immunohistochemical samples are shown in Fig. 1C and D (red arrow). Strong expression of HERV-K env protein was found in ductal carcinoma in situ (DCIS), IDC, and lymph node metastases (Fig. 1C, panel 4), in comparison with adjacent benign tissues (black arrow). There was weak positive HERV-K staining in 3 of 31 uninvolved epithelial cells adjacent to tumor cells of HERV-K–positive tumor cases (P < 0.0001, McNemar’s test). This expression correlated with lymph node involvement and not with other associations measured (P = 0.0483, Fisher’s exact test). In contrast, HERV-K staining was absent in normal breast biopsies (n = 20) obtained from patients without cancer.
Figure 1.
The expression profiles of HERV-K env protein in breast tissue arrays. A, summary of HERV-K env protein expression in arrays of 182 breast tissue samples, including normal breast tissues from healthy patients without cancer (n = 56), benign breast tissues from breast hyperplasia (n = 7), and BC tissues (n = 119). B, panels 1 to 4, representative samples from multiple breast tissue array slides stained with 6H5. The expression of HERV-K env protein was detected in two BC biopsies (panels 1 and 2) but not in normal mammary tissues (panels 3 and 4). C, detection of HERV-K env protein expression in one 48-year-old female diagnosed with infiltrating mammary carcinoma. Positive-staining tumor epithelial cells (brown color indicated with red arrow) were detected in DCIS (panel 1; magnification, ×40), IDC (panel 2), and metastases to the lymph node (panel 4). Positive-staining was greatly reduced (panel 3; black arrows indicate negative staining) in adjacent uninvolved epithelial cells. Red arrow, however, a few tumor cells were positively stained. D, detection of HERV-K env protein expression in one 59-year-old female diagnosed with IDC. All of the panels in this figure have regions of IDC, DCIS, uninvolved epithelial cells, and normal cells. Positive staining (brown color indicated with red arrows) was detected in tumor epithelial cells from DCIS (panel 1; magnification, ×20) and IDC (panels 1 to 4) but not in normal or uninvolved epithelial cells (negative staining indicated with black arrows in panels 2 and 3). More than 91% (31 of 34 cases) of biopsies containing >70% tumor cells had intermediate or strong expression of HERV-K env protein.
Thus, HERV-K env protein is expressed in >85% of human BC, with >50% of tumors exhibiting strong expression. Little or no HERV-K env protein is seen in adjacent normal tissues and no expression is seen in normal breast ductal tissues. In addition, HERV-K env protein expression is found in both ER/PR+ and HER2/neu+ BC.
Detection and specificity of IgG against HERV-K env in BC patients
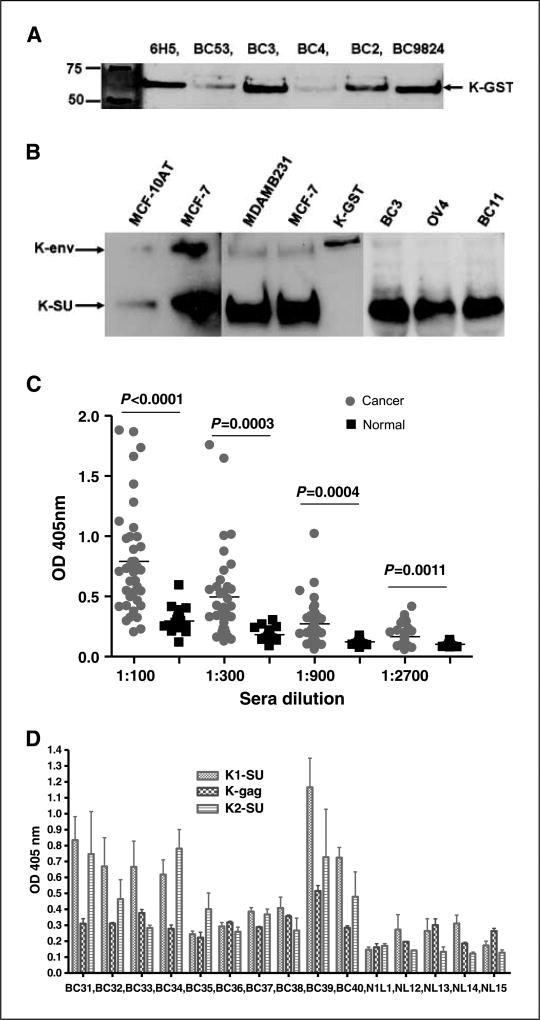
We next tested whether BC patients exhibit elevated levels of IgG reactive against HERV-K env protein over normal donors. Figure 2A shows the presence of anti–HERV-K env protein reactivity in sera from five invasive BC patients using immunoblot analysis in comparison with our 6H5 mAb. The responses varied from weak to intermediate to strong reactivity (Fig. 2A). Several BC patients (patients BC3, BC2, and BC98-24) had anti-HERV-K serum antibodies, which detected K10G17 protein (K-GST; Fig. 2A), as did 6H5 antibody. Anti–HERV-K antibody from patient BC2’s sera was affinity purified with K-GST and used as primary antibody to detect the HERV-K env SU fusion protein (K-GST) and HERV-K env proteins obtained from MDA-MB-231 and MCF-7 BC cells using immunoblot analysis as a further test for the specificity of BC patient serum IgG against HERV-K proteins (Fig. 2B, center gel). Here, the 6H5 mAb was first used to immunoprecipitate HERK-K proteins (K-env and K-SU) from lysates obtained from HERV-K–expressing BC cell lines (MCF-7 and MDA-MB-231) and the weakly expressing MCF10AT precancerous cell line. The immunoprecipitates were separated and blotted with affinity-purified IgG from patient sera (BC2 for example). As shown in Fig. 2B (center blots), purified IgG from the patient sera samples recognized either both full-length K-env (67 kDa) and the K-SU (30 kDa) proteins or the K-SU of the same molecular weights as those recognized by the 6H5 mAb (left blots). The HERV-K env fusion protein (K-GST) was used as a positive control for the IgG purified from BC2 to further show the specificity of the BC patients’ IgG against HERV-K env SU proteins. HERV-K env SU proteins immunoprecipitated with 6H5 in patient sera were detected by immunoblot using 6H5 (BC3 and BC11; right gel). Serum obtained from an ovarian cancer patient (OV4) was used as a control. In all cases, reactivity against the shorter 30-kDa protein was the strongest, suggesting that mostly the shorter HERV-K env SU protein is released in the patient circulation to trigger B-cell responses. Thus, IgG responses against HERV-K env seem to be dominated by recognition of the 30-kDa HERV-K env SU protein.
Figure 2.
Detection of HERV-K env SU protein and anti–HERV-K IgG in patient sera. A, HERV-K env SU fusion protein K-GST was used to detect anti–HERV-K IgG in patient sera by immunoblot analysis. Antibodies against HERV-K env protein were higher in sera obtained from some BC patients (BC3, BC2, and BC9824) than others (BC53 and BC4), as shown by immunoblot using a 6H5 mAb. B, HERV-K env proteins immunoprecipitated from MCF-7 and MDA-MB-231 cells with mAb 6H5 were used to detect anti–HERV-K antibody in patient sera by immunoblot. Left blot, 6H5 mAb detected HERV-K env protein and SU protein in MCF-7 and the weakly expressing MCF10AT precancerous cell line. Full-length K-env (67 kDa) and the K-SU (30 kDa) proteins were recognized by the 6H5 mAb. The amount of IgG against HERV-K env SU protein was greater than the amount of IgG against full-length env protein precipitated from MDA-MB-231 and MCF-7 cells (middle blot) in the serum of patient BC2. K-GST fusion protein, which served as a positive control, was also detected by BC2 serum. Right blot, furthermore, HERV-K env SU proteins from BC3 and BC11 sera were immunoprecipitated by 6H5. Serum obtained from an ovarian cancer patient (OV4) was used as a control. C, titration of antibodies against HERV-K env SU protein in sera from BC patients or normal female donors using ELISA. Anti–HERV-K SU antibody titers were significantly higher in BC patients (n = 39) than in normal donors (n = 20, unpaired Student’s t test). D, proteins from type 1 HERV-K surface (K1-SU; without the 292-bp insert), gag (K-gag), and type 2 HERV-K surface (K2-SU; with the 292-bp insert) were used to detect their respective antibodies in the sera (1:200 dilution) by ELISA. Specific IgGs were detected in BC patients BC31, BC32, BC33, BC34, BC39, and BC40.
To follow-up on this observation, we screened a cohort of BC patient sera for IgG against K-SU using ELISA. ELISA results for one series of serum dilutions (Fig. 2C) revealed higher K-SU antibody titers (P < 0.001–0.005, unpaired Student’s t test) in BC patients (n = 39) than normal female controls (n = 20). Approximately 50% of the BC patient samples (n = 48) were positive for antibodies against HERV-K SU env protein (type 1, without a 292-bp insert), 15% had antibodies against HERV-K gag protein, and 35% had antibodies against HERV-K SU env protein [type 2, with a 292-bp insert, as described previously (8)]. In contrast, anti–HERV-K antibodies in normal donor serum samples (n = 50, absorbance < 0.5) were detected only below the cutoff value of absorbance < 0.5, approximately the mean absorbance of serum samples from normal controls with no history of BC, as described previously (11). One sample is shown in Fig. 2D.
Thus, a high proportion of BC exhibits IgG responses against HERV-K dominated by anti–HERV-K env SU responses with some responses against HERV-K env type 2 SU and HERV-K gag proteins also detected.
HERV-K–specific T-cell responses in BC patients
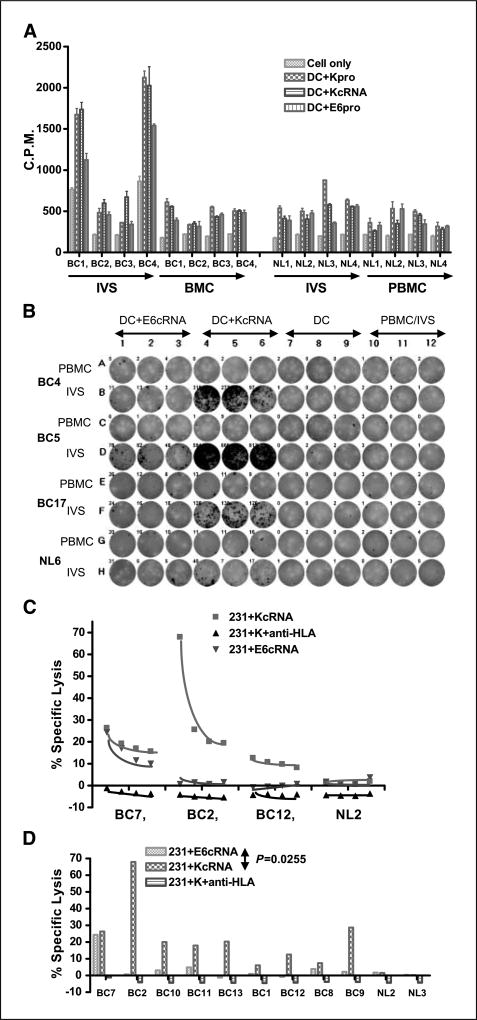
PBMCs obtained from BC patients (see Table 1 for description of patient samples), or normal age-matched female donors, were stimulated with autologous mature DC pulsed with HERV-K env antigen for 1 week. T-cell proliferation specific to HERV-K env cRNA-transfected DC or K-SU protein-treated DC as APCs was compared between PBMC and IVS cells obtained from the same donors by a [3H]thymidine incorporation assay (Fig. 3). Marked HERV-K–specific proliferation was detected after IVS of PBMC from BC patients, with no significant difference between DC pulsed with HERV-K env cRNA (Fig. 3A) or protein (Supplementary Fig. S1). The rate of HERV-K–specific proliferation in BC patients PBMC after IVS was significantly higher than in IVS cells from normal donors (NL). The proliferation index to HERV-K, calculated as the ratio of counts per minute (cpm) from IVS PBMC after HERV-K restimulation to the cpm obtained in response to HERV-K restimulation in the same donor PBMC without initial IVS, was significantly higher (P = 0.0043, unpaired Student’s t test) in BC patients than in healthy control subjects (Table 2). Overall, the proliferation index was 4.54 ± 0.84 (n = 30) for BC patients and only 1.6 ± 0.38 (n = 25) for the normal donors.
Figure 3.
Detection of HERV-K–specific T-cell responses in BC patient PBMC. A, T-cell proliferation was compared in freshly isolated (ex vivo) PBMC versus IVS cells pulsed with HERV-K cRNA from four BC patients and four normal donors. Higher T-cell proliferation was observed only in IVS cells obtained from BC patients (BC1 and BC4). There was no significant difference in proliferation between IVS performed with DC pulsed with HERV-K cRNA or HERV-K protein. B, IFN-γ ELISPOT was used to determine HERV-K–specific T-cell response. A representative ELISPOT result is shown. More spots were detected in IVS cells (1 × 105 loaded) from BC patients (BC4, BC5, and BC17) than a normal donor (NL6) stimulated with HERV-K env cRNA. C, specific lysis by CTL was measured in PBMC after IVS from HLA-A2+ BC in comparison with normal donor PBMC using MDA-MB-231 (HLA-A2.1+) as target cells. IVS cells obtained from the three BC patients (BC7, BC2, and BC12), but not a normal subject (NL2), had increased killing of MDA-MB-231 cells transfected with HERV-K cRNA than MDA-MB-231 cells transfected with control E6 cRNA. D, HERV-K–specific T-cell killing was observed only in PBMC from patients with BC after IVS, as seen by the higher lysis of MDA-MB-231 cells transfected with HERV-K cRNA than MDA-MB-231 cells transfected with control E6 cRNA (P = 0.0255, paired Student’s t test). CTL lysis was blocked by anti–HLA-A2 antibody (231+K+ anti-HLA). The percentages of CTL-specific lysis of MDA-MB-231 cells expressing various antigens were compared, and lysis of only target BC cells expressing HERV-K was observed in PBMC obtained from BC patients after IVS.
Table 2.
Enhanced immune function in IVS PBMC with HERV-K antigen
| Measurement | Cancer patients/cells | Normal controls | P | t test |
|---|---|---|---|---|
| T-cell proliferation index* | 4.540 ± 4.608 (n = 30) | 1.603 ± 1.877 (n = 25) | 0.0043 | Unpaired |
| IFN-γ spots for IVS cells† | 367.4 ± 201.3 (n = 16) | |||
| IFN-γ spots for PBMCs† | 3.771 ± 8.787 (n = 16) | <0.0001 | Paired | |
| IFN-γ spots for IVS cells† | 367.4 ± 201.3 (n = 16) | 15.96 ± 38.81 (n = 8) | <0.0001 | Unpaired |
| Percent specific lysis‡ | 18.50 ± 14.15 (n = 17) | 1.193 ± 1.478 (n = 7) | 0.0043 | Unpaired |
| IL-2 secretion by IVS cells§ | 63.24 ± 84.82 (n = 22) | |||
| IL-2 secretion by PBMCs§ | 11.89 ± 8.387 (n = 22) | 0.0109 | Paired | |
| IFN-γ secretion by IVS cells§ | 410.5 ± 494.6 (n = 22) | |||
| IFN-γ secretion by PBMCs§ | 41.81 ± 70.74 (n = 22) | 0.0031 | Paired | |
| IFN-γ secretion by PBMCs§ | 41.81 ± 70.74 (n = 22) | 145.9 ± 139.8 (n = 19) | 0.0033 | Unpaired |
| IL-6 secretion by IVS cells§ | 1,357 ± 1,838 (n = 17) | |||
| IL-6 secretion by PBMCs§ | 278.4 ± 587.9 (n = 11) | 0.0396 | Paired | |
| IP-10 secretion by IVS cells§ | 176.4 ± 84.10 (n = 17) | |||
| IP-10 secretion by PBMCs§ | 78.03 ± 62.80 (n = 11) | 0.0045 | Paired | |
| GrB spots for IVS cells‖ | 234.1 ± 166.5 (n = 13) | |||
| GrB spots for PBMCs‖ | 2.769 ± 3.938 (n = 13) | 0.0003 | Paired | |
| GrB spots for IVS cells‖ | 81.00 ± 69.56 (n = 17) | |||
| GrB spots for PBMCs‖ | 1.216 ± 1.568 (n = 17) | 0.0003 | Paired | |
| GrB spots for IVS cells‖ | 234.1 ± 166.5 (n = 13) | 81.00 ± 69.56 (n = 17) | 0.0019 | Unpaired |
| IL-8 spots for IVS cells¶ | 10,817 ± 6,401 (n = 17) | |||
| IL-8 spots for PBMCs¶ | 4,880 ± 7,504 (n = 11) | 0.0169 | Paired |
The T-cell proliferation index, measured by [3H]thymidine incorporation, was significantly increased in IVS cells obtained from breast cancer patients compared with IVS cells obtained from normal donors.
Antigen-specific IFN-γ responses, measured by ELISPOT assay, were significantly higher in PBMC from cancer patients after IVS with DCs pulsed with HERV-K cRNA than in freshly isolated ex vivo (no intervening IVS) PBMC or than in IVS cells obtained from normal female control subjects.
Higher specific lysis, measured as 4-h 51Cr-release, of MDA-MB-231 cells expressing HERV-K antigen was observed for stimulated PBMC from breast cancer patients than from normal donors.
HERV-K–specific cytokine production in breast cancer patient PBMCs using cytokine bead arrays after 72-h restimulation with K-SU cRNA or control HPV16 E6 cRNA. A statistically significant increase in cytokine/chemokine secretion was found in BC patients in response to HERV-K restimulation following IVS. No statistically significant increases in the secretion of these factors was found after IVS of normal control PBMC.
Antigen-specific GrB spot numbers were significantly higher in IVS cells than in their corresponding PBMC obtained from cancer patients or control subjects. GrB spot numbers from IVS cells (1 × 105 cells loaded) obtained from cancer patients were significantly higher than GrB spot numbers from control subjects.
IL-8 secretion was significantly elevated in 1-wk IVS from breast cancer patients, in comparison with their PBMC. No significant difference in IL-8 secretion was observed between 1-wk IVS (n = 12) and their PBMC (n = 8) obtained from normal controls.
PBMCs from BC patients after IVS with HERV-K were then assessed for antigen-specific effector cell function using IFN-γ and GrB ELISPOT as well as CTL killing assays using autologous HERV-K antigen-expressing DC as APC targets. Figure 3B shows a representative IFN-γ sample from three HLA-A2.1+ BC patients and an HLA-A2.1+ normal donor (NL6). No significant IFN-γ responses were seen after IVS in the BC patients and NL using autologous PBMC from each donor or using the DC transfected with control E6 cRNA (APC in each case negative for HERV-K). In contrast, IFN-γ production was clearly seen after IVS in the three patient samples when HERV-K–transfected DCs were used as targets, with no reactivity against this latter target in the normal donor sample. We also performed a single IVS on 15 BC PBMC with HERV-K cRNA-transduced autologous DC versus cells from 10 normal female donors followed by IFN-γ ELISPOT analysis using HERV-K antigen-pulsed DC (Table 2). A significantly greater number of IFN-γ spots were produced in BC patients than in normal female donors after a one-time IVS with HERV-K antigen-pulsed autologous DC (P < 0.0001, unpaired Student’s t test). Spots were observed in CD4+ or CD8+ depleted IVS cells, and spots were reduced if anti-CD4 or CD8+ antibody was added (data not shown). Furthermore, a significantly greater number of GrB spots were produced in BC patients (n = 13) than in normal female donors (n = 17) after a one-time IVS with HERV-K antigen-pulsed autologous DC (P = 0.0019, unpaired Student’s t test; Table 2). No spots were observed when IVS cells were depleted of CD8+ cells (data not shown). GrB ELISPOT may be a more specific indicator of CTL and natural killer cytotoxicity than IFN-γ secretion because GrB is a key mediator of target cell death via the granule-mediated pathway.
The ability of antigen-specific T cells to kill target cells expressing tumor antigens is critical for tumor control and immunotherapy. We next determined whether IVS of PBMC from BC with HERV-K–expressing APC could elicit CTL activity against antigen-expressing targets. As before, PBMCs from HLA-A2.1+ BC patients and HLA-A2.1+ normal female donors were stimulated with autologous DC pulsed with HERV-K cRNA and the PBMCs were then tested for CTL activity against HERV-K env SU-transduced or human papillomavirus 16 (HPV16) E6–transduced HLA-A2.1+ MDA-MB-231 BC cells (HERV-K transduction was needed to see consistent CTL activity). Figure 3C shows a representative HERV-K–specific CTL experiment from three HLA-A2.1+ BC patients (BC7, BC2, and BC12) and an HLA-A2.1+ normal donor (NL2). As shown in Fig. 3D, a higher CTL activity was seen against K-SU–transduced MDA-MB-231 cells than in targets transduced with control HPV16 E6 (P = 0.025, paired Student’s t test). CTL killing of the K-SU–expressing targets was completely blocked by addition of anti–HLA-A2 antibody to the assay. Using this approach (IVS followed by CTL assay using K-SU–expressing MDA-MB-231 cells), we screened 17 additional HLA-A2.1+ BC patient PBMCs in comparison with 7 HLA-A2.1+ normal donor PBMCs for HERV-K–specific CTL induction. The percentage of CTL lysis of MDA-MB-231 cells expressing K-SU antigen in BC patients (n = 17; 18.5 ± 14.2) was significantly enhanced in comparison with normal donors (n = 7; 1.19 ± 1.48; P = 0.0043, unpaired Student’s t test), as shown in Table 2.
These results show that PBMCs from BC patients contain HERV-K–specific CTL that can be restimulated in vitro to induce detectable cytolytic activity against HERV-K–expressing targets. These CTL responses in BC patients were induced by expressed HERV-K env protein and not by other viral proteins produced by the same expression vector, such as HPV16 E6, HPV16 E7, or LMP2A, which were also tested using transduced APC in our IVS experiments (data not shown). In addition, the HERV-K–specific T-cell responses were due to HERV-K env protein itself and not to bacterial contamination (e.g., endotoxin) for several reasons. First, control HPV16 E6 and HPV16 E7 proteins produced in bacteria and used as antigen with DC in the IVS experiments did not promote a considerable immune response as did HERV-K protein (data not shown). Second, the marked HERV-K responses in patient PBMC samples were induced by only a single in vitro sensitization by HERV-K cRNA or with HERV-K protein pulsing, although these responses were largely undetectable in IFN-γ and CTL assays using PBMC from normal donors. This indicates that the IVS system did not simply activate a naive T-cell response against HERV-K nor did any bacterial products lead to a nonspecific anti–HERV-K T-cell response.
HERV-K antigen-specific cytokine production in BC patient PBMC
We next determined the ability of BC patient PBMC to produce additional cytokines in response to restimulation with HERV-K in vitro. PBMCs from BC patients or normal donors were stimulated with autologous HERV-K–pulsed DC as before and incubated for 1 week to generate IVS cells. PBMCs or their IVS cells were then restimulated for 72 h with DC pulsed with no added protein, K-SU protein (or cRNA) or control E6 protein (or cRNA), at a DC to PBMC or IVS ratio of 1:30. The supernatants were harvested and tested for the secretion of T-helper 1 (Th1) and Th2 cytokines using a human cytokine bead array system (BD Biosciences). Multiple Th1 and Th2 cytokines (see Table 2), including IL-2 (P = 0.0109, paired Student’s t test), IFN-γ (P = 0.0031), IL-6 (P = 0.0396), IL-8 (P = 0.0169), and the chemokine IP-10 (P = 0.0045), showed significantly greater secretion in samples from BC patients after IVS (n = 22). In contrast, no significant change was observed in normal controls after IVS (n = 19). Of interest is our result showing that IP-10 secretion is elevated in BC patients, as IP-10 is induced in T cells by IFN-γ, which we also show to be elevated in these BC patients. A markedly increased frequency of TNF-α, IL-2, and IFN-γ–secreting CD8+ T cells was also detected in the HERV-K–stimulated cultures by intracellular cytokine staining (data not shown).
Thus, T cells isolated from BC patients exhibit HERV-K–specific proliferation, proinflammatory cytokine secretion, and cytotoxic activity against HERV-K target cells, and these changes are not found in normal healthy controls.
Discussion
The results presented here show that HERV proteins (HERV-K env) are widely expressed in human invasive BC and DCIS. These novel antigens elicit both serologic and cell-mediated immune responses against HERV-K protein and HERV-K–expressing target cells in BC patient sera and PBMC, respectively. CD8+ T-cell proliferative and effector cell responses specific for HERV-K could be readily detected in PBMC from a high proportion of BC patients after IVS, whereas little or no responses were detected in PBMC from normal control donors. The expanded CD8+ T cells could also kill HERV-K–expressing target cells, suggesting that HERV-K and other retroviral antigen products may be a useful source of antigens for active immunotherapy.
The precise mechanism by which cancer cells respond to processed peptides of HERV-K env proteins has not been elucidated. The reactivation of endogenous retroviral gene products and synthesis of mature protein products in cancer makes these a potentially valuable new pool of TAA for targeting therapeutic vaccines to BC and other solid tumors. The silencing of retroviral gene expression throughout our lifetime and their reactivation specifically in cancer cells suggests that immunologic self-tolerance mechanisms against HERV-K and other retroviral proteins may be limited or that the immune system exists in a state of ignorance toward these antigens until cancer develops. This hypothesis, however, will need to be formally addressed in future experiments. Nevertheless, the specific reactivation of retroviral protein expression in cancer cells raises the exciting possibility of prophylactic vaccination to prevent primary tumor development as well as a novel approach to therapeutic vaccination. Similar to antiviral vaccines (e.g., anti-papillomavirus vaccines) now used to prevent cervical cancer and other cancers such as liver cancer, prophylactic vaccines against hitherto nonexpressed retroviral antigens may elicit long-lived retroviral antigen-specific T-cell responses (T-central memory) in an otherwise ignorant host that can eradicate early malignancies reexpressing these retroviral gene products. A recent study in HIV-infected individuals found that the expression of endogenous HERV-K env is induced during HIV infection (14). CTL responses against HERV-K gag epitopes shared with HIV gag were also found in a significant number of infected individuals. Interestingly, an inverse correlation between these CTL response and HIV viral load began to emerge in this study, indicating that anti–HERV-K CTL responses can control HIV viral infection. In a similar fashion, vaccination against HERV-K elements expressed in cancer cells can be postulated to also induce protective CTL responses. We predict that patients with better anti–HERV-K antibody and/or CTL responses may have a better prognosis. This will need to be tested in a larger cohort of patient samples.
Optimal antitumor responses against a specific TAA have been found in cases where both CD8+ and Th1 CD4+ tumor antigen-specific responses are generated (15). The presence of these antibodies suggests that soluble retroviral envelope proteins such as HERV-K secreted by tumor cells or shed from dying tumors may circulate in the blood of cancer patients and may be a diagnostic marker for BC. The presence of anti–HERV-K IgG is indicative of the activation of CD4+ T-helper cells. CD8+ T cells have traditionally been the main focus of tumor immunologists developing anticancer vaccines. However, more recently, the critical role of antigen-specific CD4+ T-cell responses in generating more effective antitumor responses has been recognized. This appreciation for the role of CD4+ T cells stemmed from the discovery of antibodies in patients against tumor antigens, including cancer testis antigens such as MAGE-3 and NY-ESO-1 using “SEREX,” and the identification of HLA class II–binding epitopes from the same tumor antigens that are recognized by CD4+ T cells (16, 17). The activation of CD4+ T-helper cells along with CD8+ T cells against HERV-K is significant, especially in light of the growing importance of T-helper cells in driving and maintaining CTL responses through the provision of cytokines and signals activating DC antigen presentation to CD8+ T cells.
A critical issue with most BC antigens being tested in therapeutic anticancer vaccines and passive immunotherapy clinical trials is that the current armamentarium of antigen targets is mostly overexpressed tissue differentiation antigens. These antigens are expressed to different degrees in normal tissues such as HER2/neuin the heart, CEA and MUC in normal epithelial cells of the gut, thyroid and other organs, and telomerase, which is expressed by cycling T cells and T-regulatory cells. Overall, relatively few overexpressed antigens have been found in BC that show high specificity of expression in tumor cells with low or negligible expression in normal tissues to limit peripheral self-tolerance mechanisms. A few new antigens, such as NY-BR-1 of the cancer testis family and TRPS-1, have been found to be more BC specific (18, 19). There is little evidence that HERV-K protein is expressed in normal cells. In placenta, HERV-K rec is expressed at the transcriptional level. Our research has shown that even if the HERV-K gene is transcribed, that does not necessarily mean that the protein will be expressed. We previously reported that HERV-K has numerous stop codons in normal tissue that would prevent its translation. In many cancer tissues, however, we found that these stop codons are no longer present, and expression of a full-length HERV-K open reading frame is thus possible in cancer tissues. There is a need for additional targets, especially for the development of multiantigen vaccines targeting as many tumor cells in a mass as possible. HERV-K may be the basis of more specific and more potent anti-BC vaccines that can be given in the preventative or adjuvant setting.
Chronic viral infections have been shown to cause several different human cancers, such as cervical cancer (human papilloma virus), hepatocellular carcinoma (hepatitis B and C viruses), Hodgkin’s disease, Burkitt’s lymphoma, and nasopharyngeal tumors (EBV), adult T-cell leukemia (human T-cell leukemia virus), and Kaposi’s sarcoma (human herpesvirus 8; refs. 20–27). Hepatitis B virus vaccine is considered the first anticancer vaccine because it can prevent HBV-induced hepatocellular carcinoma (28). Recently, vaccine formulations targeting the HPV16 E6 and E7 antigens have undergone phase III trials for the prevention of cervical cancer, with one product, Gardasil, recently approved by the Food and Drug Administration (20, 29). The prevalence of HERV-K expression in human BC and the immunogenicity of HERV-K protein products in patients suggests that these endogenous viral antigens can be added to the growing list of viral antigen targets for therapeutic use against BC and other solid tumors, such as ovarian cancer, where HERV proteins have also been found to be expressed (11).
In summary, we have found that the HERV gene product, HERV-K env, is widely expressed in human invasive BC and DCIS and elicits both serologic and cell-mediated immune responses in BC patients recognizing HERV-K antigens and HERV-K–expressing breast tumor cells. CD8+ T-cell proliferative and effector cell responses specific for HERV-K could be readily detected in PBMC from a high proportion of BC patients after a single IVS, whereas little or no responses were detected in PBMC of normal control donors. The expanded CD8+ T cells could also kill HERV-K–expressing BC cells, suggesting that HERV-K and other retroviral antigen products may be a useful source of antigens for active immunotherapy.
Supplementary Material
Acknowledgments
Grant support: Department of Defense grant DAMD1700-1-0123, Susan G. Komen Breast Cancer Foundation (Susan G. Komen for the Cure) grants BCTR0402892 and BCTR000007888, National Institute of Environmental Health Sciences Center grant ES007784, The Cattlemen for Cancer Research, and The University of Texas M. D. Anderson Cancer Center start-up funds.
We thank Dr. Danielle W. Lu (Department of Pathology, Huntington Memorial Hospital, Pasadena, CA) for providing the immunohistochemical staining scores of tissue arrays, Dr. Caimiao Wei for some statistical analysis, and Dr. Pramod N. Nehete for advice on some immune assays.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Multiple breast tissue arrays (n = 182) that included normal breast tissues (n = 56), hyperplastic breast tissues (n = 7), and breast cancer tissues (n = 119) were obtained from Cybrdi, Inc. Characteristics and further pathologic information about normal breast and breast cancer specimens in these tissue arrays can be found at http://www.cybrdi.com (slide codes are CC08-01-002, CC08-02-002 and CC08-01-001).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Fetsch PA, Marincola FM, Abati A. The new melanoma markers: MART-1 and Melan-A (the NIH experience) Am J Surg Pathol. 1999;23:607–10. doi: 10.1097/00000478-199905000-00017. [DOI] [PubMed] [Google Scholar]
- 2.Lopes L, Fletcher K, Ikeda Y, Collins M. Lentiviral vector expression of tumour antigens in dendritic cells as an immunotherapeutic strategy. Cancer Immunol Immunother. 2006;55:1011–6. doi: 10.1007/s00262-005-0095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A. 1996;93:5177–84. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol. 1995;69:141–9. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mosch C. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69:6408–16. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etkind PR, Lumb K, Du J, Racevskis J. Type 1 HERV-K genome is spliced into subgenomic transcripts in the human breast tumor cell line T47D. Virology. 1997;234:304–8. doi: 10.1006/viro.1997.8670. [DOI] [PubMed] [Google Scholar]
- 8.Wang-Johanning F, Frost AR, Johanning GL, et al. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–60. [PubMed] [Google Scholar]
- 9.Wang-Johanning F. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–35. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 10.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62:5510–6. [PubMed] [Google Scholar]
- 11.Wang-Johanning F, Liu J, Rycaj K, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 12.Wang-Johanning F, Gillespie GY, Grim J, et al. Intracellular expression of a single-chain antibody directed against human papillomavirus type 16 E7 oncoprotein achieves targeted antineoplastic effects. Cancer Res. 1998;58:1893–900. [PubMed] [Google Scholar]
- 13.Dolbier CL, Cocke RR, Leiferman JA, et al. Differences in functional immune responses of high vs. low hardy healthy individuals. J Behav Med. 2001;24:219–29. doi: 10.1023/a:1010762606006. [DOI] [PubMed] [Google Scholar]
- 14.Garrison KE, Jones RB, Meiklejohn DA, et al. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerloni M, Zanetti M. CD4 T cells in tumor immunity. Springer Semin Immunopathol. 2005;27:37–48. doi: 10.1007/s00281-004-0193-z. [DOI] [PubMed] [Google Scholar]
- 16.Tureci O, Sahin U, Pfreundschuh M. Serological analysis of human tumor antigens: molecular definition and implications. Mol Med Today. 1997;3:342–9. doi: 10.1016/s1357-4310(97)01081-2. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann TB, Bazhin AV, Schadendorf D, Eichmuller SB. SEREX identification of new tumor antigens linked to melanoma-associated retinopathy. Int J Cancer. 2005;114:88–93. doi: 10.1002/ijc.20762. [DOI] [PubMed] [Google Scholar]
- 18.Jager D, Unkelbach M, Frei C, et al. Identification of tumor-restricted antigens NY-BR-1, SCP-1, and a new cancer/testis-like antigen NW-BR-3 by serological screening of a testicular library with breast cancer serum. Cancer Immun. 2002;2:5. [PubMed] [Google Scholar]
- 19.Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2005;102:11005–10. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadish AS, Einstein MH. Vaccine strategies for human papillomavirus-associated cancers. Curr Opin Oncol. 2005;17:456–61. doi: 10.1097/01.cco.0000174038.92526.29. [DOI] [PubMed] [Google Scholar]
- 21.Ho GY, Burk RD, Klein S, et al. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995;87:1365–71. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- 22.Comoli P, Pedrazzoli P, Maccario R, et al. Cell therapy of stage IV nasopharyngeal carcinoma with autologous Epstein-Barr virus-targeted cytotoxic T lymphocytes. J Clin Oncol. 2005;23:8942–9. doi: 10.1200/JCO.2005.02.6195. [DOI] [PubMed] [Google Scholar]
- 23.Foster AE, Rooney CM. Improving T cell therapy for cancer. Expert Opin Biol Ther. 2006;6:215–29. doi: 10.1517/14712598.6.3.215. [DOI] [PubMed] [Google Scholar]
- 24.Gottschalk S, Heslop HE, Rooney CM. Adoptive immunotherapy for EBV-associated malignancies. Leuk Lymphoma. 2005;46:1–10. doi: 10.1080/10428190400002202. [DOI] [PubMed] [Google Scholar]
- 25.Dohmen K, Shigematsu H, Irie K, Ishibashi H. Trends in clinical characteristics, treatment and prognosis of hepatocellular carcinoma. Hepatogastroenterology. 2003;50:1872–7. [PubMed] [Google Scholar]
- 26.Komori K, Hasegawa A, Kurihara K, et al. Reduction of human T-cell leukemia virus type 1 (HTLV-1) proviral loads in rats orally infected with HTLV-1 by reimmunization with HTLV-1-infected cells. J Virol. 2006;80:7375–81. doi: 10.1128/JVI.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–9. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 28.Poland GA, Jacobson RM. Clinical practice: prevention of hepatitis B with the hepatitis B vaccine. N Engl J Med. 2004;351:2832–8. doi: 10.1056/NEJMcp041507. [DOI] [PubMed] [Google Scholar]
- 29.Schiffman M, Castle PE. The promise of global cervical-cancer prevention. N Engl J Med. 2005;353:2101–4. doi: 10.1056/NEJMp058171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.