Chronic lymphocytic leukemia (CLL) is the most common leukemia in adults in the Western countries but is relatively rare in East Asia1. CLL is a disease of high heterogeneity. The clinical course ranges from indolence to rapid progression to death. Although the Rai and Binet clinical staging systems remain to be the cornerstone for CLL prognosis, the rapidly developed biological and genetic techniques enable the detection of novel prognostic factors.
Mutations in myeloid differentiation primary response gene 88 (MYD88) in CLL were first reported in 2011 with a mutation frequency of 9/310 (2.9%)2. Subsequent studies found that MYD88 mutations exist in 2.0–4.4% Caucasian patients with CLL3–7. However, subjects of Asia showed a higher MYD88 mutated rate of 8% as previously reported8. The above MYD88 mutated cases consist mainly of a p. L265P substitution.
CLL patients with MYD88 mutations were reported to be younger at diagnosis and have longer time to treatment (TTT) and overall survival (OS) than those with wild-type MYD889. However, this conclusion was controversial10. Initial studies indicated that most MYD88-mutated patients belonged to the IGHV-mutated group5,9,11, which is generally accepted as a molecular sign of favorable prognosis. These studies could be more convincing if taking IGHV mutation status and MYD88 mutations together into prognostic consideration10. In the current study, we analyzed MYD88 mutations exclusively in the IGHV-mutated CLL cases to explore its prognostic value.
Two hundred and eighty-four patients with previously untreated CLL at the First Affiliated Hospital of Nanjing Medical University between January 2000 and June 2016 were retrospectively enrolled. All cases were reviewed to confirm the diagnosis according to the 2008 International Workshop in CLL-National Cancer Institute (IWCLL-NCI)12. Clinical and biological parameters including absolute lymphocyte count, hemoglobin, platelet, cytogenetic abnormalities, mutation status of TP53, IGHV, NOTCH1 as well as surface markers of CLL cells were assessed at first presence at our center. The study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University with a reference number as 2014-SR-204. Informed consents were provided according to the Declaration of Helsinki.
Mononuclear cells from 281 peripheral blood samples and three bone marrow samples of untreated CLL patients were used for AS-PCR assay. Genomic DNA was extracted using the QIAamp DNA Blood Kits (Qiagen, Düsseldorf, Germany) according to the manufacturer’s recommendation. Two different forward primers (FW5′-GTGCCCATCAGAAGCGCCT-3′ and FM5′-GTGCCCATCAGAAGCGCCC-3′) and one reverse primer (5′-AGGAGGCAGGGCAGAAGTA-3′) were used to amplify the wild-type allele or the MYD88 L265P mutation allele as previously reported4. The sensitivity of AS-PCR was 0.625% in the present study. The Sanger sequencing was performed to confirm the AS-PCR assay and to detect MYD88 mutations other than L265P. Exon 3–5 was amplified by Sanger sequencing with a forward primer (5′- AGCGACATCCAGTTTGTGC-3′) and a reverse primer (5′- AGGCGAGTCCAGAACCAAG -3′)8. Amplified fragments were sequenced with both the forward and reverse primers. Both detecting methods were applied on all samples included in the study.
All statistical analyses were performed by SPSS for Windows (version 19.0; IBM Corporation, Armonk, NY, USA) and Graphpad Prism 6. Fisher’s exact test and the chi-square test were used to determine the correlations between MYD88 mutations and clinical characteristics. Mann-Whitney U test was applied for comparing mean fluorescence intensity (MFI) as a continuous parameter in MYD88 mutated and wild-type groups. Time to treatment (TTT) was defined as the time from initial diagnosis to first treatment. OS was defined as the time from diagnosis to death or to the last follow-up. TTT and OS curves were estimated by the Kaplan-Meier method and compared by the log-rank test. The prognostic impact of MYD88 mutations on TTT and OS was assessed using both univariate and multivariate Cox analysis. All statistical tests were two-sided, and P value < 0.05 was considered to be significant.
A total of 284 CLL patients were included in our study. Clinical and biological characteristics are summarized in Table 1. The median proportion of CD19+CD5+ cells in the samples was 65.3% (range 32.3–98.1%). Using both AS-PCR and Sanger sequencing, we detected MYD88 mutations (n = 25) in 25/284 (8.8%) patients with the hotspot L265P substitution representing 72.0% (18/25) of all mutations. Other detected mutations were all single-nucleotide substitutions including S219C (n = 3), V217F (n = 2), M232T (n = 1) and S243N (n = 1).
Table 1.
Characteristics of the CLL patients according to MYD88 mutation status
| All (n = 284) | MYD88 wild type (n = 259) | MYD88 mutated (n = 25) | |||||
|---|---|---|---|---|---|---|---|
| Characteristic | n* | % | n* | % | n* | % | P |
| Age, y (range) | 60 (54–69) | 60 (54–69) | 60 (54–66) | 0.512 | |||
| Male | 183 | 64.4 | 164 | 63.3 | 19 | 76.0 | 0.275 |
| Binet C | 86 | 32.1 | 77 | 31.3 | 9 | 40.9 | 0.351 |
| IGHV mutated | 165 | 59.1 | 143 | 56.3 | 22 | 88.0 | 0.002 |
| CD38 ≥ 30% | 52 | 18.8 | 52 | 20.6 | 0 | 0.0 | 0.011 |
| ZAP70 ≥ 20% | 71 | 28.7 | 66 | 29.1 | 5 | 25.0 | 0.802 |
| TP53 disruption | 62 | 22.5 | 58 | 23.0 | 4 | 16.7 | 0.613 |
| HBV ( + ) | 62 | 21.9 | 54 | 20.8 | 8 | 33.3 | 0.195 |
| + 12 | 42 | 18.0 | 40 | 18.9 | 2 | 9.5 | 0.383 |
| ATM deletion | 37 | 17.1 | 30 | 15.2 | 7 | 36.8 | 0.026 |
| NOTCH1 | 17 | 7.5 | 17 | 8.1 | 0 | 0.0 | 0.373 |
*Median and 25th–75th percentiles are reported for continuous variables
Patients with MYD88 mutations preferentially carried mutated IGHV genes (MYD88 mutated: 22/25 vs. MYD88 wild-type: 143/254, P = 0.001). None of the MYD88 mutated CLL patients showed CD38 positivity (defined as ≥30%) (P = 0.011). Besides, MYD88 mutated CLL were more frequently ATM-deleted (36.8%, P = 0.026). In addition, we observed lower CD200 MFI in MYD88 mutated CLL patients (P < 0.001) within both the overall cohort and CLL patients with mutated IGHV. None of the mutated patients had Ig paraproteinemia in our analysis. No difference was observed in the distribution of TP53 disruptions between MYD88 wild-type and mutated subjects in the mutated IGHV-CLL (referred as M-CLL) (19 vs. 14%, P = 0.767).
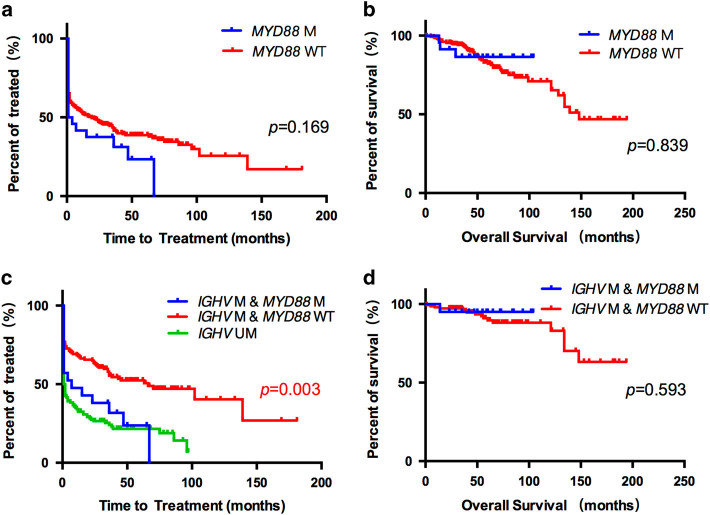
With a median follow-up of 54.5 months, MYD88 mutations showed no significant impact on either TTT or OS (Figs. 1a, b). Then we conducted survival analysis in the M-CLL patients. Variables included in the univariate analysis on TTT were: (1) conventional clinical (Binet staging system) factors; (2) widely accepted genetic (TP53 disruption, defined as TP53 mutation and/or deletion, NOTCH1 mutation ATM deletion and 12 trisomy) prognostic risk factors; 3) specific protein expression (CD38 and ZAP70). Univariate Cox analysis selected MYD88 mutation (HR 1.873; 95% CI 1.067-3.287; P = 0.029), Binet C (HR 3.617; 95% CI 2.278-5.742; P < 0.001) and TP53 disruption (HR 1.798; 95% CI 1.090-2.966; P = 0.022) as risk factors for shorter TTT, and these three parameters went for multivariate analysis in the next step. Multivariate analysis confirmed MYD88 mutations (HR 2.233; 95% CI 1.233-4.045; P = 0.008) alongside with Binet C (HR 3.653; 95% CI 2.244-5.944; P < 0.001) were independently correlated with shorter TTT (Table 2 and Fig. 1c) in M-CLL patients. However, no difference on OS was observed between MYD88-mutated and -unmutated cases in the same cohort (P = 0.593) (Fig. 1d).
Fig. 1.
a Kaplan-Meier estimates of TTT according to MYD88 mutation status among all patients. Time to treatment analysis according to MYD88 mutation status in the CLL patients (N = 284). MYD88 wild-type cases (MYD88 (-)) are represented by the red line. MYD88 mutated cases (MYD88 ( + )) are represented by the blue line. b Kaplan-Meier estimates of OS according to MYD88 mutation status among all patients. Overall survival analysis according to MYD88 mutation status in the CLL patients (N = 284). MYD88 wild-type cases (MYD88 (-)) are represented by the red line. MYD88 mutated cases (MYD88 ( + )) are represented by the blue line. c Kaplan-Meier estimates of TTT according to MYD88 mutation status and IGHV mutation status among M-CLL patients. Time to treatment analysis according to MYD88 mutation status and IGHV mutation status among all CLL patients (N = 284). Of the M-CLL cases, MYD88 wild-type cases (IGHV ( + ) MYD88 (-)) are represented by the red line, while MYD88 mutated cases (IGHV ( + ) MYD88 ( + )) are represented by the blue line. IGHV unmutated cases (IGHV(-)) are represented by the green line. d Kaplan-Meier estimates of OS according to MYD88 mutation status among M-CLL patients. Overall survival analysis according to MYD88 mutation status in the M-CLL patients (N = 165). MYD88 wild-type cases (IGHV ( + ) MYD88 (-)) are represented by the red line. MYD88 mutated cases (IGHV ( + ) MYD88 ( + )) are represented by the blue line
Table 2.
Univariate and Multivariate analysis for time to treatment in the M-CLL patients
| Risk Factors | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | |
| MYD88 mutation | 1.873 | 1.067-3.287 | 0.029 | 2.233 | 1.233-4.045 | 0.008 |
| Binet C | 3.617 | 2.278-5.742 | < 0.001 | 3.653 | 2.244-5.944 | < 0.001 |
| TP53 disruption | 1.798 | 1.090-2.966 | 0.022 | 1.454 | 0.861-2.455 | 0.162 |
| ZAP70 ≥ 30% | 1.004 | 0.616-1.637 | 0.988 | - | - | - |
| CD38 ≥ 20% | 1.139 | 0.638-2.033 | 0.659 | - | - | - |
| NOTCH1 mutation | 2.669 | 0.651-10.946 | 0.173 | - | - | - |
| + 12 | 1.021 | 0.547-1.908 | 0.948 | - | - | - |
| ATM deletion | 1.830 | 0.886-3.779 | 0.102 | - | - | - |
HR hazards ratio; 95% CI, 95% confidence interval;
We further analyzed the correlation between MYD88 mutations and 6 mostly used IGHV genes in M-CLL patients. None of the MYD88 mutated cases used IGHV4-34, the most prevalent IGHV gene in the M-CLL cohort, (P = 0.015) (Table 3), suggesting that MYD88 mutation might be IGHV gene-biased, and that certain antigen exposure might avoid the emergence of MYD88 mutations in the pathogenesis of CLL.
Table 3.
The correlation of MYD88 mutation and 6 mostly used IGHV gene in M-CLL patients in China
| All | MYD88-wild | MYD88-mutated | P value | ||
|---|---|---|---|---|---|
| n | % | n | n | ||
| VH4-34 | 0.015 | ||||
| yes | 28 | 16.7 | 28 | 0 | |
| no | 140 | 83.3 | 117 | 23 | |
| VH3-23 | 0.484 | ||||
| yes | 20 | 11.9 | 16 | 4 | |
| no | 148 | 88.1 | 129 | 19 | |
| VH3-7 | 0.484 | ||||
| yes | 20 | 11.9 | 16 | 4 | |
| no | 148 | 88.1 | 129 | 19 | |
| VH4-39 | 1.000 | ||||
| yes | 5 | 3.0 | 5 | 0 | |
| no | 163 | 97.0 | 140 | 23 | |
| VH4-59 | 0.526 | ||||
| yes | 5 | 3.0 | 4 | 1 | |
| no | 163 | 97.0 | 141 | 22 | |
| VH3-21 | 0.448 | ||||
| yes | 4 | 2.4 | 3 | 1 | |
| no | 164 | 97.6 | 142 | 22 | |
In this study, we explored the detection method and clinical relevance of MYD88 mutations in Chinese patients with CLL. We found MYD88 mutations: (1) occur in 8.8% CLL patients in our center upon diagnosis; (2) cluster with cases harboring mutated IGHV; (3) identify a group of patients with poor prognosis among M-CLL; (4) are rare, if not absent, in IGHV-4-34 users. The incidence of MYD88 mutations was 2.0–4.4% in Caucasian CLL patients3–5. However, we have detected a higher frequency of 8.8% in our cohort upon diagnosis. The disparities of ethnic groups may explain the difference in frequencies; meanwhile the application of AS-PCR assay in our study indeed improved the detection sensitivity. AS-PCR is previously used in detecting MYD88 L265P mutations in Waldenstrom macroglobulinemia and diffused large B cell lymphoma3,13,14. Our data showed that AS-PCR is capable of detecting samples with a tumor cell load as low as 0.625%, which is far beyond the sensitivity of Sanger sequencing.
The role of MYD88 mutations in determining the biological features and clinical outcome of CLL patients remains controversial. The initial study indicated that patients with MYD88 mutations were diagnosed younger and suffered a moreless advanced clinical stage9. Contradictory results, however, were observed in that MYD88 mutations showed no age and stage preference in CLL patients7,11, nor does our data do. In the subgroup analysis of M-CLL, we observed MYD88 mutations predict shorter TTT in this category with favorable outcome. Furthermore, CLL patients with MYD88 mutations had comparable prognosis with those with unmutated IGHV in our cohort, implying MYD88 mutations may counteract the survival advantage of mutated IGHV gene.
Early research has shown that CLL cells with MYD88 mutation co-immunoprecipitates with a larger amount of IRAK1&IL-1/TLR signaling pathway, and that activation of the IL-1/TLR pathway promotes proliferation in CLL cells15. Furthermore, MYD88 mutated CLL cells have higher phosphorylation and more DNA-binding activity in NF-κB subunits than CLL cells with wild-type MYD88. All these results suggests MYD88 mutation is a gain-of-function molecular change which may aberrantly activates NF-κB signaling pathway in CLL cells2,9 and offers explanation for the unfavorable prognostic impact of MYD88 mutation on the M-CLL subgroup.
We also found patients with MYD88 mutations have a relatively lower CD200 MFI compared to the wildtype cases do, consistent with a previous report16. Along with the fact that none of the MYD88 mutated CLL patients expressed positive CD38 in our study, we postulate that this subgroup of CLL patients may have a distinct immunophenotype from CLL without MYD88 mutations. This will be further explored by targeted RNA sequencing and whole genome sequencing. MYD88 mutations are mutually exclusive of IGHV 4-34 gene usage, which was not shown before to our knowledge. Unlike previously reported, we did not observe a preferable IGHV 3-23 gene usage in MYD88-mutated cases10.
In conclusion, in our cohort of newly diagnosed CLL patients, MYD88 mutations showed an incidence of 8.8%, including 6.3% on the hotspot missense mutation L265P. MYD88 mutations predict unfavorable prognosis within the M-CLL subgroup.
Acknowledgements
This study is supported by National Natural Science Foundation of China (30971296, 81170485, 81170488, 81370657, 81470328, 81500125, 81522001, and 81570141), Project of National Key Clinical Specialty, the National Science & Technology Pillar Program (2014BAI09B12), the Jiangsu Province Scientific Research Innovation Project of University graduate students (KYZZ15_0260), a project funded by Jiangsu Provincial Special Program of Medical Science (BL2014086) and National Science and Technology Major Project (2017ZX09304032).
Competing interest
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Shu-Chao Qin and Yi Xia contributed equally to this work.
Contributor Information
Jian-Yong Li, Phone: +86-25-83781120, Email: lijianyonglm@medmail.com.cn.
Wei Xu, Phone: +86-258-37-81120, Email: xuwei10000@hotmail.com.
Chun Qiao, Phone: +86-25-83781120, Email: qiaochun001004@163.com.
References
- 1.Eichhorst B, et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26:v78–v84. doi: 10.1093/annonc/mdv303. [DOI] [PubMed] [Google Scholar]
- 2.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;469:115–9. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu L, et al. MYD88 L265P in Waldenström macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood. 2013;121:2051–8. doi: 10.1182/blood-2012-09-454355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varettoni M, et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom’s macroglobulinemia and related lymphoid neoplasms. Blood. 2013;121:2522–8. doi: 10.1182/blood-2012-09-457101. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Trillos A, et al. Mutations in TLR/MYD88 pathway identify a subset of young chronic lymphocytic leukemia patients with favorable outcome. Blood. 2014;123:3790–6. doi: 10.1182/blood-2013-12-543306. [DOI] [PubMed] [Google Scholar]
- 6.Cortese D, et al. On the way towards a ‘CLL prognostic index’: focus on TP53, BIRC3, SF3B1, NOTCH1 and MYD88 in a population-based cohort. Leukemia. 2015;28:710–3. doi: 10.1038/leu.2013.333. [DOI] [PubMed] [Google Scholar]
- 7.Jeromin S, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2013;28:108–17. doi: 10.1038/leu.2013.263. [DOI] [PubMed] [Google Scholar]
- 8.Xia Y, et al. Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget. 2015;6:5426–34. doi: 10.18632/oncotarget.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baliakas P, et al. Prognostic relevance of MYD88 mutations in CLL: the jury is still out. Blood. 2015;126:1043–4. doi: 10.1182/blood-2015-05-648634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baliakas P, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29:329–36. doi: 10.1038/leu.2014.196. [DOI] [PubMed] [Google Scholar]
- 12.Hallek M, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, et al. Waldenström macroglobulinemia with extramedullary involvement at initial diagnosis portends a poorer prognosis. J Hematol Oncol. 2015;8:74. doi: 10.1186/s13045-015-0172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staiger AM, et al. Allele-specific PCR is a powerful tool for the detection of the MYD88 L265P mutation in diffuse large B cell lymphoma and decalcified bone marrow samples. Br J Haematol. 2015;171:145–8. doi: 10.1111/bjh.13369. [DOI] [PubMed] [Google Scholar]
- 15.Muzio M, et al. Expression and function of toll like receptors in chronic lymphocytic leukaemia cells. Br J Haematol. 2009;144:507–16. doi: 10.1111/j.1365-2141.2008.07475.x. [DOI] [PubMed] [Google Scholar]
- 16.Miao Y, et al. Low expression of CD200 predicts shorter time-to-treatment in chronic lymphocytic leukemia. Oncotarget. 2016;7:13551–62. doi: 10.18632/oncotarget.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]