Abstract
Despite better therapeutic options and improved survival of diffuse large B-cell lymphoma (DLBCL), 30–40% of the patients experience relapse or have primary refractory disease with a dismal prognosis. To identify biological correlates for treatment resistance, we profiled microRNAs (miRNAs) of matched primary and relapsed DLBCL by next-generation sequencing. Altogether 492 miRNAs were expressed in the DLBCL samples. Thirteen miRNAs showed significant differential expression between primary and relapse specimen pairs. Integration of the differentially expressed miRNAs with matched mRNA expression profiles identified highly anti-correlated, putative targets, which were significantly enriched in cancer-associated pathways, including phosphatidylinositol (PI)), mitogen-activated protein kinase (MAPK), and B-cell receptor (BCR) signaling. Expression data suggested activation of these pathways during disease progression, and functional analyses validated that miR-370-3p, miR-381-3p, and miR-409-3p downregulate genes on the PI, MAPK, and BCR signaling pathways, and enhance chemosensitivity of DLBCL cells in vitro. High expression of selected target genes, that is, PIP5K1 and IMPA1, was found to be associated with poor survival in two independent cohorts of chemoimmunotherapy-treated patients (n = 92 and n = 233). Taken together, our results demonstrate that differentially expressed miRNAs contribute to disease progression by regulating key cell survival pathways and by mediating chemosensitivity, thus representing potential novel therapeutic targets.
Background
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid malignancy in adults. It is a heterogeneous disease, which can be classified into activated B-cell, germinal center B-cell (GCB), and primary mediastinal B-cell subtypes according to gene expression profiling1–4. The standard therapy for DLBCL is a combination of CD20 antibody rituximab with cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP)5, 6. Despite the efficacy of this regimen, approximately one-third of the patients have primary refractory disease or relapse, which remains a major cause of morbidity and mortality. However, molecular mechanisms behind the treatment failure remain largely unknown.
MicroRNAs (miRNAs) participate in several biological processes by regulating gene expression at the posttranscriptional level. These short non-coding RNAs (20- to 22-nucleotides) bind to complementary sites in their target gene mRNAs, thereby inhibiting translation or inducing destabilization and degradation of the target mRNAs7. Moreover, dysregulation of miRNAs has been linked to the development and progression of a number of human cancers, making them ideal candidates for both predictive and prognostic biomarkers8, 9.
Recently, an 8-miRNA classifier was reported to distinguish among DLBCL molecular subgroups10. MiRNA signatures can also identify specific tumor drug resistances or drug sensitivities, and predict clinical outcome in DLBCL patients treated with chemoimmunotherapy10–12. A comprehensive profiling of the DLBCL miRNome identified miRNAs associated with patient survival independently of established indicators of outcome13. MiRNAs, such as miR-155, miR-21, and members of the miR-17-92 cluster have been demonstrated to drive lymphomagenesis in miRNA mouse models14–18, while the miR-34 family represents a tumor suppressor in DLBCL19. However, so far, the potential regulatory role of miRNAs in DLBCL progression has not been explored.
Here, we have performed miRNA and mRNA profiling of matched primary and relapsed DLBCLs. Our work establishes a landscape of miRNA expression in poor prognosis DLBCL, and highlights regulatory roles for miRNAs in signaling pathways contributing to disease progression and poor survival.
Methods
Patient samples
The discovery cohort consisted of matched primary-relapse sample pairs from seven DLBCL patients (Supplementary Table 1). The patient selection was based on availability of fresh frozen tissue containing adequate material for RNA extraction and next-generation sequencing (NGS). The patients were 62–76 years old and had a primary, high-risk (age-adjusted International Prognostic Index (aaIPI) score 2–3) disease. They had received chemoimmunotherapy (R-CHOP or R-CHOEP), but relapsed between 4 to 62 months after the treatment.
The validation cohort consisted of 13 primary-relapse sample pairs from formalin-fixed paraffin-embedded tissue containing adequate material for RNA extraction for real-time reverse transcription PCR (qRT-PCR) (Supplementary Table 1). The patients were 44–81 years old and had a primary, high-risk disease. They had received chemoimmunotherapy (R-CHOP or R-CHOEP), but relapsed between 4 to 157 months after the treatment.
The study was approved by the National Authority for Medicolegal Affairs, Finland, and Institutional Review Boards, and Ethics Committees in Helsinki, Finland, Aarhus, Denmark and Oslo, Norway. All patients gave informed consent.
Next-generation sequencing and data analysis
RNAs from seven primary-relapse DLBCL sample pairs (Supplementary Table 1) were subjected to NGS. Expression quantification was performed for all but one relapse sample (DLBCL1_R), which was filtered out during quality control. TruSeq small RNA sample preparation kit (Illumina, Inc. CA, USA) was used for library construction according to manufacturer’s protocol. Cluster generation and sequencing were performed on the Illumina HiSeqTM 2000 platform by Beijing Genomics Institute (Beijing, China), with average read depth of ~ 30 M per sample. More details on data analysis are provided in the Supplementary methods. The NGS data have been submitted to the Gene Expression Omnibus (GEO) database with accession number GSE69810.
miRNA−mRNA transcript target pair analysis
We identified highly anti-correlated (Spearman rho < −0.7, p < 0.05) transcript expression across the samples for each miRNA. Anti-correlated miRNA−mRNA pairs were then filtered for support by at least one of the selected target database resources (TargetScan Human v.5.220, DIANA microT v.1821, Microcosm v.522, PITA v.523, and a manually curated mirTarbase v.4.524). The miRNA-target gene pairs then proceeded to gene ontology (GO) and KEGG pathway enrichment analysis.
Real-time reverse transcription PCR (qRT-PCR) analysis
MiRNA expression was analyzed with the miRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Vedbaek, Denmark) using 100 ng RNA as starting material. Relative expression was determined by the 2−ΔΔC T method25 with 5S RNA as an endogenous control. The samples were run at least twice in triplicates.
For determining the target gene mRNA expression, 1 µg total RNA was reverse transcribed with iScript cDNA synthesis kit (Bio-Rad Laboratories Inc., Hercules, CA, USA). Subsequently, the cDNAs were diluted 1/10 and subjected to Taqman Fast qPCR with Gene Expression Assays from Applied Biosystems. Relative expression was determined by the 2−ΔΔC T method25 using GAPDH as an endogenous control. The samples were run three times with triplicates.
Survival analyses and statistics
For survival analyses, data from the Cancer Genome Characterization Initiative (CGCI) (n = 92) (the database of Genotypes and Phenotypes study accession: phs000532.v2.p1)26, 27 and the Lymphoma/Leukemia Molecular Profiling Project (LLMPP) (n = 233) (GEO accession: GSE10846)2 were used. CGCI level 1 RNA-seq data were downloaded and processed with SePIA transcriptomics analysis pipeline28. A web-based cutoff finder tool (http://molpath.charite.de/cutoffanalysis) was used to determine the most prognostic cutoff level for survival outcomes29. Kaplan−Meier plots were created with SPSS 22.0 (IBM, Armonk, NY, USA) and log-rank test was used for calculating the significance. Overall survival (OS) was determined from the date of diagnosis until last follow-up or death from any cause. Progression-free survival (PFS) was measured as the period between the date of diagnosis and progression or death from any cause. Multivariate analyses were performed according to the Cox proportional hazards regression model using categorical data. p-values < 0.05 were considered significant and all p-values were two-sided.
Cell culture and lentiviral transductions
SU-DHL-4 cells were from Deutsche Sammlung von Microorganismen und Zellkulturen GmbH (DSMZ) and they were cultured in RPMI 1640 Medium (Corning Life Sciences, Tewksbury, MA, USA) in the presence of 10% fetal bovine serum, 2 mm l-glutamine and 1% penicillin/streptomycin. The cells were tested and authenticated by Short Tandem Repeat profiling and checked for mycoplasma infections regularly using the MycoAlert™ Mycoplasma Detection Kit (Lonza, Basel, Switzerland).
The human shMIMIC lentiviral vectors for miR-370-3p, miR-381-3p, and miR-409-3p were obtained from GE Dharmacon (Lafayette, CO). SmartVector shMIMIC non-targeting Control 1 (Co1) and Control 10 (Co10) were used as negative controls. The lentiviral vectors had hEF1 promoter and expressed turboGFP. SU-DHL-4 cells were transduced with the lentiviral vectors and incubated for three days. Thereafter, the cells were subjected for puromycin (1 µg/ml) selection to create stable cell lines. The GFP and miRNA expression of the cell lines was confirmed by microscopy and qRT-PCR, respectively.
Cell viability assays
SU-DHL-4 cells stably expressing miR-370-3p, miR-381-3p, and miR-409-3p as well as non-targeting control Co10 were plated on black, clear-bottom 96-well plates (25,000 cells/well). Rituximab (1 µg/ml) or doxorubicin (100 nm) was added at the time of plating. The cells were incubated for 72 h, and cell viability measured with CellTiter-Blue Cell Viability assay (Promega, Madison, WI). The experiments were done in triplicates and repeated for four times.
Results
MiRNA sequencing of primary and relapsed DLBCL
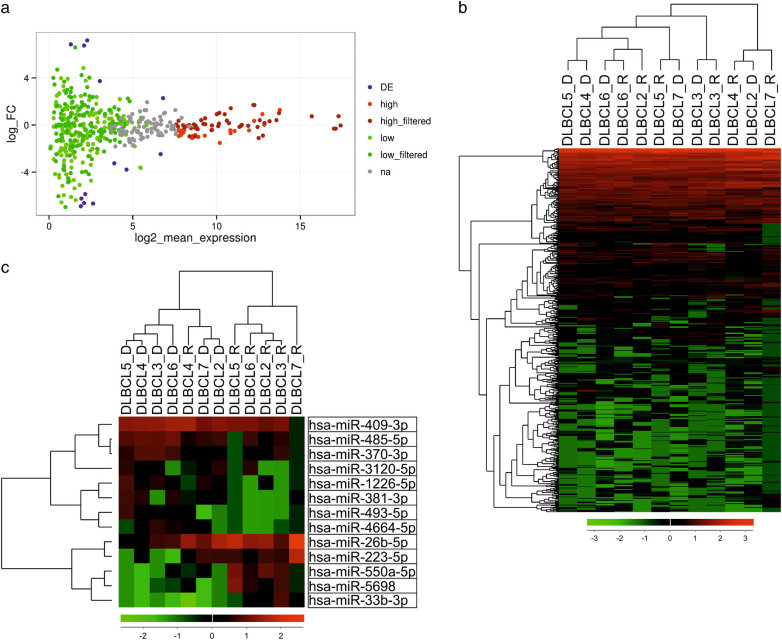
To uncover molecular mechanisms behind treatment resistance and progression in DLBCL, we searched for differentially expressed miRNA profiles between matched primary and relapsed DLBCLs. The discovery cohort consisted of seven DLBCL patients (Supplementary Table 1). A total of 492 known miRNAs were detected in the samples (Fig. 1a, b). Overall, the miRNA expression profiles in the primary and relapse samples were quite similar, suggesting that the miRNA expression remains relatively constant during the disease progression (Fig. 1a). Unsupervised clustering analysis of the miRNA expression could not distinguish primary from relapsed tumor samples (Fig. 1b).
Fig. 1. MiRNA-sequencing results.
Seven primary-relapse DLBCL sample pairs were subjected to next-generation miRNA sequencing. One relapse sample (DLBCL1_R) was exluded from the analyses because it failed the quality control. Therefore, the paired analyses included data from six pairs. a MA (Log ratio (M) vs. mean average (A) expression) plot visualizing miRNA expression in primary and relapse sample pairs. The colors denote different subgroups of miRNAs: differentially (DE), high and low expressed (high and low expressed miRNAs were filtered against a reference data set from normal and non-malignant B-cells, as described in Methods). b A heatmap visualizing clustering of the DLBCL samples based on their miRNA expression. c A heatmap of miRNAs differentially expressed between primary and relapse pairs with p-value < 0.05. FC fold change, na not assigned
We identified subsets of miRNAs with high (n = 24) and low expression (n = 177) as compared to a reference set of non-malignant B-cells (Fig. 1a; Supplementary Table 2). Among the high-expressed miRNAs were members of the miR-10 family, which has been demonstrated to have oncogenic effects for many cancer cells30. In contrast, many low-expressed miRNAs, including miR-129-5p31, 32, miR-66333, 34, and miR-203a35, 36, are known from their tumor suppressive roles. Further analysis revealed hypermethylation in the promoter regions of these tumor-suppressive miRNAs (Supplementary Fig. 1), suggesting methylation as the cause for their downregulation in DLBCL.
The analysis revealed 13 miRNAs with differential expression (p < 0.05) between primary and relapsed samples. Five miRNAs had higher and eight had lower expression in the relapse samples as compared to the primary samples (Fig. 1c and Supplementary Table 3).
MiRNA−mRNA data integration reveals potential miRNA target genes enriched in lymphoma-associated pathways
Given that miRNAs often regulate target gene expression by inducing RNA degradation37, identification of negatively correlated miRNA and mRNA transcript interactions provides functional insights to the oncogenic mechanisms of miRNAs. Therefore, we correlated miRNA expression with that of mRNAs derived from total RNA-sequencing data, and subsequently integrated these with miRNA target predictions. For the high expressed miRNAs, the analysis resulted in 243 miRNA-transcript pairs representing 186 individual genes (Supplementary Table 4). GO enrichment analysis showed these genes to be enriched for cell adhesion (not shown). Altogether 5531 miRNA−mRNA transcript pairs for the low expressed miRNAs were identified, and they represented 3064 individual genes (Supplementary Table 5). These genes were enriched for cancer-related pathways or processes, such as KEGG-pathways in cancer, MAPK signaling pathway, cell cycle and apoptosis (Supplementary Table 6), suggesting that the low-expressed miRNAs contain tumor suppressive miRNAs, and their oncogenic targets are over-expressed in the DLBCL.
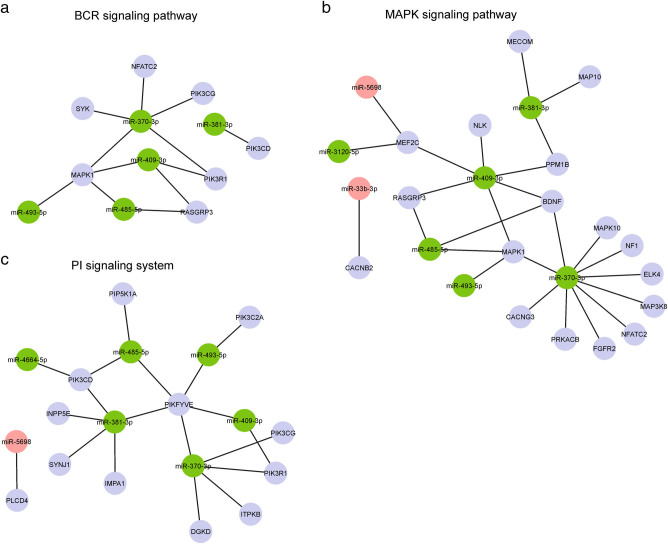
The number of miRNA−mRNA pairs for the 13 differentially expressed miRNAs was 1088 representing 787 individual genes (Supplementary Table 7). These were enriched for lymphoma-associated pathways38, 39, including PI signaling system (e.g. IMPA1, PIP5K1A, PIK3C2A, PIK3CG, PIK3R1), JAK-STAT cascade (e.g. STAT5A, STAT5B), BCR signaling (e.g. SYK, MAPK1, PIK3R1, PIK3CG, PIK3CD, RASGRP3) and MAPK signaling (e.g. MAPK1, MAPK10, MAP3K8, CACNG3) (Supplementary Table 8). Interestingly, these pathways were linked to the targets of those differentially expressed miRNAs that showed lower expression in relapse samples (Fig. 2 and Supplementary Fig. 2).
Fig. 2. Pathways regulated by the differentially expressed miRNAs.
a–c The networks visualize miRNA-target gene associations with the B-cell receptor signaling (a), MAPK signaling (b), and phosphatidylinositol signaling system (c). Green circles denote miRNAs downregulated in relapse, whereas red circles denote miRNAs upregulated in relapse as compared to primary tumor. The networks were created with Cytoscape 3.1.1
Validation of the differentially expressed miRNAs
To validate the NGS data, we performed qRT-PCR for eight differentially expressed miRNAs (miR-409-3p, miR-381-3p, miR-493-5p, miR-370-3p, miR-4664-5p, miR-485-5p, miR-26b-5p, and miR-550-5p) using an independent set (n = 13) of matched primary and relapsed DLBCL samples. Five miRNAs were excluded from the validation, because of low expression levels undetectable by qRT-PCR or because no functional primer pairs were available. In the validation set, three miRNAs (miR-409-3p, miR-381-3p, and miR-370-3p) were significantly downregulated in majority of the relapse samples (Fig. 3). The results provide support for the NGS results, and confirm the low expression of these miRNAs in relapsed DLBCL.
Fig. 3. Validation of differentially expressed miRNAs.
MiRNAs were validated in a cohort of 13 FFPE sample pairs with the miRCURY LNA™ Universal RT microRNA PCR system using 5S RNA as an endogenous control. MiRNAs which were significantly differentially expressed are shown in the figure. Mean±SD of log2 transformed relative expression values in primary vs. relapse samples are shown (negative values, down in relapse; positive values, up in relapse; zero values, no expression). All samples were run at least twice in triplicates
Overexpression of miR-370-3p, miR-381-3p, and miR-409-3p increases sensitivity to rituximab and doxorubicin
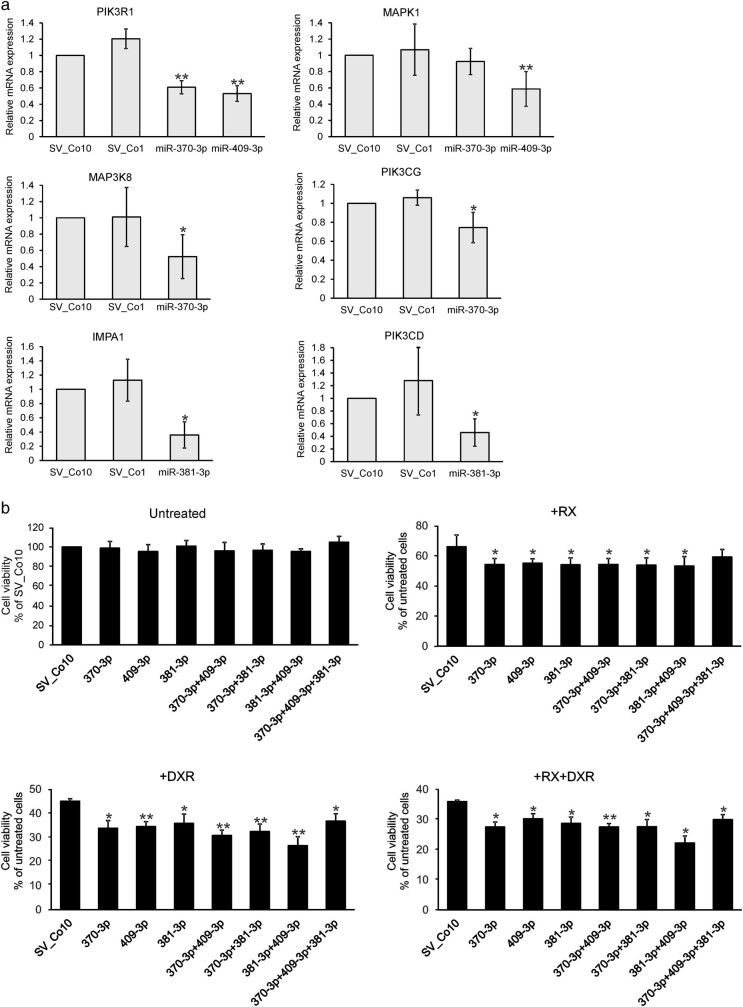
In functional analyses we chose to focus on miR-370-3p, miR-381-3p, and miR-409-3p as these differentially expressed miRNAs showed significantly lower expression in the relapsed samples, and appeared to be the most potent regulators of the MAPK, BCR, and PI signaling system. MiR-370-3p, miR-381-3p, and miR-409-3p were stably overexpressed in SU-DHL-4 cells using lentiviral vectors (Supplementary Fig. 3), and the expression of selected target genes from the MAPK, BCR, and PI signaling system was assayed by qRT-PCR. Overexpression of miR-370-3p resulted in downregulation of MAP3K8, PIK3R1, and PIK3CG mRNA (Fig. 4a). MiR-409-3p downregulated PIK3R1 and MAPK1 mRNA, whereas miR-381-3p suppressed IMPA1 and PIK3CD mRNA (Fig. 4a). MiRNA-induced regulation was detected also at the protein level, where miR-370-3p and miR-409-3p inhibited ERK1/2 protein levels (Supplementary Fig. 4). MiR-370-3p decreased PIK3CG protein levels, which is in line with the in silico and qRT-PCR results. However, also miR-409-3p and miR-381-3p affected PIK3CG, which is not predicted to be targeted by these miRNAs. This highlights the importance of experimental validation and verifies the well-known fact that miRNAs have a broad impact on protein expression40.
Fig. 4. MiRNA-mediated regulation of target gene expression and drug response.
a The expression of putative miRNA target genes was determined by qRT-PCR from the mRNAs isolated from SU-DHL-4 cells stably overexpressing miR-370-3p, miR-381-3p, or miR-409-3p. Cells expressing non-targeting controls (Co1 or Co10) were used as negative controls. The results normalized for GAPDH are shown as relative expression (2^−ΔΔCt). The samples were run three times with triplicates, and the results are shown as mean±SD. b SU-DHL-4 cells stably overexpressing miR-370-3p, miR-381-3p, or miR-409-3p were treated with rituximab (RX; 1 µg/ml), doxorubicin (DXR; 100 nm), as indicated. After 72 h incubation, the cell viability was determined by CellTiter-Blue Cell Viability assay. Non-targeting SV_Co10-expressing cells were used as controls. The experiment was performed in triplicates and repeated four times. The results are shown as mean±SEM. *p < 0.05, **p < 0.01
Next, we examined whether the miRNAs enhance the growth inhibitory effects of doxorubicin and rituximab, the key components of the R-CHOP regimen. In untreated cells, the miRNA overexpression did not affect the cell growth (Fig. 4b). However, when the cells were treated with rituximab or doxorubicin alone or in combination, the number of viable cells was lower in the miRNA-expressing cells, as compared to control SV-Co10 cells (Fig. 4b). The data suggest that miR-370-3p, miR-381-3p, and miR-409-3p favor a response of DLBCL cells to rituximab and doxorubicin, and thus are involved in the mechanisms of chemosensitivity or -resistance.
Survival association of the target genes
We investigated whether the expression of the target genes of the differentially expressed miRNAs is associated with survival. First, the prognostic impact was assessed in the CGCI cohort of 92 patients treated with R-CHOP-like regimen26. Clinical characteristics of the patients are shown in Table 1. Twenty-eight genes from the PI signaling system, BCR, and MAPK signaling pathways targeted by the differentially expressed miRNAs were used in the survival analyses. In Kaplan−Meier analyses, six genes were associated with survival (Table 2 and Supplementary Fig. 4). The patients with high expression of SYK, MAPK1, CACGN3, IMPA1, PIP5K1A, and RASGRP3 genes had a shorter survival in comparison to the remaining patients with lower mRNA levels. In Cox multivariate analyses with IPI, PIP5K1A expression remained an independent negative prognostic factor for both PFS (RR = 3.431, CI95 = 1.407–8.366, p = 0.007) and OS (RR = 3.897, CI95 = 1558–9.744, p = 0.004), whereas IMPA1 and RASGRP3 had adverse prognostic impact for OS (RR = 2.850, CI95 = 1.202–6.757, p = 0.017 and RR = 3.270, CI95 = 1.255–8.520, p = 0.015, respectively), and SYK for PFS (RR = 2.925, CI95 = 1.152–7.425, p = 0.024) (Supplementary Fig. 5). When target gene-related PFS and OS were analyzed separately for the patients in different molecular subtypes, high expression of PIP5K1A and RASGRP3 was associated with shorter survival in the non-GCB subtype, whereas high expression of IMPA1 was associated with shorter survival in the GCB subtype. SYK was associated with shorter PFS in the GCB subtype, and MAPK1 and CACNG3 were associated with shorter PFS in the non-GCB subtype (data not shown). When clinical characteristics and gene expression were compared according to molecular subtypes, no differences were observed between the subgroups (Table 1).
Table 1.
Patient characteristics of the CGCI and LLMPP cohorts
| CGCI | LLMPP | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients n (%) | GCB n (%) | Non-GCB n (%) | p -val | Patients n (%) | GCB n (%) | Non-GCB n (%) | p -val | ||
| All n (%) | 92 (100) | 51 (55) | 41 (45) | 233 (100) | 107 (46) | 126 (54) | |||
| Gender | Female | 31 (34) | 19 (37) | 12 (29) | 0.51 | 99 (42) | 49 (46) | 50 (40) | 0.356 |
| Male | 61 (66) | 32 (63) | 29 (71) | 134 (58) | 58 (54) | 76 (60) | |||
| Age | <60 | 39 (42) | 20 (39) | 19 (46) | 0.78 | 109 (47) | 56 (52) | 53 (42) | 0.264 |
| 60–65 | 15 (16) | 9 (18) | 6 (15) | 29 (12) | 13 (12) | 16 (13) | |||
| >60 | 38 42) | 22 (43) | 16 (39) | 95 (41) | 38 (36) | 57 (45) | 0.109 | ||
| Stage | I−II | 44 (48) | 28 (55) | 16 (39) | 0.15 | 105 (45) | 54 (52) | 51 (42) | |
| III−IV | 48 (52) | 23 (45) | 25 (61) | 121 (55) | 49 (48) | 72 (58) | |||
| IPI | 0–2 | 62 (67) | 35 (69) | 27 (66) | 0.83 | 112 (48) | 56 (76) | 56 (62) | 0.091 |
| 3–5 | 30 (33) | 16 (31) | 14 (34) | 52 (52) | 18 (24) | 34 (38) | |||
| IMPA1 | Low | 69 (75) | 42 (82) | 27 (66) | 0.09 | 218 (94) | 104 (97) | 114 (90) | 0.058 |
| High | 13 (25) | 9 (18) | 14 (34) | 15 (6) | 3 (3) | 12 (10) | |||
| MAPK1 | Low | 27 (29) | 12 (24) | 15 (37) | 0.25 | 50 (21) | 27 (25) | 23 (18) | 0.205 |
| High | 65 (71) | 39 (76) | 26 (63) | 183 (79) | 80 (75) | 103 (82) | |||
| PIP5K1A | Low | 78 (85) | 46 (90) | 32 (78) | 0.15 | 223 (96) | 104 (97) | 119 (94) | 0.35 |
| High | 14 (15) | 5 (10) | 9 (22) | 10 (4) | 3 (3) | 7 (6) | |||
| CACNG3 | Low | 77 (84) | 46 (90) | 31 (76) | 0.09 | 131 (56) | 63 (59) | 68 (54) | 0.508 |
| High | 15 (16) | 5 (10) | 10 (24) | 102 (44) | 44 (41) | 58 (46) | |||
| RASGRP3 | Low | 82 (89) | 47 (92) | 35 (85) | 0.33 | 183 (79) | 77 (72) | 106 (84) | 0.026 |
| High | 10 (11) | 4 (8) | 6 (15) | 50 (21) | 30 (28) | 20 (16) | |||
| SYK | Low | 81 (88) | 44 (86) | 37 (90) | 0.75 | 219 (94) | 98 (92) | 121 (96) | 0.176 |
| High | 11 (12) | 7 (14) | 4 (10) | 14 (6) | 9 (8) | 5 (4) | |||
Table 2.
DE-miRNA target gene expression is associated with survival in DLBCL patients (n = 92) (log-rank test)
| Gene | Pathway | Targeting miRNA | OS a ( p -val ) | PFS ( p -val) |
|---|---|---|---|---|
| PIP5K1A | PI | miR-485-5p | <0.001 | <0.001 |
| IMPA1 | PI | miR-381-3p | 0.004 | 0.016 |
| SYK | BCR | miR-370-3p | 0.231 | 0.002 |
| RASGRP3 | BCR, MAPK | miR-409-3p, miR-485-5p | 0.001 | <0.001 |
| MAPK1 | MAPK | miR-370-3p, miR-409-3p, miR-493-5p, miR-485-5p | 0.248 | 0.038 |
| CACNG3 | MAPK | miR-370-3p | 0.224 | 0.007 |
a OS overall survival, PFS progression-free survival, PI phosphatidylinositol, BCR B-cell receptor, MAPK mitogen-activated protein kinase
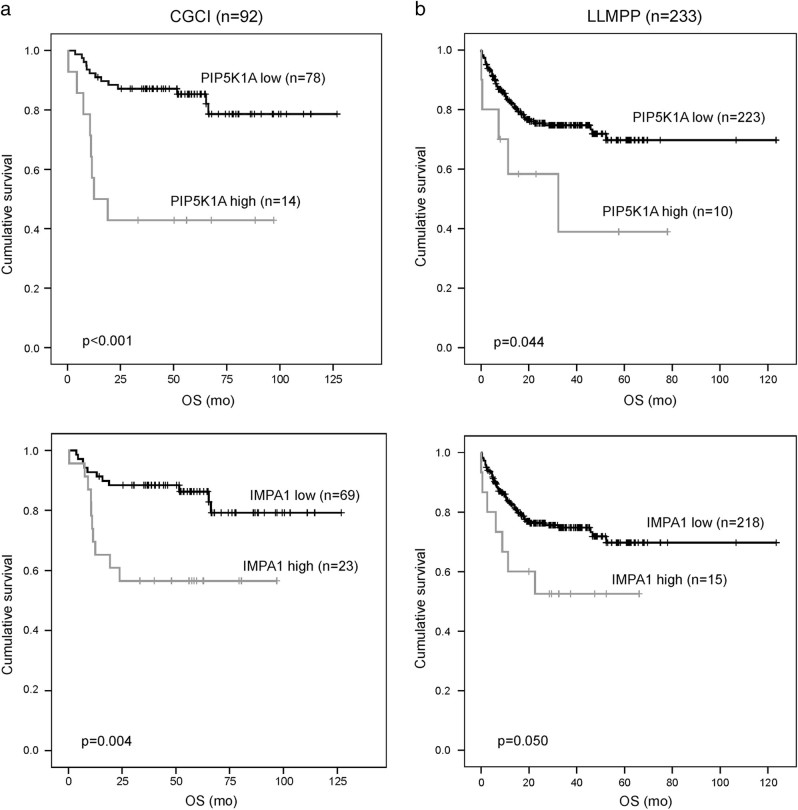
To validate the survival data, LLMPP data set was exploited (n = 233) (Table 1)3. In this cohort, high expression of IMPA1 and PIP5K1A was found to have adverse impact on OS (Fig. 5). Association of PIP5K1A with poor survival was restricted to samples of non-GCB subtype (not shown). Together, our clinical, molecular, and functional data show that differentially expressed miRNAs target gene expression and thereby regulate key cell survival pathways, proliferation of lymphoma cells and survival of patients with DLBCL progression.
Fig. 5. PIP5K1A and IMPA1 are significantly associated with survival in two DLBCL patient cohorts.
a, b The chemoimmunotherapy-treated patients from two independent cohorts (CGCI n = 92 (a) and LLMPP n = 233 (b)) were divided into two groups with high and low gene expression. The ideal cut-off values were calculated using Cutoff Finder. Kaplan–Meier curves depict overall survival (OS) of patients whose tumors contained low or high levels of the selected genes. p-values were obtained using a log-rank test
Discussion
Genomic profiling of paired primary-relapse samples enables the discovery of the biologic pathways and mechanisms that drive therapy resistance. Here, we have utilized NGS and profiled the miRNAome of matched primary and relapsed DLBCL. MiRNAs are important regulators of both normal and pathological cellular processes controlling gene expression through complex regulatory networks. Recently, it was reported that miRNAs are associated with DLBCL outcome and can be used as predictors of treatment effectiveness10, 13. To our knowledge, the present study is the first one characterizing the changes that occur in miRNA expression during the treatment course of DLBCL and validating these observations with functional studies. Our results show that miRNA expression profiles remain relatively constant as the disease progresses with only a few miRNAs that were differentially expressed between the primary-relapse pairs. The limitation of our study is the small sample size, which may hamper the identification of differences between primary and relapse tumors. However, the small set of differentially expressed miRNAs participates in the control of central lymphoma-associated pathways, and represents a set of potential molecular targets for therapeutic intervention. Putative targets for the differentially expressed miRNAs were enriched for important lymphoma-associated pathways, such as PI signaling system, BCR, and MAPK signaling. Specifically, they were targeted by miRNAs downregulated in relapse tissue, suggesting that the pathways are activated during DLBCL progression. Our data further demonstrated that high expression of selected genes from these signaling pathways in the pretreatment samples was associated with poor survival. Importantly, the expression of PIP5K1A and IMPA1 genes had negative prognostic impact on survival in two independent DLBCL cohorts. PIP5K1A and IMPA1 are components of the PI signaling pathway, which is an important intracellular second-messenger signaling system linking to the PI3K/AKT pathway. The PI3K/AKT pathway in turn plays an important role in controlling proliferation and survival of tumor cells, and therefore represents a promising therapeutic target in DLBCL39 . IMPA1 (Inositol monophosphatase 1) is responsible for the provision of inositol required for synthesis of PI and polyphosphoinositides, whereas PIP5K1A (Phosphatidylinositol-4-Phosphate 5-Kinase, Type I, Alpha) catalyzes the phosphorylation of phosphatidylinositol 4-phosphate (PIP) to form phosphatidylinositol 4,5-bisphosphate (PIP2), implicated in a wide variety of cellular signaling pathways.
BCR signaling has also been suggested to act as a driver of lymphoma development41. It is essential for normal B-cell development and maturation, but has emerged as an important target for the treatment of B-cell malignancies, including DLBCL. Central hubs in the BCR signaling pathway include SYK, Bruton’s tyrosine kinase (BTK), and PI3K. Novel therapies targeting the BCR signaling in lymphoma are currently under investigation. For instance, ibrutinib, a small molecule inhibiting BTK has shown significant anti-tumor activity in clinical studies on B-cell lymphomas42. Additionally, tonic BCR signaling and lymphoma cell survival can be selectively targeted with a SYK inhibitor fostamatinib, which has shown activity against relapsed DLBCL in a phase I/II study43.
MiR-409-3p, miR-381-3p, and miR-370-3p were validated as being downregulated in the relapse samples of an independent patient cohort. Interestingly, overexpression of these miRNAs enhanced the chemosensitivity of DLBCL cells in vitro. Several studies have suggested that miRNAs are novel players in mediating drug resistance. Overexpression of miR-34a sensitized DLBCL cells to doxorubicin in vitro44. MiR-17~92 cluster mediated chemoresistance in mantle cell lymphoma45, and miR-331-5p and miR-27a were downregulated in doxorubicin-resistant leukemia cells46. MiR-381 may play a role in regulating the drug resistance in leukemia cells47.
In addition to differentially expressed miRNAs, we characterized miRNAs with high or low expression in the DLBCL samples compared to a control data set of non-malignant B-cells. Tumor suppressive miRNAs miR-129-5p, miR-663a, and miR-203a were hypermethylated at their promoter region, suggesting methylation as a potential mechanism for downregulation. This concurs with previous studies demonstrating hypermethylation of miR-129 and miR-203 in hematological cancers, including non-Hodgkin lymphoma48, 49. Indeed, lower miR-129-5p expression was associated with shorter survival in DLBCL patients both with and without R-CHOP treatment31. In our integrated miRNA−mRNA analysis miR-129-5p was predicted to target several oncogenes that included IRF4, PIM1, FOXP1, SYK, BCL10, RUNX1, and ABL1. These were enriched for cancer-associated pathways, such as regulation of I-kappaB kinase/NF-kappaB cascade, regulation of protein kinase cascade, cell death, and apoptosis (not shown).
In conclusion, our analysis of the miRNAome in matched primary refractory and relapsed DLBCL uncovers biological processes underlying relapse/progression. In addition, the data imply that miRNAs mediate the chemoresistance of DLBCL. The results are novel and promising and emphasize that the molecular mechanisms of DLBCL progression are complex, involve multiple pathways, and are heterogeneous between the patients.
Electronic supplementary material
Acknowledgements
The authors would like to thank Anne Aarnio, Kristina Lystlund, and Marika Tuukkanen for expert technical assistance. Dr. Marko Hyytiäinen is acknowledged for providing lentiviral packaging plasmids and for advising in lentiviral transduction experiments. The diffuse large B-cell lymphoma (DLBCL) data set is part of the Cancer Genomics Characterization Initiative (CGCI), supported by NCI contract N01-C0-12400 (http://cgap.nci.nih.gov/cgci.html).
Authors’ contributions
S-KL designed and conceived the study, analyzed data, and wrote the manuscript. KI analyzed the NGS data, performed bioinformatics analyses, and participated in writing the manuscript. KJ designed and performed experiments. IS performed experiments. CL performed pathway analyses. AA analyzed methylation data. AC analyzed the total RNA-Seq data and preprocessed the CGCI data. ML, SJH-D, FA, M-LK-L, JD and HH provided clinical samples. RL and SH designed the study, supervised bioinformatics analyses, and assisted in interpretation. SL designed and supervised the study and revised the manuscript. All authors have read and approved the manuscript.
Funding
The study was supported by grants from the Academy of Finland (SH), Finnish Cancer Societies (SL, SH), Sigrid Jusélius Foundation (SL, SH), University of Helsinki (SL), and Helsinki University Hospital (SL).
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Suvi-Katri Leivonen and Katherine Icay contributed equally to this work.
Supplementary information
The online version of this article (10.1038/s41408-017-0033-8) contains supplementary material, which is available to authorized users.
References
- 1.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Lenz G, et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coiffier B, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002;346:235–242. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 6.Pfreundschuh M, et al. CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in young patients with good-prognosis diffuse large-B-cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) Group. Lancet Oncol. 2006;7:379–391. doi: 10.1016/S1470-2045(06)70664-7. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iqbal J, et al. Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. Blood. 2015;125:1137–1145. doi: 10.1182/blood-2014-04-566778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alencar AJ, et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin. Cancer Res. 2011;17:4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montes-Moreno S, et al. miRNA expression in diffuse large B-cell lymphoma treated with chemoimmunotherapy. Blood. 2011;118:1034–1040. doi: 10.1182/blood-2010-11-321554. [DOI] [PubMed] [Google Scholar]
- 13.Lim EL, et al. Comprehensive miRNA sequence analysis reveals survival differences in diffuse large B-cell lymphoma patients. Genome Biol. 2015;16:18. doi: 10.1186/s13059-014-0568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costinean S, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin HY, et al. MicroRNA-17~92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J. 2013;32:2377–2391. doi: 10.1038/emboj.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 18.Sandhu SK, et al. B-cell malignancies in microRNA Emu-miR-17~92 transgenic mice. Proc. Natl. Acad. Sci. USA. 2013;110:18208–18213. doi: 10.1073/pnas.1315365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig VJ, et al. Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia. 2012;26:2421–2424. doi: 10.1038/leu.2012.110. [DOI] [PubMed] [Google Scholar]
- 20.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maragkakis M, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 24.Hsu SD, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Morin RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Icay K, et al. SePIA: RNA and small RNA sequence processing, integration, and analysis. BioData Min. 2016;9:20. doi: 10.1186/s13040-016-0099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budczies J, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tehler D, Hoyland-Kroghsbo NM, Lund AH. The miR-10 microRNA precursor family. Rna Biol. 2011;8:728–734. doi: 10.4161/rna.8.5.16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedstrom G, et al. Low expression of microRNA-129-5p predicts poor clinical outcome in diffuse large B cell lymphoma (DLBCL) Int. J. Hematol. 2013;97:465–471. doi: 10.1007/s12185-013-1303-2. [DOI] [PubMed] [Google Scholar]
- 32.Wu Q, et al. Methylation of miR-129-5p CpG island modulates multi-drug resistance in gastric cancer by targeting ABC transporters. Oncotarget. 2014;5:11552–11563. doi: 10.18632/oncotarget.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y, et al. Primate-specific miR-663 functions as a tumor suppressor by targeting PIK3CD and predicts the prognosis of human glioblastoma. Clin. Cancer Res. 2014;20:1803–1813. doi: 10.1158/1078-0432.CCR-13-2284. [DOI] [PubMed] [Google Scholar]
- 34.Zang W, et al. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Mol. Cancer. 2015;14:37. doi: 10.1186/s12943-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Diao Y, et al. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in rhabdomyosarcoma. J. Biol. Chem. 2014;289:529–539. doi: 10.1074/jbc.M113.494716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hailer A, et al. Loss of tumor suppressor mir-203 mediates overexpression of LIM and SH3 Protein 1 (LASP1) in high-risk prostate cancer thereby increasing cell proliferation and migration. Oncotarget. 2014;5:4144–4153. doi: 10.18632/oncotarget.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi D, Ciardullo C, Gaidano G. Genetic aberrations of signaling pathways in lymphomagenesis: revelations from next generation sequencing studies. Semin. Cancer Biol. 2013;23:422–430. doi: 10.1016/j.semcancer.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat. Rev. Clin. Oncol. 2014;11:12–23. doi: 10.1038/nrclinonc.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niemann CU, Wiestner A. B-cell receptor signaling as a driver of lymphoma development and evolution. Semin. Cancer Biol. 2013;23:410–421. doi: 10.1016/j.semcancer.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Advani RH, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J. Clin. Oncol. 2013;31:88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedberg JW, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 2010;115:2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marques SC, et al. High miR-34a expression improves response to doxorubicin in diffuse large B-cell lymphoma. Exp. Hematol. 2016;44:238–246 e232. doi: 10.1016/j.exphem.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 45.Rao E, et al. The miRNA-17-92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia. 2012;26:1064–1072. doi: 10.1038/leu.2011.305. [DOI] [PubMed] [Google Scholar]
- 46.Feng DD, et al. Down-regulated miR-331-5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J. Cell. Mol. Med. 2011;15:2164–2175. doi: 10.1111/j.1582-4934.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, et al. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS One. 2013;8:e82062. doi: 10.1371/journal.pone.0082062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chim CS, et al. Epigenetic inactivation of the hsa-miR-203 in haematological malignancies. J. Cell. Mol. Med. 2011;15:2760–2767. doi: 10.1111/j.1582-4934.2011.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong KY, et al. Epigenetic inactivation of the MIR129-2 in hematological malignancies. J. Hematol. Oncol. 2013;6:16. doi: 10.1186/1756-8722-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.