Abstract
Hospital-acquired infections pose a significant risk to patient health, while their surveillance is an additional workload for hospital staff. Our overall aim is to build a surveillance system that reliably detects all patient records that potentially include hospital-acquired infections. This is to reduce the burden of having the hospital staff manually check patient records. This study focuses on the application of text classification using support vector machines and gradient tree boosting to the problem. Support vector machines and gradient tree boosting have never been applied to the problem of detecting hospital-acquired infections in Swedish patient records, and according to our experiments, they lead to encouraging results. The best result is yielded by gradient tree boosting, at 93.7 percent recall, 79.7 percent precision and 85.7 percent F1 score when using stemming. We can show that simple preprocessing techniques and parameter tuning can lead to high recall (which we aim for in screening patient records) with appropriate precision for this task.
Keywords: clinical decision-making, databases and data mining, ehealth, electronic health records, secondary care
Introduction
Patient security in hospitals is crucial. Various risk factors for patients can be found within clinical settings, including hospital-acquired infections (HAIs). HAI is defined as
[a]n infection occurring in a patient in a hospital or other healthcare facility in whom the infection was not present or incubating at the time of admission. This includes infections acquired in the hospital but appearing after discharge, and also occupational infections among staff of the facility.1
HAIs may be caused by medical procedures, for instance, during the implantation of contaminated urinary tract catheters. HAI might also develop in wounds after surgery or occur when micro-organisms spread from person to person, such as during winter vomiting diseases. HAIs pose a public health problem worldwide. A survey conducted under the patronage of World Health Organization (WHO) in 2002 found that for 55 hospitals in 14 countries, an average of 8.5 percent of all hospital patients suffer from HAI.1
Many attempts have been made to confine HAIs, for example, better hygiene or manual surveillance performed by infection control professionals, constituting an additional workload for hospital medical staff and hospital management. Nevertheless, the presence of HAIs remains unvaried in modern health facilities. Hospital Information Systems, which are standard in most health facilities today, in combination with the increasing amount of digital data, has pioneered the way for automatic surveillance systems. In the course of this development, research that focuses on the automatic detection of HAI has emerged throughout the past years. The exact approaches vary, ranging from numerous attempts that implement rule-based systems to fewer machine learning–based approaches.
Our study is of an experimental nature and focuses on applying machine-learning techniques to the problem of detecting HAIs. For our task, two well-known learning algorithms, support vector machines (SVMs) and gradient tree boosting (GTB), were applied to the data. The data used in this study comprise patient records provided by Karolinska University Hospital.
The focus of our study lies on the recall values obtained using different classifiers. We aim at approaching 100 percent recall with the highest precision possible, which is a reasonable overall performance in terms of F1. As presented in the literature,2 we obtained encouraging results when applying Naive Bayes, SVM and a C4.5 Decision Tree to the problem in an initial approach. Therefore, SVM, in particular, revealed its potential application for our task, as it tendentiously yielded the best results. Thus, we decided to apply SVM once again, this time with tuned parameters. We further applied GTB since it has good classification abilities and interesting data-mining capabilities.3 The data-mining capability of interest is the ability to interpret the trained classifier. It makes it possible to get a measurement of how important each feature is. This is of interest since it enables us to assess if the features used by the classifier are plausible indicators of HAI. In combination with each of the classifiers, we applied different data preprocessing and feature selection methods, namely, term frequency (TF), lemmatization, stemming, stop word removal, infection-specific terms, term frequency–inverse document frequency (TF-IDF), a combination of lemmatization, respectively, stemming, stop word removal and TF-IDF. The study focused on answering the question regarding whether or not any preprocessing method or parameter tuning would help to increase performance.
Algorithms with high recall are especially suitable for the screening of infections.4 Thus, this study is an important step toward implementing a system that is expected to constantly screen patient records and determine whether they contain HAI. Automatic HAI screening is especially valuable for medical staff and hospital management, since it would significantly reduce the burden of manually checking patient records for HAI, which is a time-consuming task even for highly trained experts.5 Instead of analyzing all records, the hospital staff would only have to check those patient records that the system preselected as containing HAI.
Related work
During the past decade, multiple studies have utilized machine learning in the medical domain. See Claster et al.6 for an overview of some of the more recent papers. The following section presents recent studies that adapt machine-learning approaches to the problem of detecting HAI.
Researchers have aimed at developing a monitoring system that predicts potential HAIs.7 In that particular study, six classifiers were applied to the problem: Alternating Decision Tree (ADTree), C4.5, ID3, RNA, Decision Tables and nearest neighbor with generalization (NNge). Their data comprise 1520 patient records from the intensive care unit (ICU) of the University Hospital of Oron, Algeria. From this, 17 features were derived, some of which are sex of patient, age of patient, reason for the hospitalization or catheter. They solely measure accuracy, obtaining the highest one of 100 percent with the NNge classifier.
In a study conducted in Taiwan,8 linear regression (LR) and artificial neural networks (ANNs) were used to predict HAI. The system used structured data. A total of 16 features were extracted from patient records, ranging from demographic, procedural and therapeutic features to features concerning the general health status of the patient. ANNs are trained using back-propagation and conjugate gradient descent. Evaluation of the system was done using an internal test set from the same hospital, as well as an external test set from a different hospital. For the internal test set of 461 hospitalizations, the best result was produced using the ANN approach, reaching a recall of 96.64 percent and a specificity of 85.96 percent. For the external test set consisting of 2500 hospitalizations from different hospitals, LR gave the best result with 82.76 percent recall and 80.90 percent specificity.
In a series of papers,9–12 researchers around Gilles Cohen addressed the task of monitoring and detecting HAI using data from the University Hospital of Geneva, Switzerland. Their focus lied on the class imbalance, a problem that can be observed in many real-world classifications, especially in the medical domain. They used data from 683 patients, out of which 11 percent were positive cases (contracted HAI) and 89 percent were negative (did not contract HAI). From these records, the researchers collected features, such as demographic characteristics, admission date or admission diagnosis, and applied various techniques in order to detect patients with HAI.
In another study,9 the researchers tested (1) random and agglomerative-hierarchical-clustering (AHC) oversampling, (2) K-means subsampling and random subsampling and (3) combined AHC oversampling and K-Means subsampling. They compared them using five different classifiers: IB1, Naive Bayes, C4.5, AdaBoost and a symmetrical-margin SVM. They obtained a recall ranging from 49 percent (IB1) to 87 percent (NB) for the five different classifiers when applying combined AHC oversampling and K-Means subsampling. Specificity ranged from 74 percent (NB) to 86 percent (IB1).
In yet another study,10 the researchers compared a symmetrical SVM against an asymmetrical one. The experiments showed the inadequacy of the symmetrical SVM when dealing with a skewed class distribution. They obtained the highest recall, at 92 percent, with a specificity of 72.2 percent when using the asymmetrical SVM.
In the follow-up paper,11 the researchers applied one-class SVMs to the problem. This adaption of SVM can be trained to distinguish two classes by ignoring one of the two classes and learning from one class only. Their best results yielded a recall of 92.6 percent at the cost of a very low specificity of 43.73 percent.
In a study from 2006,12 the researchers compared the resampling strategy that had yielded the best results in a prior study,9 that is, they combined AHC oversampling and K-means subsampling, to the asymmetrical soft-margin SVM, which had been proven to be suitable for an imbalanced data, as shown in an earlier study.10 The asymmetrical soft-margin SVM obtained a recall of 92 percent and a specificity of 72 percent, thus clearly outperforming their resampling method that obtained the highest recall at 87 percent with a 74 percent specificity for Naive Bayes.
In two additional studies,13,14 researchers presented results from a retrospective analysis of data that were collected during the 2006 HAI prevalence survey at the University Hospital of Geneva. The objective of their study, which encompassed both papers, was to define the minimal set of features needed for automated case reporting of HAI. Their dataset comprised 1384 cases, with 166 positive cases (11.99%) and 1218 negative cases (88.01%). The data contained four categories of interest: demographic information, admission diagnosis, patient information on the study date and 6 days before and information related to the infection. They used information gain and SVM recursive feature elimination, combined with chi-squared filtering, to select the most important features. They built two datasets: S1, which contained the most significant features retained by both feature selection algorithms; and S2, which also contained the most important features, but the features that were not well-documented in the patient record were removed. They then applied Fisher’s Linear Discriminant for classification. As a result, they obtained 65.37 percent recall and 41.5 percent precision for S1 and 82.56 percent recall and 43.54 percent precision for S2.
Method
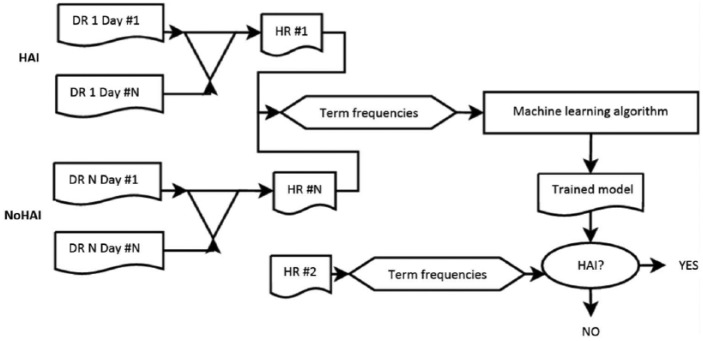
The method used in text classification using machine learning, a high-level flow chart, can be seen in Figure 1. An explanation of each part of the flow chart is given in the sections below.
Figure 1.
A high-level flow chart describing this study’s text-classification approach for automatically detecting HAI. DR stands for daily patient record. In this study, a patient’s DR comprises data from four modules. All DRs of a patient together amount to the patient’s HR.
Data
The dataset (This research has been approved by the Regional Ethical Review Board in Stockholm (Etikprövningsnämnden i Stockholm), permission number 2012/1838-31/3.) encompasses data from the electronic health records (EHRs) of 120 inpatients at a major university hospital in Sweden and was collected during a Point Prevalence Survey (PPS) (In Sweden, PPSs are performed twice a year to estimate the occurrence of HAI by counting existing cases of HAI at one specific time) in spring 2012. Not all information stored in the patients’ EHRs was considered valuable by the physicians for detecting HAI. Thus, a subset of information from the EHRs was retrieved: Journalanteckning (Engl.: record notes), Läkemedelsmodul (Engl.: drug module), Mikrobiologiska Svar (Engl.: microbiological result) and Kroppstemperatur (Engl.: body temperature). The information extracted from these modules consists of structured and unstructured data. Structured data refer to data that are stored in predefined fields, such as International Classification of Diseases–10th Revision (ICD-10) diagnosis codes, medication or body temperature. Unstructured data refer to textual notes written by physicians, such as daily notes or microbiological results.
For each of the 120 patients, information from all four modules was extracted for the patient’s entire hospitalization. The physicians defined one hospitalization as the stay of a patient at a health facility for one care process. If the patient is discharged from one department of the hospital and admitted to another within 24 h, this was regarded as the same hospitalization. Moreover, any noted event occurring within 24 h after discharge was included in the hospitalization. From this point on, we will refer to the file that contains the data of a patient’s entire hospitalization as the hospitalization record (HR). Since some of the 120 patients were hospitalized multiple times during the 5-month period of records we received, our dataset comprises 213 HRs. Hospitalizations of less than 48 h are not represented in the final dataset as they were considered to carry too little information. (This time frame is based on international definitions of HAI and the incubation period of infections and is estimated to be less than 48 h for a multitude of disorders.15) Table 1 depicts the characteristics of the HRs that were used as input for the classifiers.
Table 1.
The characteristics of the HRs used in our study.
| HAI | NoHAI | Total | |
|---|---|---|---|
| Number of HRs | 128 | 85 | 213 |
| Length of hospitalization in days | 2–144 | 3–93 | 2–144 |
| Total number of tokens | 22,528,102 | 2,598,036 | 25,126,138 |
HR: hospitalization record; HAI: hospital-acquired infection.
All 120 patients had experienced HAI according to the PPS results. We had access to a 5-month period of records. As a result, the physicians in this study, unlike the physicians who carried out the PPSs, obtained information on how the health status of the patient progressed and which assessment he or she received during the time after the PPSs had been conducted. The physicians in this study could therefore give a more accurate answer on whether or not HAI occurred. Only 128 of 213 HRs contained HAI diagnoses (positive examples). Those records represent the HAI class. According to the physician’s assessment, the remaining 85 HRs contained no HAI diagnoses (negative examples), thus representing the NoHAI class. The dataset was not balanced, but instead, it was skewed toward the positive class. We only used the class containing HAI for prediction and not the class without HAI.
Machine learning
There are a large number of different learning algorithms and classifier models that could be applied in our classification task. We decided to apply SVMs and GTB to the problem. Instead of exhaustively testing different learning strategies, we focused on a problem that is common for all supervised learning techniques—feature selection and the optimization of parameters. The authors in previous studies16,17 stated that preprocessing, feature selection and parameter tuning have a large impact on performance—more than the actual choice of the classification model. For a more detailed description of GTB, see Hastie et al.,18 and for SVM, see Dalal and Zaveri16 and Noble.19 The two classifiers are part of the scikit-learn environment (available via http://scikit-learn.org).
SVM
SVMs use the concept of representing the documents that are to be classified as points in a high-dimensional space and finding the hyperplane that separates them. This concept, in fact, is not unique to SVM. However, the difference between SVM and other classifiers using this concept is how the hyperplane is selected. SVM tries to find the hyperplane with the maximum margin, where margin refers to the distance between the hyperplane and the nearest data points.19 Using SVM is, among others, motivated by the statement that SVM is very effective for two-class classification problems.16
We used an optimized and non-optimized SVM on our dataset. For the non-optimized SVM classifier, a radial basis function (RBF) kernel with degree = 3, C = 1, epsilon = 0.001 and gamma = 1/1000 was used. Usually, an RBF kernel is preferred unless the number of features is huge. In that case, a linear kernel is appropriate.20
GTB
GTB utilizes the power of a forest of weak tree learners to approximate the sought-after classification function. By training a number of tree classifiers on different parts of the training data and then weighting their collective decision, a strong classifier is produced. The weak learner is a learner that may only have slightly better classification abilities than random guessing, but the combined strong learner will be an approximation of the true classification function. Using decision trees as the weak learner has the advantage of being able to handle different data types without conversion. Inherent when using trees with a maximum depth is feature selection, as only the most important features will be used when constructing the trees. Using trees also makes it possible to interpret the trained model by examining which variables are used most commonly to branch in each individual decision tree.18 We used GTB both with and without parameter optimization. When used without parameter optimization, the default parameters used were v = 0.1, J = 3, M = 100 and subsample = 1.0.
Preprocessing techniques and parameter optimization
According to previous researchers,17,21 the high-dimensional feature space, that is, the amount of unique terms that occur in the text documents to be classified, marks a major characteristic and difficulty in text classification, making it a non-trivial task for automatic classifiers. It is thus desirable to reduce the dimensionality of the data to be processed by the classifier, in addition to reducing execution time and improving predictive accuracy. In our study, we used well-known preprocessing and filter methods in order to optimize and reduce the feature space. The preprocessing techniques are depicted in Table 2.
Table 2.
Different combinations of applied text-classification techniques and feature selection methods as well as the name chosen for each combination.
| Name | Text-classification method | Feature selection method |
|---|---|---|
| TF 1000 | Data not processed | TF 1000 |
| Lemma | Data lemmatized | TF 1000 |
| Stem | Data stemmed | TF 1000 |
| Stop | Stop words removed from data | TF 1000 |
| IST | Data not processed | Infection-specific terms used |
| TF-IDF 1000 | Data not processed | TF-IDF 1000 |
| LS-TFIDF 1000 | Data lemmatized + stop words removed | TF-IDF 1000 |
| SS-TFIDF 1000 | Data stemmed + stop words removed | TF-IDF 1000 |
TF: term frequency; IST: infection-specific terms; TF-IDF: term frequency–inverse document frequency.
Term frequency (TF)
In this method, TF 1000, the 1000 most frequent terms, was chosen based on their TF. TF refers to the simplest weighting scheme, where the weight of a term is equal to the number of times the term occurs in a document.22,23
Lemmatization and stemming
In machine learning, lemmatization and stemming are the frequently used methods when preprocessing data.16 In our study, we use the CST lemmatize (http://cst.dk/online/lemmatiser/uk/) in order to perform lemmatization. Lemmatization describes the process of reducing a word to a common base form, normally its dictionary form (lemma). This is achieved by removing inflectional forms and sometimes derivationally related forms of the word, by means of vocabulary usage and morphological analysis, for instance, am, are, is, be, or hospitals, hospital’s → hospital.22,23 For the Swedish language, which is highly inflectional, lemmatization is more important than it is for English.
We further use stemming, which is a simpler form of lemmatization, where the produced stemmed words do not need to be real words but the minimal set of characters that distinguish the different stemmed words, for example, hospitals, hospital’s → hospit. We used the Snowball stemmer (http://snowball.tartarus.org/algorithms/swedish/stemmer.html) for the Swedish language. The patient records were lemmatized and stemmed separately, before then being given as input for the classifiers.
Stop word removal
Stop words are terms that are regarded as not conveying any significant semantics to the texts or phrases they appear in and are consequently discarded.24 The filter was configured to use the Swedish stop list, which is available via Snowball (http://snowball.tartarus.org/algorithms/swedish/stop.txt) and comprises 113 words, such as och (Engl.: and), att (Engl.: to) or i (Engl.: in).
Infection-specific terms
In the course of the Detect-HAI project (Detection of HAIs through language technology project—conducted in collaboration between Karolinska University Hospital and the Department of Computer and System Science (DSV) at Stockholm University during 2012 and 2013; the aim of the project was to ultimately build a system that can automatically detect HAI in Swedish patient records), a terminology database containing infection-specific terms was built using a semi-automatic approach. Infection-specific terms, such as kateter (Engl.: catheter), ultraljud (Engl.: ultrasound), operation (Engl.: surgery) or feber (Engl.: fever), are expected to be contained in patient records in case an infection occurs. In order to build the terminology database, the medical experts involved in that project supplied a seed set of about 30 infection-specific terms, which were based on frequent observations in the above-mentioned data and their knowledge about infections. The seed set was then extended by giving each term of it as input for an automatic synonym extractor. The synonym generator used was implemented in-house and was based on random indexing.25 For each input term, a table holding related terms, which could include synonyms or misspellings, was generated as an output by the synonym generator. One medical expert then manually analyzed all proposed terms with respect to whether or not they could be regarded as applicable infection-specific terms. All relevant terms were added to the terminology database. The final infection-specific term (IST) terminology database comprised a total of 1045 terms. When using the terminology database as a feature reduction technique, we removed all terms from the HRs except for those that occurred in the terminology database. By means of this procedure, the feature space was decreased to 374.
Term frequency–inverse document frequency (TF-IDF)
In a final approach, we assigned a TF-IDF weight to all terms. TF is defined in section “Term frequency (TF).” IDF is, according to previous research,22,23 a mechanism used in combination with TF to attenuate the effect of words that occur too often in the set of documents, as they could be important in order to discriminate between those. IDF is calculated as follows: idft = log N / dft, where N is the number of documents in a collection and dft is the document frequency of term t, that is, the number of documents in the collection that contain t. TF-IDF for a term is calculated using: tf-idft,d = tft,d × idft. Thus, TF-IDF for a t is large if t occurs many times within a small number of documents. We reduced the number of features to a maximum of 1000 terms with the highest TF-IDF scores. For more information on TF-IDF and different weighting schemes, see Manning et al.22 and Van Rijsbergen.23
Combination of preprocessing techniques
In an additional preprocessing step, lemmatization, stop word removal and TF-IDF 1000 were combined. This preprocessing step is named LS-TFIDF 1000 in Table 2. The corresponding step with stemming is named SS-TFIDF. (For more examples of different preprocessing and filtering techniques, see Dalal and Zaveri,16 Yang and Pedersen21 and Doraisamy et al.26)
Parameter optimization
The chosen machine-learning algorithms have a number of parameters that can be fine-tuned to better adapt to the problem and data they are applied on. For SVM using the RBF kernel, there are two main parameters: C and gamma.20 C controls the number of misclassified examples tolerated in the training set, while the gamma value affects the number of support vectors used.
In the case of GTB, the important parameters are J, v and M.18 J refers to the number of terminal nodes in each tree and reflects the number of variable interactions that are possible, v is the learning rate and M is the number of trees. It is usually beneficial to use subsampling with GTB. When using subsampling, a random sample of a predefined size is used to train each tree. This reduces the risk of overfitting. We chose to use 0.5 subsampling, as it is a commonly chosen strategy and has been proven to work well for the task. The learning rate v was fixed to 0.01 after some initial experiments.
In order to find good combinations of parameter values, a grid-search was conducted using fivefold cross-validation on the training data in each fold. Using fivefold cross-validation instead of a higher value of K avoids overfitting, given the small amount of available data. The parameters searched for GTB were as follows:
J ◊ {1, 3, 6, 8};
M ∈ {25, 50, 100, 200, 500, 1000}.
The parameters searched for SVM were as follows:
Gamma ∈ {1.0, 0.1, 0.01, 0.001, 0.0001, 0.00001, 0.00001, 0.000001};
C ∈ {1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, 9.5, 10}.
Evaluation
10-fold cross-validation
For evaluation, we use stratified 10-fold cross-validation, which is one of the best known and most commonly used evaluation techniques. Cross-validation is especially useful if the dataset is small, such as in our case, as it maximizes the amount of training data.27
Statistical tests
When comparing the classifiers’ results, statistical testing is necessary in order to verify the significance of the results. In this study, the non-parametric sign test was used. The choice was motivated by the fact that the authors in previous research27 presented this statistical test as being simple to calculate and yet appropriate when wanting to compare the performance of multiple classifiers on a single domain. Just like the researchers27 did in their example calculations, the sign test was one-tailed and performed at 5 percent significance level.
Results
Table 3 depicts the results of SVM, GTB and their optimized counterparts, given the different preprocessing and feature selection methods. The best precision, recall and F1 scores for each preprocessing method are highlighted. For both classifiers, we built the models using both classes, that is, the 128 HRs containing HAI and 85 HRs not containing HAI. However, since the focus of this study lies on obtaining high recall for HRs that contain HAI, we only present performance measures of the classifiers for those records. Precision, recall and F1 scores for HRs not containing HAI are thus neither depicted nor analyzed.
Table 3.
Precision, recall and F1 score (in %) for detecting HAIs using GTB, optimized GTB, SVM and optimized SVM given the different preprocessing methods.
| GTB |
GTB optimized |
SVM |
SVM optimized |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| TF 1000 | 83.4 | 90.6 | 86.7 | 79.6 | 92.2 | 85.2 | 76.3 | 79.8 | 78.0 | 80.2 | 88.1 | 83.7 |
| Lemma | 79.3 | 87.6 | 83.0 | 76.5 | 92.2 | 83.1 | 60.1 | 100.0 | 75.1 | 78.9 | 88.2 | 83.1 |
| Stem | 82.4 | 88.3 | 85.0 | 79.7 | 93.7 | 85.7 | 60.1 | 100.0 | 75.1 | 80.7 | 89.8 | 84.8 |
| Stop | 79.0 | 83.6 | 80.6 | 79.0 | 93.0 | 85.0 | 76.5 | 78.3 | 77.4 | 83.1 | 89.8 | 84.8 |
| IST | 79.0 | 86.0 | 81.7 | 76.7 | 89.1 | 81.9 | 73.0 | 65.1 | 68.9 | 72.9 | 84.5 | 78.0 |
| TF-IDF 1000 | 81.7 | 91.2 | 86.0 | 79.5 | 92.1 | 84.9 | 60.1 | 100.0 | 75.1 | 78.1 | 89.7 | 82.8 |
| LS-TFIDF 1000 | 80.2 | 84.4 | 81.9 | 78.9 | 91.3 | 84.2 | 60.1 | 100.0 | 75.1 | 72.7 | 88.9 | 79.3 |
| SS-TFIDF 1000 | 78.6 | 85.8 | 81.6 | 78.8 | 93.0 | 85.0 | 60.1 | 100.0 | 75.1 | 75.3 | 86.6 | 79.8 |
GTB: gradient tree boosting; SVM: support vector machine; TF: term frequency; IST: infection-specific term; TF-IDF: term frequency–inverse document frequency.
In total, the material comprised 213 HRs of which 128 contained HAI giving a baseline precision of 60 percent, recall of 100 percent and F-score of 75 percent.
When considering the recall score, one needs to take the F1 score into consideration. A baseline majority classifier would classify all instances as HAI, yielding a recall of 100 percent, precision of 60 percent and F1 score of 75 percent. Hence, if the F1 score for a classifier is close to the baseline of 75 percent, the result is not of interest, even if the recall value is high, as the performance is not better than is the baseline majority classifier. This means that we can disregard all of the results with a recall value of 100 percent since these do not have an F1 score larger than 75 percent. The optimized GTB yields the highest recall for all preprocessing techniques, with a maximum recall value of 93.7 percent and an F1 score of 85.7 percent when using stemming as the preprocessing technique.
When comparing the unoptimized SVM with the optimized SVM approach, it becomes clear that it is very important to perform parameter optimization when using SVM; the optimized SVM obtains a higher F1 score than does the unoptimized SVM for all preprocessing techniques. It is also worth noting that the F1 scores of the unoptimized SVM only differ slightly from the baseline. In the case of GTB, however, the results did not differ much when parameter optimization was applied, and the default parameters used produced a result as good as, slightly better or slightly worse than its optimized counterpart.
To statistically verify the classifiers’ different performance, we applied the non-parametric sign test, as mentioned earlier when using stemming as preprocessing, since it yielded the highest recall score, in combination with a high F1 score for GTB, optimized GTB and optimized SVM. The conclusion was that the performance results obtained using the GTB, optimized GTB and optimized SVM, respectively, are not significantly different. However, they are significantly better compared to the unoptimized SVM that does not perform significantly better than the baseline classifier.
When comparing the recall, precision and F1 scores that the optimized GTB and optimized SVM obtained for the different preprocessing methods, it became apparent that the techniques did not generate a significant difference in the results. For the optimized GTB, the obtained recall values ranged from 89.1 percent (GTB-IST) at the lowest to 93.7 percent (optimized GTB-Stem) at the highest, indicating significant improvement from using one or the other technique. Likewise, the precision and F1 score values did not show any significant difference. The same can be stated for the performance results of the optimized SVM. The recall values varied between the minimum recall of 83.7 percent and the maximum recall of 89.8 percent, not differing significantly.
To summarize our observations, GTB obtained the highest recall and F1 score for all preprocessing methods when results too close to the baseline are disregarded. The difference between the best recall value, 93.7 percent (Stem), and the second best, 93.0 percent (SS-TFIDF 1000 or Stop), was only 0.7 percentage points, and thus was not statistically significant. The difference in the third best recall value, 92.2 percent (TF 1000), amounted to 1.5 percentage points. Compared to this spread, the respective F1 scores remained quite close: 85.7 percent (Stem), 85.0 percent (stop word removal/SS-TFIDF 1000) and 85.2 percent (TF 1000), indicating a comparable overall performance. The highest F1 score, 85.7 percent, was obtained when only stemming was applied. Since we aimed for the highest recall with the highest precision possible, that is, a reasonable overall performance in terms of F1, we concluded that the performance of optimized GTB-Stem came closest to our objective.
Decision features
It is interesting to look at the features upon which the classifiers base their decision. Using GTB, a measure of the relative importance of each feature used can be obtained by examining and scoring features that are most frequently used to branch off in each tree.18 The way to visualize and interpret the results happens in the form of a relative importance plot: the value of each feature is calculated as feature value = 100×(feature score / max score), giving each feature a value relative to the most important feature.
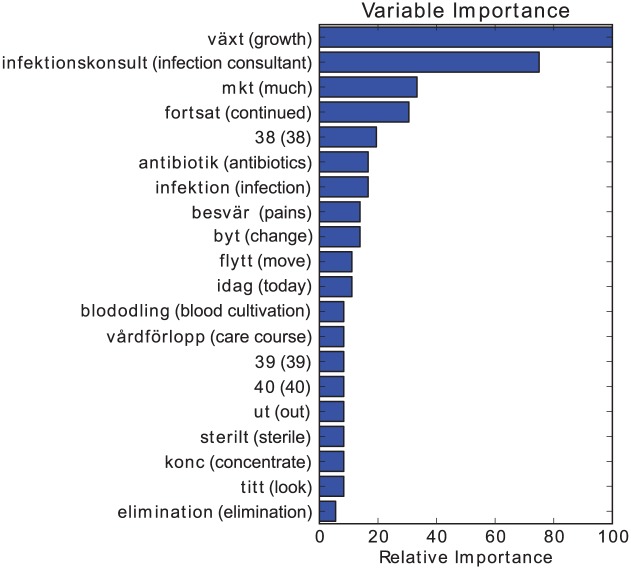
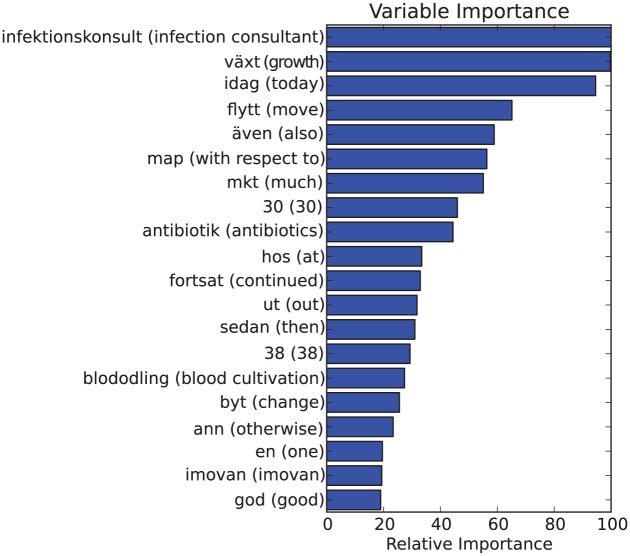
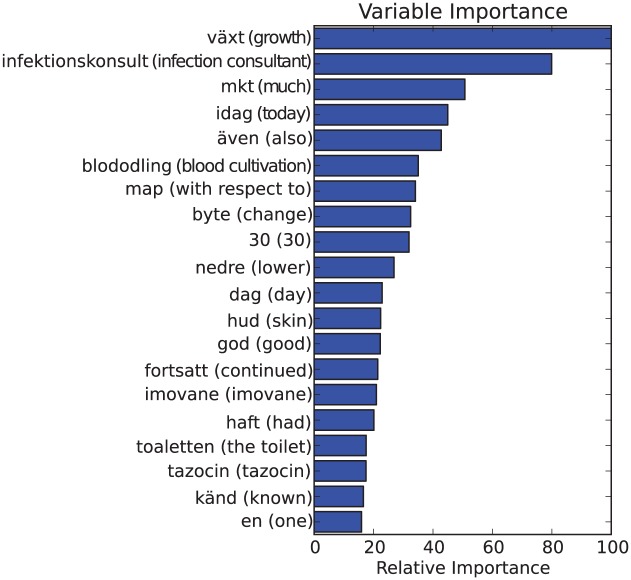
Doing this for a GTB classifier trained on the whole stemmed point prevalence measurement (PPM) dataset yielded Figure 2. This can be compared to Figures 3 and 4, which show relative feature importance for unoptimized GTB-Stem and unoptimized GTB using TF1000 without stemming. Based on these figures, some important observations can be made: (1) among the most important features are words that are plausible HAI indicators, such as växt (Eng.: growth), infektionskonsult (Eng.: infection consultant), antibiotik (stemmed Swedish word, Eng.: antibiotics) and infektion (Eng.: infection). This is good as it hints that the approach was not building a model randomly. The features should be important indicators, even for larger datasets. (2) We can, however, also observe that idag (Eng.: today) and the abbreviation mkt (Eng. much), which are considered to be Swedish stop words, were seen as important features. This observation asks for a more thorough analysis of the terminological structure of patient records in order to optimize feature selection. (3) The most important features were independent of parameter optimization and preprocessing: the top two features in all of the figures were växt (Eng.: growth) and infektionskonsult (Eng.: infection consultant). This observation strengthens the case that the application of different preprocessing techniques may not be very significant. (4) Furthermore, the two most important features were not present in the database of ISTs in section “Infection-specific terms.” In other words, there were terms that were either overlooked or deemed to not be indicators of HAI that were, in fact, important. This indicates that feature selection should either be automatic or semi-automatic since there may be important terms that may be left out otherwise.
Figure 2.
Top 20 feature importances for optimized GTB TF1000+stemming trained on the whole dataset. English translation within parenthesis. Note that since stemming is used the english translation is an approximation as directly translating a stem is not always possible.
Figure 3.
Top 20 feature importances for un-optimized GTB TF1000+stemming trained on the whole dataset. English translation within parenthesis.
Figure 4.
Top 20 feature importances for unoptimized GTB using TF1000 without stemming. English translation within parenthesis.
Classifier errors
As the optimized GTB-Stem produced the overall best results for our objective, it is interesting to analyze the types of errors the classifier made. To do this, all misclassified examples from each of the 10-folds were examined. It is observable from Table 3 that a recall of 100 percent can be achieved, yet at the cost of very low precision. However, as stated earlier, we emphasized recall (aiming at 100 percent) with the highest precision possible. If we considered the obtained recall of above 90 percent as being sufficiently high, it would be interesting to look at what keeps the precision low. In order to do so, we must evaluate what type of errors were made in the NoHAI class since misclassifications for this class appeared as false positives in the predicted HAI class, keeping the precision score below 80 percent in almost all cases.
As visible in Table 4, the class NoHAI can be divided into two disjoint subclasses: hospitalizations with no infections at all (NoINF) and hospitalizations with community-acquired infections (CAIs), the latter of which we defined as infections that were not acquired in the hospital. Furthermore, some of the hospitalizations were, at the time the PPMs were carried out, considered to contain HAI, but in retrospect did not contain any HAI, but rather, some other type of infection or no infection at all. These are referred to as “HAI suspects.”
Table 4.
Classifier errors (optimized GTB-Stem) for the classes HAI and NoHAI, the latter being divided into four disjoint subclasses.
| Class structure | Errors | Dataset | ||
|---|---|---|---|---|
| HAI | 11 | 128 | ||
| NoHAI | CAI | Suspected HAI | 4 | 5 |
| Not suspected HAI | 8 | 18 | ||
| NoINF | Suspected HAI | 7 | 9 | |
| Not suspected HAI | 9 | 53 | ||
| Total | 39 | 213 |
GTB: gradient tree boosting; HAI: hospital-acquired infection; CAI: community-acquired infection; NoINF: no infections at all.
If we examine the type of errors made in the NoHAI class, we can make the following observations: 11/14 of all the “suspected HAI” cases were misclassified in comparison to the non-suspects, which were only misclassified in 17/70 of the cases. Furthermore, of all hospitalizations that contained CAI, 12/22 were misclassified, while only 16/62 of all hospitalizations not containing infections were misclassified. Based on this, it seems like it is difficult for the classifier to distinguish between a HAI and CAI, and the cases misclassified by the medical staff during the PPM study are indeed hard to classify.
Compared to the fairly high recall, the precision stayed below 80 percent in almost all cases. Error analysis revealed that 42.8 percent of the false positives were patient records that contained a CAI. Another 25 percent of the false positives were “suspected HAI.” Handling these false positives in a future approach is crucial for increasing precision. Excluding records containing CAI from classification is one option. Another idea is to, as a first step, train a classifier to differentiate between patient records containing an infection (including HAIs and CAIs) and those not containing an infection. In the second step, HAIs are then detected from the records that were predicted as infections.
Table 5 depicts the classifier errors for each label in class HAI. All hospitalizations that contained ventilator-associated pneumonia (VAP) were classified correctly. Likewise, the classifier performed well for the hospitalizations containing pneumonia and sepsis: only 3 and 2 percent, respectively, of the hospitalizations containing these types of HAI were classified incorrectly. On the other hand, 20 percent of all hospitalizations containing urinary tract infections (UTI) and Clostridium difficile were classified into the wrong class. The analysis gives an indication that some types of HAI are easier to classify compared to others.
Table 5.
Classifier errors (optimized GTB-Stem) for the different types of HAIs.
| Label | Errors | Dataset |
|---|---|---|
| Ventilator-associated pneumonia | 0 | 8 |
| Sepsis | 1 | 46 |
| Pneumonia | 1 | 33 |
| Other HAI | 1 | 15 |
| Fungus/virus | 1 | 15 |
| Central venous catheter–related HAI | 1 | 10 |
| Wound infection | 2 | 25 |
| Urinary tract infection | 4 | 20 |
| Clostridium difficile | 2 | 10 |
GTB: gradient tree boosting.
A hospitalization marked with HAI may have one or more types of HAI. Hence, a misclassified HAI hospitalization may contribute to the number of errors for multiple labels.
Yet, the count of certain types, for example, VAP, central venous catheter–related HAI or Clostridium difficile, is quite low. It is therefore difficult to say whether or not the error rate would be the same for a larger number of cases. Looking at how the different types of HAIs were classified, it is evident that some types, such as VAP, sepsis and pneumonia, have a lower error rate compared to, for instance, UTI or Clostridium difficile. This observation demands a more thorough analysis of the records and how they have been classified with regard to which type of HAI they contain. This is to ultimately find out whether there are any indications in the records that keep the classification error rate for some types of HAI low, while others remain high.
Discussion
To our knowledge, our project group is the first or only one that has applied machine-learning techniques to Swedish patient records in order to detect HAIs. Compared to our previous approach, which was previously presented,2 we increased performance while using the same input data, that is, from 89.84 percent recall, 66.9 percent precision and 76.7 percent F1 score when applying SVM-tfidf50 in our previous paper to 93.7 percent recall, 79.7 percent precision and 85.7 percent F1 score when applying optimized GTB-Stem in our present approach. Although the recall values differed by only 3.86 percentage points, we could increase the precision significantly by 12.8 percentage points. This yielded a considerably better F1 score, bringing us an important step toward our aim of approaching a 100 percent recall with the highest precision possible. Moreover, our experiments suggested that we can achieve better results than can some of the approaches presented in section “Related work,” even though they are not directly comparable due to different datasets and variant languages used.
Limitations
One limitation of this study was the size of the dataset. The task of manually classifying patient records as to whether or not they contained HAI was difficult and time consuming. Manually analyzing a larger amount of patient records than the one we had and marking them as HAI or NoHAI have therefore not been possible in the amount of time available.
Another limitation was the distribution of positive and false cases in our dataset. The number of records that contain HAI (61%) and NoHAI (39%) in our dataset does not relate to the real-life distribution, which is approximately 10 percent HAI and 90 percent NoHAI. Furthermore, the majority of the NoHAI records were from patients who had a HAI at some point or another (there also exists a HAI record for the same patient). This, as well as the fact that several of the NoHAI records were incorrectly classified as HAI by the medical staff at some point, gives us a dataset where the NoHAI class is harder to distinguish from the HAI class that what we thought would be the case.
Importance of preprocessing methods and choice of classifier
Even though the optimized GTB-Stem yielded the best performance results, it became apparent that the results yielded by a classifier, given the different preprocessing and feature selection techniques, were only marginal. This means that, given our data, it does not make a significant difference whether we choose, for instance, stemming, stop word removal or SS-TFIDF 1000 as a preprocessing technique since the performance results are nearly the same. In our case, it made a bigger difference as to which classifier was used and whether the parameters were tuned. In terms of recall and F1 score, the optimized GTB generally yielded better results than did the (un)optimized SVM. Moreover, the improvement in results obtained using the optimized SVM compared to the unoptimized SVM were clearly visible.
Text classification
A major difference between our approach and the similar work mentioned in Ehrentraut et al.2 is the fact that we treated all available data—free-text and lab results were treated as a single unstructured text document. This allowed us to apply standard text-classification methods, namely, applying TFs, such as features and standard machine-learning algorithms, to the problem. This has a big advantage compared to methods that rely on structured data: the amount and type of structured data available are different between hospitals and journal systems. The approach does not rely on the availability of such data and it does not rely on the data being available in a certain format. Furthermore, our approach was able to detect HAI indicators that were not known ahead of time, as shown in section Decision Features.
Comparing the text-classification approach with an approach based on structured data requires further research—evaluating and comparing the text-classification approach with a structured data approach using the same dataset. The results of this study showed that in terms of recall, the text-classification approach was proven to produce results that were as good as using structured data (see Table 6). However, the specificity was lower than were the best scores found in the studies using structured data, but this may be due to the characteristics of the dataset used. If the text-classification approach was able to produce the results seen in Table 3 on larger datasets, it might be a good candidate for application in real-world scenarios, with minimal changes to the journal systems used and minimal additional work for the medical staff.
Table 6.
Recall, specificity and precision for optimized GTB-Stem compared with the results found in the “Related work” section.
| Recall | Specificity | Precision | |
|---|---|---|---|
| GTB-Stem optimized | 93.7 | 64.1 | 79.7 |
| [7] ANN Internal | 96.64 | 85.96 | – |
| [7] LR external | 82.76 | 80.90 | – |
| [10] SVM | 92.6 | 43.73 | – |
| [11] SVM | 92.0 | 72.0 | – |
| [11] NB | 87 | 74.0 | – |
| [13] FLD S2 | 82.56 | – | 43.54 |
GTB: gradient tree boosting; ANN: artificial neural network; LR: linear regression; SVM: support vector machine; NB: Naïve Bayes classifiers; FLD: Fisher’s linear discriminant.
Note that the evaluation methods and datasets are not the same.
Cost analysis
In a report by the Swedish National Board of Health and Welfare,28 it is estimated that HAIs prolong the length of a patient’s stay in the hospital by an average of 4 days. Given all of the patients suffering from HAI, this is estimated to be about 500,000 extra hospital days per year. With a daily average cost of SEK7.373 (US$860) per day of care, HAIs generate an additional cost of approximately SEK3.7b (US$0.43b) per year except for the labor-intensive cost for carrying out manual PPM twice a year. However, if we can automatize this process, we would save labor, as well as improve the quality of the controls by carrying them out automatically and continuously, 24 h a day, all year long.12,29
Future work
We are well aware of the facts that
Our dataset is small, containing only 213 instances;
The distribution of positive and negative cases, that is, 128 HAI and 85 NoHAI instances, does not correlate with the real-life distribution;
The differences in the results of optimized GTB and optimized SVM are marginal and are not significantly different.
However, the result is significantly better compared to a majority baseline classifiers and unoptimized SVM, and we are convinced that the results reveal the potential for applying text-classification techniques to patient records, including the structured as well as unstructured parts. This is further motivated by the fact that, so far, we have used no particularly elaborate preprocessing and feature reduction methods. Future research will thus have to focus on improving the scores by, for instance, using wrapper techniques for feature reduction that are optimized on a specific learning algorithm and, therefore, yield better results according to previous research.30
Moreover, the medical experts involved in this project will manually analyze 292 additional HRs from the rheumatic clinic at Karolinska University Hospital. Thus, we will be able to train the classifiers on about twice as many data as we did for the current project, leading us to expect an improvement in performance. In addition, we aim at training the classifiers on a more realistic dataset, with a real-life distribution of about 10 percent positive and 90 percent negative cases.
Conclusion
This article focuses on applying SVM and GTB to the problem of detecting HAI in digital patient records. By means of applying different preprocessing, as well as feature selection, methods, we tried to increase recall. The results of the machine-learning algorithms were all in all very encouraging. Optimized GTB-Stem came closest to the objective of obtaining high recall with the highest precision possible, that is, yielding a recall of 93.7 percent, precision of 79.7 percent and F1 score of 85.7 percent.
This revealed the applicability of GTB to the task. The increased recall value obtained with the optimized SVM compared to the unoptimized SVM confirmed the assumption that SVM seemed to be suitable for the task and, more importantly, revealed the importance of parameter tuning, leading to significantly better results. Applying stemming yielded high performance results for all three classifiers, yet the difference in the results yielded by the classifiers when other preprocessing techniques (especially stop word removal) are applied were marginal.
Finally, the overall goal will continue to be obtaining high recall (approaching 100%) with the highest precision possible for HRs. This will enable us to implement a system that can screen all HRs and filter out all HRs that contain HAI. This would reduce the workload for hospital staff tremendously as they only need to analyze those HRs that were preselected by the system.
Acknowledgments
We would like to thank our excellent physicians, Maria Kvist and Elda Sparrelid, for their insightful classifications of hospital-acquired infections, as well as Martin Duneld for suggestions regarding preprocessing and computing methods. C.E. did the main work, together with M.E. C.E. wrote the main part of the article and contributed to the data analysis, while M.E. ran the experiments, did the data analysis and contributed to the writing. H.T. extracted and prepared the datasets from the patient record system. J.T. and H.D. gave feedback on the writing and the analysis of the results.
Footnotes
Declaration of conflicting interests: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by VINNOVA’s (Swedish Governmental Agency for Innovation Systems) project Innovation mot Infektion (grant number 2012-01252, 2012) and HIPPA (Hospital Intelligence for Better Patient Security) (grant number 2013-00677, 2013).
Contributor Information
Claudia Ehrentraut, Stockholm University, Sweden.
Markus Ekholm, KTH Royal Institute of Technology, Sweden.
Hideyuki Tanushi, Stockholm University, Sweden.
Jörg Tiedemann, University of Helsinki, Finland.
Hercules Dalianis, Stockholm University, Sweden.
References
- 1. Ducel G, Fabry J, Nicolle L. Prevention of hospital-acquired infections: a practical guide. 2nd ed. Geneva: World Health Organization, 2002, p. 1. [Google Scholar]
- 2. Ehrentraut C, Tiedemann J, Dalianis H, et al. Detection of hospital acquired infections in sparse and noisy Swedish patient records. In: Proceedings of the sixth workshop on analytics for noisy unstructured text data (AND 2012), Mumbai, India, 9 December 2012, pp. 1–8. New York: ACM. [Google Scholar]
- 3. Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference and prediction. 2nd ed. New York: Springer, 2008, p. 758. [Google Scholar]
- 4. Klompas M, Yokoe DS. Automated surveillance of health care-associated infections. Clin Infect Dis 2009; 48(9): 1268–1275. [DOI] [PubMed] [Google Scholar]
- 5. Blacky A, Mandl H, Adlassnig KP, et al. Fully automated surveillance of healthcare-associated infections with MONI-ICU: a breakthrough in clinical infection surveillance. Appl Clin Inform 2011; 2(3): 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Claster WB, Shanmuganathan S, Ghotbi N, et al. Text classification for medical informatics: a comparison of models for data mining radiological medical records. Asia Pac World 2011; 2(1): 121–137. [Google Scholar]
- 7. Benhaddouche D, Benyettou A. Control of nosocomial infections by data mining. World Appl Program 2012; 2(4): 216–219. [Google Scholar]
- 8. Chang YJ, Yeh ML, Li YC, et al. Predicting hospital-acquired infections by scoring system with simple parameters. PLoS ONE 2011; 6(8): e23137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cohen G, Hilario M, Sax H, et al. Data imbalance in surveillance of nosocomial infections. In: Proceedings of the medical data analysis: 4th international symposium (ISMDA 2003) (ed Perner P, Brause R, Holzhütter HG.), Berlin, 9–10 October 2003, pp. 109–117. Berlin, Heidelberg: Springer. [Google Scholar]
- 10. Cohen G, Hilario M, Hugonnet S, et al. Asymmetrical margin approach to surveillance of nosocomial infections using support vector classification. In: Proceedings of the intelligent data analysis in medicine and pharmacology (IDAMAP 2003), Protaras, 19–22 October 2003, pp. 1–13. [Google Scholar]
- 11. Cohen G, Hilario M, Sax H, et al. An application of one-class support vector machine to nosocomial infection detection. In: Proceedings of the 11th world congress on medical informatics (MedInfo 2004) (ed Fieschi M, Coiera E, Li YCJ.), San Francisco, CA, 7–14 September 2004, pp. 716–720. Amsterdam: IOS Press. [PubMed] [Google Scholar]
- 12. Cohen G, Hilario M, Sax H, et al. Learning from imbalanced data in surveillance of nosocomial infection. Artif Intell Med 2006; 37(1): 7–18. [DOI] [PubMed] [Google Scholar]
- 13. Iavindrasana J, Cohen G, Depeursinge A, et al. Minimal set of attributes required to report hospital-acquired infection cases. In: Proceedings of the workshop on intelligent data analysis in biomedicine and pharmacology (IDAMAP 2008) (ed Holmes J, Tucker A.), Washington, DC, 7 November 2008, pp. 23–28. [Google Scholar]
- 14. Iavindrasana J, Cohen G, Depeursinge A, et al. Towards an automated nosocomial infection case reporting-framework to build a computer-aided detection of nosocomial infection. In: Proceedings of the second international conference on health informatics (HEALTHINF 2009) (ed Azevedo L, Londral AR.), Porto, 14–17 January 2009, pp. 317–322. Porto: INSTICC Press. [Google Scholar]
- 15. Kelly KN, Monson JRT. Hospital-acquired infections. Surgery 2012; 30(12): 640–644. [Google Scholar]
- 16. Dalal MK, Zaveri MA. Automatic text classification: a technical review. Int J Comput Appl 2011; 28(2): 37–40. [Google Scholar]
- 17. Colas F, Brazdil P. Comparison of SVM and some older classification algorithms in text classification tasks. In: Bramer M. (ed.) IFIP international federation for information processing, vol. 217: artificial intelligence in theory and practice Boston, MA: Springer, 2006, pp. 169–178. [Google Scholar]
- 18. Hastie T, Tibshirani R, Friedman J, et al. The elements of statistical learning: data mining, inference and prediction. Math Intell 2005; 27(2): 83–85. [Google Scholar]
- 19. Noble WS. What is a support vector machine? Nat Biotechnol 2006; 24(12): 1565–1567. [DOI] [PubMed] [Google Scholar]
- 20. Hsu CW, Chang CC, Lin CJ. A practical guide to support vector classification, https://www.cs.sfu.ca/people/Faculty/teaching/726/spring11/svmguide.pdf (2000, accessed 17 January 2014).
- 21. Yang Y, Pedersen JO. A comparative study on feature selection in text categorization. In: Proceedings of the fourteenth international conference on machine learning (ICML’97) (ed Fisher DH.), Nashville, TN, 8–12 July 1997, pp. 412–420. San Francisco, CA: Morgan Kaufmann Publishers Inc. [Google Scholar]
- 22. Manning CD, Raghavan P, Schütze H. Introduction to information retrieval (online edition). Cambridge: Cambridge University Press, 2009, p. 27. [Google Scholar]
- 23. Van Rijsbergen CJ. Information retrieval. 2nd ed. London: Butterworth, 1979, p. 208. [Google Scholar]
- 24. Dragut E, Fang F, Sistla P, et al. Stop word and related problems in web interface integration. In: Proceedings of the VLDB endowment (ed Jagadish HV.), Lyon, 24–28 August 2009, vol. 2, pp. 349–360. New York: ACM. [Google Scholar]
- 25. Hassel M. JavaSDM: a Java package for working with Random Indexing and Granska, http://www.nada.kth.se/~xmartin/java/ (2006, accessed 17 January 2014).
- 26. Doraisamy S, Golzari S, Norowi NM, et al. A Study on feature selection and classification techniques for automatic genre classification of traditional malay music. In: Proceedings of the 9th international conference of music information retrieval (ISMIR 2008) (ed Bello JP, Chew E, Turnbull D.), Philadelphia, PA, 14–18 September 2008, pp. 331–336. Drexel University. [Google Scholar]
- 27. Japkowicz N, Shah M. Evaluating learning algorithms: a classification perspective. 1st ed Cambridge: Cambridge University Press, 2011, p. 231. [Google Scholar]
- 28. Tegnell A, Carlson J. Att förebygga vårdrelaterade infektioner. Ett kunskapsunderlag, https://www.folkhalsomyndigheten.se/pagefiles/20412/att-forebygga-vardrelaterade-infektioner-ett-kunskapsunderlag-2006-123-12.pdf (2006, accessed 21 March 2015).
- 29. Bouzbid S, Gicquel Q, Gerbier S, et al. Automated detection of nosocomial infections: evaluation of different strategies in an intensive care unit 2000–2006. J Hosp Infect 2011; 79(1): 38–43. [DOI] [PubMed] [Google Scholar]
- 30. Hall MA. Correlation-based feature selection for machine learning. PhD Thesis, The University of Waikato, Hamilton, New Zealand, 1999. [Google Scholar]