The 2016 revision of the World Health Organization classification of lymphoid neoplasms introduces the umbrella category “nodal T-cell lymphomas with T-follicular helper (TFH) phenotype”, which includes angioimmunoblastic T-cell lymphoma (AITL), follicular T-cell lymphoma and nodal peripheral T-cell lymphoma (PTCL) with a TFH phenotype1. One of the genetic features clustering TFH cell-derived lymphomas is a recurrent RHOA G17V mutation, which is present in approximately 60% of investigated cases2,3. RHOA is a member of the Rho family of GTPases which function as molecular regulators of diverse cellular functions4. Mutant RHOA acts as a dominant-negative signaling protein sequestering guanine nucleotide exchange factors (GEFs) thereby inhibiting wildtype RHOA and potentially other GEF-dependent proteins3. In vivo, mutant RHOA has recently been shown to skew CD4+ T-cell differentiation towards the TFH lineage and promote AITL lymphomagenesis5. Thus, the RHOA G17V mutation can be viewed as pivotal genetic aberration in AITL and potentially other TFH cell-derived lymphomas. Mutations contributing to lymphomagenesis in wildtype RHOA AITL cases remain largely unknown. This mutational heterogeneity points towards the existence of distinct AITL lymphomagenic pathways. In this report, we explore the mutational landscape of AITL by assessing the data from large sequencing studies focusing on the association between RHOA mutational status and recurrent mutations in other genes to provide evidence for the existence of distinct lymphomagenic pathways in AITL.
Sequencing studies of AITL and/or PTCL published between 01-01-2014 and 28-02-2017 using an English language restriction were identified with PubMed. In total, 117 abstracts were screened. Only 34 articles were eligible for full text review. Studies were included in our analysis if they contained ten or more AITL cases and used targeted deep sequencing of RHOA, TET2, DNMT3A, IDH2, CD28 and multiple other genes or whole genome/exome/transcriptome approaches. Also, the original dataset had to be available to the authors. Five of the 34 articles met the prespecified inclusion criteria and were included in our analysis6–10. The article selection process was performed by two authors.
In total, these studies analyzed 239 AITL cases using various sequencing techniques. Of interest, in 13.8% (33/239) of investigated AITL cases no detectable mutations were reported. RHOA was mutated in 61.1% (146/239) of the investigated AITL cases. The remaining 25.1% (60/239) of cases were wildtype, but carried mutations in other genes (Table 1a).
Table 1.
RHOA mutational status of included studies (A) and association with recurrent mutations in wildtype RHOA AITL cases (B)
| (A) Author | Sequencing method | AITL cases | No detectable mutations | Wildtype RHOA | Mutant RHOAa |
|---|---|---|---|---|---|
| Abate et al.6 | RNA-seq, TDS (VAV1) | 60 | 15 | 20 | 25 |
| Nguyen et al.7 2017 | TDS (71 genes) | 48 | 6 | 9 | 33 |
| Vallois et al.9 | TDS (69 genes) | 72 | 8 | 18 | 46 |
| Yoo et al.10b | TDS (70 genes) | 29 | 3 | 5 | 21 |
| Palomero et al.8c | RNA-seq, TDS (13 genes), allele-specific PCR (RHOA) | 30 | 1 | 8 | 21 |
| Total | 239 | 33 (13.8%) | 60 (25.1%) | 146 (61.1%) | |
| (B) Gene | Mutant RHOA AITL cases/totala | Wildtype RHOA AITL cases/totald | Odds ratio | 95% CI | p-value |
| TET2 c, e | 101/146 (69.2%) | 33/93 (35.5%) | 3.46 | 1.92, 6.22 | <0.001 |
| DNMT3A c, e | 34/146 (23.3%) | 10/93 (10.8%) | 2.14 | 0.99, 4.66 | 0.076 |
| IDH2 c, e | 51/146 (34.9%) | 9/93 (9.7%) | 6.68 | 2.89, 15.45 | <0.001 |
| CD28 | 14/92 (15.2%) | 7/69 (10.1%) | 1.73 | 0.64, 4.73 | 0.399 |
| CD28 f | 24/92 (26.1%) | 8/69 (11.6%) | 2.60 | 0.98, 6.86 | 0.093 |
| FYN c | 5/113 (4.4%) | 3/78 (3.8%) | 1.38 | 0.30, 6.29 | 0.972 |
| PLCG1 | 6/92 (6.5%) | 6/69 (8.7%) | 0.85 | 0.26, 2.85 | 0.960 |
| STAT3 | 2/92 (2.2%) | 3/69 (4.3%) | 0.68 | 0.13, 3.69 | 0.979 |
| VAV1 | 2/92 (2.2%) | 5/69 (7.2%) | 0.27 | 0.046, 1.56 | 0.268 |
TDS targeted deep sequencing, CI confidence interval
aIncluding non-G17V RHOA mutations
bOnly 29/45 AITL cases were analyzed using TDS
cOnly 30/35 AITL cases were analyzed by allele-specific PCR and targeted deep sequencing. Only one case showed no mutations in both techniques (no detectable mutations). RNA sequencing data not included due to uncertainty of diagnosis
dIncluding cases with no detectable mutations
e Missing data regarding mutational status of TET2 (n = 14), DNMT3A (n = 14) and IDH2 (n = 10) from Vallois et al. included as not mutated
fIncluding cases with CTLA4–CD28 gene fusion
We focused on the data extract of all wildtype RHOA AITL cases to identify potentially recurrent mutations contributing to AITL lymphomagenesis other than RHOA. Only mutations occurring in more than 5% of targeted cases and identified in two or more studies were classified as recurrent. TET2, CD28, DNTM3A, PLCG1, IDH2, VAV1, FYN and STAT3 were mutated in 60.7% (34/56), 18.6% (8/43), 17.9% (10/56), 14.0% (6/43), 13.8% (8/58), 11.6% (5/43), 7.8% (4/51) and 7.0% (3/43) of targeted wildtype RHOA AITL cases, respectively (Supplementary Data set 1). As these mutations also frequently occur in mutant RHOA AITL cases, we performed Mantel–Haenszel statistics to assess the association between these mutations and RHOA mutational status across different studies (SPSS v21 IBM Corp., Armonk, NY, USA). A p-value ≤0.05 was considered significant.
Statistical analysis showed that mutations in TET2 and IDH2 were associated with mutant RHOA status (p < 0.001 for both genes). Mutations in DNMT3A and CD28, including CTLA4–CD28 fusion, also tend to show this association (p = 0.076 and 0.093). Interestingly, despite being mutated in a low number of cases, mutations in VAV1 tend to associate with wildtype RHOA status (p = 0.268). Mutations in FYN, PLCG1 and STAT3 showed no significant association with RHOA mutational status (p = 0.972, 0.960 and 0.979) (Table 1b).
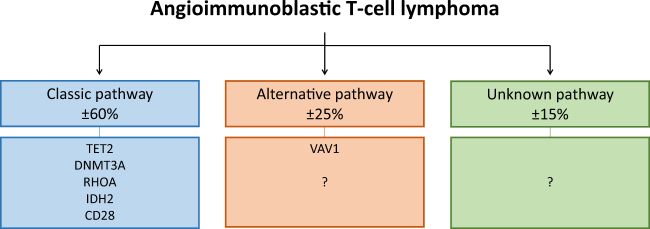
This study reports on the association between RHOA mutational status and other recurrent mutations in AITL. We found an association between mutant RHOA and mutations in TET2 and IDH2. Despite being mutually exclusive in acute myeloid leukemia, mutations in IDH2 and TET2 tend to co-occur in AITL11. Gene expression profiling and promoter methylation analysis of double mutant AITL cases showed upregulation of genes associated with TFH phenotype and downregulation of genes associated with TH1 phenotype11. Mutant IDH2 and TET2 potentially cooperate with mutant RHOA to induce a potent TFH phenotype in vivo. This mechanism would explain the association found between these mutations in the present study. We also identified a strong tendency towards association between mutant RHOA and mutations in DNMT3A. The exact mechanism by which mutations in epigenetic modifiers contribute to lymphomagenesis remain to be elucidated, but alterations in hematopoietic stem cell differentiation is an attractive theory. The present study also identified a strong tendency towards association between mutant RHOA and mutations in CD28, including CTLA4–CD28 gene fusion. CD28 mutations in AITL are confined to hotspot residues D124 and T195 and render CD28 constitutively active10,12. The CTLA4–CD28 fusion gene has only been reported in an Asian cohort10. Therefore, validation of this fusion gene in other cohorts is essential to confirm the association between mutant RHOA and mutations in CD28. Altogether, these findings point towards a classic AITL lymphomagenic pathway (Fig. 1). Several therapeutic approaches targeting epigenetic modifiers, IDH2 or CD28 are currently in clinical trials or have already been approved for other diseases13,14. The tendency of these mutations to cluster will potentially help to develop novel combinatorial therapeutic regimens.
Fig. 1. Three distinct lymphomagenic pathways in AITL.
Our statistical analysis has identified three potentially distinct lymphomagenic pathways in AITL. The classic pathway accounts for approximately 60% of AITL cases and is characterized by the RHOA G17V mutation in association with several other recurrent mutations. The alternative pathway accounts for approximately 25% of AITL cases and is characterized by mutations in VAV1 or potentially other members of the Rho family of GTPases or their regulatory proteins. The mutations that drive the remaining approximately 15% of AITL cases are unknown. It is possible that mutations in signaling pathways directly regulating TFH differentiation contribute to these cases
Despite being mutated in rather a low number of AITL cases, this study identified the tendency of mutations in VAV1 to associate with wildtype RHOA. VAV1 encodes a Rho GTPase family-specific GEF which is primarily expressed in the hematopoietic system15. The studies that targeted VAV1 identified three missense mutations (E524D, E556D and D797G), two frameshift deletions (151_158del and 778_783del), one fusion gene (VAV1–S100A7) and one in-frame deletion (778_786del)6,9,10. Abate et al. found the 778_786 in-frame deletion and VAV1–S100A7 fusion gene to be locked in a constitutively active conformation, indicated by high levels of Tyr174 phosphorylation6. Both genetic aberrations resulted in increased VAV1 catalytic-dependent functions downstream of RAC1, another member of the Rho family of GTPases6. These findings are in accordance with previous experiments showing that constitutively active VAV1 predominantly increases nucleotide exchange of RAC1 and to a lesser extend of RHOA16. Interestingly, the RAC1 pathway is upregulated in mutant RHOA compared to wildtype RHOA AITL cases, providing evidence that both mutations have similar effects on VAV1 catalytic-dependent pathways17. Additionally, Abate et al. found that the VAV1–S100A7 fusion gene resulted in increased NFAT activity, a functional readout of VAV1 non-catalytic activity, whereas both the 778_786 in-frame deletion and VAV1–S100A7 fusion gene increased expression of NFAT target genes6. A recently published study, not yet indexed by PubMed at the time of our search, identified activating VAV1 mutations in 8.2% (7/85) of wildtype RHOA AITL cases, compared to 0% (0/41) in mutant RHOA AITL cases, respectively18. They also showed that mutant RHOA enhances the non-catalytic functions of VAV1 through increased Tyr174 phosphorylation, thereby increasing NFAT activity and expression of NFAT target genes. Together, these data not only strengthen the association between mutant VAV1 and wildtype RHOA, but also provide evidence that mutant RHOA and mutant VAV1 have similar effects on catalytic and non-catalytic signaling pathways downstream of VAV1. Therefore, we deduce from these data that mutant RHOA and mutant VAV1 contribute to AITL lymphomagenesis in a similar manner. This would mean that VAV1 is part of an alternative AITL lymphomagenic pathway (Fig. 1). Previous clinicopathological studies have shown that mutant RHOA AITL cases have worse performance status, more frequent B-symptoms and splenomegaly and a more potent TFH immunophenotype compared to wildtype RHOA AITL cases19,20. These data provide additional justification for separating AITL subgroups.
According to our analysis, no mutations were detectable in approximately 15% (range 3–25%) of AITL cases (Fig. 1). Exploring the mutational landscape of AITL using targeted deep sequencing panels enriched with members of the Rho family of GTPases and their regulatory proteins might identify driver mutations in this subgroup. It is also possible that other lymphomagenic mechanisms contribute to some AITL cases, for example mutations in signaling pathways directly regulating TFH differentiation.
We are aware that there are some limitations to our study. Our findings are entirely based on retrospective data from a relatively small sample size. Furthermore, there is significant technical heterogeneity between the sequencing studies from which the data is derived. The individual studies use different sequencing techniques, bioinformatics pipelines for data processing and mutation calling methods. Despite these limitations, this study remains noteworthy as it provides a unique perspective on associations and possible collaborations between the most common genetic aberrations in AITL as well as providing a rationale for future research.
In short, using data from large sequencing studies this study reports on varying associations between RHOA mutational status and other recurrent mutations in AITL. These findings enable us to identify three potentially distinct AITL lymphomagenic pathways. First, the classic pathway with the RHOA G17V mutation which is associated with mutations in TET2, DNMT3A, IDH2 and CD28. Secondly, the alternative pathway with mutations in VAV1 or potentially yet unidentified mutations in members of the Rho family of GTPases or their regulatory proteins. Third, AITL cases with unknown mutations which might arise from direct mutations in pathways regulating TFH differentiation. To what extend these different lymhpomagenic pathways result in different clinical behavior of AITL is largely unknown. Additional evidence on the mutational landscape of AITL, especially wildtype RHOA AITL cases, is needed to either confirm or refute our findings. Furthermore, prospective data is needed to identify potential clinical differences between the distinct lymphomagenic pathways of AITL proposed in this manuscript.
Electronic supplementary material
Acknowledgments
Authors’ contributions
M.W. developed the initial hypothesis that lead to this work and wrote the manuscript. M.A.H. and A.Z.H. were involved in critical reviewing and revising the manuscript. B.W. supervised the statistical analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at (10.1038/s41408-017-0047-2).
References
- 1.Swerdlow SH, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobay MP, et al. Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica. 2017;102:e148–e151. doi: 10.3324/haematol.2016.158428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cools J. RHOA mutations in peripheral T cell lymphoma. Nat. Genet. 2014;46:320–321. doi: 10.1038/ng.2937. [DOI] [PubMed] [Google Scholar]
- 4.Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell. Biol. 2016;17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 5.Cortes JR, et al. Role and Mechanisms of Rhoa G17V in the Pathogenesis of AITL. Blood. 2016;128:621. [Google Scholar]
- 6.Abate F, et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2017;114:764–769. doi: 10.1073/pnas.1608839114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen T, et al. Identification of cell-type-specific mutations in nodal T-cell lymphomas. Blood Cancer J. 2017;7:e516. doi: 10.1038/bcj.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014;46:166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallois D, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell–derived lymphomas. Blood. 2016;128:1490–1502. doi: 10.1182/blood-2016-02-698977. [DOI] [PubMed] [Google Scholar]
- 10.Yoo HY, et al. Frequent CTLA4-CD28 gene fusion in diverse types of T cell lymphoma. Haematologica. 2016;101:757–763. doi: 10.3324/haematol.2015.139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126:1741–1752. doi: 10.1182/blood-2015-05-644591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohr J, et al. Recurrent activating mutations of CD28 in peripheral T-cell lymphomas. Leukemia. 2016;30:1062–1070. doi: 10.1038/leu.2015.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willemsen M, Schouten HC. Inappropriate costimulation and aberrant DNA methylation as therapeutic targets in angioimmunoblastic T-cell lymphoma. Biomark. Res. 2017;5:6. doi: 10.1186/s40364-017-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz N, Leval L. How I manage peripheral T‐cell lymphoma, not otherwise specified and angioimmunoblastic T‐cell lymphoma: current practice and a glimpse into the future. Br. J. Haematol. 2017;176:851–866. doi: 10.1111/bjh.14473. [DOI] [PubMed] [Google Scholar]
- 15.Bustelo XR. Vav family exchange factors: an integrated regulatory and functional view. Small GTPases. 2014;5:e973757. doi: 10.4161/21541248.2014.973757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crespo P, Schuebel KE, Ostrom AA, Gutkind JS, Bustelo XR. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 17.Manso R, et al. The RHOA G17V gene mutation occurs frequently in peripheral T-cell lymphoma and is associated with a characteristic molecular signature. Blood. 2014;123:2893–2894. doi: 10.1182/blood-2014-02-555946. [DOI] [PubMed] [Google Scholar]
- 18.Fujisawa M. et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia.10.1038/leu.2017.273 (2017). [DOI] [PMC free article] [PubMed]
- 19.Nagao R, et al. Clinicopathologic analysis of angioimmunoblastic T-cell lymphoma with or without RHOA G17V mutation using formalin-fixed paraffin-embedded sections. Am. J. Surg. Pathol. 2016;40:1041–1050. doi: 10.1097/PAS.0000000000000651. [DOI] [PubMed] [Google Scholar]
- 20.Ondrejka SL, et al. Angioimmunoblastic T-cell lymphomas with the RHOA p. Gly17Val mutation have classic clinical and pathologic features. Am. J. Surg. Pathol. 2016;40:335–341. doi: 10.1097/PAS.0000000000000555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.