Abstract
T-cell prolymphocytic leukemia (T-PLL) is an aggressive malignancy with a median survival of the patients of less than two years. Besides characteristic chromosomal translocations, frequent mutations affect the ATM gene, JAK/STAT pathway members, and epigenetic regulators. We here performed a targeted mutation analysis for 40 genes selected from a RNA sequencing of 10 T-PLL in a collection of 28 T-PLL, and an exome analysis of five further cases. Nonsynonymous mutations were identified in 30 of the 40 genes, 18 being recurrently mutated. We identified recurrently mutated genes previously unknown to be mutated in T-PLL, which are SAMHD1, HERC1, HERC2, PRDM2, PARP10, PTPRC, and FOXP1. SAMHD1 regulates cellular deoxynucleotide levels and acts as a potential tumor suppressor in other leukemias. We observed destructive mutations in 18% of cases as well as deletions in two further cases. Taken together, we identified additional genes involved in JAK/STAT signaling (PTPRC), epigenetic regulation (PRDM2), or DNA damage repair (SAMHD1, PARP10, HERC1, and HERC2) as being recurrently mutated in T-PLL. Thus, our study considerably extends the picture of pathways involved in molecular pathogenesis of T-PLL and identifies the tumor suppressor gene SAMHD1 with ~20% of T-PLL affected by destructive lesions likely as major player in T-PLL pathogenesis.
Introduction
T-cell prolymphocytic leukemia (T-PLL) is a rare leukemia with an aggressive disease course and a median survival of the patients of less than 2 years. Leukemic cells are characterized by expression of pan-T-cell markers with the unique feature of CD4 and CD8 co-expression in 25% of cases. A CD4+CD8− phenotype is observed in 60% of patients, whereas a CD4−CD8+ phenotype is rare (~15%)1,2. Most T-PLL carry typical genetic alterations, namely inv(14)(q11q32), t(14;14)(q11;q32), or, less often, t(X;14)(q28;q11). These alterations, involving the TCRAD locus on chromosome 14q11, cause overexpression of the oncogenes TCL1A on chromosome 14q32 or MTCP1 on chromosome Xq283–6. Other frequent genetic lesions involve chromosome 8 (idic(8p), t(8;8)(p21;q11), trisomy 8q), and the ATM gene on chromosome 11 (11q2.23). ATM is deleted or mutated in up to 70% of cases7–9. Further recurrent deletions or losses occur on chromosomes 12p13 (CDKN1B), 6q, 17p13.1 (TP53 locus), and 22q10. Sequencing analyses identified recurrent mutations in members of the JAK/STAT signaling pathway, as well as in epigenetic regulators7,11–14. Recent next-generation sequencing studies included whole-genome and exome sequencing7 as well as targeted deep sequencing13,14.
We characterized T-PLL by RNA sequencing, targeted capture sequencing, and whole-exome sequencing (WES) for somatic mutations, and by single-nucleotide polymorphism (SNP) arrays for detection of genomic imbalances in candidate regions. We identified recurrent mutations in SAMHD1 in 6/33 cases (18%). Copy number losses were observed in two more patients. Other genes that exhibited recurrent mutations and/or copy number alterations were HERC2, HERC1, PRDM2, PARP10, PTPRC, and FOXP1.
Materials and methods
Patients and samples
Patient samples were obtained from archived material of the participating institutions. Patients were diagnosed between 2005 and 2012 in accordance with the WHO 2008 classification2. The study was approved by the ethical review committees of the Universities of Duisburg-Essen and Kiel (14-6080-BO and B295/11). All patients provided written informed consents according to the Declaration of Helsinki. The detection of an inv(14)/t(14;14) or t(X;14) by cytogenetic analysis and/or detection of TCL1 or MTCP1 breakpoints by FISH was required for inclusion into the study. Clinical data of 33 study patients are summarized in Table 1. Standard clinical criteria were applied for initiation of therapy15. RNA sequencing data and copy number analyses were assessed for 10 patients. As control, we sequenced T-cell RNA from five healthy donors. CD3+ T cells were enriched by magnetic cell separation (Miltenyi Biotech, Bergisch Gladbach). For 28 samples, including the 10 with RNA-sequencing analysis, DNA capture sequencing was performed. WES was carried out for five additional T-PLL. The assignment of samples and experiments is given in Supplementary Table 1.
Table 1.
Clinical patient data
| Patient ID | Sex | Age at diagnosis (years) | Genetic group | T-PLL Immunophenotype | SAMHD1 status | Absolute WBC count at diagnosis (10 9 /L) | Absolute lymph. count at diagnosis (10 9 /L) | Other sites of involvement (except blood and BM) | Type of Treatment* | Allogenic transplant | CR at any time | Overall survival (months) | Death | Comorbidity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 74 | inv(14)/t(14;14) | CD4+ | Loss | 719 | 62.7 | None | 1; 2; 3; 4 | No | No | 34 | Yes | Prior T-PLL: Cervical carcinoma (radiotherapy, CR); immunoblastic lymphoma (CTX, CR) |
| 2 | F | 64 | t(X;14) | CD4+ | Mutated | 17.3 | 8.5 | Skin | 1; 2; 4 | No | No | 18 | Yes | Rheumatoid arthritis; myociadial inf.; renal insuff, Std. III |
| 3 | M | 62 | inv(14)/t(14;14) | CD4+ | Mutated | 148 | 121 | Spleen, liver, lymph nodes, skin | 1; 2; 3; 4 | No | No | 7 | Yes | None |
| 4 | M | 69 | inv(14) | CD4+ | WT | n.a. | n.a. | n.a. | n.a. | No | No | 0 | Yes | n.a. |
| 5 | M | 75 | inv(14) | CD8+ | WT | 830 | 730 | Spleen, liver, lymph nodes | 1; 2; 3; 4 | No | Yes | 33 | Yes | Ponsinfarction right; vasculare leukencephalopathie; hypertension; kidney-surgery right; polyneuropathy |
| 6 | F | 41 | t(X;?) | CD4+ | WT | 102 | 91 | Spleen, lymph nodes, mediastinal | 1; 2; 4 | yes | no | 12 | Yes | None |
| 7 | F | 51 | t(14;14) | CD8+ | WT | 11.8 | 8 | Spleen, liver, lymph nodes | 1; 2; 3; 4 | Yes | Yes | 16 | Yes | Diabetes mellitus; hypertension; adiposity; bronchial asthma |
| 8 | M | n.a. | t(X;14) | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | 33 | Yes | n.a. | |
| 10 | M | 82 | inv(14) | CD4+ | WT | 32 | 28 | Spleen | 1; 2; 4 | No | No | 5 | Yes | Hypertension; renal insuff.; depression; prior to T-PLL: Prostate cancer |
| 11 | F | 62 | inv(14) | CD4+/CD8+ | Loss | 34 | 30 | Spleen | 1; 2; 3; 4 | Yes | No | 21 | Yes | Hypertension; diabetes mellitus; ulcerative colitis |
| 12 | M | 57 | inv(14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 13 | F | 53 | inv(14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 15 | F | 78 | t(X;14) | CD4+ | Mutated | n.a. | n.a. | n.a. | n.a. | No | n.a. | 21 | Yes | n.a. |
| 16 | M | 47 | inv(14) | n.a. | Mutated | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 17 | M | 48 | inv(14) | CD4+ | WT | 22 | 17.6 | None | 1; 2; 3; 4 | No | No | 20 | n.a. | n.a. |
| 18 | M | n.a. | n.a. | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 19 | F | 54 | n.a. | n.a. | WT | 181 | n.a. | Lymph nodes | 1; 2; 3; 4 | Yes | Yes | 158 | No | None |
| 20 | F | 55 | n.a. | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 21 | M | n.a | n.a. | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 22 | F | 69 | inv(14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 24 | M | n.a | t(14;14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 25 | F | 76 | inv(14) | CD4+ | WT | 118 | 106 | Spleen, lymph nodes | 3 | No | Yes | 30 | No | CLL |
| 26 | M | n.a. | inv(14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 28 | F | n.a. | inv(14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 29 | M | n.a. | inv(14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 30 | F | n.a. | t(X;14) | n.a. | WT | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 31 | F | n.a. | inv(14) | CD4+ | Mutated | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| 33 | F | 64 | inv(14) | CD4+ | WT | 30.2 | 28.4 | Spleen, liver | 3 | No | No | 17 | No | Chronic HepC infection, liver cirrhosis; diabetes mellitus; renal insuff.; hypertension; heart insuff.; pulmonary hypertension; atrial fibrillation |
| 34 | M | 71 | n.a. | CD4+ | WT | 76 | 68 | Skin | 3 | Yes | Yes | 13 | Yes | Hypertension |
| 35 | M | 76 | inv(14) | CD4+ | WT | 407 | 387 | Spleen | 3 | No | No | 19 | No | Hypertension; hyperuricemia |
| 36 | M | 70 | t(X;14) | CD8+ | UPD | 185 | 180 | Spleen, liver, lymph nodes, | 3; 4 | Yes | Yes | 8 | Yes | Hypertension; diabetes mellitus; renal insuff. |
| 37 | M | 74 | inv(14) | CD4+ | Mutated | 53 | 44 | Lymph nodes, skin | 4 | No | No | 51 | No | M. Bechterew, psoriasis |
| 38 | F | 74 | inv(14) | CD4+ | WT | 79 | 72 | Skin | 1; 2; 3; 4 | No | Yes | 29 | Yes | Hypertension |
WT wildtype, WBC white blood cell, Lymph. lymphocyte, BM bone marrow, CR complete remission, n.a. not available.
*Type of treatment:
1 alkylators (chlorambucile, etc.),
2 purine analogs (Fludarabine, Pentostatin, etc.),
3 Anti-CD52 antibody (Alemtuzumab),
4 others
Tumor cell enrichment
Details are given in the supplementary methods.
RNA and DNA isolation
RNA and DNA were extracted from 1–2 × 107 enriched tumor cells per sample. Details are given in the supplementary methods.
Transcriptome sequencing
Sample libraries were prepared from RNA of isolated cells of 10 patients and five healthy blood donors. RNA sequencing (RNA-Seq) was performed on the HiSeq 2500 system with 2 × 101 bp paired-end reads (Illumina). Duplicate reads were removed and reads were quality filtered. Mutations were considered only if the particular position was covered at least 20-fold. For exclusion of polymorphisms, the dbSNP database was used. In general, single nucleotide variants were excluded if they matched i) a 1000 genomes entry and/or ii) exhibited an annotated variant allele frequency (VAF) above 1% and/or iii) occurred in one or more healthy donor samples. Filtering against healthy donor samples was performed to exclude sequencing artefacts. Database version dbSNP137 was used for data evaluation. Expression analysis of RNA-Seq data was performed with Partek Genomics Suite software, version 6.6; 2016 (Partek Inc., St. Louis, MO, USA)16. Further details are given in the supplementary methods. Data are available under GEO accession number GSE100882.
Targeted capture sequencing
To validate candidate mutations in genes identified by RNA-Seq, we selected 40 genes for which capture oligonucleotides for all coding exons of the respective genes were designed (Fig. 1). Further information is given in the supplementary methods. All variant calls originating from positions covered with less than 20 reads were removed. We excluded variants with less than 20% VAF and analyzed non-synonymous variants only. Polymorphisms were excluded as indicated above. Database version dbSNP137 was used for evaluation of capture sequencing data. Data are available under SRA accession number SRP111041.
Fig. 1. Distribution of mutated genes in the T-PLL cohort analyzed by targeted capture sequencing and WES.
Mutated genes are indicated as black fields for the 33 T-PLL that carried at least one mutated gene. * indicate the five cases for which results are generated by WES. Not listed are 10 genes without any mutations, which are BCL11B, CBL, CUX1, ETV6, JAK1, MTOR, RUNX1, SUZ12, VOPP1, and ZRSR2
Whole-exome sequencing
For five T-PLL samples WES was performed. SNPs with entries in the 1000 genomes project17 were removed. We eliminated mutations if the respective position was covered with less than 20 reads. Only non-synonymous mutations were considered. We excluded variants with a VAF below 20%. Database version dbSNP147 was used for data evaluation. Data are available under SRA accession number SRP111041.
Amplification and sequence analysis of mutations
To verify candidate mutations detected in SAMHD1 by targeted capture sequencing or WES, we selected the respective mutated positions in this gene for three SAMHD1-mutated T-PLL. For all three cases, non-tumor DNA extracted from CD14+ and CD19+ cells was available. After PCR, amplicons were analyzed by Sanger sequencing (ABI3130 Genetic Analyzer; Applied Biosystems, Life Technologies). Primer sequences are available from the authors upon request.
Copy number analysis
Copy number variation (CNV) analysis was carried out on Affymetrix SNP 6.0 microarrays (n = 10) and CytoScan HD arrays (n = 4; Affymetrix, Santa Clara, CA, USA). Further details are given in the supplementary information.
Western blotting
For analysis of SAMHD1, equal amounts of protein lysates were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Details are found in the supplementary information.
Quantitative reverse transcription PCR
RNA was transcribed into cDNA with the high-capacity cDNA reverse transcription kit (Applied Biosystems). Details are given in the supplementary methods.
Cell viability assay
For determination of the number of metabolically active cells, the CellTiter-Glo LuminescenT-cell Viability Assay (Promega, Fitchburg, WI, USA), which is based on quantification of ATP, was used. Details are described in the supplementary methods.
Determination of cellular dNTP content
To obtain intracellular dNTPs, cell pellets prepared from 2 × 106 cells of seven T-PLL samples comprising four SAMHD1 mutated and three wild-type samples as well as three CD3+ MACS-enriched healthy donor samples were lysed. The extraction and quantitative measurement of intracellular dNTPs were conducted as reported previously18.
Results
Identification of mutations in T-PLL by RNA and DNA sequencing
Outline of the experimental design
For identification of recurrent mutations in T-PLL, we first performed RNA-Seq analysis of isolated tumor cells of 10 cases. We then selected 40 candidate genes from this analysis and studied them for somatic mutations in an extended cohort of 28 patients with a targeted DNA capture sequencing approach. Five additional T-PLL were analyzed by WES. For both DNA-sequencing analyses, we excluded mutations with low coverage (read counts < 20) at a respective position and occurring only at subclonal levels, as mutation detection is less reliable in these instances. The cutoff for subclonality was set to 20%. Polymorphisms were excluded (see Materials and Methods section).
Transcriptome sequencing
In the RNA-Seq analysis of 10 T-PLL, we obtained an average number of 9.5 million reads on the target region after quality filtering and duplicate removal. Focusing on variants with high sequence quality scores, we selected all genes carrying non-synonymous mutations and fulfilling the following criteria: (i) mutations in a gene in at least two T-PLL and (ii) a quality score of 1000, which is the highest possible value. The score is calculated as the −10log10 of the p value given by the AVADIS software. The 40 genes fulfilling the criteria (Fig. 1) were subjected to further analyses on the DNA level.
Capture sequencing
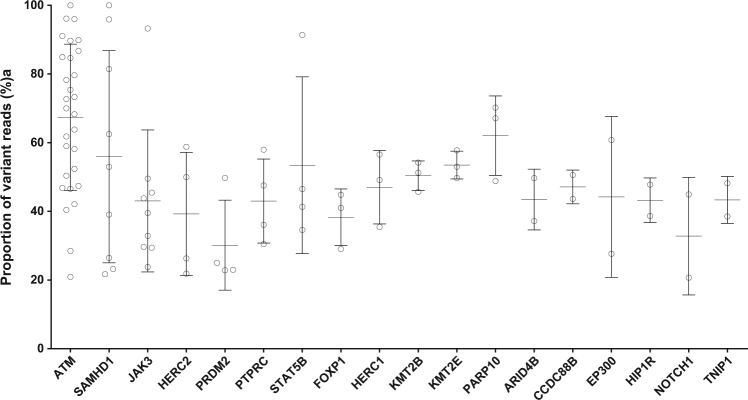
All coding exons of the 40 candidate genes selected from the RNA-Seq analysis were studied by a targeted deep sequencing approach in a total of 28 T-PLL, including the 10 samples on which RNA-Seq was performed. The average sequence coverage for the target region in the capture approach was 151, with 97% of target-region nucleotides covered at least 20 times. Twenty-nine of the forty genes (73%) showed non-synonymous mutations (Fig. 1). Most mutations appeared with VAF of 40–60%, indicating that they are clonal heterozygous mutations (Fig. 2; but note that SNVs with VAF below 20% were filtered out). Two genes (ATM and SAMHD1) showed several mutations with VAF between 90 and 100%, suggesting either homozygous or hemizygous clonal mutations. Ten of the forty genes selected from the RNA-Seq analysis were not found to be mutated in the targeted sequencing (and also not in a WES of five further cases, see below). The discrepancy to the RNA-Seq data is mainly due to the fact that we applied more stringent selection criteria in the DNA analysis (e.g., VAF ≥ 20%). Moreover, not all mutations identified by RNA-Seq were validated in the targeted sequencing approach, indicating some false-positive results in the RNA-Seq analysis, or that some alleles with subclonal mutations were preferentially transcribed.
Fig. 2. Variant allele frequencies of 18 mutated genes carrying at least two mutations.
Variant allele frequencies below 20% are not considered. Depicted are all mutations per gene. Bars indicate mean and S.D.
WES
Five further T-PLL were analyzed by WES. On average, the target region of the exome was covered 75 times across all samples. Overall 88% of nucleotides were covered at least 20 times. After applying the above-mentioned filtering criteria, we first focused on the 40 genes selected for targeted sequencing. Ten of the forty genes (25%), namely ATM, FOXP1, HERC1, HERC2, JAK3, NOTCH1, PARP10, PRMT2, SAMHD1, and STAT5B, showed non-synonymous mutations (Fig. 1). Analyzing the complete WES data, we identified several previously unreported recurrently mutated genes (Supplementary Table 2). Three T-PLL displayed mutations in the ryanodine receptor 3 gene, RYR3, namely two replacement mutations and one splice site mutation. RYR3 can release calcium from the endoplasmic reticulum. PARN was mutated in two T-PLL. It encodes a poly(A)-specific ribonuclease, which degrades poly(A) tails of mRNAs. PCLO, coding for the piccolo presynaptic cytomatrix protein, was mutated in two T-PLL. This gene is also frequently mutated in diffuse large B-cell lymphomas19,20. One T-PLL also carried a mutation in the IL2RG gene, which is known to be recurrently mutated in T-PLL7.
Combining capture (n = 28) and WES data (n = 5), we observed in total mutations in 30 of the 40 genes (75%). ATM was the most frequently mutated gene, with 76% of cases harboring ATM mutations (Fig. 1), in line with prior studies9. Other genes known to be mutated in T-PLL7,13,14 were also recurrently mutated in our cohort, including JAK3 (27% of cases mutated) and STAT5B (12%). Mutations in genes encoding epigenetic regulators, including four members of the KMT2 lysine-methyltransferase family, were observed in 3–9% of cases, and overall in 6 of the 33 cases. Mutations in other epigenetic regulators occurred at lower frequencies compared to previous reports, e.g., BCOR and TET2 each in only 3% of cases (Fig. 1), whereas in prior studies, they were mutated in up to 9% and up to 17% of T-PLL, respectively7,13,14.
We also identified recurrently mutated genes known to be involved in tumorigenesis of various hematologic malignancies, but which have not been described in T-PLL yet. These are SAMHD1, HERC1, HERC2, PRDM2, PARP10, PTPRC, and FOXP1, which could be assigned to distinct functional categories (Fig. 1).
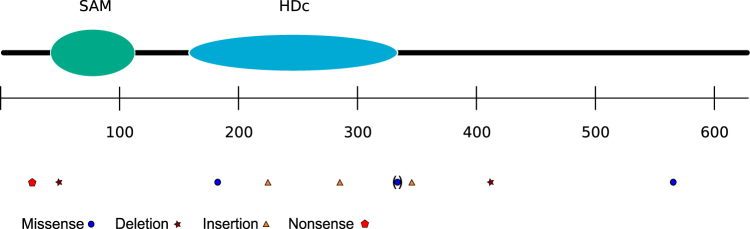
Besides ATM, the group of mutated genes related to DNA damage/repair included SAMHD1 as the second most frequently mutated gene, which was mutated in 18% of cases. The pattern and distribution of these mutations are depicted in Figs. 1 and 3. SAMHD1 encodes a dGTP-activated triphosphohydrolase, which regulates the cellular dNTP pool. Inactivating mutations in SAMHD1, which lead to increased dNTP levels, promote tumor cell survival. In our cohort, the majority of SAMHD1 mutations are frameshift or nonsense mutations. Three of six mutated cases with four SAMHD1 mutations could be used for verification studies by PCR and Sanger sequencing. All SAMHD1 mutations observed in the next-generation sequencing approaches were confirmed. The pattern and distribution of these mutations are depicted in Fig. 3. An analysis of corresponding non-tumor DNA of these patients confirmed the somatic origin of three of the four mutations, whereas in the patient with two SNVs in the gene one of these was also detected in the non-tumor DNA, indicating that this is a germline variant. To test for mutual exclusivity of SAMHD1 mutations with other recurrent mutations, we performed a contingency analysis, which revealed a significant negative association between ATM and SAMHD1 mutations (Fisher’s exact test, p = 0.02).
Fig. 3. Pattern and distribution of mutations in SAMHD1.
Depicted are all mutations. The missense mutation in brackets was also observed in hematopoietic non-tumor cells of a female patient carrying two SAMHD1 mutations. The scale indicates the numbering of the amino acids. SAM sterile alpha motif, HDc Histidine (H)-Aspartate (D) containing
The genes HERC2 and HERC1, which are mutated in 12 and 9% of T-PLL, respectively, encode for E3 ubiquitin protein ligases. In both genes, we observed replacement mutations distributed along the whole length of the gene. HERC2 and HERC1 function as DNA repair genes21. HERC1 is recurrently mutated in T-cell acute lymphoblastic leukemias (T-ALL)22.
PARP10, encoding the mono-ADP-ribosyltransferase PARP10, is mutated in 3/33 cases (9%). Two mutations lead most likely to loss of the protein or impaired protein function as they are a nonsense mutation and a missense mutation in the ubiquitin-interacting motif, which is part of the catalytic domain of the protein23. PARP10 is recruited to stalled replication forks and mediates cellular resistance to DNA damage, thereby promoting genomic stability24.
The most frequently mutated gene in the group of epigenetic regulators is PRDM2, with 12% of T-PLL harboring mutations. It is a member of a nuclear histone/protein methyltransferase superfamily and described as tumor suppressor gene, mutated in several types of cancers25,26. Microsatellite-instable tumors show frequent frameshift mutations of PRDM227. Three of four mutations are predicted to change the amino-acid sequence in the coiled-coil domain of the protein. The lysine-methyltransferase genes KMT2B and KMT2E are mutated in three cases each (9%). All mutations are missense mutations, dispersed along the gene. KMT2B, mutated in several cancers28, is a methyltransferase, while KMT2E, which was initially grouped to the KMT2 family and is also mutated in various cancers29, seems to lack intrinsic histone methyltransferase activity.
PTPRC, encoding CD45, is expressed on all human hematopoietic cells, and has, among other functions, a role in inhibiting JAK/STAT signaling. A downregulation of CD45 is reported in T-ALL carrying PTPRC mutations30. We observed replacement mutations in PTPRC in 9% of cases. Two of three mutations were located in the catalytic protein tyrosine phosphatase domain. CD45 protein expression measured by FACS was slightly reduced in two of the three mutated cases (not shown).
Finally, 9% of T-PLL in our cohort carried mutations in FOXP1, encoding a ubiquitously expressed transcription factor and essential regulator in human CD4+ T cells31,32. All mutations, including missense and nonsense mutations, are located in the forkhead domain, which mediates monomeric DNA binding33.
Copy number variations
To identify gains and losses in this T-PLL cohort, we performed genome-wide CNV analyses for 14 patients (Table 2). Besides already known CNV, we identified several previously unknown gains and losses as well as uniparental disomies (UPDs) for the 40 genes of interest (Table 2). Two of fourteen (14%) patients displayed losses of the SAMHD1 locus at 20q11. In addition, we identified two further patients carrying UPDs involving SAMHD1. None of these four patients had a point mutation or small indel in SAMHD1. For HERC2, HERC1, FOXP1, and PRDM2, no copy number alterations were observed.
Table 2.
Copy number variations and UPDs for 40 selected genes
| Array | SNP 6.0 | CytoHD | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Chr | T-PLL 1 | T-PLL 8 | T-PLL 2 | T-PLL 7 | T-PLL 10 | T-PLL 4 | T-PLL 38 | T-PLL 11 | T-PLL 5 | T-PLL 3 | T-PLL 33 | T-PLL 34 | T-PLL 35 | T-PLL 36 |
| ARID4B | 1 | ||||||||||||||
| JAK1 | 1 | ||||||||||||||
| MTOR | 1 | ||||||||||||||
| PRDM2 | 1 | ||||||||||||||
| PTPRC | 1 | UPD 4.7 Mb | |||||||||||||
| CEBPZ | 2 | Gain 46.6 Mb | |||||||||||||
| FOXP1 | 3 | ||||||||||||||
| FBXW7 | 4 | Gain 7.6 Mb | |||||||||||||
| TET2 | 4 | ||||||||||||||
| WHSC1 | 4 | Loss 1.8 Mb | |||||||||||||
| TNIP1 | 5 | ||||||||||||||
| BCLAF1 | 6 | Loss 66 Mb | UPD 104 Mb | Loss 4.1 Mb | |||||||||||
| CUX1 | 7 | Gain 2.8 Mb | Gain 10.2 Mb | Gain 0.8 Mb | |||||||||||
| KMT2C | 7 | Loss 4 Mb | Loss 1.3 Mb | UPD 8.1 Mb | Gain 5.8 Mb | Loss 3.9 Mb | Loss 14.7 Mb | ||||||||
| KMT2E | 7 | Gain 0.6 Mb | Gain 1.6 Mb | Gain 3.5 Mb | Loss 0.4 Mb | ||||||||||
| VOPP1 | 7 | Gain 9.2 Mb | |||||||||||||
| PARP10 | 8 | Gain 2.2 Mb | Gain 5.2 Mb | Gain 4.3 Mb | Gain 34 Mb | Gain 7.1 Mb | Gain 2.2 Mb | Gain 93.1 Mb | Gain 27.2 Mb | Gain 88.3 Mb | Gain 58 Mb | ||||
| NOTCH1 | 9 | ||||||||||||||
| USP20 | 9 | ||||||||||||||
| ATM | 11 | Loss 8.8 Mb | Loss 2.2 Mb | Loss 7.1 Mb | Loss 7.2 Mb | Loss 7.1 Mb | UPD 74.4 Mb | Loss 25.4 Mb | Loss 9 Mb | Gain mosaic 51.3 Mb | |||||
| CBL | 11 | Loss 6.2 Mb | Loss 10.4 Mb | UPD 74.4 Mb | Loss 25.4 Mb | Gain mosaic 51.3 Mb | |||||||||
| CCDC88B | 11 | UPD 74.4 Mb | |||||||||||||
| DDB2 | 11 | Loss 2.5 Mb | Loss 1 Mb | Loss 3.5 Mb | |||||||||||
| ETV6 | 12 | Loss 2.8 Mb | Loss 3.3 Mb | Loss 2.3 Mb | Loss mosaic 4.5 Mb | ||||||||||
| HIP1R | 12 | UPD 84.4 Mb | |||||||||||||
| KMT2D | 12 | UPD 84.4 Mb | |||||||||||||
| RPLP0 | 12 | UPD 84.4 Mb | |||||||||||||
| BCL11B | 14 | Gain 3.4 Mb | Gain 4.2 Mb | Gain 18.5 Mb | Ggain 66.7 Mb | ||||||||||
| HERC1 | 15 | ||||||||||||||
| HERC2 | 15 | ||||||||||||||
| STAT5B | 17 | UPD 50.7 Mb | Gain 0.2 Mb | ||||||||||||
| SUZ12 | 17 | UPD 50.7 Mb | |||||||||||||
| JAK3 | 19 | UPD 19.5 Mb | |||||||||||||
| KMT2B | 19 | ||||||||||||||
| SAMHD1 | 20 | Loss 1.9 Mb | Loss 2 Mb | ||||||||||||
| PRMT2 | 21 | Gain 0.1 Mb | |||||||||||||
| RUNX1 | 21 | ||||||||||||||
| EP300 | 22 | Loss 1.5 Mb | Gain 21.9 Mb | ||||||||||||
| BCOR | X | Loss mosaic 56.8 Mb | |||||||||||||
| ZRSR2 | X | Loss mosaic 56.8 Mb | |||||||||||||
Chr chromosome, UPD uniparental disomy, Mb megabases
In the region containing PTPRC, UPDs were observed in 3/14 patients (21%). None of these patients displayed other PTPRC mutations. Recurrent gains were observed in BCL11B in 4/14 patients (29%). The role of BCL11B, which is a main maturation factor of T cells34,35, in hematologic malignancies is still discussed. In T-ALL it is reported as either a tumor suppressor or oncogene36,37.
SAMHD1 protein expression is reduced in T-PLL carrying mutations or gene losses
As most mutations in SAMHD1 were destructive or caused loss of one allele, we analyzed whether these genetic lesions affected RNA and/or protein expression of SAMHD1. Quantitative real-time reverse transcription PCR showed variable mRNA levels of SAMHD1 in nine T-PLL analyzed (Fig. 4a). Three T-PLL with SAMHD1 point mutations or a deletion were among the five T-PLL with lowest transcript levels. However, there was no strict correlation between SAMHD1 mRNA levels and presence of mutations. More importantly, western blot analysis of 12 T-PLL revealed that T-PLL without genetic lesions in SAMHD1 had strong protein expression, comparable to normal peripheral blood CD4+ and CD8+ T cells, whereas three of the four mutated cases had hardly detectable levels, and one case slightly reduced levels (Fig. 4b). A similarly low expression was observed for cases with losses and even for one case with a UPD (Fig. 4b).
Fig. 4. SAMHD1 mRNA and protein expression in T-PLL.
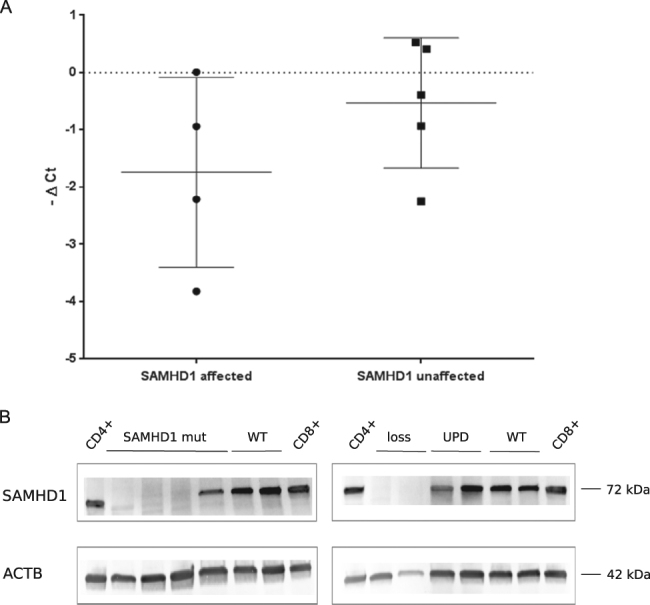
a mRNA expression levels (−ΔCT values) for SAMHD1 of T-PLL with mutated or deleted SAMHD1 (affected) compared to T-PLL with wild-type SAMHD1 (unaffected). Samples with mutated or deleted SAMHD1 have a lower SAMHD1 mRNA expression. GAPDH was used as internal reference. Bars indicate mean and S.D. b. Protein expression levels of 12 T-PLL samples. Samples with mutated SAMHD1 or losses show lower SAMHD1 protein expression compared to WT samples. β-actin served as loading control. mut mutated, WT wild type, UPD uniparental disomy, ACTB Actin beta
T-PLL cells carrying SAMHD1 mutations do not respond differently to cytarabine in vitro
SAMHD1 reduces cytarabine (Ara-C) toxicity in acute myeloid leukemia cells38. Ara-CTP, which is the active triphosphate metabolite of Ara-C, is hydrolyzed by SAMHD1 as its direct substrate, which results in reduction of Ara-C levels. Hence, in cells with inactive SAMHD1, the cytotoxic activity of Ara-C is potentiated38. On the other hand, SAMHD1 may potentially also decrease cell sensitivity to AraC, namely due to the increase of cellular dCTP as competitor of AraCTP. The net outcome may depend on cell types. Although Ara-C is not particularly effective in T-PLL39, we tested drug toxicity to determine a potential effect of Ara-C in cases with SAMHD1 mutations. We performed cell viability assays on thawed viably cryopreserved T-PLL cells after treatment with Ara-C and a number of other cytotoxic agents in different concentrations (Table 3). However, we observed no difference in the IC50 values between cells with low or high SAMHD1 protein expression, indicating that Ara-C is not cytotoxic for T-PLL cells, even not in those with low SAMHD1 expression (Fig. 5). The response of SAMHD1 mutant cases was not improved for any of the drugs tested. When analyzing the cases for the five most frequent mutations occurring in this cohort, we observed next to Ara-C non-responsiveness that SAMHD1 mutant cases neither responded to fludarabine nor clofarabine. By contrast, four of the ATM mutated cases responded well to fludarabine and clofarabine (Table 3). Each of the four cases carried non-synonymous mutations that were predicted to be destructive for ATM protein function by at least 5/7 prediction tools. Two of the cases additionally showed deletions of the second ATM allele. Hence, although the functional impairment of ATM in these cases could not be experimentally validated, it seems that in these T-PLL, ATM mutations are not linked to resistance against fludarabine or clofarabine.
Table 3.
Cell viability assay
| IC50 (µM) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound name | T-PLL_2 | T-PLL_3 | T-PLL_10 | T-PLL_11 | T-PLL_7 | T-PLL_36 | T-PLL_5 | T-PLL_25 | T-PLL_34 | T-PLL_37 | T-PLL_35 | Mean # |
| Cytarabine | 16.095 | 1.875 | 3.212 | 29.91 | 14.727 | 3.897 | 29.91 | 7.761 | 2.034 | 24.826 | 6.233 | 8.3 |
| Fludarabine | 29.91 | 29.91 | 0.468 | 29.91 | 29.91 | 0.76 | 25.828 | 39.91 | 0.241 | 29.91 | 0.363 | 6.4 |
| Clofarabine | 29.91 | 29.91 | 0.046 | 29.91 | 29.91 | 0.225 | 29.91 | 29.91 | 0.372 | 29.91 | NA | 6.2 |
| Cyclophosphamide | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.9 |
| Prednisone | ND | ND | ND | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.91 | 29.9 |
| Doxorubicine | 0.226 | 2.523 | 3.535 | 4.011 | 4.208 | 4.532 | 5.645 | 1.935 | 2.061 | 0.553 | 5.439 | 2.3 |
| SAMHD1 status | Mutated | Mutated | WT | Loss | WT | Loss (UPD) | WT | WT | WT | Mutated | WT | |
| ATM status | WT | WT | Mutated/loss | Mutated | Mutated | Mutated/gain | Mutated | WT* | Mutated | WT* | Mutated/loss | |
| JAK3 status | WT | Mutated | WT | WT | WT | Mutated | Mutated/UPD | * | WT | Mutated* | WT | |
| HERC2 status | Mutated | WT | WT | WT | WT | WT | WT | * | WT | * | WT | |
| FOXP1 status | WT | WT | WT | WT | WT | WT | WT | * | Mutated | * | WT | |
NA not available, ND not determined, WT wild type, UPD uniparental disomy.
*No copy number status available;
#calculated as geometric mean
Fig. 5. Lack of toxic effects of cytarabine on T-PLL cells.
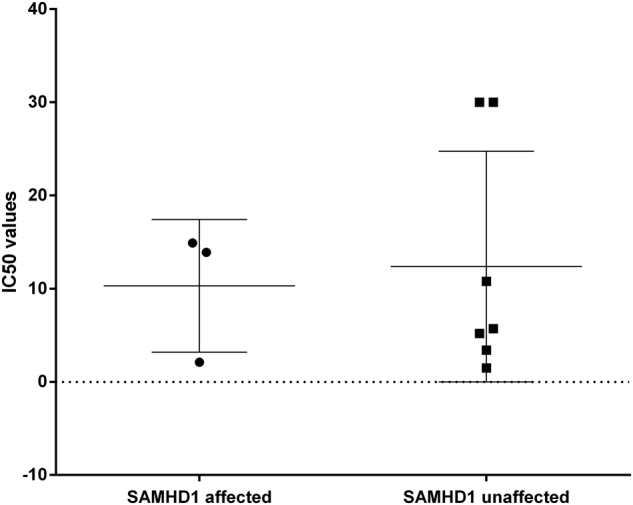
IC50 values for cytarabine of 12 T-PLL samples separated into SAMHD1 affected (n = 4) and SAMHD1 unaffected (n = 7) cases. Bars indicate mean and S.D.
Correlating SAMHD1 mutations to clinical data, we observed no differences between mutated and unmutated cases in terms of age at diagnosis, genetic group, T-PLL immunophenotype, absolute white blood cell count, and absolute lymphocyte count at diagnosis, sites of involvement or response to therapy (Table 1).
Intracellular dNTP levels of SAMHD1-mutated T-PLL are elevated compared to SAMHD1 wild-type and healthy blood donor-derived CD3+ samples
Intracellular dNTP levels measured from 2 × 106 cells of four SAMHD1-mutated samples revealed detectable levels, whereas no dNTPs were detected in three T-PLL SAMHD1 wild-type samples and three healthy donor-derived CD3+ samples, using the same cell numbers for analysis (Fig. 6). The dNTP levels did not correlate with the size of the SAMHD1 mutant clone. The contingency analysis revealed a significant difference between SAMHD1-mutated and unmutated samples (p = 0.0048, two-tailed Fisher’s exact test).
Fig. 6. dATP levels of T-PLL and healthy donor samples.
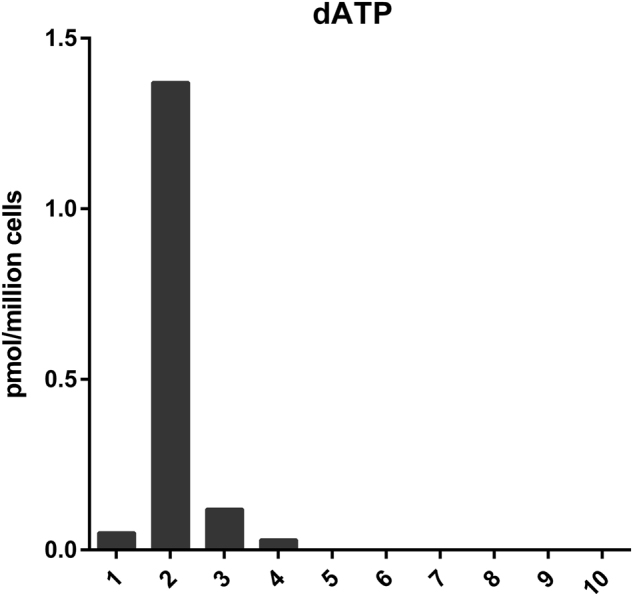
Display of dATP levels from dNTP measurement from 2 × 106 cells. Samples 1–4: T-PLL with SAMHD1 mutations. Samples 5–7: T-PLL with wild-type SAMHD1. Samples 8–10: healthy donor-derived CD3+ T cells. In samples 5–10 the dNTP levels were not detectable
Discussion
T-PLL is characterized by inversions or translocations involving the TCL1 gene or the MTCP1 gene, along with one of the TCR loci3–6. Further genetic lesions involved in T-PLL pathogenesis involve ATM7–9, members of the JAK/STAT signaling pathway, and epigenetic regulators7,11,12,40.
We focused here on 40 candidate genes selected from an exploratory RNA-Seq analysis. Formally, we cannot exclude that a few alterations identified in the RNA or DNA analyses are not somatic mutations but represent germline variants, because for most cases non-tumor RNA or DNA was not available. However, our stringent filtering against known polymorphisms, the occurrence of distinct non-synonymous mutations for recurrently mutated genes, and the direct experimental verification for the somatic origin of three of four tested SAMHD1 mutations together argue that the vast majority of events described here are indeed somatic mutations. Overall, our results confirm previously reported data from our12 and other groups7 and furthermore reveal novel recurrently altered genes in T-PLL (Fig. 1).
Most recurrently altered genes encode proteins with functions in DNA damage/repair. This category included HERC2 and HERC1, the latter of which has been previously reported as recurrently mutated in up to 13% of patients with T-ALL22. We observed nonsense and missense mutations in the ubiquitin-interacting motif of PARP10. As PARP10 deficiency leads to severe DNA repair defects41, these mutations may disturb DNA repair in T-PLL. Ten of fourteen patients (71%) displayed gains involving the PARP10 gene. Such gains are in a considerable number of cases occurring due to formation of an isochromosome i(8q). While the point mutations observed in PARP10 likely have functional consequences, it remains unclear whether these gains translate into higher protein levels and hence functional alterations.
A further gene in the category of DNA damage/repair is SAMHD1, which is mutated in 6/33 (18%) of cases and, moreover, is affected by deletions in two further cases. SAMHD1 encodes a dGTP-activated triphosphohydrolase, which regulates the cellular dNTP pool42. Inactivating mutations in SAMHD1 can promote tumor cell survival as they lead to increased dNTP levels42. SAMHD1 is recurrently mutated in CLL, and the protein is supposed to act as a tumor suppressor43. Most mutations we observed in SAMHD1 are frameshift or nonsense mutations. In several cases, the high VAF of mutations indicates biallelic destruction of the gene. SAMHD1 is highly expressed in monocytes, macrophages, dendritic cells, and resting T cells44,45. In T-PLL, we observed variable SAMHD1 protein expression levels, but in general protein expression of wild-type SAMHD1 was comparable to resting normal CD4+ and CD8+ T cells (Fig. 4b). By contrast, T-PLL cases carrying SAMHD1 mutations or losses showed hardly any SAMHD1 protein, further validating the destructive nature of the mutations (Fig. 4b). We also observed downregulation of SAMHD1 protein in four T-PLL cases not carrying mutations or losses in the coding region of this gene (data not shown). In these cases, epigenetic silencing might occur, although at the group level we did not observe hypermethylation of the SAMHD1 promotor by comparing over 50 T-PLL to normal T-cell subsets (data not shown; to be published elsewhere). Thus, the situation in T-PLL is different from that of CD4 T cells of patients with Sézary syndrome in which the SAMHD1 promotor is hypermethylated46,47. We did not observe a stringent correlation between SAMHD1 protein and mRNA expression (Fig. 4a), which hints toward a regulation of gene expression on different levels. Notably, in the present cohort SAMHD1 mutations are significantly negatively associated with ATM mutations (Fisher’s exact test, p = 0.02). Hence, SAMHD1 inactivation may be an alternative lesion to loss of ATM function to impede proper DNA repair in T-PLL cells. The observation that SAMHD1 mutant cases did not show differences to wild-type cases regarding clinical data might be explained by the relatively low number of cases analyzed, but even more by the aggressive nature of T-PLL and short survival of patients with this disease, so that moderate alterations of tumor cell physiology do not translate into a significant change of clinical outcome.
SAMHD1 regulates dNTP homeostasis by decreasing intracellular dNTP levels. In T-PLL, we observed elevated dNTP levels in SAMHD1-mutated cases compared to SAMHD1 wild-type cases and CD3+ T cells from healthy donors, confirming that mutant SAMHD1 is not able to regulate dNTP levels in T-PLL (Fig. 6). Nucleoside analogs, which are used as therapeutic agents against leukemias, are incorporated into the DNA by a competitive mechanism. It was supposed that in leukemias SAMHD1 mutations causing loss of the proteins’ tumor-suppressive function could lead to enhanced drug efficacy48,49. However, for patients with acute myeloid leukemia it was observed that Ara-CTP, the active metabolite of cytarabine, is a direct substrate of SAMHD1. Hence, intact SAMHD1 reduces Ara-C levels, whereas impaired SAMHD1 protein leads to more effective drug responses38.
Although Ara-C is not generally effective in T-PLL, we analyzed whether T-PLL with SAMHD1 mutations or deletions show an increased sensitivity toward cytarabine. However, this was not observed (Fig. 5). Testing of further nucleoside analogs including Fludarabine and other drugs showing cytotoxic effects in T-PLL also revealed no increased sensitivity in SAMHD1 mutant cases toward any of the drugs (Table 3). Therefore, we conclude that the disturbance of the dNTP pool caused by SAMHD1 mutations in T-PLL leads to a benefit for the tumor cells, since increased intracellular dNTPs promote cell cycle progression, proliferation, and survival of the cells. Regarding the non-responsiveness to drugs, in T-PLL further regulatory mechanisms are likely playing a major role in this context, reflecting the overall drug resistance of these tumor cells.
In the present cohort, PRDM2 is the most frequently mutated gene among those involved in epigenetic regulation. PRDM2 is frequently inactivated by mutations in colorectal cancer cell lines and in relapsed bladder cancer25,26. The amino-acid changing mutations we observed, occurring in the coiled-coil domain of the protein, lead most likely to an impaired function, potentially resulting in tumor cell advantage. We also validate the occurrence of mutations in other epigenetic regulators, namely members of the KMT2 family, BCOR, and TET2.
PTPRC, encoding CD45, is recurrently mutated in T-ALL, leading to downregulation of the protein30. Two of three mutations we observed were located in the catalytic protein tyrosine phosphatase domain, suggesting functional consequences. CD45 negatively regulates JAK/STAT signaling by dephosphorylation of all four JAKs50. Therefore, mutations in PTPRC can enhance the already constitutively activated JAK/STAT signaling in T-PLL12.
FOXP1 is a ubiquitously expressed transcription factor and essential regulator in human CD4+ T cells32. Main mechanisms leading to FOXP1 protein overexpression in cancers are translocations, amplifications, or the repression of miRNAs normally downregulating FOXP1 translation51. All three replacement mutations observed in our T-PLL cohort are located in the DNA-binding forkhead domain, with unclear consequences. Notably, however, in other T-cell lymphoproliferative disorders FOXP1 expression is constitutionally repressed32. Clearly, the role of FOXP1 alterations in T-PLL requires further investigations.
Novel recurrently mutated genes identified by WES included the ryanodine receptor 3 gene RYR3, PARN, coding for a poly(A)-specific ribonuclease and PCLO, encoding the piccolo presynaptic cytomatrix protein. RYR3 releases calcium from the endoplasmic reticulum. Intracellular calcium homeostasis plays an important role in cell metabolism, therefore a disturbed calcium metabolism caused by mutations in RYR3 or overexpression of RYR3 may be oncogenic, as reported for several cancers52. As the point mutations we observed are not located in already described domains, their impact is unclear. However, the observation of a fourfold higher expression of the RYR3 gene in T-PLL compared to normal CD3+ T cells (p = 0.026) is in line with altered RYR3 expression in several cancer types52. PARN was already described as a potential tumor-suppressor gene53. Two missense mutations are located directly in front and behind a coiled-coil region of PARN, which may alter its function. PCLO is recurrently mutated or targeted by copy number gains in diffuse large B-cell lymphomas19,20. We identified one missense and one frameshift mutation, and two copy number gains in two further patients. These seemingly contradictory findings (inactivating frameshift mutation vs. increased gene dosage by copy gain) warrant further investigations.
In conclusion, we identified novel recurrently mutated genes in T-PLL, including PTPRC, regulating the JAK/STAT pathway, epigenetic regulators like PRDM2 and HERC1/2, and genes involved in DNA damage response and DNA repair like SAMHD1, which has most likely a tumor-suppressor function in T-PLL.
Electronic supplementary material
Acknowledgements
We thank Michael Möllmann and Sabine Senkel for expert technical assistance. We thank the Imaging Center Essen (IMCES). We thank the technicians in the Baek Kim Laboratory. This work was supported by a research grant provided from the Dr. Werner Jackstädt-Stiftung, by the Deutsche Krebshilfe (70112112), by National Institutes of Health to B. Kim (GM104198 and AI049781), and by the Deutsche Forschungsgemeinschaft (KU1315/9-2). R.S. is supported through SFB 1074/2 B09*. P.J. is supported by IFORES.
Authors’ contributions
P.J. and L.K.H. performed the sequencing experiments. A.C. and P.H. performed the cell viability assays. A.B. und R.Sie provided samples and performed genetic and epigenetic analyses. B.M. and B.K. performed dNTP measurements. R.Sch and L.K.H. processed all sequencing data. P.J., M.P. prepared cells from healthy donors and patients. P.J. and A.L. performed the protein analyses. T.Z., J.D., and P.J. designed the study. P.J. and R.K. interpreted data and wrote the manuscript. All authors read, edited, and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41408-017-0036-5.
References
- 1.Dungarwalla M, Matutes E, Dearden CE. Prolymphocytic leukaemia of B- and T-cell subtype: a state-of-the-art paper. Eur. J. Haematol. 2008;80:469–476. doi: 10.1111/j.1600-0609.2008.01069.x. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow S. H. et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 4th edn (IARC Press, Lyon, 2008).
- 3.de Oliveira FM, et al. Translocations t(X;14)(q28; q11) and t(Y;14)(q12;q11) in T-cell prolymphocytic leukemia. Int. J. Lab. Hematol. 2009;31:453–456. doi: 10.1111/j.1751-553X.2008.01036.x. [DOI] [PubMed] [Google Scholar]
- 4.Fisch P, Forster A, Sherrington PD, Dyer MJ, Rabbitts TH. The chromosomal translocation t(X;14)(q28; q11) in T-cell pro-lymphocytic leukaemia breaks within one gene and activates another. Oncogene. 1993;8:3271–3276. [PubMed] [Google Scholar]
- 5.Madani A, et al. Expression of p13MTCP1 is restricted to mature T-cell proliferations with t(X;14) translocations. Blood. 1996;87:1923–1927. [PubMed] [Google Scholar]
- 6.Stern MH, et al. MTCP-1: a novel gene on the human chromosome Xq28 translocated to the T-cell receptor alpha/delta locus in mature T-cell proliferations. Oncogene. 1993;8:2475–2483. [PubMed] [Google Scholar]
- 7.Kiel MJ, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124:1460–1472. doi: 10.1182/blood-2014-03-559542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stankovic T, Taylor AM, Yuille MR, Vorechovsky I. Recurrent ATM mutations in T-PLL on diverse haplotypes: no support for their germline origin. Blood. 2001;97:1517–1518. doi: 10.1182/blood.V97.5.1517. [DOI] [PubMed] [Google Scholar]
- 9.Stilgenbauer S, et al. Biallelic mutations in the ATM gene in T-prolymphocytic leukemia. Nat. Med. 1997;3:1155–1159. doi: 10.1038/nm1097-1155. [DOI] [PubMed] [Google Scholar]
- 10.Dürig J, et al. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32) Leukemia. 2007;21:2153–2163. doi: 10.1038/sj.leu.2404877. [DOI] [PubMed] [Google Scholar]
- 11.Bellanger D, et al. Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia. 2014;28:417–419. doi: 10.1038/leu.2013.271. [DOI] [PubMed] [Google Scholar]
- 12.Bergmann AK, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes. Chromosomes Cancer. 2014;53:309–316. doi: 10.1002/gcc.22141. [DOI] [PubMed] [Google Scholar]
- 13.Lopez C, et al. Genes encoding members of the JAK-STAT pathway or epigenetic regulators are recurrently mutated in T-cell prolymphocytic leukaemia. Br. J. Haematol. 2016;173:265–273. doi: 10.1111/bjh.13952. [DOI] [PubMed] [Google Scholar]
- 14.Stengel A, et al. Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker. Genes. Chromosomes Cancer. 2016;55:82–94. doi: 10.1002/gcc.22313. [DOI] [PubMed] [Google Scholar]
- 15.Dearden C. Management of prolymphocytic leukemia. Hematol. Am. Soc. Hematol. Educ. Program. 2015;2015:361–367. doi: 10.1182/asheducation-2015.1.361. [DOI] [PubMed] [Google Scholar]
- 16.PartekInc. Partek® Genomics Suite® Revision 5.0 (Partek Inc, St. Louis, 2016).
- 17.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diamond TL, et al. Macrophage tropism of HIV-1 depends on efficienT-cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mareschal S, et al. Whole exome sequencing of relapsed/refractory patients expands the repertoire of somatic mutations in diffuse large B-cell lymphoma. Genes. Chromosomes Cancer. 2016;55:251–267. doi: 10.1002/gcc.22328. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Tena S, Cubillos-Rojas M, Schneider T, Rosa JL. Functional and pathological relevance of HERC family proteins: a decade later. Cell. Mol. Life Sci. 2016;73:1955–1968. doi: 10.1007/s00018-016-2139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann M, et al. Mutational spectrum of adult T-ALL. Oncotarget. 2015;6:2754–2766. doi: 10.18632/oncotarget.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 24.Nicolae CM, et al. The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J. Biol. Chem. 2014;289:13627–13637. doi: 10.1074/jbc.M114.556340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouradov D, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, et al. Novel variants in MLL confer to bladder cancer recurrence identified by whole-exome sequencing. Oncotarget. 2016;7:2629–2645. doi: 10.18632/oncotarget.6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piao Z, et al. Frequent frameshift mutations of RIZ in sporadic gastrointestinal and endometrial carcinomas with microsatellite instability. Cancer Res. 2000;60:4701–4704. [PubMed] [Google Scholar]
- 28.Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Novera W., Zhang Y., Deng L. W. MLL5 (KMT2E): structure, function, and clinical relevance. Cell Mol. Life Sci. (2017). [DOI] [PMC free article] [PubMed]
- 30.Porcu M, et al. Mutation of the receptor tyrosine phosphatase PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood. 2012;119:4476–4479. doi: 10.1182/blood-2011-09-379958. [DOI] [PubMed] [Google Scholar]
- 31.Durek P, et al. Epigenomic profiling of human CD4+ T-cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity. 2016;45:1148–1161. doi: 10.1016/j.immuni.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Garaud S, et al. FOXP1 is a regulator of quiescence in healthy human CD4+ T-cells and is constitutively repressed in T-cells from patients with lymphoproliferative disorders. Eur. J. Immunol. 2017;47:168–179. doi: 10.1002/eji.201646373. [DOI] [PubMed] [Google Scholar]
- 33.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 34.Avram D, Califano D. The multifaceted roles of Bcl11b in thymic and peripheral T-cells: impact on immune diseases. J. Immunol. 2014;193:2059–2065. doi: 10.4049/jimmunol.1400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satterwhite E, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.V98.12.3413. [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez A, et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood. 2011;118:4169–4173. doi: 10.1182/blood-2010-11-318873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraszewska MD, et al. BCL11B, FLT3, NOTCH1 and FBXW7 mutation status in T-cell acute lymphoblastic leukemia patients. Blood Cells Mol. Dis. 2013;50:33–38. doi: 10.1016/j.bcmd.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Schneider C, et al. SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat. Med. 2017;23:250–255. doi: 10.1038/nm.4255. [DOI] [PubMed] [Google Scholar]
- 39.Dearden C. How I treat prolymphocytic leukemia. Blood. 2012;120:538–551. doi: 10.1182/blood-2012-01-380139. [DOI] [PubMed] [Google Scholar]
- 40.Bergmann A. K. et al. DNA methylation profiling of pediatric B-cell lymphoblastic leukemia with KMT2A rearrangement identifies hypomethylation at enhancer sites. Pediatr. Blood Cancer64(2017). [DOI] [PubMed]
- 41.Shahrour MA, et al. PARP10 deficiency manifests by severe developmental delay and DNA repair defect. Neurogenetics. 2016;17:227–232. doi: 10.1007/s10048-016-0493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballana E, Este JA. SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends Microbiol. 2015;23:680–692. doi: 10.1016/j.tim.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Rossi D. SAMHD1: a new gene for CLL. Blood. 2014;123:951–952. doi: 10.1182/blood-2013-12-545384. [DOI] [PubMed] [Google Scholar]
- 44.Descours B, et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Silva S, et al. Promoter methylation regulates SAMHD1 gene expression in human CD4+ T-cells. J. Biol. Chem. 2013;288:9284–9292. doi: 10.1074/jbc.M112.447201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Silva S, et al. Downregulation of SAMHD1 expression correlates with promoter DNA methylation in Sezary syndrome patients. J. Invest. Dermatol. 2014;134:562–565. doi: 10.1038/jid.2013.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clifford R, et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 2014;123:1021–1031. doi: 10.1182/blood-2013-04-490847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ballana E, et al. SAMHD1 specifically affects the antiviral potency of thymidine analog HIV reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 2014;58:4804–4813. doi: 10.1128/AAC.03145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irie-Sasaki J, et al. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 51.Katoh M, Katoh M. Human FOX gene family (Review) Int. J. Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 52.Cui C, Merritt R, Fu L, Pan Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B. 2017;7:3–17. doi: 10.1016/j.apsb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maragozidis P, et al. Alterations of deadenylase expression in acute leukemias: evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol. 2012;128:39–46. doi: 10.1159/000337418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.