Myelodysplastic syndromes (MDS) are a heterogeneous group of myeloid neoplasms with extremely variable clinical outcome1. The latest update of the World Health Organization (WHO) classification (2016) has defined various MDS subtypes on the basis of dysplastic and cytopenic lineages, the prevalence of blasts, the percentage of ring sideroblasts (RS), and the presence of cytogenetic and genetic abnormalities. In this assessment, the category defined by ring sideroblasts, MDS-RS, is subdivided into cases with single-lineage dysplasia (MDS-RS-SLD) and cases with multilineage dysplasia (MDS-RS-MLD). Moreover, in the presence of cytopenias, dysplasia, and as few as 5% of RS, the MDS-RS category can be established by the identification of a mutation in the SF3B1 gene2. Therefore, genetic data like SF3B1 mutation provide diagnostic utility in MDS and probably also relevant prognosis information. In recent years, next-generation sequencing (NGS) studies in MDS have found new recurrently mutated genes3, 4. Thus, we have utilized these findings for a specific mutational analysis of the MDS-RS subgroup.
We studied 122 patients diagnosed of MDS-RS according to the 2016 WHO classification. From them, 80 patients (66%) had been diagnosed by morphology of MDS-RS-SLD and 42 patients (34%) corresponded to MDS-RS-MLD (Supplementary Table 1). The International Prognostic Scoring System (IPSS) was low for 105 patients (86%) and intermediate-1 for 17 patients (14%). According to the Revised IPSS (IPSS-R), 53 patients (43%) were classified as very low risk, 60 patients (50%) as low risk, 8 patients (6%) as intermediate risk, and only 1 patient (1%) as high risk (Supplementary Table 1). The median follow-up was 35 months (95% confidence interval, 6–204 months), and 5 patients (4%) progressed to acute myeloid leukemia (AML, Supplementary Table 2).
DNA samples were isolated from bone marrow samples at diagnosis and were obtained with written informed consent in accordance with the Declaration of Helsinki, and the approval of the internal review of Bioethics and Medical Research at the University Hospital La Fe. NGS was based on AmpliSeq chemistry and was performed on an Ion Proton instrument. A custom panel of 39 genes with 659 amplicons was designed and allowed a 98% detection rate for 5% variant frequency at positions with an average sequencing coverage from 1000× to 5000× (Supplementary Methods; Supplementary Table 3). Mutations of CALR exon 9, not included in the original panel, were analyzed by Sanger sequencing5. In the statistical analyses P < 0.05 values were considered as statistically significant.
With this approach, up to 97% patients (118 out of 122) have been found to have a somatic mutation in at least one gene (Supplementary Tables 4 and 5). As it was expected, the majority of cases carried SF3B1 mutations (106/122, 86.9%), but other mutations were also detected: TET2 (38/122, 31.1%); DNMT3A (21/122, 17.2%); JAK2 (8/122, 6.6%); SRSF2 (6/122, 4.9%); SETBP1 (6/122, 4.9%); EZH2 (5/122, 4.1%); and ZRSR2 (5/122, 4.1%) genes (Fig. 1).
Fig. 1. Distribution of genetic alterations of significantly mutated genes in 122 MDS-RS cases.
Each column represents an individual sample. Gray cells indicate a mutation in the gene described in that row on the left
In the overall series, patients carrying SF3B1 mutations showed a lower proportion of poor prognosis chromosomal abnormalities compared with SF3B1 ɯt cases (6% vs. 31%, P= 0.038; Supplementary Table 1). In addition, SF3B1 mutations favorably influenced the overall survival (OS) of patients with single-lineage dysplasia (median OS, 88 vs. 22 months, P = 0.010; Supplementary Fig. 1A and B). Although in previous reports MDS-RS cases carrying SF3B1 mutation appear to have a favorable prognosis compared to those without the mutation, the role of multilineage dysplasia vs. the SF3B1 mutation remains controversial6, 7.
Of note, we found a significant positive correlation between the MDS-RS-MLD category and the median number of mutations per patient (2, range 0–5 vs. 1, range 0–5, for MDS-RS-MLD and MDS-RS-SLD; P = 0.003).
It was also observed that SF3B1 mut patients with the K700E mutation showed a significant higher level of RS than SF3B1 mut patients with other SF3B1 mutations (median, 44% vs. 27%, P = 0.012). SF3B1 mutations affect the gene expression of the iron transporter ABCB7 and determine the accumulation of aberrant mitochondrial ferritin in the erythroblasts8. It is likely that the distinct SF3B1 mutations cause a different degree of ABCB7 downregulation and therefore a phenotype with more or less RS.
Mutations in other important splicing gene, SRSF2, were generally found in SF3B1 ɯt patients suggesting that they would play a redundant role in disease pathogenesis9. Most SRSF2 mut patients (83%) were clustered in lower-risk categories of the IPSS-R however showed a significantly lower platelet count (median, 93 × 109 vs. 262 × 109/L, P < 0.001) a higher red blood cell (RBC) transfusion-dependency (100% vs. 59%, P = 0.046), and an inferior OS (hazard ratio, HR = 10.89; P = 0.001) than SRSF2 ɯt patients (Supplementary Tables 1 and 6, Supplementary Figure 1C).
In methylation categories, TET2 mutations were found in a high number of MDS-RS patients although no differences in the clinical features were found according to their mutation status. Nevertheless, TET2 mut patients showed a higher median number of mutations compared with TET2 ɯt patients (3, range 0–5 vs. 1, range 0–5, P < 0.001). TET2 mutations are currently considered as drivers of “clonal hematopoiesis of indeterminate potencial” and would lead to a primary permissive environment for subsequent genetic alterations10.
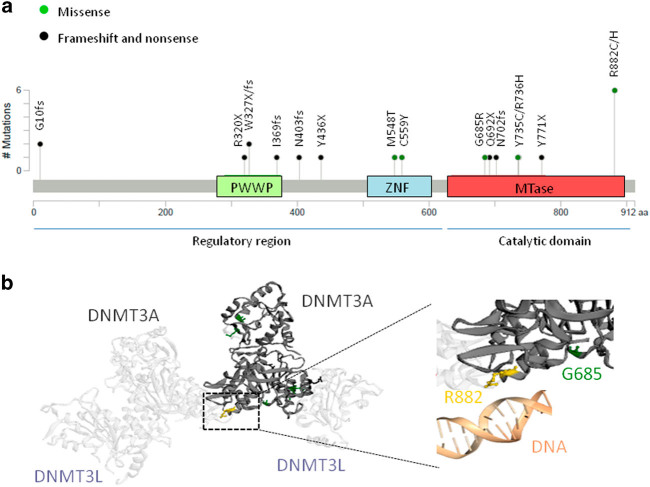
The other methylator gene frequently mutated was DNMT3A. Patients with DNMT3A mutations showed a more adverse clinical status with a significant higher RBC transfusion-dependency compared with DNMT3A ɯt patients (81% vs. 56%, P = 0.029; Supplementary Table 1). Furthermore, when DNMT3A mutations were considered according to their occurrence in the protein domain important differences were observed. In the regulatory domain (RG, exon 1–15, n = 9), frameshift and nonsense mutations predominantly occurred (80%; Fig. 2a) and were likely to be loss-of-function mutations and therefore would not exhibit significant changes in DNA methylation11. DNMT3A_RG mutations did not show any influence on OS or AML transformation. Conversely, in the DNMT3A methyltransferase domain (MT, exon 16–23, n = 12), missense mutations involving highly conserved residues were especially found (75%; Fig. 2a), suggesting that they may not be simple loss-of-function mutations and may confer a novel protein function12, 13. Several studies, especially based on R882 hotspot mutations, demonstrate profound loss of de novo methyltransferase activity resulting from the dominant negative consequences of the missense alterations. The mutant DNMT3A protein interacts with wild-type DNMT3A and DNMT3L proteins to form functionally deficient complexes that change the normal methylation patterns in the cell (Figure 2b)12, 14. Accordingly, the presence of DNMT3A_MT mutations in our series determined a more adverse clinical outcome with a very prominent RBC transfusion-dependence (92% vs. 57%, P = 0.017), a shorter OS, and a higher risk of AML progression (OS: HR = 4.99, P < 0.001; AML transformation: HR = 9.84, P = 0.047; Supplementary Table 6, Supplementary Fig. 1E and F).
Fig. 2. Locations of DNMT3A mutations and structure of DNMT3A protein.
Plot a reports the number of DNMT3A mutations found in the MDS-RS series and their locations with respect to the protein functional domain (MTase, methyltransferase; PWWP, chromatin targeting; ZNF, zinc finger). Plot b reports the interaction of two DNMT3L and two DNMT3A proteins to construct a tetramer with DNA methylating function. The R882 residue (in yellow color) is critical in the correct binding of the DNMT3A protein with the DNA molecule12
Finally, when analyzing other minority mutated genes, the negative clinical impact of the EZH2 mutations should also be noted (OS, HR = 7.06, P = 0.004; Supplementary Table 6, Supplementary Fig. 1D). All EZH2 mut patients were clustered in lower-risk categories of the IPSS and IPSS-R however had a high RBC transfusion-dependency at diagnosis and showed a median OS of 30 months, near to the OS observed in MDS patients within the intermediate IPSS-R risk category15.
In summary, the stratification of risk remains the essential step before treatment decision- making. Nevertheless, clinical behaviors differ from what expected on the basis of calculated prognostic indexes. In this context, our findings highlight the potential utility of SF3B1, SRSF2, EZH2, and DNMT3A gene mutations on prognostic risk stratification and treatment decisions in MDS-RS patients.
Electronic supplementary material
Acknowledgements
This work was supported in part by the following grants: PI12/01047; RD12/0036/0014; PIE13/00046; PT13/0010/0026; PI14/01649; PI16/01113; and CIBERONC Consortium grant CB16/12/00284 by the Instituto de Salud Carlos III and PROMETEOII/2015/008 from the “Consellería de Educación, Cultura y Deporte” of the Generalitat Valenciana. We thank Ana Belén Martín for her excellent assistance in optimizing DNA extraction.
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
José Cervera and Guillermo Sanz contributed equally to this work.
Supplementary information
The online version of this article (10.1038/s41408-017-0016-9) contains supplementary material.
References
- 1.Jädersten M, Hellström-Lindberg E. Myelodysplastic syndromes: biology and treatment. J. Intern. Med. 2009;265:307–328. doi: 10.1111/j.1365-2796.2008.02052.x. [DOI] [PubMed] [Google Scholar]
- 2.Arber DA, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klampfl T, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N. Engl. J. Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- 6.Malcovati L, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233–241. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patnaik MM, et al. Prognostic irrelevance of ring sideroblast percentage in World Health Organization-defined myelodysplastic syndromes without excess blasts. Blood. 2012;119:5674–5677. doi: 10.1182/blood-2012-03-415356. [DOI] [PubMed] [Google Scholar]
- 8.Nikpour M, et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia. 2013;27:889–896. doi: 10.1038/leu.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy JA, Ebert BL. Clinical implications of genetic mutations in myelodysplastic syndrome. J. Clin. Oncol. 2017;35:968–974. doi: 10.1200/JCO.2016.71.0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 13.Im AP, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014;28:1774–1783. doi: 10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russler-Germain DA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bejar R, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.