Abstract
Somatic or de novo mutations of Additional sex combs-like 1 (ASXL1) frequently occur in patients with myeloid malignancies or Bohring-Opitz syndrome, respectively. We have reported that global loss of Asxl1 leads to the development of myeloid malignancies and impairs bone marrow stromal cell (BMSC) fates in mice. However, the impact of Asxl1 deletion in the BM niche on hematopoiesis remains unclear. Here, we showed that BMSCs derived from chronic myelomonocytic leukemia patients had reduced expression of ASXL1, which impaired the maintaining cord blood CD34+ cell colony-forming capacity with a myeloid differentiation bias. Furthermore, Asxl1 deletion in the mouse BMSCs altered hematopoietic stem and progenitor cell (HSC/HPC) pool and a preferential myeloid lineage increment. Immunoprecipitation and ChIP-seq analyses demonstrated a novel interaction of ASXL1 with the core subunits of RNA polymerase II (RNAPII) complex. Convergent analyses of RNA-seq and ChIP-seq data revealed that loss of Asxl1 deregulated RNAPII transcriptional function and altered the expression of genes critical for HSC/HPC maintenance, such as Vcam1. Altogether, our study provides a mechanistic insight into the function of ASXL1 in the niche to maintain normal hematopoiesis; and ASXL1 alteration in, at least, a subset of the niche cells induces myeloid differentiation bias, thus, contributes the progression of myeloid malignancies.
Introduction
The Drosophila Asx protein belongs to the enhancer of Trithorax and Polycomb group and functions in both transcriptional activation and repression1,2. Trithorax and Polycomb proteins have significant impacts on various biological processes by modifying chromatin structures to control the active/repressive transcriptional states, respectively3. There are three Asx homologs in mammals, additional sex combs-like 1 (ASXL1), ASXL2, and ASXL34. Three ASXL members share conserved domains, including N-terminal ASXN, ASXH domains, and a C-terminal plant homeodomain4. As a chromatin regulator, ASXL1 plays an important role in epigenetic regulation by activating or repressing the transcription of genes involved in either differentiation or proliferation through its effect on histone methylation marks5,6. ASXL1 has been shown as an essential cofactor for the histone H2A deubiquitinase BAP16, as well as a critical mediator of the function of polycomb repressive complex 2 (PRC2)5. Recently, we reported that ASXL1-cohesin interaction functions as a novel way to maintain normal sister chromatid separation and to regulate gene expression in hematopoietic cells7. These studies demonstrate multifaceted functions of ASXL1 in gene regulation by assembling epigenetic regulators and transcription factors to specific gene loci.
Genomic sequencing studies have uncovered an array of distinct genomic driver mutations in various cancers, including myeloid malignancies. ASXL1 mutations are often found in a wide range of myeloid malignancies8–11, and its alterations are associated with poor prognosis12. Hoischen et al.13 reported that de novo ASXL1 mutations occur in patients with Bohring-Opitz syndrome (BOS) and some of these patients develop Wilms tumors14. We and others have established mouse models and verified that loss of Asxl1 leads to myelodysplastic syndrome (MDS)-like disease15,16 and BOS-like phenotypes17. We also showed that ASXL1 regulates the self-renewal and differentiation of bone marrow stromal cells (BMSCs)17 and hematopoietic stem/progenitor cells (HSC/HPCs)15,16.
HSC/HPCs reside in the bone marrow (BM), known as BM “niche”. The normal function of the BM niche is critical for the maintenance of cellular function of HSC/HPCs18–23. BMSCs are the major component of the BM niche that maintain and regulate the HSC/HPC pool throughout life24,25. Two independent studies using different mouse models revealed that systemic deletion of Asxl1 (Asxl1−/−) leads to severer hematologic phenotypes16 than the conditional loss of Asxl1 in hematopoietic cells alone15. This led us to hypothesize that Asxl1 loss in the niche of Asxl1−/− mice contributes to the hematopoietic phenotypes in vivo. Biased myeloid differentiation prerequisites leukemia formation26. Furthermore, preferential expansion of the granulocyte-macrophage progenitor (GMP) population is associated with a high risk of leukemic transformation in MDS patients27,28. Given the fact that global deletion of Asxl1 results in biased myeloid differentiation, we questioned that Asxl1-deficient niche may alter the cell fates of HSC/HPCs, contributing to the pathogenesis of myeloid malignancies.
In the current study, we found that the expression of ASXL1 significantly decreased in the BMSCs of chronic myelomonocytic leukemia patients (CMML-BMSCs) compared with healthy donors (HD-BMSCs). In addition, CMML-BMSCs displayed a reduced hematopoietic supportive activity and induced a skewed HSC/HPC differentiation toward granulocytic/monocytic lineage. Furthermore, utilizing OsxCre;Asxl1fl/fl mouse model, we showed that deletion of Asxl1 in the BM niche impaired HSC/HPC pool and skewed cell differentiation with a bias to granulocytic/monocytic lineage. Interestingly, immunoprecipitation assays showed that ASXL1 interacted with the core subunit of RNA polymerase II (RNAPII) complex, POLR2A, in BMSCs. Chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) analyses identified a co-occupancy of ASXL1 and RNAPII at the gene promoter regions. Loss of Asxl1 reduced RNAPII enrichment genome-wide accompanied by altered expression of genes critical for BMSC self-renewal, differentiation, and biological functions. Our study provides a further mechanistic insight into ASXL1 functions in the BM niche, and how ASXL1 alteration-associated defective niche works in concert with an intrinsic effect of ASXL1 alteration-mediated HSC/HPC defects to promote the pathogenesis of myeloid malignancies.
Results
Reduced CFU-F frequency and decreased proliferative capacity in CMML-BMSCs
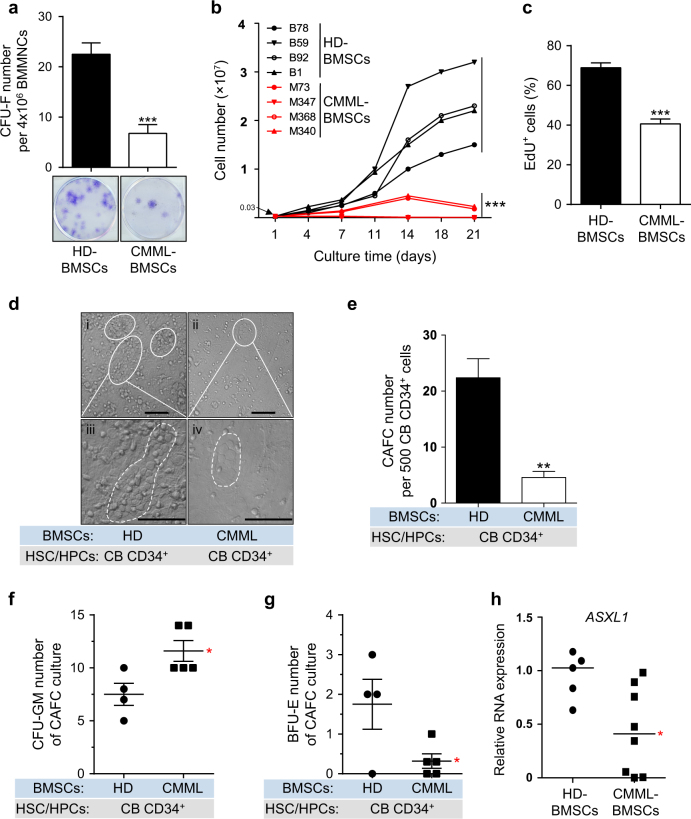
BMSCs from thirteen CMML patients and ten healthy donors were isolated and cultured in vitro. The clinical characteristics of CMML patients were listed in Supplementary Table S1. CMML-BMSCs exhibited similar morphology and expression pattern of cell surface markers as in HD-BMSCs (Supplementary Fig. S1a, b). Colony-forming unit-fibroblast (CFU-F) assay revealed a significant reduction in the frequency of CFU-F in the BM of CMML patients compared with that in the HD-BM (Fig. 1a), indicating a reduced BMSC pool in the BM of CMML patients.
Fig. 1. CMML-BMSCs exhibit decreased proliferative capacity and reduced hematopoietic supportive activity.
a The frequency of CFU-F from CMML patients shows a dramatically decreased BMSC pool compared with HD controls after 10 days of culture (n = 5 individual samples for each group). Representative photographs of CFU-F are shown. b Growth curves for 21 days shows reduced proliferation ability of CMML-BMSCs (red lines, n = 4) compared with HD-BMSCs (black lines, n = 4). Each dot represents one passage. c Bar graph shows the percentage of EdU+ cells (n = 4 individual samples for HD, 3 for CMML). d Long-term co-culture of CB CD34+ cells with CMML-BMSCs or HD-BMSCs indicates impaired hematopoietic supportive ability of CMML-BMSCs. Representative micrographs of CAFCs after 5 weeks of co-culture with HD-BMSCs (i, iii) or CMML-BMSCs (ii, iv) are shown. Scale bar, 50 µm. e The numbers of CAFC are reduced in CB CD34+ cells co-cultured with CMML-BMSCs. Quantitative evaluation of CAFC number per 500 CB CD34+ cells is shown (n = 4 individual samples for HD, 5 for CMML). f, g After co-culture of CB CD34+ cells with HD-BMSCs or CMML-BMSCs, the CFU-C assays were performed. The colony numbers of CFU-GM (f) and BFU-E (g) are shown (n = 4 individual samples for HD, 5 for CMML). h qPCR analysis shows that ASXL1 transcripts were significantly decreased in CMML-BMSCs compared with HD-BMSCs (n = 5 individual samples for HD, 8 for CMML). Bars represent the median. Data represent mean ± s.e.m., * P < 0.05. **P < 0.01, ***P < 0.001
To determine the growth characteristics of CMML-BMSCs and HD-BMSCs, cell numbers were assessed dynamically in the cultures of BMSCs. CMML-BMSCs exhibited limited proliferative capacity during passaging in vitro compared with HD-BMSCs (Fig. 1b). EdU staining further confirmed the impaired proliferation in the CMML-BMSCs. After incubation with EdU for 36 h, ~70% of HD-BMSCs were EdU-positive (EdU+), while only 40% CMML-BMSCs were EdU+ (Fig. 1c; Supplementary Fig. S1c). Cell cycle analysis revealed a marked reduction in S-phase of CMML-BMSCs compared with HD-BMSCs (Supplementary Fig. S1d, e). In addition, senescence-associated β-galactosidase staining showed a threefold increase in the percentage of senescent cells in CMML-BMSCs compared with HD-BMSCs (Supplementary Fig. S1f). Collectively, these data indicate that CMML-BMSCs have a decreased proliferative capacity, an increased senescent activity, and reduced DNA replication.
BMSCs are capable of differentiating into osteoblasts29. CMML-BMSCs exhibited a less alkaline phosphatase-positive expression (Supplementary Fig. S1g), as well as decreased mRNA expression of multiple genes (ALPL, BGLAP, and RUNX2) critical for osteoblast differentiation compared with HD-BMSCs (Supplementary Fig. S1h). These data suggest an impaired osteoblast differentiation in CMML-BMSCs.
Reduced hematopoietic supportive activity and increased granulomonocytic lineage differentiation
BMSCs are widely recognized to be important in HSC maintenance within the BM microenvironment24. Given that CMML-BMSCs exhibited impaired proliferative capacity and cell fate determination, we next examined the hematopoietic supportive activity of CMML-BMSCs using cobblestone-area-forming cell (CAFC) assays in vitro. A significant reduction of CAFC number and smaller CAFC sizes were observed in the co-culture of cord blood (CB) CD34+ cells with CMML-BMSCs compared with HD-BMSCs (Fig. 1d, e). These data indicate that CMML-BMSCs have an impaired hematopoietic supportive activity.
To examine the effect of CMML-BMSCs on the differentiation of normal HSC/HPCs in vitro, we performed long-term culture-initiating cell (LTC-IC) assays. An increased number of CFU-granulocytes/macrophages (CFU-GM) was observed in the co-culture of CB CD34+ cells with CMML-BMSCs compared with HD-BMSCs (Fig. 1f). In contrast, the number of burst forming unit-erythrocytes (BFU-E) was significantly decreased in the co-culture of CB CD34+ cells with CMML-BMSCs compared with HD-BMSCs (Fig. 1g). These results demonstrate that the co-culture of CB CD34+ cells with CMML-BMSCs preferentially promotes myeloid differentiation.
Decreased ASXL1 expression in CMML-BMSCs
Mutations of ASXL1 frequently occurred in the hematopoietic cells of CMML patients12. To examine whether the CMML-BMSCs carry ASXL1 mutation, targeted PCR followed by Sanger sequencing was performed on genomic DNA extracted from the CMML-BMSCs. No ASXL1 mutation was identified in the BMSCs of these 13 CMML patients who have ASXL1 mutations in the hematopoietic cells (data not shown). However, the mRNA expression levels of ASXL1 were significantly decreased in the cohort of CMML-BMSCs (Fig. 1h).
To determine whether the decreased ASXL1 expression is a direct cause of the functional alteration of the patient BMSCs, we next introduced WT ASXL1 into BMSCs derived from myeloproliferative neoplasms (MPN) patients. Quantitative PCR (qPCR) confirmed that ASXL1 mRNA was overexpressed in MPN-BMSCs compared with the empty vector (EV) controls (Supplementary Fig. S1i). After 7 days of co-culture of MPN-BMSCs with CB CD34+ cells, ASXL1 overexpression in MPN-BMSCs restored their effect on CB CD34+ cell differentiation as evidenced by the reduced percentage of myeloid cells (CD45+/CD33+) and increased percentage of erythroid cells (CD71+/CD235a+) compared with EV-transduced MPN-BMSCs (Supplementary Fig. S1j, k).
Altered cell fates of HSC/HPCs in OsxCre;Asxl1fl/fl mice
We and others reported that loss of Asxl1 in mice led to MDS-like disease15,16. Interestingly, global deletion of Asxl1 leads to severer hematologic phenotypes16 than the conditional loss of Asxl1 in hematopoietic cells alone15. Our previous study showed that loss of Asxl1 in mice led to a marked decrease in the number of BMSCs compared with WT mice; and Asxl1−/− BMSCs exhibited impaired self-renewal and skewed differentiation17. We argued that deletion of Asxl1 in the BM niche could cooperate with Asxl1-null HSC/HPCs to accelerate the pathogenesis of myeloid malignancies in vivo. To test this hypothesis, we performed hematopoietic phenotypic analyses in OsxCre;Asxl1fl/fl mice, in which Asxl1 is deleted in the BMSCs and preosteoblasts30,31. Of note, BMSCs and preosteoblasts have been considered as important components of BM niche32. The successful Asxl1 deletion in BMSCs was shown by PCR (Supplementary Fig. S2a), and the expression of Asxl1 was not affected in lineage negative/cKit+ (LK) cells (Supplementary Fig. S2a, b). The von Kossa/McNeal staining of the OsxCre;Asxl1fl/fl femur revealed a decreased trabecular bone surface and an increased adipocyte number (Supplementary Fig. S2c–e).
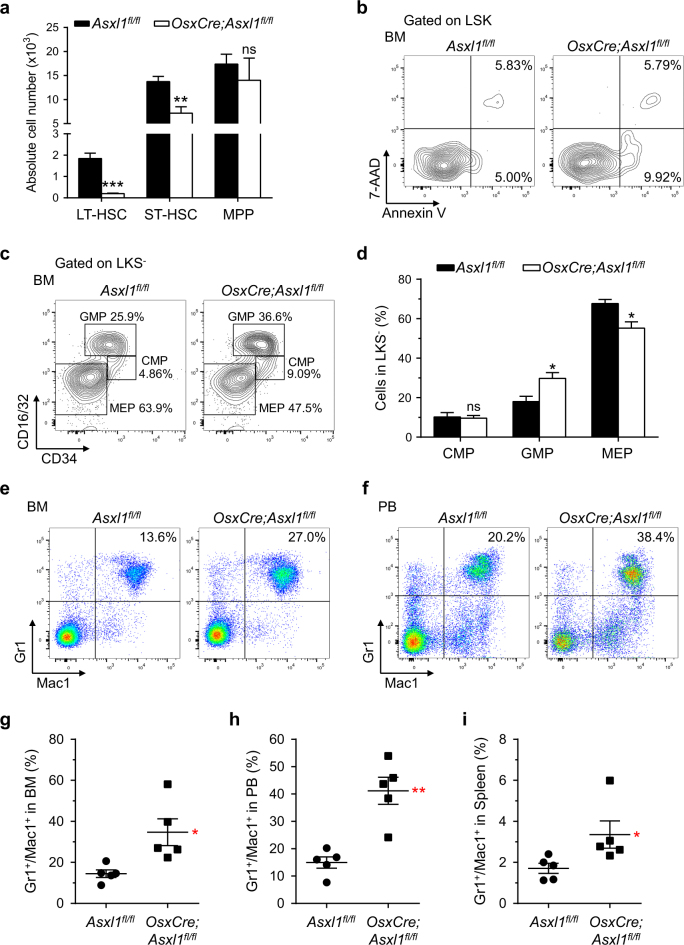
To assess whether deletion of Asxl1 in the BM niche affects the HSC/HPC pool, we performed phenotypic analyses on the BM cells of Asxl1fl/fl (control) and OsxCre;Asxl1fl/fl mice by flow cytometry. The number of long-term HSCs (LT-HSCs) and short-term HSCs (ST-HSCs) per femur was dramatically reduced in OsxCre;Asxl1fl/fl mice compared with Asxl1fl/fl control mice (Fig. 2a). OsxCre;Asxl1fl/fl mice had a significantly lower proportion of LT-HSCs in BM Lin−Sca1+cKit+ (LSK) cells compared with control mice (Supplementary Fig. S2f, g). The ST-HSCs and multipotential progenitor (MPP) cell populations in the BM LSK cells of OsxCre;Asxl1fl/fl mice were similar to that of Asxl1fl/fl mice (Supplementary Fig. S2f, g).
Fig. 2. Altered HSC/HPC and myeloid populations in OsxCre;Asxl1fl/fl mice.
a Absolute numbers of LT-HSCs, ST-HSCs, and MPP cells are shown (n = 4 mice per genotype). b Apoptosis analysis (Annexin V/7-AAD staining) on freshly isolated LSK cells from BM of representative Asxl1fl/fl and OsxCre;Asxl1fl/fl mice. c Flow cytometric analysis of CMP, GMP, and MEP populations in the Lin−cKit+Scal1− (LKS−) cells from BM of representative Asxl1fl/fl and OsxCre;Asxl1fl/fl mice. d Quantification of the percent GMP and MEP populations in LKS− cells of OsxCre;Asxl1fl/fl mice are shown (n = 4 mice per genotype). e, f Flow cytometric analysis of Gr1+/Mac1+ cell populations in BM (e) and PB (f) of representative Asxl1fl/fl and OsxCre;Asxl1fl/fl mice. g–i Quantification of Gr1+/Mac1+ cell populations in BM (g), PB (h), and spleen (i) revealed increased granulocytic/monocytic cells in OsxCre;Asxl1fl/fl mice (n = 5 mice per genotype). Data represent mean ± s.e.m., ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001
The changes in the HSC/HPC frequencies in vivo can be associated with altered apoptosis. We then examined whether Asxl1 loss in the BM niche affects the survival of HSC/HPCs by flow cytometric analyses following Annexin V and 7-amino-actinomycin D (7-AAD) staining. OsxCre;Asxl1fl/fl LSK cells had a significantly higher proportion of Annexin V+/7-AAD− cells than Asxl1fl/fl cells (Fig. 2b; Supplementary Fig. S2h). These results indicate that loss of Asxl1 in the BM niche increases HSC/HPC apoptosis, which may contribute to the lower numbers of LT-HSCs and ST-HSCs in vivo.
Further analyses showed that deletion of Asxl1 in the BM niche significantly increased GMP population and decreased megakaryocyte-erythrocyte progenitor (MEP) population, whereas common myeloid progenitor (CMP) population was similar in the BM Lin−cKit+Sca1− (LKS−) cells of OsxCre;Asxl1fl/fl mice compared with control mice (Fig. 2c, d). However, the number of GMP, MEP, and CMP cell populations was similar between the two genotypes of mice (Supplementary Fig. S2i). To further confirm the effect of Asxl1 loss in BM niche on HSC/HPCs, we performed the CFU-C assays using the BM of Asxl1fl/fl and OsxCre;Asxl1fl/fl mice. The frequency of CFU-GM was significantly higher in the BM of OsxCre;Asxl1fl/fl mice than in the BM of Asxl1fl/fl control mice (Supplementary Fig. S2j), suggesting a preferential myeloid progenitor increment.
To further characterize the Asxl1 loss in BM niche-mediated dysregulation of hematopoiesis, flow cytometric analyses were performed on peripheral blood (PB), BM, and spleen cells of Asxl1fl/fl and OsxCre;Asxl1fl/fl mice. An increased proportion of granulocytic/monocytic cells (Gr1+/Mac1+) was observed in the PB, BM, and spleens of OsxCre;Asxl1fl/fl mice compared with control mice (Fig. 2e–i).
Given that Asxl1−/− BMSCs exhibit impaired self-renewal and skewed differentiation17, we sought to explore the role of Asxl1 in BMSCs in the maintenance of HSC/HPC activity. We began by performing CAFC assay to evaluate the hematopoietic supportive activity of Asxl1−/− BMSCs. When LK cells were co-cultured for 4 weeks on BMSC feeder layers, the numbers of CAFC and CFU-C were significantly reduced in the co-culture of Asxl1−/− BMSCs plus WT LK cells compared with WT co-cultures (Supplementary Fig. S3a, b). This result suggests that Asxl1−/− BMSCs had an impaired hematopoietic supportive activity. To further assess the effect of Asxl1−/− BMSCs in hematopoietic cell differentiation, the percentage of Gr1+/Mac1+ cells in the co-cultures was analyzed by flow cytometry. After 2 weeks of co-culture, Asxl1−/− BMSCs induced a biased Gr1+/Mac1+ differentiation of WT LK cells compared with WT BMSCs (Supplementary Fig. S3c), verifying that Asxl1−/− BMSCs mediates a biased myeloid differentiation of HSC/HPCs.
Loss of Asxl1 in the BM niche alters HSC/HPC fate
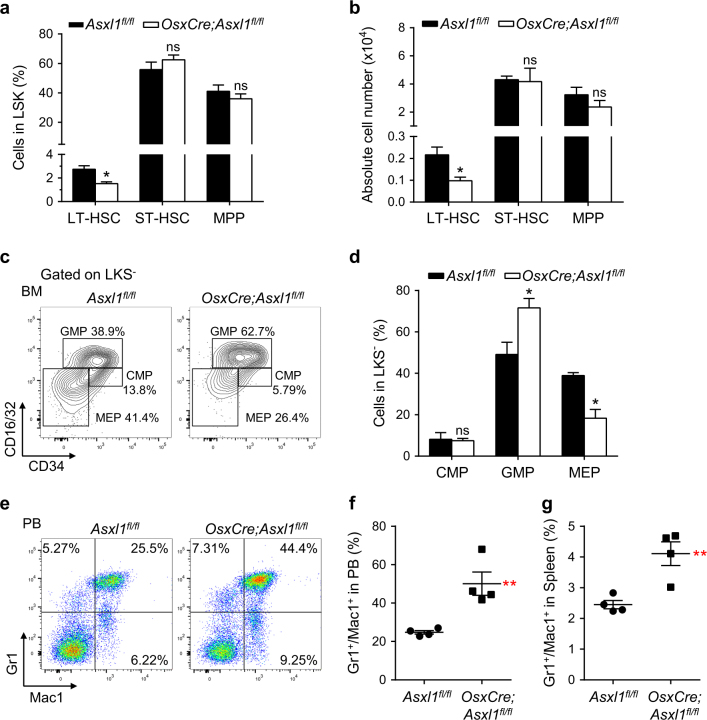
To assess the contribution of the microenvironment to the hematopoietic phenotypes in OsxCre;Asxl1fl/fl mice, we next transplanted WT BM (CD45.1+) cells into lethally irradiated Asxl1fl/fl and OsxCre;Asxl1fl/fl recipient mice, respectively. Five months after the transplantation, we performed flow cytometric analyses to determine the frequencies of HSC/HPCs and the myeloid cell population of reconstituted cells (CD45.1+) (Supplementary Fig. S3d). Interestingly, the frequency and number of LT-HSCs were significantly decreased in OsxCre;Asxl1fl/fl recipients compared with the control recipients (Fig. 3a, b; Supplementary Fig. S3e). Furthermore, significantly increased GMP and decreased MEP populations were observed in BM LKS− cells of OsxCre;Asxl1fl/fl recipients compared with that of control recipients (Fig. 3c, d). However, the number of GMP population in OsxCre;Asxl1fl/fl recipients was comparable to that in the control recipients, whereas the number of MEP was significantly decreased (Supplementary Fig. S3f). OsxCre;Asxl1fl/fl recipients had a significantly high proportion of Gr1+/Mac1+ cells in the PB and spleens (Fig. 3e–g; Supplementary Fig. S3g). The results reinforce the impact of OsxCre;Asxl1fl/fl microenvironment in altering the HSC/HPC pool and promoting myeloid differentiation bias in vivo.
Fig. 3. Loss of Asxl1 in the BM niche alters HSC/HPC pool.
a Frequencies of LT-HSC, ST-HSC, and MPP cell populations in BM reconstituted CD45.1+/LSK cells of OsxCre;Asxl1fl/fl recipients are shown (n = 3 mice per genotype). b Absolute numbers of LT-HSCs, ST-HSCs, and MPP cells from Asxl1fl/fl and OsxCre;Asxl1fl/fl recipients are shown (n = 3 mice per genotype). c Flow cytometric analyses of CMP, GMP, and MEP populations in the reconstituted CD45.1+/LKS− cells from BM of representative Asxl1fl/fl and OsxCre;Asxl1fl/fl recipients. d Quantification of the percent CMP, GMP, and MEP populations in BM reconstituted CD45.1+/LKS− cells of OsxCre;Asxl1fl/fl recipients are shown (n = 3 mice per genotype). e Flow cytometric analysis of Gr1+/Mac1+ cell populations in PB of representative Asxl1fl/fl and OsxCre;Asxl1fl/fl recipients. f, g Quantification of Gr1+/Mac1+ cell populations in PB (f) and spleen (g) shows increased myeloid cells in OsxCre;Asxl1fl/fl recipients (n = 4 mice per genotype). Data represent mean ± s.e.m., ns, not significant, *P < 0.05, **P < 0.01
To further examine whether the impaired HSC/HPCs in OsxCre;Asxl1fl/fl mice are transplantable, we transplanted Asxl1fl/fl and OsxCre;Asxl1fl/fl BM cells into lethally irradiated WT-recipient mice, respectively. Histological analyses of the femur at 6 months post transplantation revealed an accumulation of myeloid cells in the recipients transplanted with OsxCre;Asxl1fl/fl BM cells compared with the recipients transplanted with Asxl1fl/fl BM cells (Supplementary Fig. S3h). In addition, the area with adipocytes was larger in OsxCre;Asxl1fl/fl BM transplanted recipients compared with Asxl1fl/fl BM transplanted recipients (Supplementary Fig. S3h). The increase in adipocyte expansion may be caused by the following reasons: (1) a less proliferative potential of donor HSC/HPCs (OsxCre;Asxl1fl/fl BM) may reduce the marrow cellularity, resulting in an expansion of adipocytes in the BM; (2) the impaired HSC/HPCs of OsxCre;Asxl1fl/fl donor mice produce altered growth factors, which dysregulate cell fates of BMSCs, leading to adipocyte accumulation. Collectively, these results suggest that the Asxl1-deficient niche may mediate permanent marks in hematopoietic cells.
Loss of Asxl1 in BMSCs dysregulates genes required for HSC/HPC maintenance
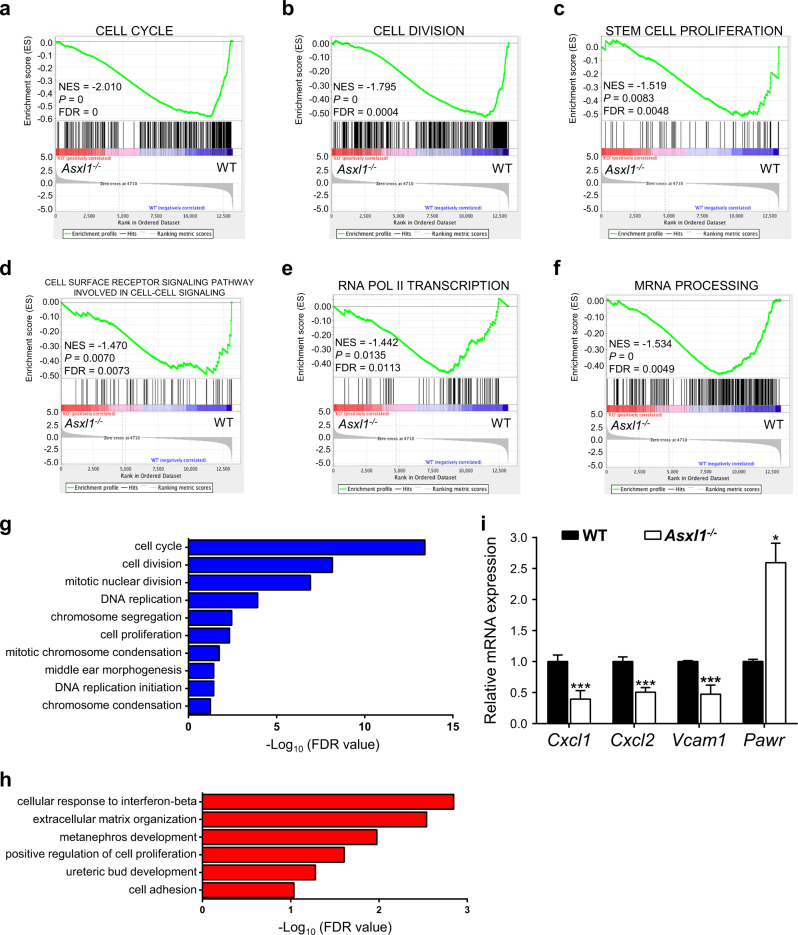
We have previously reported that Asxl1 loss dysregulated transcriptional programs to induce lineage commitment of BMSCs17. To examine if the dysregulated genes in Asxl1−/− BMSCs are associated with the HSC/HPC maintenance, we re-analyzed the RNA-seq data (GSE75787)17. Gene set enrichment analysis (GSEA) showed that the downregulated genes in Asxl1−/− BMSCs were enriched for cell cycle, cell division, stem cell proliferation, cell surface receptor signaling pathway involved in cell–cell signaling, and mRNA transcription signatures (Fig. 4a–f). Gene ontology (GO) analyses revealed that the downregulated genes were associated with cell cycle, cell division, and cell proliferation (Fig. 4g). In contrast, the upregulated genes were associated with cellular response to interferon-beta, extracellular matrix organization, and cell adhesion (Fig. 4h). Interestingly, the genes critical for the function of the BM niche on hematopoiesis, such as Vcam1, Cxcl133, and Cxcl234, were downregulated in Asxl1−/− BMSCs (Fig. 4i; Supplementary Fig. S4a). These data suggest that Asxl1 is required for normal BM niche activity to maintain normal hematopoiesis. Further studies are warranted to investigate the underlying mechanisms by which ASXL1 alteration dysregulates the BM niche, contributing to the impaired HSC/HPC function.
Fig. 4. Loss of Asxl1 in BMSCs alters the expression of genes associated with the HSC/HPC maintenance.
a–f GSEA plots showing downregulated genes in Asxl1−/− BMSCs significantly enriched in the cell cycle (a), cell division (b), stem cell proliferation (c), cell surface receptor signaling pathway involved in cell–cell signaling (d), and mRNA transcription (e, f) signature. The normalized enrichment score (NES), P-value and false discovery rate (FDR) are shown. g, h GO analysis of the down- (g, blue) and up- (h, red) regulated genes in Asxl1−/− BMSCs compared with WT cells (FDR < 0.1). i qPCR shows dysregulated genes expression in Asxl1−/− BMSCs compared with WT BMSCs (n = 3~5 mice for WT, n = 4~6 mice for Asxl1−/−). Data represent mean ± s.e.m., *P < 0.05, ***P < 0.005
ASXL1 functionally associates with RNAPII complex
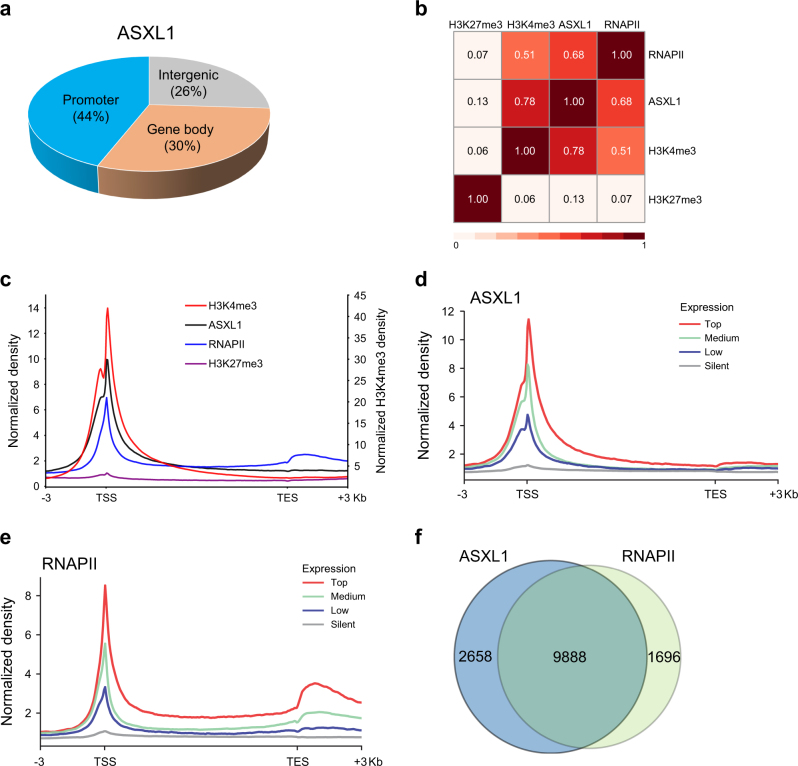
We and others have reported that ASXL1 is mainly located at transcription start sites (TSSs) on the genome of hematopoietic cells7,15. To determine the ASXL1-associated regions on the genome of BMSCs, ChIP-seq assay was performed using anti-ASXL1 antibody. We identified 24,347 ASXL1-binding sites in WT BMSCs. Genome-wide mapping of the ASXL1-binding regions showed forty-four percent of ASXL1-binding sites were localized at the promoter regions (± 3 kilobase (kb) of TSSs, Fig. 5a), suggesting an association of ASXL1 in gene regulation. ASXL1 was reported to bind to PRC2 complex and loss of Asxl1 reduced the global levels of tri-methylation of histone H3 lysine 27 (H3K27me3) and H3K4me3 in myeloid cells5,15,16. To determine the putative impact of ASXL1-mediated H3K27me3 and H3K4me3 on BMSC cell fates, we then performed H3K27me3, H3K4me3, and RNAPII ChIP-seq using WT and Asxl1−/− BMSCs. Surprisingly, we did not find a strong association (r = 0.13) between ASXL1 binding and H3K27me3 enrichment on the genome of BMSCs (Fig. 5b, c; Supplementary Fig. S4b, c). Furthermore, a minimal correlation (r = 0.07) was observed between H3K27me3 enrichment and RNAPII occupancy (Fig. 5b). In contrast, a significant positive association (r = 0.78) was observed between ASXL1 binding and H3K4me3 enrichment (Fig. 5b, c; Supplementary Fig. S4c). However, loss of Asxl1 in BMSCs did not affect the H3K4me3 enrichment on the genome of BMSCs (Supplementary Fig. S4d, e). These results suggest ASXL1 regulates gene expression in an H3K27me3/H3K4me3-independent manner in BMSCs.
Fig. 5. ASXL1 co-localizes with RNAPII in BMSCs.
a Genomic distribution of ASXL1 peaks in WT BMSCs. b Heatmap showing a pairwise correlation between ASXL1, H3K4me3, H3K27me3, and RNAPII in WT BMSCs. Pearson correlation coefficients are given for each comparison. c Average genome-wide occupancies of H3K4me3 (red, right y-axis), ASXL1 (black), RNAPII (blue), and H3K27me3 (purple) on ASXL1-bound genes along the transcription unit in WT BMSCs. d, e ASXL1 (d) and RNAPII (e) occupancies across genes classified by expression. The gene body length is aligned by percentage from the TSS to transcription end site (TES). 3 kb upstream of TSS and 3 kb downstream of TES are also included. f Venn-diagram shows the overlap among target genes of ASXL1 with RNAPII in WT BMSCs
It is recognized that increased gene transcription correlates with increase dynamic supercoiling and demands more recruitment of specific genes35,36. It is possible that more ASXL1 can be found in the promoter regions of active genes. At genes with high (75–100%), medium (25–75%), low (bottom 25% of genes with significant output), or silent (genes with nonsignificant output) expression as measured by RNA-seq, the density of ASXL1 peaks was indeed associated with gene expression levels (Fig. 5d). The distribution of ASXL1 at promoter regions was comparable to that of RNAPII in BMSCs (Fig. 5e). To examine whether ASXL1 and RNAPII co-localize at the same genomic regions, integrative analyses were performed to assess the genome-wide distribution in BMSCs. The results showed a significant positive correlation between ASXL1 binding and RNAPII occupancy (r = 0.68, Fig. 5b). ASXL1 promoter binding was highly overlapped with RNAPII loading (Fig. 5c). Importantly, analyses of the coverage between ASXL1 and RNAPII bound genes showed a high degree (85.36%) of overlay (Fig. 5f; Supplementary Fig S4f, g). These data suggest a role for ASXL1 involving in gene regulation.
ASXL1 interacts with RNAPII in BMSCs
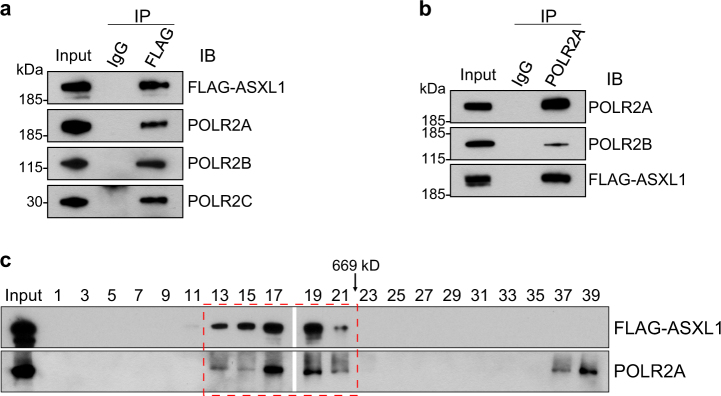
To test whether ASXL1 and RNAPII interact and co-localize in the nucleus of BMSCs, we generated a stable BMSC line that constitutively expresses a FLAG-tagged ASXL1 (FLAG-ASXL1) to overcome the limitation of commercially available ASXL1 antibodies (Supplementary Fig. S4h) and then performed immunoprecipitation (IP) assays. We used anti-FLAG antibody to pull down ASXL1 in WT BMSCs, followed by western blots using antibodies against POLR2A, POLR2B, and POLR2C, the subunits of RNA polymerase II complex. Indeed, ASXL1 associated with endogenous POLR2A, POLR2B, and POLR2C (Fig. 6a). In addition, the reciprocal co-IP using POLR2A antibody further verified the interaction between ASXL1 and RNAPII family members (Fig. 6b). When the fractionated BMSC nuclear extracts by gel filtration chromatography were subjected to western blot analysis, we observed a co-elution of ASXL1 and RNAPII in the high-molecular-weight fractions (Fig. 6c). These data indicate that ASXL1 interacts with RNAPII complex in the nucleus of BMSCs.
Fig. 6. ASXL1 interacts with RNAPII complex in BMSCs.
a Nuclear extracts of WT BMSCs transduced with FLAG-tagged ASXL1 immunoprecipitated (IP) with anti-FLAG or non-immune IgG and probed for FLAG, POLR2A, POLR2B, and POLR2C. b Reciprocal IP and western blotting confirmed interaction of ASXL1 with POLR2A, and POLR2B. Nuclear extracts were subjected to IP using POLR2A antibody. c Gel filtration analysis of nuclear extracts from FLAG-ASXL1 overexpressing cells. ASXL1 and RNAPII complex were co-eluted from a Superose 6 column as analyzed by western blotting. The numbers over the lanes represent the eluted fraction numbers
Loss of Asxl1 alters the target gene expression by deregulating RNAPII transcriptional activity
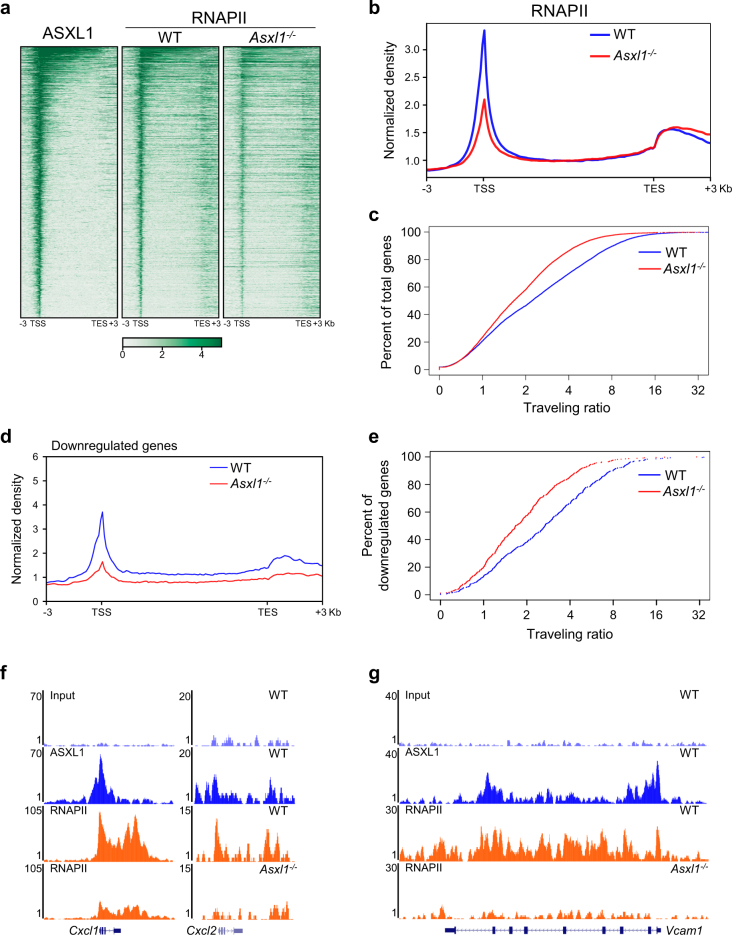
To examine whether loss of Asxl1 affects RNAPII transcriptional activity, we generated heatmaps using ASXL1 and RNAPII ChIP-seq data (Fig. 7a; Supplementary Fig. S5a). The density of RNAPII peaks was significantly decreased, especially on ASXL1-binding genes in Asxl1−/− BMSCs compared with WT BMSCs (Fig. 7a, b; Supplementary Fig. S5a, b). The traveling ratio (TR) or the pausing index represents the relative ratio of RNAPII density in the promoter region and the gene body37–39. To further characterize RNAPII occupancy on the genome of WT and Asxl1−/− BMSCs, we calculated the relative ratio of RNAPII density in the promoter region and the gene body (Supplementary Fig. S5c). A substantial shift in TR was observed in Asxl1−/− BMSCs compared with WT BMSCs, suggesting a decrease in RNAPII density at the transcribed region (Fig. 7c). These results indicate an impact of ASXL1 in regulating RNAPII transcription in BMSCs.
Fig. 7. Loss of Asxl1 dysregulates transcriptional program through RNAPII transcriptional activity.
a Heatmaps of ASXL1 and RNAPII on ASXL1-bound genes in WT and Asxl1−/− BMSCs. The genes ranked from highest to lowest ASXL1 level. b Asxl1 deletion reduces genome-wide RNAPII occupancy. c RNAPII traveling ratio (TR) shows that many genes become less paused in Asxl1−/− BMSCs compared with WT controls. Lower TR values indicate a lower degree of pausing. d Average genome-wide occupancies of RNAPII in WT (blue) and Asxl1−/− (red) BMSCs on downregulated genes along the transcription unit. e RNAPII TR calculations of downregulated genes in WT and Asxl1−/− BMSCs. f, g Representative genome browser tracks showing ASXL1 and RNAPII occupancies on regions of Cxcl1, Cxcl2, and Vcam1
We have previously shown that loss of Asxl1 in BMSCs altered BMSC self-renewal capacity and lineage commitment with a dysregulated transcriptional program17. To correlate the RNAPII status with the dysregulated genes in Asxl1−/− BMSCs, we next performed convergent analyses on RNA-seq and ChIP-seq data. Decreased RNAPII occupancy at transcribing regions was strongly associated with changes in the mRNA expression for the set of downregulated genes in Asxl1−/− BMSCs (Fig. 7d, e). Importantly, ~ 51% of the downregulated genes were ASXL1-binding genes (Supplementary Fig. S5d), which had significant changes of RNAPII occupancy compared with the genes without ASXL1 binding (Supplementary Fig. S5e, f). GO analysis revealed that these ASXL1-binding genes were associated with cell cycle, mRNA transcription, cell division, and stem cell maintenance (Supplementary Fig. S5g). Interestingly, the downregulated genes identified by RNA-seq and qPCR, such as Vcam1, Cxcl1, and Cxcl2, were ASXL1 target genes and had decreased RNAPII signals in Asxl1−/− BMSCs (Fig. 7f, g).
Similarly, the upregulated genes were also associated with the RNAPII transcriptional activity in Asxl1−/− BMSCs (Supplementary Fig. S6a). About 44% of the upregulated genes were ASXL1-binding genes, which had significant changes of RNAPII enrichment compared with the genes without ASXL1 binding (Supplementary Fig. S6b). The upregulated Pawr, an ASXL1 target, had increased RNAPII density in Asxl1−/− BMSCs (Supplementary Fig. S6c). Of note, Runx2 and Cebpa, two key genes controlling osteoblast and adipocyte differentiation were ASXL1 target genes, and their expressions were strongly associated with RNAPII occupancy (Supplementary Fig. S6d, e). Collectively, these data suggest that ASXL1 regulates gene expression by regulating RNAPII transcriptional activity.
Discussion
Somatic mutations of ASXL1 are frequently found in myeloid malignancies40, and loss of Asxl1 in mice leads to MDS-like diseases15,16. Interestingly, systemic deletion of Asxl1 leads to severer hematologic phenotypes than the conditional loss of Asxl1 in hematopoietic cells alone. This led us to hypothesize that loss of Asxl1 in BM niche cooperated with Asxl1-deleted HSC/HPCs to contribute the pathogenesis of myeloid malignancies in vivo. Here, we show that ASXL1 in niche cells is required to maintain HSC/HPC functions, and Asxl1 loss in BMSCs alters the HSC/HPC pool and induces a biased differentiation toward granulocytic/monocytic lineages.
Over the past decade, mutations in epigenetic regulators in the hematopoietic cells have been discovered to be broadly involved in the pathogenesis of myeloid malignancies40,41. However, initiation and progression of myeloid malignancies can also be accelerated by the disruption of the architecture and cellular components in the BM niche32,42–46. As a vital component of the hematopoietic microenvironment, BMSCs are essential for HSC maintenance and regulation of blood production. Geyh et al.47 reported that impaired BMSCs could contribute to hematopoietic insufficiency. In addition, several groups have reported that the BMSCs generated from patients with different myeloid malignancies displayed cellular and functional alterations48,49. For example, BMSCs derived from MDS patients appeared to be impaired in immunomodulatory and supporting hematopoiesis function50. Recently, Arranz et al.51 have demonstrated that depletion of Nestin+ BMSCs or their production of CXCL12 may accelerate the progression of MPNs both in vivo and in vitro.
Inoue et al.52 have reported that the truncated ASXL1 protein can be detected in MDS cells. They also found that C-terminal deletion mutants of ASXL1 induced an MDS-like disease in the transplanted mice53. In the current study, although CMML-BMSCs displayed impaired cellular functions, we did not identify ASXL1 mutation (a cohort of 13 patients) using targeted PCR followed by Sanger sequencing. Further studies to screen the ASXL1 mutations with a larger cohort of patients are warranted. ASXL1, TET2, and NF1 are known to be important in hematopoiesis54,55. We next performed qPCR to compare mRNA levels of ASXL1, TET2, and NF1 between CMML-BMSC and HD-BMSC. Interestingly, significant downregulation of ASXL1 was found in CMML-BMSCs compared with HD-BMSCs, while the expression of TET2 was identical between these two types of BMSCs. It has been reported that SOX2 directly binds to ASXL1 promoter regions and positively regulates ASXL1 expression56. Inoue et al.57demonstrated that USP7, a deubiquitinating enzyme, plays an important role in maintaining stabilization of ASXL1. Knockdown of USP7 reduced ASXL1 expression in HL60 cells. However, the mechanism of dysregulated ASXL1 expression in CMML-BMSCs requires further investigation.
Deletion of Asxl1 in the BM niche (OsxCre;Asxl1fl/fl) significantly increased the GMP population with skewed differentiation favoring the granulocytic/monocytic lineages. Ye and colleagues26 recently showed that skewed myeloid differentiation is a prerequisite for leukemic stem cell formation and leukemia development. Consistently, skewed expansion of GMP population is associated with higher risks of leukemic transformation in MDS patients27,28. The hematopoietic phenotypes observed in OsxCre;Asxl1fl/fl mice indicate that ASXL1 is required for the maintenance of normal function of the BM niche, and Asxl1 loss in the niche may contribute to the pathogenesis of myeloid malignancies.
The phenotype and cell fate of a given cell rely on the precise control of gene expression by complex transcriptional and epigenetic networks, which is essential for proper differentiation, cellular function, development, and homeostasis58,59. Although extensive studies have focused on the identification of intrinsic targets of transcription factors regulating BMSC cell fate and functions, the epigenetic events that control BMSC identity and/or functions remain largely unknown. Here, our data demonstrate that ASXL1 works in concert with RNAPII complex to maintain normal gene expression, and loss of Asxl1 impaired the genome-wide occupancy of RNAPII. We found that deletion of Asxl1 in BMSCs dramatically deregulated RNAPII transcriptional activity and resulted in the up- and downregulation of genes critical for BMSC cell fates and biological functions. The interaction between ASXL1 and the core subunits of RNAPII complex supports the notion of ASXL1 involving in gene regulation. We and others have previously reported that loss of Asxl1 reduced global H3K27me3 levels in myeloid progenitor cells5,15,16. To our surprise, we did not observe a significant positive correlation between ASXL1 and H3K27me3 enrichment on the genome of BMSCs, suggesting a cell-lineage dependent association between ASXL1 and interacting partners in the nucleus.
Collectively, our results demonstrate that ASXL1 in the BM niche is required for normal hematopoiesis and Asxl1 deletion in BMSCs alters HSC/HPC pool and leads to a myeloid differentiation bias. These data provide a mechanistic basis for ASXL1 functions in the BM niche to maintain normal hematopoiesis; and ASXL1 alteration in, at least, a subset of the niche cells induces myeloid differentiation bias, thus, contributes the progression of myeloid malignancies. This work identifies a novel ASXL1-binding partner, RNAPII protein complex, through which ASXL1 loss leads to dysregulated gene expression. Future efforts are warranted to investigate if ASXL1 mutation/dysregulation in the BM niche of patients contributes to the pathogenesis of myeloid malignancies, which will shed light on identifying novel therapeutic strategies for patients with ASXL1 alterations.
Materials and methods
Mouse models and reagents
The generation of Asxl1fl/fl mice have been previously described15. Osx-Cre transgenic mice were purchased from Jackson Laboratory. All of the murine models were bred on a C57BL/6 genetic background. All studies were conducted in accordance with the regulatory guidelines by the Institutional Animal Care and Use Committee at the University of Miami Miller School of Medicine. Chemicals were obtained from Sigma unless otherwise indicated.
Patients
Thirteen patients with CMML (ten males and three females) and ten healthy donors (seven males and three females) were included in this study. The study was approved by the Ethics Committee of Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences according to guidelines of the 1975 Helsinki Declaration, and informed consent was received according to the institute’s guidelines on the use of human subjects. All patients were reevaluated and met the 2008 WHO diagnostic criteria.
Isolation and expansion of BMSCs
Whole BM cells from human CMML patients and healthy donors were cultured at 4 × 106 cells/well in 6-well plate at 37 °C, 5% CO2, 5% O2 in a fully humidified atmosphere in expansion medium Dulbecco’s modified Eagle medium (DMEM)/F12 (Gibco), supplemented with 10% fetal bovine serum (FBS, Hyclone), 1x insulin transferrin selenium-A (Life Technologies), 10 ng/mL human epidermal growth factor (EGF, Peprotech), and 10 ng/mL human platelet derived growth factor-BB (PDGF-BB, Peprotech). After 10 days’ culture, CFU-fibroblasts (CFU-F) were counted to measure the frequency of BMSCs in the BM following Wright-Giemsa staining as previously reported17 and photographs were taken by a Fujifilm digital camera (FinePix 2400Zoom, Fujifilm, Tokyo, Japan). For functional analysis, cells were trypsinized and replated upon reaching 80% confluence and BMSCs at passage three to five were used for the following experiments. Generation of full-length ASXL1 complementary DNA overexpressing BMSCs were performed as previously described17.
Phenotypic analysis of BMSC surface markers
The phenotypic analyses of BMSCs were performed by evaluating the expression of surface markers on a BD LSR II flow cytometer (BD Biosciences). In brief, BMSCs were incubated with antibodies against CD45 (FITC), CD34 (APC), CD73 (PE), CD105 (APC), CD44 (APC-H7), and CD29 (PE-Cy5) (BD PharMingen) at 4 °C for 30 min. The cells were then washed three times with PBS containing 0.1% bovine serum albumin (BSA) and analyzed by flow cytometry.
Differentiation assay
To induce osteogenic differentiation, BMSCs were plated at 5 × 104/well in osteogenic differentiation medium, which is DMEM/F12 supplemented with 10% FBS, 10−8 M dexamethasone, 10 mM β-glycerophosphoric acid, and 100 μM ascorbic acid in 6-well plate for 1 week. Cells were stained with Alkaline Phosphatase Kit according to the manufacturer’s instruction.
Cell proliferation assays
To study the proliferative ability of the BMSCs, cell growth was evaluated by manually counting at each passage. Briefly, 3 × 105 BMSCs were seeded in the 60 mm2 dish and passaged once the culture reached 80–90% confluence. Cell numbers were recorded and cells were replated at the same density each time. Cell proliferation was also examined by EdU assay. In brief, 1.5 × 105 BMSCs were plated in 20 mm glass bottom dish (NEST Biotechnology, China) and cultured overnight. After incubation with EdU for another 36 h, the cells were fixed in 4% paraformaldehyde for 15 min at room temperature, washed twice with 3% BSA in PBS, incubated with 0.5% Triton X-100 in PBS for 20 min and stained with the Click-iT EdU Imaging Kit (Life Technologies) according to the manufacturer’s instruction. Nuclear staining was performed with 4′, 6-diamidino-2-phenylindole (DAPI). Photomicrographs were taken by Laser confocal fluorescence microscopy (Leica, Germany). The percentage of proliferating cells was calculated based on counting EdU-positive cells/total cells in five individual visual fields.
Senescence assay
Histochemical staining for senescence-associated β-galactosidase activity was used to measure the senescence of BMSCs. BMSCs were plated in 6-well plate at 5 × 104 cells/well and cultured for 12 h at 37 °C, 5% CO2, and 5% O2. Cells were then stained with the Senescence Staining Kit (Beyotime Institute of Biotechnology, China) according to the manufacturer’s instructions. Senescent cells displayed a blue staining in the cytoplasm. The percentage of senescent cells was calculated based on counting β-galactosidase-positive cells/total cells in five individual visual fields.
Cell cycle analysis
Cell cycle analysis was performed using an APC BrdU Flow Kit (BD PharMingen). In brief, 2 × 105 cells were seeded in a T25 flask and cultured for 12 h, BrdU was added 2 h before culture termination, and then nuclear staining was performed with 7-amino-actinomycin D (7-AAD) according to the manufacturer's instruction. Data were acquired on BD LSR II and analyzed by FlowJo.
Long-term co-culture of HSC/HPCs with BMSCs
The function of BMSCs to support cord blood (CB) CD34+ cells was measured by cobblestone-area-forming cell (CAFC) assay and LTC-IC assay. In brief, mononuclear cells of CB were separated by Ficoll-Hypaque density gradient centrifugation. CD34+ cells were purified by using magnetic microbeads following the manufacturer’s instructions (Miltenyi, Germany). BMSCs (7000 cells/well) were irradiated with 20 Gy after reaching 90% confluence, and 500 CB CD34+ cells were inoculated on top of BMSCs in Iscove modified Dulbecco medium (IMDM, Gibco) supplemented with 10% horse serum (Gibco), 10% FBS, 10−6 M hydrocortisone, incubating at 33 °C and 5% CO2. After 5 weeks of weekly half-media replacement, the areas containing phase-dim hematopoietic clones (at least five cells) beneath the BMSCs layer were counted as CAFCs. LTC-IC assay was performed by plating all the cells from one co-culture well in H4435 methylcellulose (STEMCELL Technologies, Canada) in one well of a 24-well plate for 14 days.
Phenotypic analyses of the hematologic system in mice
PB was collected by retro-orbital bleeding and total white blood cells were obtained after lysis of PB with red cell lysis buffer. Single-cell suspensions from BM, spleen, and PB were stained with panels of fluorochrome-conjugated antibodies (Supplementary Table S2). Flow cytometric analysis of HSC/HPCs was performed as previously described60. Dead cells were excluded by DAPI staining. The analyses were performed using a BD FACS Canto II or LSR Fortessa flow cytometer. For cell apoptosis analysis, freshly isolated BM cells were stained with the PE-Annexin V/7-AAD Apoptotic Kit (BD Biosciences) according to the protocol and analyzed with LSK subpopulation. All data were analyzed by FlowJo-V10 software. For CFU assays, BM cells were plated in triplicate in methylcellulose medium (Methocult M3134, STEMCELL Technologies) supplemented with mouse stem cell factor (100 ng/mL), human interleukin 6 (50 ng/mL), mouse interleukin 3 (5 ng/mL), erythropoietin (4 U/mL), thrombopoietin (100 ng/mL), granulocyte-macrophage-colony-stimulating factor (10 ng/mL, Peprotech) and scored on day 7 of the cultures61.
Gene expression analysis
Total RNA was extracted with TRIzol reagent (Invitrogen), and 1 µg RNA was used for each reverse transcription PCR amplifications with the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. qPCR was performed in triplicate using an ABI 7500 with the Fast SYBR green master mix (Applied Biosystems). Mouse β-actin and human GAPDH were used as the reference genes. Primers used are listed in Supplementary Table S3 and gene expression was calculated by the 2−ΔΔCt method.
Immunoprecipitation (IP) and western blot
IP was performed using nuclear fraction buffer and antibodies (monoclonal mouse anti-FLAG, Sigma, F3165, clone M2, 1:200; and monoclonal mouse anti-POLR2A, ThermoFisher Scientific, MA1-46093, clone 4H8, 1:200). After washing with IP buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 2 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 2 mM NaF, and protease inhibitor cocktail) four times, the associated proteins were collected for western blot analysis. Antibodies against the following proteins were used: anti-FLAG (1:1000); anti-POLR2A (1:1000); polyclonal rabbit anti-POLR2B (ThermoFisher Scientific, PA5-21446, 1:500); and polyclonal rabbit anti-POLR2C (Abcam, ab138436, 1:2000).
Gel filtration chromatography
The nuclear extracts were prepared and concentrated to ~ 0.2 mL, and then loaded onto a superose 6 GL 10/300GL column (GE Healthcare) equilibrated at 4 °C with buffer A (20 mM Tris-HCl, 75 mM NaCl, pH 7.8) using an AKTApurifer machine (GE Healthcare). The sample was eluted with buffer A. The column fractions (1 mL each) were subjected to concentrate with Amicon Ultra-0.5 mL Centrifugal Filters (Millipore) and analyzed by western blot.
RNA-seq analysis
Cuffdiff62 was used to detect the differentially expressed genes with a cutoff of P < 0.05 and fold change >2. The identified differentially expressed genes were used for pathway enrichment analysis and functional annotation of the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources63. GSEA64 was performed with gene signatures in GSEA/MSigDB v6.0, including KEGG pathway signatures and GO signatures. Enriched gene sets or pathways were selected using a cutoff of P < 0.05 and FDR < 0.25.
Chromatin immunoprecipitation assays
BMSCs were fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated and sonicated to an average length of 300 to 500 bp. An aliquot of chromatin was precleared with protein A agarose beads (Invitrogen). Genomic DNA regions of interest were isolated using ASXL1 (Santa Cruz #sc-85283), RNAPII (Abcam #ab5095), H3K4me3 (Active Motif #39159), and H3K27me3 (Millipore #07-449) antibodies. Complexes were eluted from the beads and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65 °C, and ChIP DNA was purified by phenol–chloroform extraction and ethanol precipitation. Illumina sequencing libraries were prepared and sequenced on NextSeq 500. The sequence reads were aligned to the mouse genome (mm9) using the BWA algorithm (default settings). ASXL1, RNAPII, H3K4me3, and H3K27me3 peaks were called using the MACS2 program65 with default settings. The resulting histograms (genomic “signal maps”) were stored in bigWig files. The UCSC genome browser was used for data visualization. Heatmap and profile plot were generated by deepTools66. The accession number for the ChIP-seq data is NCBI GEO: GSE99103.
Statistical analysis
Differences between experimental groups were determined by Student’s t-test or analysis of variance followed by Newman-Keuls multiple comparison tests as appropriate. P-values of <0.05 were considered significant.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Institutes of Health (CA172408 and CA185751 to F.-C.Y. and M.X., and HL112294 to M.X.), the Ministry of Science and Technology of China (2016YFA0100600 to T.C.), the National Natural Science Foundation of China (81421002 to T.C, and 81570113 to Y.Z.), and Tianjin Municipal Science and Technology Commission (16JCZDJC34500 to J.B.). We thank the services provided by the Satellite Histological Core and Flow Cytometry Core Facilities of Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine.
Authors' contributions
F.-C.Y. conceived the project. P.Z., Y.Z., and F.-C.Y. designed the study. P.Z., R.L., Y.G., H.S., H.Y., M.S., Z.L., Z.L., J.L., and S.C. performed the experiments. P.Z. and Z.C. performed the RNA-seq and ChIP-seq analyses. P.Z., Z.C., R.L., J.B., W.Y., T.C., M.X., Y.Z., and F.-C.Y. discussed and analyzed the data. P.Z., Y.Z., and F.-C.Y. wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
Footnotes
Peng Zhang, Zizhen Chen and Rong Li contributed equally to this work
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Information accompanies the paper at (10.1038/s41421-017-0004-z).
Contributor Information
Peng Zhang, Email: pxz36@med.miami.edu.
Yuan Zhou, Email: yuanzhou@ihcams.ac.cn.
Feng-Chun Yang, Phone: +1 305 243 7836, Email: fxy37@med.miami.edu.
References
- 1.Milne TA, Sinclair DA, Brock HW. The Additional sex combs gene of Drosophila is required for activation and repression of homeotic loci, and interacts specifically with Polycomb and super sex combs. Mol. General Genet.: MGG. 1999;261:753–761. doi: 10.1007/s004380050018. [DOI] [PubMed] [Google Scholar]
- 2.Sinclair DA, Milne TA, Hodgson JW, et al. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development. 1998;125:1207–1216. doi: 10.1242/dev.125.7.1207. [DOI] [PubMed] [Google Scholar]
- 3.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Katoh M. Functional and cancer genomics of ASXL family members. Br. J. Cancer. 2013;109:299–306. doi: 10.1038/bjc.2013.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Wahab O, Adli M, LaFave LM, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Zhang P, Yan A, et al. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Sci. Adv. 2017;3:e1601602. doi: 10.1126/sciadv.1601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbuccia N, Murati A, Trouplin V, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–2186. doi: 10.1038/leu.2009.141. [DOI] [PubMed] [Google Scholar]
- 9.Boultwood J, Perry J, Pellagatti A, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24:1062–1065. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- 10.Chou WC, Huang HH, Hou HA, et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. 2010;116:4086–4094. doi: 10.1182/blood-2010-05-283291. [DOI] [PubMed] [Google Scholar]
- 11.Gelsi-Boyer V, Trouplin V, Adelaide J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 12.Gelsi-Boyer V, Trouplin V, Roquain J, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br. J. Haematol. 2010;151:365–375. doi: 10.1111/j.1365-2141.2010.08381.x. [DOI] [PubMed] [Google Scholar]
- 13.Hoischen A, van Bon BW, Rodriguez-Santiago B, et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 2011;43:729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- 14.Russell B, Johnston JJ, Biesecker LG, et al. Clinical management of patients with ASXL1 mutations and Bohring-Opitz syndrome, emphasizing the need for Wilms tumor surveillance. Am. J. Med. Genet. A. 2015;167A:2122–2131. doi: 10.1002/ajmg.a.37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O, Gao J, Adli M, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Li Z, He Y, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123:541–553. doi: 10.1182/blood-2013-05-500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, Xing C, Rhodes SD, et al. Loss of asxl1 alters self-renewal and cell fate of bone marrow stromal cell, leading to bohring-opitz-like syndrome in mice. Stem Cell Rep. 2016;6:914–925. doi: 10.1016/j.stemcr.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 19.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 20.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 22.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 23.Naveiras O, et al. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dazzi F, Ramasamy R, Glennie S, Jones SP, Roberts I. The role of mesenchymal stem cells in haemopoiesis. Blood Rev. 2006;20:161–171. doi: 10.1016/j.blre.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Kfoury Y, Scadden DT. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Ye M, Zhang H, Yang H, et al. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell. 2015;17:611–623. doi: 10.1016/j.stem.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goardon N, Marchi E, Atzberger A, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Will B, Zhou L, Vogler TO, et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120:2076–2086. doi: 10.1182/blood-2011-12-399683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, et al. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS ONE. 2014;9:e85161. doi: 10.1371/journal.pone.0085161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizoguchi T, Pinho S, Ahmed J, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev. Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinclair A, Park L, Shah M, et al. CXCR2 and CXCL4 regulate survival and self-renewal of hematopoietic stem/progenitor cells. Blood. 2016;128:371–383. doi: 10.1182/blood-2015-08-661785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmadzadeh A, Kast RE, Ketabchi N, et al. Regulatory effect of chemokines in bone marrow niche. Cell Tissue Res. 2015;361:401–410. doi: 10.1007/s00441-015-2129-4. [DOI] [PubMed] [Google Scholar]
- 35.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baranello L, Wojtowicz D, Cui K, et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Zeitlinger J, Stark A, Kellis M, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahl PB, Lin CY, Seila AC, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 41.Woods BA, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Immunol. Rev. 2015;263:22–35. doi: 10.1111/imr.12246. [DOI] [PubMed] [Google Scholar]
- 42.Dong L, Yu WM, Zheng H, et al. Leukaemogenic effects of Ptpn11 activating mutations in the stem cell microenvironment. Nature. 2016;539:304–308. doi: 10.1038/nature20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiozawa Y, Taichman RS. Dysfunctional niches as a root of hematopoietic malignancy. Cell Stem Cell. 2010;6:399–400. doi: 10.1016/j.stem.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Walkley CR, Olsen GH, Dworkin S, et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129:1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B, Ho YW, Huang Q, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geyh S, Rodriguez-Paredes M, Jager P, et al. Functional inhibition of mesenchymal stromal cells in acute myeloid leukemia. Leukemia. 2016;30:683–691. doi: 10.1038/leu.2015.325. [DOI] [PubMed] [Google Scholar]
- 48.Borojevic R, Roela RA, Rodarte RS, et al. Bone marrow stroma in childhood myelodysplastic syndrome: composition, ability to sustain hematopoiesis in vitro, and altered gene expression. Leuk. Res. 2004;28:831–844. doi: 10.1016/j.leukres.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Zhao ZG, Liang Y, Li K, et al. Phenotypic and functional comparison of mesenchymal stem cells derived from the bone marrow of normal adults and patients with hematologic malignant diseases. Stem Cells Dev. 2007;16:637–648. doi: 10.1089/scd.2007.0008. [DOI] [PubMed] [Google Scholar]
- 50.Zhao ZG, Xu W, Yu HP, et al. Functional characteristics of mesenchymal stem cells derived from bone marrow of patients with myelodysplastic syndromes. Cancer Lett. 2012;317:136–143. doi: 10.1016/j.canlet.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 51.Arranz L, Sanchez-Aguilera A, Martin-Perez D, et al. Neuropathy of haematopoietic stem cell niche is essential for myeloproliferative neoplasms. Nature. 2014;512:78–81. doi: 10.1038/nature13383. [DOI] [PubMed] [Google Scholar]
- 52.Inoue D, Matsumoto M, Nagase R, et al. Truncation mutants of ASXL1 observed in myeloid malignancies are expressed at detectable protein levels. Exp. Hematol. 2016;44:172–176. doi: 10.1016/j.exphem.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Inoue D, Kitaura J, Togami K, et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J. Clin. Invest. 2013;123:4627–4640. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The Ten-eleven translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28:485–496. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- 55.Shannon KM, O'Connell P, Martin GA, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N. Eng. J. Med. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 56.Kang M, Lee SW, Kim EJ, Um SJ. Positive regulation of additional sex comb-like 1 gene expression by the pluripotency factor SOX2. Biochem. Biophys. Res. Commun. 2012;421:621–626. doi: 10.1016/j.bbrc.2012.04.072. [DOI] [PubMed] [Google Scholar]
- 57.Inoue D, Nishimura K, Kozuka-Hata H, Oyama M, Kitamura T. The stability of epigenetic factor ASXL1 is regulated through ubiquitination and USP7-mediated deubiquitination. Leukemia. 2015;29:2257–2260. doi: 10.1038/leu.2015.90. [DOI] [PubMed] [Google Scholar]
- 58.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 59.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, He F, Zhang P, et al. Loss of Asxl2 leads to myeloid malignancies in mice. Nat. Commun. 2017;8:15456. doi: 10.1038/ncomms15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi H, Yamamoto S, Sheng M, et al. ASXL1 plays an important role in erythropoiesis. Sci. Rep. 2016;6:28789. doi: 10.1038/srep28789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 64.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramirez F, Ryan DP, Gruning B, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.