Abstract
The use of defined conditions for derivation, maintenance, and differentiation of human-induced pluripotent stem cells (hiPSCs) provides a superior experimental platform to discover culture responses to differentiation cues and elucidate the basic requirements for cell differentiation and fate restriction. Adoption of defined systems for reprogramming, undifferentiated growth, and differentiation of hiPSCs was found to significantly influence early stage differentiation signaling requirements and temporal kinetics for the production of primitive neuroectoderm. The bone morphogenic protein receptor agonist LDN-193189 was found to be necessary and sufficient for neural induction in a monolayer system with landmark antigens paired box 6 and sex-determining region Y-box 1 appearing within 72 h. Preliminary evidence suggests this neuroepithelium was further differentiated to generate ventral spinal neural progenitors that produced electrophysiologically active neurons in vitro, maintaining viability posttransplantation in an immunocompromised host. Our findings support current developments in the field, demonstrating that adoption of defined reagents for the culture and manipulation of pluripotent stem cells is advantages in terms of simplification and acceleration of differentiation protocols, which will be critical for future clinical translation.
Keywords: human-induced pluripotent stem cells, ventral spinal neurons, neural differentiation
Derivation and differentiation of human-induced pluripotent stem cells (hiPSCs)1,2 provide the opportunity to study otherwise inaccessible developmentally transient progenitor cells useful for understanding of human embryogenesis and also potential of therapeutic intervention in cases of injury or disease. Evidence suggests that iPSC-derived progenitors function similarly to their fetal-derived counterparts3,4 and possess the ability to engraft and survive in a human host, making it perhaps possible to alleviate various pathological conditions caused by cell loss or dysfunction. Terminal maturation of iPSC-derived progenitors in vitro can also support chemical screening for identification of novel therapeutic drugs. These efforts are especially important for conditions of the central nervous system where tissue acquisition from living patients is problematic.
Platforms for cell reprogramming and differentiation are a central aspect of both research and potential translation and have matured in recent years to incorporate increasingly defined reagents. Undefined reagents used in medium supplementation and for cellular anchorage are now being replaced with defined compositions. While these changes may help mitigate unpredictable and inconsistent outcomes that negatively impact the construction and reproducibility of stem cell differentiation protocols, it is important to establish material equivalence. Maintenance and loss of self-renewal,5 differentiation kinetics, and trajectory may be impacted6 by the elimination of unknown factors present in commonly used undefined reagents.
Co-inhibition of receptors to bone morphogenetic protein (BMP) and transforming growth factor β (TGFβ) has been shown to result in diminished downstream sma (a drosophila gene name that cannot be further elucidated) mothers against decapentaplegic (SMAD) signaling and has been widely utilized to promote formation of primitive neural epithelium from human pluripotent stem cells (hPSCs).7 Termed “dual-SMAD inhibition,” defining characteristics of this strategy are the efficient production of neural progenitors from hPSCs in an anchorage-dependent manner, eliminating the need for 3-dimentional multicellular aggregation techniques. A commonly preferred substrate during this process is a murine tumor extract consisting of undefined mixtures of growth factors and basement membrane proteins.8 Recent advances in undifferentiated hPSC growth systems now provide defined anchorage motifs,5,9 but the extent to which these systems influence differentiation protocols requires examination.
We evaluated defined media and substrates for reprogramming and growth of undifferentiated hiPSCs10 as well as their anchorage-dependent differentiation into neural progenitors. We found that recombinant human vitronectin (rhVTN) is effective for both growth and neural differentiation of hiPSCs and discovered unexpectedly that inhibition of BMP receptors alone with the selective activin-like kinase 2 and 3 inhibitor LDN-193189 proved sufficient for neural induction. Furthermore, acquisition of primitive neuroepithelial markers paired box 6 (PAX6)/sex-determining region Y-box 1 (SOX1) was seen to be accelerated relative to previous reports.7 This neural population could then be further directed to acquire a ventral spinal fate based on transcriptome data and survived following transplantation into an athymic nude rat injury/engraftment assay. Our results highlight the potential challenges involved with translating long-established protocols to defined systems. Utilization of alternative materials may require reevaluation of both signaling requirements and differentiation kinetics during the process of differentiation.
Materials and Methods
Reprogramming of Human Dermal Fibroblasts into iPSCs
Neonatal human dermal fibroblasts isolated from the foreskin of a single, male donor (Lonza CC-2509, Basel, Switzerland) were grown in FibroGRO Defined Fibroblast Growth Medium (Millipore SCM044, Billerica, MA, USA). Fibroblasts were dissociated with TryPLE (12563011; Thermo Fisher Scientific, Waltham, MA, USA) and seeded at a density of 50,000 cells per 10 cm2 well. After 24 h, fibroblasts were transduced with CytoTune 2.0 Sendai (SeV) vectors (A16518; Thermo Fisher Scientific), bearing transgenes for pituitary-specific octamer unc-86 class 5 homeobox 1 (POU5F1), SOX2, Kruppel-like factor 4 (KLF4), and myc proto-oncogene according to manufacturer recommendations. After 24 h, viral supernatants were aspirated and medium replaced with FibroGRO. Fibroblasts became dense by the sixth day and were passaged 1:6 onto tissue culture–treated plates (Fischer Scientific, Hampton, NH, USA) coated with rhVTN (AF-140-09; PeproTech, Rocky Hill, NJ, USA). The following day, media was exchanged for Essential 8 Flex (A2858501, Flex; Thermo Fisher Scientific). Cultures were monitored for the emergence of hiPSC clones over the next 2 wk. iPSC colonies were identified morphologically and dissected using a pipet tip. Individual colonies were scraped into culture medium, and the medium transferred to tissue culture–treated 48-well plates (Fischer Scientific) coated with rhVTN (AF-140-09; PeproTech) at a concentration of 0.5 µg/cm2 at room temperature for 1 h. Cultures were fed daily. Every third day, cultures were passaged with an in-house formulation of hypertonic citrate buffer10 of 1:2 to 1:3 while expanding into T25 flasks (Fischer Scientific). Cultures were maintained in T25 flasks for characterization and differentiation and expanded into T75 flasks (Fischer Scientific) for banking.
hiPSC Culture
Tissue culture–treated plates (Fischer Scientific) were coated with rhVTN (AF-140-09; PeproTech) at a concentration of 0.5 µg/cm2 at room temperature for 1 h. The iPSCs were washed twice and then incubated with hypertonic citrate buffer10 at 37 °C for 10 min until colony detachment occurred. The colonies were processed into smaller fragments by trituration and the dissociation quenched with Essential 6 Medium (Thermo Fisher Scientific A1516401). Suspensions were collected into 15 mL conical tubes and centrifuged for 1 min at 200× g. Supernatants were aspirated and pellets resuspended into an appropriate volume of Essential 8 Medium (A2858501; Thermo Fisher Scientific) according to split ratio, generally 1:5 to 1:6. Cultures were carefully redistributed onto rhVTN-coated plates. Cultures were maintained in Essential 8 Medium with daily media changes and passaged every 3 d.
Quantitative Real-time Polymerase Chain Reaction (qRT-PCR) for Sendai Loss
At confluence, RNA was extracted from hiPSCs using RNeasy kit according to manufacturer instructions (Qiagen, 74104, Hilden, Germany). cDNA was produced using SuperScript™ III Reverse Transcriptase (18080093; Thermo Fisher Scientific). Prime-time quantitative polymerase chain reaction (qPCR) assays (Integrated DNA Technologies, Coralville, IA, USA) were ordered using the following sequences: SeV ALL (5′-TGG CTA AGA ACA TCG GAA GG-3′, 5′-GTT TTG CAA CCA AGC ACT CA-3′, probe: ACA CCA CCT GGC AGT CGG AGC),11 glyceraldehyde-3-phosphate dehydrogenase (GAPDH; (5′-ACA TCG CTC AGA CAC CAT G-3′, 5′-GGG AAG TAA CTG GAG TTG ATG T-3′, probe: AAG GTC GGA GTC AAC GGA TTT GGT C). cDNA was combined with PrimeTime Master Mix (Integrated DNA Technologies), and amplification took place using an Mastercycler RealPlex2 (Eppendorf, Hamburg, Germany) with the following program: 50 °C 2 min, 95 °C 10 min, and 40 cycles of 95 °C 15 s, 59 °C 1 min, and 72 °C 15 s. Raw cycle threshold (CT) values were compared to control samples with a sample determined to have cleared SeV vector, when no SeV-specific amplification was seen following 40 cycles.
qRT-PCR for Gene Expression Analysis
At confluence, RNA was extracted from hiPSCs using RNeasy kit according to manufacturer instructions (Qiagen, 74104). cDNA was produced using SuperScript™ III Reverse Transcriptase (18080093; Thermo Fisher Scientific). qPCR was performed with FastStart Universal SYBR Green Master (Rox; 4913850001; MilliporeSigma, Billerica, MA, USA) with the following primers: POU5F1 (5′-CCT CAC TTC ACT GCA CTG TA-3′,5′-CAG GTT TTC TTT CCC TAG CT-3′), SOX2 (5′-GGG AAA TGG GAG GGG TGC AAA AGA GG-3′,5′-TTG CGT GAG TGT GGA TGG GAT TGG TG-3′), Nanog (5′-GAA ATA CCT CAG CCT CCA GC-3′,5′- GCG TCA CAC CAT TGC TAT TC-3′), lin-28 homolog A (LIN28) (5′-GCA GAA GCG CAG ATC AAA AG-3′,5′-CGG ACA TGA GGC TAC CAT ATG-3′), and GAPDH (5′-GTG GAC CTG ACC TGC CGT CT-3′,5′-GGA GGA GTG GGTGTC GCT GT-3′). An Mastercycler RealPlex2 was used for amplification (Eppendorf), with the following program: 50 °C 2 min, 95 °C 10 min, and 40 cycles of 95 °C 15 s, 59 °C 1 min, and 72 °C 15 s. Delta–delta CT method was used with an established reference hiPSC line for comparison.
hiPSC Karyotyping
Karyotyping was performed at the University of Minnesota, Masonic Cancer Center, Cytogenomics Core Laboratory, according to standard cytogenetic protocol. Briefly, adherent iPSCs cultured in three 30-mm wells of a 6-well plate were treated for 3.5 h with colcemid. Cells were harvested, and 20 metaphases were completely analyzed by G-banding at a 400 to 425 band-level resolution. Two metaphases were karyotyped and examined for gross chromosomal abnormalities.
hiPSC Teratoma Formation Assay
hiPSCs were single-cell dissociated with Accutase (A1110501; Thermo Fisher Scientific) and resuspended into Flex medium at a concentration of 20 million cells per mL. This suspension was mixed 1:1 into undiluted Matrigel™ (08-774-552; Fisher Scientific). Cell suspensions were loaded into a sterile 1-cc syringe with 23-gauge, ½-in. needle kept on ice. Three male nonobese diabetic severe-combined immunodeficient (NOD-SCID) mice were anesthetized with isoflurane. Cell suspension of 150 μL was injected subcutaneously into the lower hind leg. Mice were maintained 6 to 12 wk until palpable tumors measuring 5 mm in diameter were grown. Mice were killed and tumors extracted. Samples were submitted to the University of Minnesota, Masonic Cancer Center, Comparative Pathology Shared Resource for histology, where they were embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Complex teratomas comprising tissues originating from the 3 embryonic germ layers were verified by a clinical pathologist.
Differentiation of hiPSCs into human VSNPs (hVSNPs)
For differentiation, iPSC cultures were passaged onto rhVTN at half the colony density used for maintenance (1:6 ratio). After 18 to 24 h, culture media was aspirated and exchanged for stage 1 differentiation medium consisting of Essential 6 supplemented with LDN-193189 (250 nM, S7507; Selleckchem, Houston, TX, USA). Cultures were maintained 3 d in this medium with daily media exchanges and passaged 1:10 onto rhVTN on the third day. Following passage, medium was exchanged for stage 2 medium consisting of Essential 6 supplemented with 250 nM LDN-193189, retinoic acid (RA, 100 nM, Sigma R2625-50MG, Billerica, MA, USA), and CHIR99021 (3 mM, Bio-Techne 4423, Minneapolis, MN, USA). Medium was changed daily. Following 3 d in culture, colonies were seen to proliferate from their centers, resulting in a dome-like appearance (stage 3). By day 11, the centers detached as spherical cell aggregates, were collected, and resuspended into stage 4 medium consisting of Dulbecco’s modified Eagle’s medium (DMEM) F/12 basal (11039-047; Thermo Fisher Scientific) containing 1× N2 ( A13707-01; Thermo Fisher Scientific), 1× B27 (17504-044; Thermo Fisher Scientific), 100 nM RA, and smoothened agonist (SAG, 1 μM, 11914; CaymanChem, Ann Arbor, MI, USA). The cell spheres were placed into suspension culture in ultralow attachment plates (Fisher Scientific 3471) for an additional 6 d. Tissue culture–treated dishes were incubated with a poly-ornithine hydrobromide solution (50 μg/mL, P3655-10MG; Sigma) for 2 h, the solution aspirated, washed twice with water, and then coated with laminin (20 μg/mL, Sigma L2020) for 1 h. The laminin solution was aspirated, and the aggregates were plated for attachment into stage 5 medium comprising DMEM F/12 supplemented with N2, B27, and neurotrophin 3 (20 ng/mL, 450-03; PeproTech), brain-derived neurotrophic factor (BDNF, 20 ng/mL, 450-02; PeproTech), ciliary neurotrophic factor (CNTF, 20 ng/mL, 450-13; PeproTech), and glial-derived neurotrophic factor (GDNF, 20 ng/mL, 450-10; PeproTech). Cultures were maintained as such for later analysis.
Cryopreservation
iPSCs were cryopreserved by centrifugation of passaged colonies to form cell pellets. Pellets were resuspended in Flex medium with 10% v/v dimethyl sulfoxide (DMSO; Sigma). Stage 4 differentiations were pelleted and resuspended into stage 4 medium + 10% v/v DMSO. Cultures were vialed, placed into isopropanol baths, and frozen overnight at −80 C before being placed into long-term liquid nitrogen storage.
Immunocytochemistry
All steps were performed at room temperature. Cultures were fixed in 10% buffered formalin (Protocol 23-305-510) for 10 min, permeabilized in phosphate-buffered saline (PBS) + 0.2% v/v Triton X-100, blocked in PBS + 1% bovine serum albumin (BSA), 0.1% v/v Tween-20 (PBS-T) for 30 min, and incubated with primary antibodies for 2 h. Cultures were washed twice with blocking solution and incubated 1 h with secondary antibodies. 4′,6-diamidino-2-phenylindole dilactate (DAPI, Thermo Fisher) was added for 5 min before final washing in PBS-T. Antibodies: OCT4 (Millipore, MAB4401; 1:250), Nanog homeobox (NANOG, Bio-Techne, AF1997; 1:100), tumor-related antigen (TRA-1-81, Millipore, MAB4381; 1:100), PAX6 (PAX6; 1:25; Developmental Studies Hybridoma Bank, Iowa City, IA, USA), SOX1 (AF3369; 1:250; Bio-Techne Systems), β-III tubulin (TUJ1, MAB1637; 1:500; Millipore). IgG secondary antibodies: donkey anti-mouse, goat, or rabbit antigens each conjugated to either 488 or 555 fluorophores were used for visualization (A-31572, A-21206, A-21432, A-11055, A-31570, and A-21202; Thermo Fisher Scientific).
Microarray Analysis
hiPSC line UMN05272014JBx7 (UMNx7) and sister clone UMN05272014JBx11 (UMNx11) were differentiated in 3 independent experiments and cultures sacrificed on days 0, 11, and 17. RNA was extracted using RNeasy kit according to manufacturer instructions (74104; Qiagen). RNA was submitted to University of Minnesota Genomics Center for microarray analysis. RNA was hybridized to using the HumanHT-12 v4 Expression BeadChip kit (Illumina BD-103-0204). Expression analysis was performed in GenomeStudio. Samples extracted from the same time points were treated as replicates and grouped for analysis. Differential analysis was performed with the stage 0 group used as a reference with average normalization. Results were filtered with a diffscore cutoff of ±90 to limit analysis to the most differentially expressed genes. Euclidian distance was used to cluster samples and genes to produce heat maps. RNA from the days 0 and 11 samples was also used for qRT-PCR with the following primers to validate the microarray data: neurogenin 2 (NEUROG2; 5′-TCA GAC ATG GAC TAT TGG CAG-3′, 5′-GGG ACA GGA AAG GGA ACC-3′) and homeobox D3 (HOXD3; 5′- CGA CAG AAC TCC AAG CAG AAG-3′, 5′-CCT TTT CCA ATT CCA CCA GC-3′). 2▴▴CT method was used to demonstrate fold change gene expression.
Electrophysiology
Twenty days after differentiation, hiPSC-derived VSNPs were maintained in N2/B27 medium supplemented with of CNTF, BDNF, GDNF, and NT3 (20 ng/mL; PeproTech). After 10 d, cells were used for whole-cell patch clamp recordings. Before recording, an individual culture slide was transferred to a recording chamber and continuously superfused (2.0 ± 0.25 mL/min) with mammalian Ringer’s solution at room temperature. The mammalian Ringer’s solution contained in mM: 146.0 NaCl, 3.0 KCl, 2.0 MgCl2, 2.0 CaCl2, 1.25 NaH2PO4, 1.0 Na pyruvate, 10.0 HEPES, and 10.0 glucose (pH 7.4, 290–300 mOsm). Individual neurons were visualized using an Olympus (Shinjuku, Tokyo) BX-51WI fixed-stage microscope equipped with differential interference contrast optics and a water immersion objective (40, 0.9 numerical aperture; Olympus). Whole-cell recordings were performed using MultiClamp (Axon Instruments; Molecular Devices, Sunnyvale, CA, USA), and data were acquired and analyzed with pCLAMP 9 (Axon Instruments). For whole-cell current and voltage clamp recordings, patch pipettes had a tip resistance of 3 to 4.5 MΩ when filled with an internal solution containing (in milli molar) 140.0 KCl, 1.0 MgCl2, 1.0 CaCl2, 11.0 ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 5.0 HEPES, 2.0 glucose, 1.0 glutathione, 3.0 Mg–adenosine triphosphate (ATP), and 0.5 Na-GTP (pH adjusted to 7.2 with potassium hydroxide(KOH), 310–314 mOsm). During recordings, the pipette capacitance was neutralized and access resistance was regularly monitored. In current clamp recording mode, action potentials were evoked by current step injections (of 60–480 Pa, 100 ms duration). Voltage-gated currents were recorded in voltage clamp mode by stepping the membrane potential from a holding potential of −80 to +60 mV in 10 mV increments (20 ms duration steps). Traces and current–voltage plots were made using GraphPad Prism, version 6.
Injury and Cell Engraftment Assay
Animal use was approved by the University of Minnesota, Institutional Animal Care and Use Committee. Three adult (201–220 g) female athymic nude (Hsd: RH-rnu/rnu) rats (Charles River Laboratories, Wilmington, MA, USA) received moderate contusion injury using the Infinite Horizon Impactor (Precision Systems, Lexington, KY, USA). Female rats were utilized due to their amenability to postinjury care, specifically bladder expression. Briefly, the rats were anesthetized with isoflurane and underwent T8/T9 laminectomy followed by spinal cord impact at a force of 200 kdyn. Rats then received buprenorphine (0.05 mg/kg SC q12 h) for 3 d after surgery to moderate acute pain and amoxicillin (14 g/L in drinking water) for 7 d after surgery to prevent urinary tract infection as part of standard postoperative care. Urine was expressed manually twice daily for the first 3 d after surgery and daily thereafter until the return of independent urination. Nine days postinjury, the injury site was reexposed and hVSNPs were prepared and grafted at a concentration of 30,000 cells per μL. A 2.5 μL cell suspension was transplanted at the midline of the spinal cord 1 mm rostral and caudal to the injury site and 5 μL in the epicenter for a total of 10 μL using a stainless steel needle (26S, Hamilton 7804-04) attached to a 10-μL syringe (Hamilton 701-RN, Reno, Nevada, USA). The muscles and skin were then re-approximated and closed in layers.
Immunohistochemistry
At 2 wk postinjection, rats were anesthetized with ketamine (100 mg/kg IP) and xylazine (10 mg/kg IP), then underwent transcardial perfusion with 4% paraformaldehyde (PFA). The spinal cord was removed and cryoprotected in 30% w/v sucrose. Spinal cord tissue was frozen in optimal cutting temperature compound (OCT) compound (Sakura Finetek, Torrance, CA, USA) and cryosectioned in the parasagittal plane at a thickness of 20 μm using a cryotome (CM3050S; Leica, Wetzlar, Germany). Representative serial sections for each spinal cord were selected from the mid-sagittal area for immunohistochemical analysis. Negative controls were obtained by omission of the primary antibody. Immunofluorescent photomicrographs were obtained from a 1-mm segment centered on the lesion epicenter. Antibodies identifying human nuclear antigen (hNA, MAB1281; 1:250; Millipore, Billerica, MA, USA), TUJ1 (1:500; Millipore), Stem121 (, AB-121-U-050, 1:500; SC Proven, Newark, CA, USA), neuronal nuclei (NeuN, Abcam, , EPR12763; 1:1,000; Cambridge, United Kingdom), and microtubule-associated protein 2 (MAP2, Abcam, ab32454; 1:200) were used to characterize engraftment of injected human cells.
Results
Reprogramming of Human Dermal Fibroblasts into iPSCs Under Defined Conditions
We sought to test defined reprogramming and pluripotent stem cell culture practices as an upstream requirement for reproducible differentiation. After surveying the available culture methods, we used Essential 8 Flex medium and the truncated rhVTN produced in prokaryotic expression systems n-terminal cleaved vitronectin (VTN-N)5 to promote undifferentiated growth for preexisting lines. In addition, we evaluated a full-length rhVTN produced from eukaryotic expression systems. We titrated quantities of each side by side and observed undifferentiated growth following seeding. Gross morphological assessment and growth kinetics across our various conditions were observed, and the decision was made to proceed with the full-length vitronectin variant (data not shown). As an alternative to ethylenediaminetetraacetic acid (EDTA) passaging methods previously described,12 we adopted hypertonic citrate passaging methodology due to its compatibility with flask-based manufacture.13 Best performance was observed when cultures were processed into approximately 50-cell colonies for seeding at a ratio of 1:3 and 1:8, reaching 70% to 100% surface utilization after 3 d. As recommended by the manufacturer, feeding was skipped the day following passage, while daily feeding resumed thereafter to combat excessive media acidification on days 2 and 3.
Reprogramming of Human Dermal Fibroblasts into iPSCs
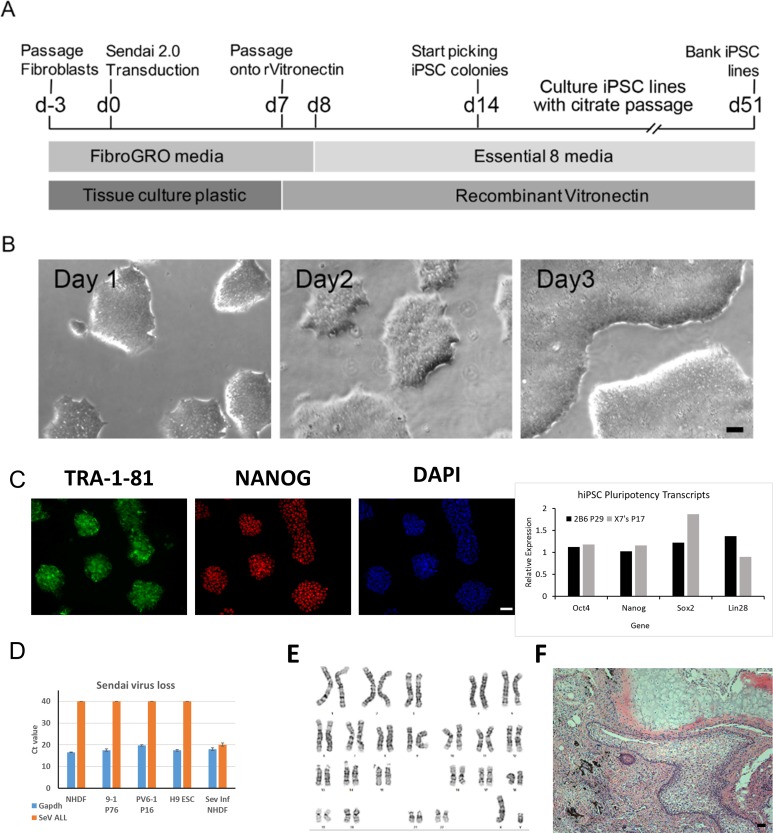
Together with our modified undifferentiated growth conditions, we adapted an SeV-vector-based reprogramming scheme using dermal fibroblasts11 (Fig. 1A). Fibroblasts were cultured prior to reprogramming in the defined FibroGro fibroblast growth medium that contains only BSA as a xenogeneic component. Twenty-four hours posttransduction with CytoTune 2.0 vectors, medium was exchanged for Flex. Cultures were fed daily before TryPLE passage onto rhVTN on the sixth day posttransduction. Two weeks posttransduction, colonies were identified morphologically, transferred to VTN-coated 48-well plates, and culture expanded into T25 flasks with hypertonic citrate as previously described10 (Fig. 1B). Cultures were passaged for analysis to confirm the expression of the pluripotency markers TRA-1-81 and NANOG by immunocytochemistry as well as OCT4, SOX2, NANOG, and LIN28 by qRT-PCR (Fig. 1C). Cultures were screened for vector loss between passages 5 and 20. Vector loss was variable, occurring as early as passage 6 (Fig. 1D) but sometimes requiring greater than 20 passages (data not shown). Clones were subsequently culture expanded into multiple T75 flasks and cryo-banked with each vial containing approximately 4 million cells, sufficient to seed a T25 flask upon thaw. Postthaw, cultures were further characterized, demonstrating a normal G-banded karyotype (Fig. 1E) and the ability to form a complex teratoma when injected intramuscularly into NOD-SCID mice (Fig. 1F).
Fig. 1.
Defined derivation and culture of human-induced pluripotent stem cell (hiPSC) line UMN05272014JBx7 (UMNx7). (A) General scheme for reprogramming dermal fibroblasts and banking of hiPSCs under defined conditions. Fibroblasts were thawed and cultured in defined FibroGro before being reprogrammed with CytoTune 2.0 Sendai (SeV) vectors. iPSC colonies were picked and iPSC lines expanded for characterization and banking. (B) Phase-contrast morphology during a standardized 3-d passage cycle. (C) Cultures demonstrate an undifferentiated phenotype via immunocytochemistry for surface antigen tumor-related antigen-1-81, nuclear expression of pluripotency-associated transcription factor Nanog homeobox (NANOG), and quantitative real-time polymerase chain reaction (qRT-PCR) for pluripotency-associated transcripts octamer-binding transcription factor 4 (OCT4), sex-determining region Y-box 2, NANOG, and LIN28. (D) Loss of exogenous SeV vectors detected by qRT-PCR. Positive-control samples contained RNA from 24-h Sendai-infected fibroblasts. Negative-control samples contained RNA from late-passage hiPSCs. (E) Passage-8 karyotype for UMNx7 showed no structural abnormalities by g-banding. (F) hiPSC UMNx7 generated a complex teratoma when injected into immunocompromised mice (all scale bars are 50 µm).
Defined Conditions Promote Accelerated Formation of Primitive Neuroectoderm
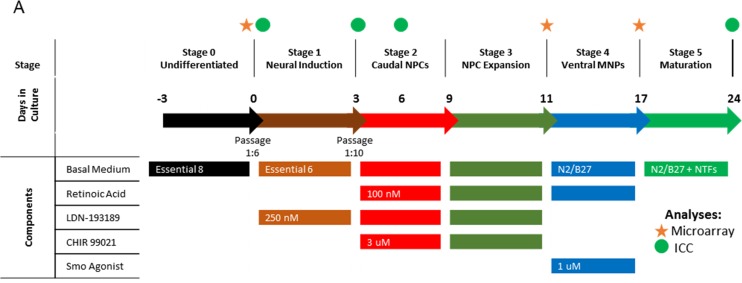
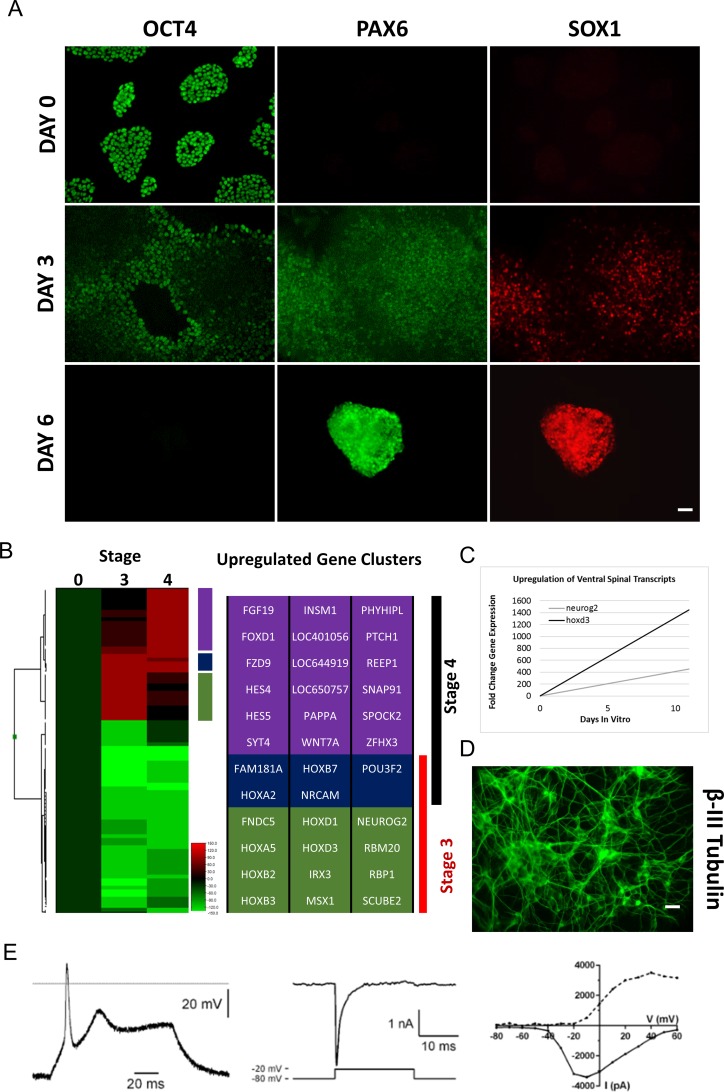
We utilized clones UMNx7 and UMNx11 for differentiation. We adopted for refinement a previously published protocol for directed differentiation of human embryonic stem cells toward the ventral spinal cord14 (Fig. 2) with a focus to mitigate variables that could negatively impact manufacturability. We identified cellular stratification and size irregularity inherent to embryoid body culture as barriers to reproducibility and purity, as diffusional limitations have the potential to establish morphogen gradients during early inductive and patterning events.15 As such, we evaluated the use of monolayer differentiation methods. To initiate differentiation, we passaged cultures identically to the process used for normal propagation so as to minimize the introduction of new reagents and techniques. After 18 to 24 h, medium was exchanged for Essential 6 medium (E6) that contains the same basal salts and additives as Flex while lacking both FGF2 and TGFβ1 responsible for promoting undifferentiated growth. The use of Noggin alone in adherent monolayer culture has previously been shown to produce anterior neuroectoderm from hPSCs.16 We therefore supplemented the medium with the chemical Noggin analog, LDN-193189. We assayed for the loss of OCT4 as well as the gain of PAX6 and SOX1 to monitor neural induction (Fig. 3A, top and middle rows). After 3 d, substantial loss of OCT4 was seen from the center of colonies, whose expression was replaced by PAX6 and SOX1. Edges of colonies maintained OCT4 expression without expression of PAX6/SOX1. Differentiating cultures maintained similar growth kinetics as undifferentiated hiPSCs (not shown), and cultures were passaged on the third day at a 1:6 split ratio.
Fig. 2.
Generation of human ventral spinal neural progenitors (hVSNPs) from human-induced pluripotent stem cell UMN05272014JBx7 (UMNx7). General scheme for differentiation of hVSNPs from undifferentiated hiPSC line UMNx7. Circles above the time line indicate time points where immunocytochemistry were performed, while stars indicate where microarray gene expression was performed. NTFs, neurotrophins, BDNF, brain-derived neurotrophic factor; CNTF, ciliary neurotrophic factor; GDNF, glial-derived neurotrophic factor; NT3, neurotrophin-3.
Fig. 3.
Characterization of human ventral spinal neural progenitors differentiation protocol. (A) Early time point immunohistochemistry demonstrating neural induction. OCT4 expression is absent from colony centers outward toward colony edges over 6 d. Cells in the colony centers gain expression of both paired box 6 (PAX6) and sex-determining region Y-box 1 (SOX1). Following passage, cultures maintain high PAX6/SOX1 expression, with undetectable OCT4. (B) Microarray analysis of stages 3 and 4 cultures. Heat map analysis highlighting caudal HOX gene activation in stage 3 cultures and hedgehog- and synapse-related transcription in stage 4 cultures, suggestive of ventral, spinal Fig. 3. (continued) neurogenesis. (C) Quantitative real-time polymerase chain reaction validates gene expression changes demonstrated by microarray analysis. (D) Cultures express the neuron-specific markers β-III tubulin (scale bar is 50 μm). (E) Electrophysiological recordings of stage 4 cultures matured an additional 2 wk with neurotrophins. Whole-cell current and voltage clamp recordings. Left: current clamp recording showing an action potential generated by injection of positive current (70 Pa). Horizontal line represents 0 mV. Middle: Voltage clamp recording showing a fast inward current evoked by a voltage step from −80 mV to −20 mV. Right: plot of the peak inward (solid line) and outward (dashed line) current evoked by sequential voltage steps from −80 mV to −70 through +60 mV.
Caudal–Ventral Patterning of hiPSC-derived Neuroectoderm
Once expression of neural tube lineage markers was established, 100 nM RA and 3 µM CHIR99021 were used to induce retinoid and WNT signaling, promoting caudalization. Over the course of 3 d under these conditions, colonies failed to spread as they grew, instead proliferating from the center of each colony. Immunostaining demonstrated complete absence of OCT4 expression, with highly evident PAX6/SOX1 expression. Cells could be seen to align radially within colonies, reminiscent of collections of small neural rosettes (Fig. 3A, bottom row). Cultures maintained under these conditions continued to proliferate without spreading, resulting in delamination from the culture matrix to form floating aggregates by day 11. Immunocytochemical analysis became unhelpful due to the nonadherent, 3-dimensional structure of the aggregates, and therefore RNA was extracted from the cell spheres at this time point for gene expression analysis by microarray (Fig. 3B) validated by qRT-PCR (Fig. 3C).
Using diffscore cutoff values showing most logical heat map clustering, gene lists were generated from the microarray data. Thirty-six genes were most differentially upregulated compared to undifferentiated stage 0 cultures. Twelve genes were specific to stage 3 differentiation, 18 genes were specific to stage 4 differentiation, and 5 genes were shared by stages 3 and 4. Among stage 3 genes specifically upregulated was retinol-binding protein 1, evidence of a transcriptional response to the RA present in the culture medium.17 Numerous caudal homeobox (HOX) genes were expressed at this time, with the most anterior and posterior limits represented by homeobox D1 and homeobox B7, respectively, which encompass neural tube segments eventually giving rise to structures spanning from the hindbrain to cervical spinal cord. Signal peptide, complement C1r/C1s, u epidermal growth factor, BMP 1 domain, and epidermal growth factor–like domain containing 2 expression were also seen, which has been implicated in the sonic hedgehog (SHH) signaling cascade.18 Ventral patterning was evident by differential expression of iroquois homeobox 3, msh homeobox 1, and NEUROG2, demonstrating regionalization to the V2 domain.19,20 It is possible that this population has lost patterning flexibility and is fully restricted to the V2 domain due to the expression of NEUROG2, which in addition to serving as a patterning landmark drives cell cycle exit and neuronal commitment.21 The myokine fibronectin type III domain containing 5 and neuronal cell adhesion molecule (NRCAM) were also expressed, demonstrating commitment to terminal neural differentiation.22,23 Taken together, the gene expression data suggest stage 3 cultures are fated toward caudal ventral spinal progenitors of the P2 domain with early commitment toward V2 interneuron production.
Stage 4 (d17) cultures were exposed to a potent SHH analog SAG for an additional 6 d, serving to mimic the function of the developing floor plate to ventralize neural progenitors. As expected, microarray analysis of RNA extracted from spheres at this time point shows highly differentially expressed mediators of hedgehog signaling represented by patched 1. Fibroblast growth factor 19 (FGF19) was also upregulated, which has been shown to be responsive to SHH signaling, albeit within the context of interneuron specification at the caudal ganglionic eminence.24 Upregulated stage 4 genes also suggest evidence of motor neurogenesis. Receptor accessory protein 1 has been found to be mutated in upper motor neuron diseases, while wingless-related integration site family member 7A differentiates the γ-motor neuron subtype from either α- or β-motor neurons.25,26 NRCAM, shared with stage 3 cultures, and insulinoma-associated transcriptional repressor 1 have been implicated in commitment to an immature neuronal fate,23,27 while synaptosome-associated protein 91 kDa and synaptotagmin 4 have been shown to be involved in synaptic vesicle formation.28,29 These data are suggestive of commitment to an immature motor neuron phenotype, specifically to the γ-subtype.
Stage 4 cultures also expressed genes associated with neural stem cell maintenance. Hairy and enhancer of split 4 and 5 serve as Notch effectors in promoting neuronal diversity through asymmetric division of neural stem/progenitor cells (NSPCs).30 Additionally, pituitary-specific octamer unc-86 class 3 homeobox 2 has been shown to bind the Nestin promoter, another NSPC marker.31 The frizzled proteins have been shown to be involved in self-renewal as receptors to WNT proteins, and frizzled class receptor 9 specifically has been found in the ependymal layer of the ventricular zone where NSCs reside.32 Finally, zinc finger homeobox 3 has been shown to upregulate the expression of platelet-derived growth factor receptor α (PDGF-α),33 which has been shown to be important for the maintenance of radial glia in the developing neocortex.34 Interpretation of our stages 3 and 4 microarray data suggests our cell population consists of a mixture of immature caudal ventral spinal neurons and restricted progenitors based on HOX gene expression, ventral domain markers, motor neuron-related transcripts, and general pathways affiliated with stemness.
Stage 4 cultures were cryopreserved and, upon thaw and maturation on laminin substrates in the presence of neurotrophins, expressed the neuron-specific protein TUJ1 (Fig. 3D). After an additional 2 wk, electrophysiological recordings were taken. Neurons generated action potentials with an overshoot upon depolarization and demonstrated voltage-gated currents whose current–voltage relationship were consistent with a fast sodium inward current and a slow outward current (n = 3; Fig. 3E).
Transplantation of hIPSC-derived hVSNPs
To investigate potential for in vivo engraftment in preparation for future functional studies, stage 4 cultures were thawed and maintained for 24 h in stage 4 media conditions before transplantation into immunocompromised rats that had received moderate SCI as described above (Fig. 4A). We optimized our cell injection parameters by assessing the impact of needle bore size on multicellular aggregate injection in vitro (Fig. 4B). Using trypan exclusion, we discovered the importance of using a wide-gauge needle to avoid compromised cell aggregate viability (data not shown). Injected animals (n = 3) were recovered for 2 wk to allow for engraftment before killing and histological sectioning. Human cells were identified in the injured spinal cords of all 3 rats at 2 wk after transplantation. Human cells were detected with either human nuclear antigen (hNA) or human-specific cytoplasmic antibody SC121.
Fig. 4.
Transplantation of human ventral spinal neural progenitors into injured rat spinal cord. (A) Time line for rat injury, injection, and analysis. (B) Schematic for injury model, anatomical orientation, injection parameters, and tissue processing planes for analysis. (C) Immunohistochemistry of sectioned rat cord. Dotted lines demarcates white matter/gray matter boundary. Dashed circle represents tissue deficit. White arrows indicate injection site. Grafted cell identity is confirmed with human nuclear antigen (hNA)-positive nuclei densely packed within the gray matter around a tissue deficit. SC121-positive cells are shown to co-localize with β-III tubulin expression (TUJ1; inset, white arrows), indicating the graft is composed of neurons. SC121 TUJ1 co-expressing axons extend caudal to the injection site. (D) Engrafted human neurons are postmitotic as shown by co-expression of NeuN and MAP2 with hNA (scale bars are 50 μm).
Nuclei-dense regions of tissue sections visualized with DAPI were used to delineate white matter from gray matter, the latter demonstrating greater neuronal cell body density. Grafted cells were located primarily within gray matter, restricted to a dense patch at the injury epicenter surrounding a tissue deficit identified by lack of DAPI-positive nuclear staining (Fig. 4C). Human-specific cytoplasmic staining co-localized with TUJ1 (Fig. 4C, inset), suggesting that the graft was composed primarily of neurons. Axons associated with the transplanted cells were observed to extend mainly in a caudal direction within the gray matter. These axons extended distances at least 780 µM from the injection bolus (Fig. 4C). Axon tracing to determine the full length of these axons was not within the scope of this study but will be included in future work. NeuN and MAP2 co-localized with hNA, indicating that engrafted human neurons were postmitotic (Fig. 4D).
Discussion
In the present study, we described a method for the reprogramming of human dermal fibroblasts into iPSCs and their subsequent differentiation into ventral spinal neural progenitors using defined reagents. Many research groups target differentiation toward the ventral spine,35 and our protocol adheres to similar developmentally guided stages. However, Matrigel™, commonly used as a substrate for both reprogramming and differentiation, has been replaced by recombinant vitronectin. We further adopted the fully defined Flex medium for reprogramming and stem cell maintenance as well as Essential 6 for differentiation. These changes significantly influence the differentiation kinetics of our populations, requiring protocol refinements and uncovering novel information regarding the induction of neural identity of hiPSCs under defined conditions.
It has been previously indicated that monolayer neural induction requires combined antagonism of both BMP and TGFβ pathways.7 We found TGFβ antagonism to be superfluous in our conditions. This mirrors similar findings by others, demonstrating fewer requirements for active pathway modulation for the production of primitive neuroectoderm under defined conditions.6 We find that the combination of defined conditions with active pathway modulation provides advantages with respect to differentiation speed, with the initial induction to PAX6/SOX1 progenitors occurring in less than 72 h. Opportunities for further stage-specific refinements to reduce time in culture may also exist during stages 3 and 4 differentiation.
Our preliminary in vivo transplantation data are included as a proof of principle that the neural progenitors generated by our accelerated induction protocol using defined reagents survive freezing, thawing, and injection and are maintained in a preclinical animal xenotransplant model,36–38 critical parameters for testing the potential application of this technology in clinical translation. Future studies investigating aspects of cell identity posttransplant and injury parameters such as cavitation, tissue sparing, and immediate and long-term functional deficits, and recovery will be required to determine the clinical application of this novel technology. Combination transplants with glia would also be an interesting research avenue to explore.39
Conclusion
In summary, our reprogramming and differentiation protocols provide an improved platform for the production of neural progenitors for use in vitro to understand cell fate decisions of the ventral spinal cord. The fully defined reagents and improved temporal kinetics in our protocol also provide advantages for eventual translation into a clinical cell manufacturing process.
Footnotes
Authors’ Note: James R. Dutton and Ann M Parr were co-senior authors. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Ethical Approval: Ethical approval is not applicable.
Statement of Human and Animal Rights: Procedures involving animal subjects were be approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Minnesota #1607-34013.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: The Wings for Life Spinal Cord Research Foundation, private philanthropy through the University of Minnesota Foundation (UMF), and the KL2 Scholar Program (A. M. Parr) from the Clinical and Translational Science Institute KL2TR113 (NIH:8UL1TR000114).
References
- 1. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. [DOI] [PubMed] [Google Scholar]
- 2. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz–Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. [DOI] [PubMed] [Google Scholar]
- 3. Lindvall O, Bjorklund A. Cell therapeutics in Parkinson’s disease. Neurotherapeutics. 2011;8(4):539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bjorklund A, Lindvall O. Replacing dopamine neurons in Parkinson’s disease: how did it happen? J Parkinson’s Dis. 2017;7(s1):S23–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lippmann ES, Estevez-Silva MC, Ashton RS. Defined human pluripotent stem cell culture enables highly efficient neuroepithelium derivation without small molecule inhibitors. Stem Cells. 2014;32(4):1032–1042. [DOI] [PubMed] [Google Scholar]
- 7. Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human es and ips cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kibbey MC. Maintenance of the EHS sarcoma and Matrigel preparation. J Tissue Cult Methods. 1994;16(3):227–230. [Google Scholar]
- 9. Jin S, Yao H, Weber JL, Melkoumian ZK, Ye K. A synthetic, xeno-free peptide surface for expansion and directed differentiation of human induced pluripotent stem cells. PLoS One. 2012;7(11):e50880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parr AM, Walsh PJ, Truong V, Dutton JR. cGMP-Compliant Expansion of Human iPSC Cultures as Adherent Monolayers. Methods Mol Biol 2016;1357:221–9. [DOI] [PubMed] [Google Scholar]
- 11. Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108(34):14234–14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7(11):2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nie Y, Walsh P, Clarke DL, Rowley JA, Fellner T. Scalable passaging of adherent human pluripotent stem cells. PLoS One. 2014;9(1):e88012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu BY, Zhang SC. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat Protoc. 2009;4(9):1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3(5):508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerrard L, Rodgers L, Cui W. Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem Cells. 2005;23(9):1234–1241. [DOI] [PubMed] [Google Scholar]
- 17. Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8(10):755–765. [DOI] [PubMed] [Google Scholar]
- 18. Jakobs P, Schulz P, Ortmann C, Schurmann S, Exner S, Rebollido-Rios R, Dreier R, Seidler DG, Grobe K. Bridging the gap: heparan sulfate and Scube2 assemble Sonic hedgehog release complexes at the surface of producing cells. Scientific reports 2016;6:26435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takahashi M, Osumi N. Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development. 2002;129(6):1327–1338. [DOI] [PubMed] [Google Scholar]
- 20. Misra K, Luo H, Li S, Matise M, Xiang M. Asymmetric activation of dll4-notch signaling by foxn4 and proneural factors activates BMP/TGFβ signaling to specify v2b interneurons in the spinal cord. Development. 2014;141(1):187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lacomme M, Liaubet L, Pituello F, Bel-Vialar S. Neurog2 drives cell cycle exit of neuronal precursors by specifically repressing a subset of cyclins acting at the g1 and s phases of the cell cycle. Mol Cell Biol. 2012;32(13):2596–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forouzanfar M, Rabiee F, Ghaedi K, Beheshti S, Tanhaei S, Shoaraye Nejati A, Jodeiri Farshbaf M, Baharvand H, Nasr-Esfahani MH. Fndc5 overexpression facilitated neural differentiation of mouse embryonic stem cells. Cell Biol Int. 2015;39(5):629–637. [DOI] [PubMed] [Google Scholar]
- 23. Sakurai T. The role of NrCAM in neural development and disorders--beyond a simple glue in the brain. Mol Cell Neurosci. 2012;49(3):351–363. [DOI] [PubMed] [Google Scholar]
- 24. Kim TG, Yao R, Monnell T, Cho JH, Vasudevan A, Koh A, Peeyush KT, Moon M, Datta D, Bolshakov VY, et al. Efficient specification of interneurons from human pluripotent stem cells by dorsoventral and rostrocaudal modulation. Stem Cells. 2014;32(7):1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Bot ST, Veldink JH, Vermeer S, Mensenkamp AR, Brugman F, Scheffer H, van den Berg LH, Kremer HP, Kamsteeg EJ, van de Warrenburg BP. Atl1 and reep1 mutations in hereditary and sporadic upper motor neuron syndromes. J Neurol. 2013;260(3):869–875. [DOI] [PubMed] [Google Scholar]
- 26. Ashrafi S, Lalancette-Hebert M, Friese A, Sigrist M, Arber S, Shneider NA, Kaltschmidt JA. Wnt7a identifies embryonic gamma-motor neurons and reveals early postnatal dependence of gamma-motor neurons on a muscle spindle-derived signal. J Neurosci. 2012;32(25):8725–8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duggan A, Madathany T, de Castro SC, Gerrelli D, Guddati K, Garcia-Anoveros J. Transient expression of the conserved zinc finger gene insm1 in progenitors and nascent neurons throughout embryonic and adult neurogenesis. J Comp Neurol. 2008;507(4):1497–1520. [DOI] [PubMed] [Google Scholar]
- 28. Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, Stachowiak JC. Intrinsically disordered proteins drive membrane curvature. Nat Commun. 2015;6:7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berton F, Iborra C, Boudier JA, Seagar MJ, Marqueze B. Developmental regulation of synaptotagmin i, ii, iii, and iv mRNAs in the rat CNS. J Neurosci. 1997;17(4):1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30(9):3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the nestin gene in neural primordial cells. Mol Cell Biol. 2004;24(20):8834–8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gonzalez-Fernandez C, Arevalo-Martin A, Paniagua-Torija B, Ferrer I, Rodriguez FJ, Garcia-Ovejero D. Wnts are expressed in the ependymal region of the adult spinal cord. Molecular Neurobiology 2017;54(8):6342–6355. [DOI] [PubMed] [Google Scholar]
- 33. Kim TS, Kawaguchi M, Suzuki M, Jung CG, Asai K, Shibamoto Y, Lavin MF, Khanna KK, Miura Y. The zfhx3 (atbf1) transcription factor induces PDGFRB, which activates atm in the cytoplasm to protect cerebellar neurons from oxidative stress. Dis Model Mech. 2010;3(11–12):752–762. [DOI] [PubMed] [Google Scholar]
- 34. Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, Oldham MC. Radial glia require PDGFD-PDGFRβ signalling in human but not mouse neocortex. Nature. 2014;515(7526):264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sances S, Bruijn LI, Chandran S, Eggan K, Ho R, Klim JR, Livesey MR, Lowry E, Macklis JD, Rushton D, et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci. 2016;19(4):542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matyas JJ, Stewart AN, Goldsmith A, Nan Z, Skeel RL, Rossignol J, Dunbar GL. Effects of bone-marrow -derived MSC transplantation on functional recovery in a rat model of spinal cord injury. Cell Transplantation 2017;26(8):1472–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruzicka J, Machova-Urdzikova L, Gillick J, Amemori T, Romanyuk N, Karova K, Zaviskova K, Dubisova J, Kubinova S, Murali R, et al. A comparative study of three different types of stem cells for treatment of rat spinal cord injury. Cell Transplant. 2017;26(4):585–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao Y, Tang F, Xiao Z, Han G, Wang N, Yin N, Chen B, Jiang X, Yun C, Han W, et al. Clinical study of NeuroRegen scaffold combined with human mesenchymal stem cells for the repair of chronic complete spinal cord injury. Cell Transplant. 2017;26(5):891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Terzic D, Maxon JR, Krevitt L, DiBartolomeo C, Goyal T, Low WC, Dutton JR, Parr AM. Directed differentiation of oligodendrocyte progenitor cells from mouse induced pluripotent stem cells. Cell Transplant. 2016;25(2):411–424. [DOI] [PubMed] [Google Scholar]