Abstract
Biologic substrates, prepared by decellularizing and solubilizing tissues, have been of great interest in the tissue engineering field because of the preservation of complex biochemical constituents found in the native extracellular matrix (ECM). The integrity of the ECM is critical for cell behavior, adhesion, migration, differentiation, and proliferation that in turn affect homeostasis and tissue regeneration. Previous studies have shown that various processing methods have a distinctive way of affecting the composition of the decellularized ECM. In this study, we developed a bioactive substrate for hepatocytes in vitro, made of decellularized and solubilized liver tissue. The present work is a comparative approach of 2 different methods. First, we decellularized porcine liver tissue with ammonium hydroxide versus a sodium deoxycholate method, then characterized the decellularized tissue using various methods including double stranded DNA (dsDNA) content, DNA size, immunogenicity, and mass spectrometry. Second, we solubilized the decellularized porcine liver with hydrochloric acid versus acetic acid (AA) and characterized the resultant solubilized tissues using relevant methodologies including protein yield, immunogenicity, and bioactivity. Finally, we isolated primary porcine hepatocytes, cultured, and evaluated their bioactivity on the optimized decellularized–solubilized liver substrate. The decellularized porcine liver ECM processed by the ammonium hydroxide method and solubilized with AA displayed higher ECM integrity, low dsDNA, no evidence of intact nuclei, low human monocyte chemoattraction, and the presence of key molecules typically found in the native liver, a very important element for normal cell function. In addition, primary porcine hepatocytes showed enhanced functionality including albumin and urea production and bile canaliculi formation when cultured on the developed liver substrate compared to type I collagen.
Keywords: liver, decellularization, solubilization, ECM, hepatocytes
Cell-based therapies have been an active area of investigation as an alternative to whole organ transplantation. Hepatocyte transplantations have been successfully performed for a variety of indications, including acute liver failure, end-stage liver disease, and inborn errors of metabolism, and have shown positive therapeutic outcomes.1–4 However, hepatocyte transplantation has been hindered by the inability to secure a reliable and healthy cell source, shortage of organ donors, problems related to isolation of high-quality hepatocytes, and the inability of hepatocytes to proliferate in vitro. Moreover, hepatocytes undergo an epithelial to mesenchymal transition and have a drastic loss of their functionality and polarity when cultured in vitro;5–10 hence, the inability to obtain a large enough number of hepatocytes for therapeutic uses.11,12 Current cell culture models include the addition of cytokines, proteins, different media, coatings, gels, and sandwich methods as well as 2-dimensional (2-D) or 3-dimensional (3-D) cultures in an effort to further improve in vitro conditions for primary hepatocytes.13–20 Furthermore, matrices used for the culture of hepatocytes, such as type I collagen, fibronectin, and matrigel, have not demonstrated sufficient long-term success, possibly due to the absence of biochemical cues only found in the native cellular environment, critical for hepatocyte function.13,15,18,21–25 To emulate native biochemical cues in vitro, the use of decellularized extracellular matrix (ECM) has emerged as a key biomaterial to produce a tissue-specific scaffold that closely mimics the complex physical and chemical profile found in the native ECM microenvironment in vivo.18,26,27 However, there are many factors to consider with decellularization and solubilization methods to achieve maximum preservation of the ECM biochemical profile created by and benefited the former resident cells in the native tissue.26,28–30
Liver ECM was obtained by removing endogenous cellular components from a native porcine liver. Porcine organ decellularization has high relevance in translational medicine, compared to rat or other subspecies, due to the high similarities of its protein biochemical profile with that of humans, in addition to regulatory advantages31,32 that allow a practical clinical transition into humans.33–35 The purpose of developing a decellularized liver matrix was to create a liver-like environment that will ultimately be exposed to human cells or a human physiological milieu, with the intention of repairing, substituting, or enhancing liver pertinent characteristics. We used 2 different decellularization methods followed by 2 different solubilizing methods to obtain an optimized matrix with a biochemical profile closest to that of the in vivo environment. Structural and biochemical analyses of the decellularized and solubilized tissue matrix provided valuable information to address current concerns in hepatocyte tissue culture and to further understand the necessary biochemical cues to preserve hepatocyte functions.
Materials and Methods
Decellularization of Porcine liver
Porcine livers were used to develop a decellularized liver ECM. Livers were obtained from deceased pigs under a material transfer agreement that were part of other surgical studies approved by the Institutional Animal Care and Use Committee, US Army Institute of Surgical Research. The procurement of the liver was done by clamping and cutting both venous ends: suprahepatic and infrahepatic of the inferior vena cava (IVC), the hepatic portal vein, the intrahepatic artery, and the bile duct. Prewarmed 0.9% saline solution containing heparin (2,000 U/L) was used to perfuse the liver through the portal vein into the suprahepatic IVC, then through the hepatic artery to the suprahepatic IVC while all other vessels were clamped. The perfusion was terminated when most of the native red color was depleted and the liver appeared pale. The flushed porcine liver was then vacuum sealed (Food Saver® Model V3425, Atlanta, GA, USA) and stored at −80 °C until use. Prior to decellularization, the liver was thawed, sliced using a deli slicer (Berkel® 827A-PLUS, Troy, OH, USA) into ∼3-mm thick slices, and rinsed with distilled water for 30 min to remove any residual blood clots and debris. The sliced tissue was then decellularized under agitation (100 to 125 rpm; New Brunswick Scientific, Incubator Shaker, Edison, NJ, USA) using the methods discussed below.
Decellularizing methods
Ammonium hydroxide (method A) and deoxycolate (method B) were used to decellularize the porcine liver slices. In both methods, the liver slices were initially incubated in 0.025% trypsin-ethylenediaminetetraacetic acid (EDTA) (Gibco, 25200056, Waltham, MA, USA) at 37 °C for 1 h, then in a solution of 1% triton X-100 (Thermo Scientific, 215680010, Waltham, MA, USA) containing 0.1% ammonium hydroxide (Ricca Chemical, RABA002025D1, Arlington, TX, USA), agitated for 8 h at room temperature (method A), or in a solution of 3% triton X-100 for 1 h followed by a solution of 4% sodium deoxycholate (Sigma-Aldrich, D6750, St. Louis, MO, USA) for another 1 h under agitation (method B). Finally, the slices in both methods were thoroughly rinsed, incubated overnight in deionized distilled water, and chemically sterilized using 0.1% peracetic acid (PAA Sigma-Aldrich, 77240) in 4% ethanol (Sigma-Aldrich, 24102) for 2 h and rinsed twice with sterile phosphate-buffered saline pH 7.4 (PBS Gibco, 10010023) followed by deionized distilled water. Under sterile conditions, the decellularized liver slices were lyophilized, cut into small pieces, and milled using a rotary blade (Thomas Wiley® Mini-Mill Cutting Mill, 3383L10, Swedesboro, NJ, USA). The obtained powder of porcine liver ECM (pLECM) was sieved (mesh sieve designation #40) to achieve a particle size of less than 0.420 mm, was collected in sterile tubes, and stored in −20 °C until further use.
Solubilization of pLECM
Solubilization of pLECM was optimized using different concentrations of hydrochloric acid (HCl) or acetic acid (AA). Briefly, pLECM powder was mixed with 0.001 to 1 N HCl (Fluka, 38272, Morris Plains, NJ, USA) and porcine pepsin (Sigma-Aldrich, P7000) or 0.001 to 1 M AA (Sigma-Aldrich, 27225) and porcine pepsin keeping a 5:1:2 (mg:mL:mg) proportion, under constant stirring at 4 °C until no traces of powder were visible and a homogenous solution was formed (∼72 h). The solution was then filtered using a sterile 100 µm sieve, centrifuged at 3,000× g for 10 min, aliquoted, and stored at −20 °C until further use.
Deoxyribonucleic Acid (DNA) Analysis
Lyophilized pLECM was analyzed for DNA content before and after decellularization using Quant-iT PicoGreen (Qiagen/Invitrogen, P7589, Hilden, Germany) according to manufacturer’s instructions. Briefly, 2 mg of pLECM or native liver tissue was digested with proteinase K for 6 h at 56 °C, then it was processed with DNeasy Blood & Tissue Kit (Qiagen, 69504) to extract the DNA following purification using a QIAprep 2.0 Spin Miniprep Columns (Qiagen, 27115). Equal volumes of eluted DNA and PicoGreen dye solution were mixed and fluorescence was measured at 520 nm (Biotek® Synergy HTX Multi-Mode Reader, VT, USA) to determine DNA amount. Known concentrations of DNA were used as a standard curve. The DNA values were normalized by the dry weight of the original pLECM sample. Southern blotting: 320 ng DNA were analyzed in a FlashGel DNA Cassette, 2.2% (Lonza, 57031) at 100 V for 1 h, stained with Syto 60® (Thermo, Waltham, MA USA; S11342), and then visualized with a Laser 700 nm (LI-COR Odyssey® Fc Imager) using a 1 Kb Plus DNA Ladder (Invitrogen, 10787018).
Protein Extraction and Analysis
Resulting pLECM (10 mg) was processed (N = 3) to extract native proteins for analysis and quantification. One milliliter of Bio-Rad Ready Prep 2-D Rehydration/Sample Buffer (BioRad, Hercules, CA, USA; 1632106) was added to each sample consisting of 8 M urea, 2% 3-([3-Cholamidopropyl]dimethylammonio)-1-propanesulfonate hydrate (CHAPS), 50 mM Dithiothreitol (DTT), 0.2% Bio-Lyte® 3/10 ampholyte, and 0.001% bromophenol blue solution then sonicated for 30 min on ice, warmed at 32 °C for 5 min, briefly snap frozen in liquid nitrogen, and thawed at 32 °C. The freeze-thaw process was repeated twice. The samples were centrifuged at 21,000× g for 10 min and the supernatant was transferred to a new centrifuge tube. The remaining precipitates were processed with 100 µL dimethyl sulfoxide (DMSO), sonicated for 30 min on ice, and centrifuged at 21,000× g for 10 min. The supernatant was combined with the supernatant from the previous step and 200 µL was transferred to a new tube, mixed with 1 mL acetone, and stored at 4 °C overnight to precipitate proteins. The samples were centrifuged at 21,000× g for 1 min and the resulting precipitates were air dried to remove the acetone.
Bicinchoninic Acid (BCA) Protein Quantification
Protein concentrations were determined using a BCA™ protein assay kit (Thermo, 23225), according to manufacturer’s instructions. Extracted protein (25 µL) was mixed with 200 µL BCA working reagent and incubated at 37 °C for 30 min. Absorbance was measured at 562 nm using a Synergy HTX Multi-Mode Reader (Biotek). Known concentrations of bovine serum albumin (BSA, Sigma-Aldrich, 1076192 USP) were used as standard curve.
Mass Spectroscopy
Proteins extracted from the pLECM (see Protein Extraction and Analysis section) were separated by 1-D sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a Criterion XT 12% gel and then stained with Coomassie blue. The protein-containing region of each gel lane was divided into 7 slices which were individually reduced in situ with tris (2-carboxyethyl) phosphine and alkylated in the dark with iodoacetamide prior to treatment with trypsin (Promega, V5111, Madison, WI, USA). The digests were analyzed by capillary high-performance liquid chromatography (HPLC)-electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) on a Thermo Fisher LTQ Orbitrap Velos Pro mass spectrometer fitted with a New Objective Digital PicoView 550 NanoESI source. Online HPLC separation of the digests was accomplished with an Eksigent/AB Sciex NanoLC-Ultra 2-D HPLC system (column, PicoFrit™ [New Objective; 75 µm internal diameter]) packed to 15 cm with C18 adsorbent (Vydac, Columbia, MD, USA; 218MS 5 µm, 300 Å). Precursor ions were acquired in the Orbitrap from 300 to 2,000 m/z in profile mode at 60,000 resolutions (400 m/z); data-dependent collision-induced dissociation spectra of the 6 most intense ions in each precursor scan were acquired at the same time in the linear trap (MS2 isolation width = 3; unassigned charge states rejected; normalized collision energy = 30; dynamic exclusion repeat count = 1; dynamic exclusion repeat duration = 30 s). Mascot (Matrix Science, Boston, MA, USA) was used to search the spectra against the mammal’s subset of the National Center for Biotechnology Information (NCBI) nr database. Cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation and deamidation of glutamine and asparagine were considered as variable modifications; trypsin was specified as the proteolytic enzyme, with 1 missed cleavage allowed. The Mascot results files were combined for subset searching of the identified proteins by X! Tandem cross-correlation with the Mascot results, and determination of protein and peptide identity probabilities was accomplished by Scaffold (Proteome Software, Inc., Portland, OR, USA). The thresholds for acceptance of peptide and protein assignments in Scaffold were 95% and 99%, respectively, 2 peptides minimum per protein.
Peptide Analysis
Scaffold (Proteome Software) was used to validate protein identifications derived from MS/MS sequencing results using the X! Tandem36 and ProteinProphet computer algorithms.37 Scaffold verified peptide identifications assigned by SEQUEST, Mascot, or other search engines (NCBI, Pantherdb) using the X! Tandem database searching program36,38 Scaffold then probabilistically validated these peptide identifications and derived corresponding protein probabilities using ProteinProphet.37–39 All protein sequences were filtered by taxonomy (Sus scrofa). Common and statistically different proteins were analyzed in protein analysis through evolutionary relationships. Proteins were classified using comprehensive gene ontology (GO)40,41 annotations. Details of the methods can be found elsewhere.42,43
Biochemical Characterization of pLECM
The amounts of hydroxyproline, elastin, and glycosaminoglycans (GAGs) present in native and decellularized tissues were quantified with Hydroxyproline Assay (BioVision, K555-100, Milpitas, CA, USA), Fastin Elastin Assay (Biocolor, Carrickfergus, UK; F2000), and Blyscan Sulfated GAG Assay (Biocolor, B1000), respectively, according to manufacturer’s instructions. The concentrations of hydroxyproline, elastin, and GAGs in each sample were determined using a standard curve of known concentrations of each protein at absorbance 560, 513, and 656 nm, respectively. All absorbance measurements were performed using a Synergy HTX Multi-Mode Reader (Biotek) and the resulting values were normalized by original dry weight of tissue.
Enzyme-linked Immunosorbent Assay (ELISA) to Assess Growth Factor Content
The amount of growth factors present in the pLECM was quantified according to manufacturer’s protocols using a commercially available ELISA kit. Hepatocyte growth factor (HGF; Abcam, Cambridge, MA, USA), basic fibroblast growth factor (bFGF; Abcam), and epidermal growth factor (EGF; Abcam) were analyzed, and the values were normalized by total protein quantified by BCA. All measurements were performed in triplicates in a Synergy HTX Multi-Mode Reader (Biotek).
Histology and Immunohistochemistry
Samples of liver tissue before and after decellularization were analyzed using standard histological and immunohistological techniques. Picrosirius red (PSR) under polarized light and 4′,6-diamidino-2-phenylindole (DAPI) to stain intact nuclei before and after decellularization were used. Tissue samples were fixed with 4% buffered formaldehyde (Fisher Scientific, F79-4) for 20 min, washed with Hank’s balanced salt solution (HBSS; Invitrogen, Carlsbad, CA, USA), treated serially with increasing concentrations of sucrose (from 5% to 20%), and then incubated overnight with 20% sucrose (Sigma-Aldrich, St. Louis, MO, USA). The sucrose-treated biopsies were embedded in a 20% sucrose:histoprep mixture (2:1; Fisher, Pittsburgh, PA, USA), frozen by immersing them in isopentane, and cooled by liquid nitrogen and stored at −80 °C for immunohistochemical analysis. Paraffin embedded or cryopreserved slides were processed, stained, and visualized using an Olympus BX60 microscope (Center Valley, PA, USA) equipped with the appropriate filters for polarized light.
Chemotaxis Assay
To evaluate the immunological response of human monocytes in the presence of pLECM in vitro, a chemoattractant assay of primary human blood monocytes was performed. The peripheral blood mononuclear cells (PBMCs) were isolated from whole peripheral blood of healthy human donors by density gradient centrifugation. Each vial (BD® Hemogard™, BD Vacutainer®, 367654, Franklin Lakes, NJ, USA) of whole blood collected in purple top vials ethylenediaminetetraacetic acid (EDTA) was diluted with an equal volume of phosphate-buffered saline (PBS) + 2% heat inactivated fetal bovine serum (HIFBS; Thermo, 10438026), then added to 15 mL of Ficoll-Paque™ PLUS (GE Healthcare Bio-Sciences) and centrifuged at 400× g without breaks for 30 min at room temperature. The buffy coat at the interface was carefully removed, added to 30 mL of PBS + 2% HIFBS, and centrifuged at 400× g for 10 min. The resulting cell pellet was washed twice with PBS and resuspended in complete Roswell Park Memorial Institute (RPMI) 1604 media (Thermo Cat. 11875119) before seeding in a T-75 tissue culture flask. After 3 h, the media was replaced with fresh media, and the adherent cells (predominantly monocytes) were cultured for 48 h. Chemotaxis was evaluated in a transmigration 96-well plate using BD Falcon FluoroBlok multiwell inserts with 3 µm pores. Freshly isolated PBMCs were stained using cell tracker CM-DiI (Molecular Probes®, Eugene, OR, USA; C7001) according to manufacturer’s specifications. Stained PBMCs were resuspended in DMEM (Gibco 0567-014) media at a density of 2 × 106 cells/mL and added onto upper compartment (50 µL/well) and set aside. In a separate BD Falcon™ 96-square well, flat-bottom plate, 200 µL of chemoattractant solution, consisting of 300 µg of total protein, from decellularized and native liver tissue, in DMEM, was added to the bottom compartment; 50 nM monocyte chemotactic protein-1 (R&D Systems, Minneapolis, MN, USA; 279-MC) was used as positive migration control per manufacturer’s recommendation.44 The multiwell insert containing the cells was gently lowered into the plate containing chemoattractants and immediately placed into an incubator at 37 °C for 1 h. After 1 h of incubation, cells that had migrated from the top to the bottom chamber were quantified by reading the fluorescence of migrated cells at 545/575 nm wavelength (Biotek).
Cytotoxicity/Attachment/Proliferation of HepG2/C3A
Cytotoxicity, attachment, and proliferation of a human cell line HepG2/C3A (ATCC® CRL-10741™) were measured by culturing cells with DMEM and 10% HIFBS on the pLECM substrate. To coat TC plates, solubilized pLECM (5 mg/mL) was mixed 1:1 with DMEM. Then, 200 µL was added per well (6-well plate) and incubated in a 5% CO2 humidified incubator at 37 °C overnight. Excess solubilized pLECM solution was then removed and the wells were washed twice with PBS before culturing cells. All cell culture experiments were performed in a 5% CO2 humidified incubator at 37 °C for 7 d. To assess cell viability, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used.
System Equipment Setup for Hepatocyte Isolation from Porcine Liver
Porcine primary hepatocytes were isolated to evaluate the effect of pLECM substrate on hepatocytes and their native functions in vitro. The perfusion apparatus to isolate porcine hepatocytes from livers included 2 perfusion pumps (Masterflex L/S 16® for inflow and Fisher Scientific™ Variable-Flow—medium–high flow for outflow), in conjunction with an oxygenator (made with Silastic® medical oxygen permeable tubing) and a bubble trap to create a constant flow of the various solutions used in the protocol. All equipment was previously sterilized using 70% ethanol or autoclaved.
Preparation of Perfusion Solutions
All perfusion solutions were prepared fresh, under sterile conditions, filtered with a 0.22 µm filter, and prewarmed in a water bath (41 °C) before use. Freshly excised porcine liver tissue was placed in cold solution 0 (Table 1) and transferred to a biological safety cabinet with the perfusion apparatus. The liver section was placed in the appropriate size container and partially filled with enough solution 0 to cover tissue when submerged. Perfusion lines were purged prior to initiating perfusion to ensure that all lines and bubble trap were free of air bubbles prior to initiating perfusion.
Table 1.
Hepatocyte Isolation Solutions.
| Solution | Constituent | Final Concentration |
|---|---|---|
| Solution 0 | Lactate ringers | 250 mL |
| Heparin | 500 U | |
| Mannitol | 50 cc (20%) | |
| Glucose and insulin transferrin sodium selenite | 25 nM | |
| Solution 1 | Hank’s balanced salt solution (HBSS), Ca/Mg free | 250 mL |
| ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) | 0.5 mM | |
| Bovine serum albumin (BSA) | 0.5% w/v | |
| Ascorbic acid | 50 µg/mL | |
| Solution 2 | HBSS with Ca/Mg | 250 mL |
| Solution 3 | Dulbecco’s modified eagle medium | 300 mL |
| BSA | 0.5% w/v | |
| Collagenase P | 0.05% w/v | |
| Soybean trypsin inhibitor | 1.25 mL/mL | |
| Solution 4 | William’s E Medium | 500 mL |
| Penicillin-streptomycin-glutamine (100×) | 5 mL | |
| Sodium pyruvate | 1 g | |
| Solution 5 | HBM™ Basal Medium Lonza HCM™ SingleQuots™ Kit Lonza |
Hepatocyte Isolation
Hepatocytes were isolated from a porcine liver section using a 2-step collagenase perfusion. The liver section (∼10 g) was excised with a single cut from the tip of the middle lobe to include as much intact Glisson’s capsule as possible. Before starting the isolation, the perfusion system was primed with solution 1 (Table 1). An irrigation cannula tip was utilized to flush out any remaining blood in the liver section using 250 mL of solution 1 at a low flow rate (10 to 20 mL/min) until the tissue became lighter, an indication of good perfusion. The liver section was kept moist with gauze soaked in 0.9% saline solution (Hospira, Lake Forest, IL, USA; 0409-7983-03). Next, 250 mL of solution 2 (Table 1) was used to perfuse the liver section followed by 300 mL of solution 3 (Table 1). The flow rate was kept between 10 and 100 mL/min. Solution 3 was reused until the liver was sufficiently digested. At the end of the perfusion, the liver tissue disintegrated under the Glisson’s capsule indicating the end of the perfusion step. The liver section was then placed in a dish containing cold (4 °C) 50 mL of solution 4 (Table 1). The Glisson’s capsule was carefully removed and the cells were gently shaken out into the dish. The cell suspension collected in solution 4 was filtered twice through a sterile 100-µm nylon mesh and centrifuged at 50× g for 5 min at 4 °C. The supernatant was aspirated and the cell pellet was resuspended in 50 mL of solution 4. The centrifugation of the cell pellet in solution 4 was repeated 3 times, with a final resuspension of the cells in 50 mL of cold solution 5 (Table 1). Cell viability was evaluated using a trypan blue exclusion assay.
Cell Culture of Porcine Hepatocytes on pLECM
To evaluate the effect of solubilized pLECM on hepatocytes, porcine primary hepatocytes were cultured on pLECM-coated multiwell plates. Type I collagen–coated plates (Cat. A11428-01, Gibco) were used as control. Various concentrations of pLECM were used to optimize the assay based on initial seeding attachment to obtain a confluent monolayer (data not shown).
Viability and Proliferation
MTT (3-(4,5-dimethylthiozole-2-yl)-2,5-diphenyltetrazolium bromide, Molecular Probes M6494) assay was used to evaluate cell number/viability. A standard curve with known cell number was used at each time point evaluated. Briefly, MTT solution (25 µL, 5 mg/mL) was added to each well and incubated for 5 h in a 5% CO2, 37 °C humidified incubator. After incubation, the formazan complex was extracted with DMSO and the absorbance was measured at 570 nm.
Albumin Production by Porcine Hepatocytes
Albumin secreted by porcine hepatocytes was measured using a commercially available porcine-specific ELISA kit (Abcam, ab108794) designed for the quantitative measurement of albumin levels in cell culture conditioned media. Known concentrations of porcine albumin were tested in parallel with the experimental samples, and absorbance measurements were performed in a Synergy HTX Multi-Mode Reader (Biotek). Sample values were normalized by cell number quantified by MTT as described above.
Colorimetric Assay for Ureogenesis
Urea secretion was measured in supernatant of cultured cells collected at various time points using a colorimetric assay kit (QuantiChrom™ Urea Assay Kit, Hayward, CA, USA) according to the manufacturer’s instructions. Known concentrations of urea were tested in parallel with the experimental samples. All measurements were performed in a Synergy HTX Multi-Mode Reader (Biotek). Sample values were normalized by cell number quantified by MTT as described above.
Fluorescence Microscopy for Bile Canaliculi Formation and Low-density Lipoprotein Uptake
Bile function formation was assessed by fluorescence microscopy, using cholyl-L-lysyl-fluorescein (CLF, Corning 451041, Corning, NY, USA), a fluorescent bile salt derivative. CLF was added (5 µM) to cells in culture, incubated at 37 °C, 5% CO2, for 30 min, and followed by 2 washes with HBSS. Hepatocyte morphology and accumulated CLF were imaged using a fluorescent microscope (Invitrogen® EVOS™ FL Auto Imaging System, Carlsbad, CA, USA).
Results
Decellularization and Characterization of Porcine Liver Tissue
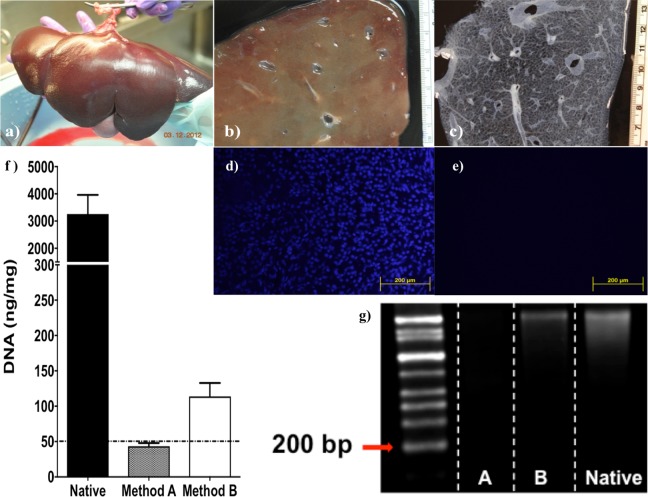
Porcine livers were successfully procured, using a surgical procedure similar to those used in human liver procurements for transplants45 (Fig. 1a). Effective cell lyses with minor disruptions of the ECM ultrastructure46 and uniform shape of the liver tissue was accomplished by freeze-thaw cycles and slicing while the tissue was still frozen, respectively (Fig. 1b). Both decellularization protocols (methods A and B) resulted in clear tissues (Fig. 1c). DAPI staining of intact nuclei and picogreen quantification of dsDNA revealed that both methods A and B significantly reduced the amount of cellular dsDNA (P < 0.01) from native porcine liver with high efficiency. Specifically, method A removed 98.67% and method B removed 96.53% of dsDNA compared to native tissue (N = 5). However, only method A resulted in a concentration of <50 ng/mg dry weight of dsDNA (Fig. 1f). Both methods A and B completely removed intact cell nuclei as seen by DAPI histological section staining (Fig. 1d and e). Interestingly, only method A had DNA fragments <200 bp (Fig. 1g). Based on the above data, method A fulfilled all minimal criteria for successful decellularization according to Badylak et al.47, (a) <50 ng dsDNA per mg ECM dry weight, (b) lack of visible nuclear material in tissue sections, and (c) <200 bp DNA fragment length, and method B did not.
Fig. 1.
(a) Porcine liver was (b) sliced and (c) decellularized. Nuclei staining (4′,6-diamidino-2-phenylindole) of histological section of (d) native porcine tissue and (e) decellularized porcine liver. (f) Quantification of dsDNA (PicoGreen stain) of native porcine liver and decellularized porcine liver using methods A and B (n = 5). (g) Southern blot of DNA from native porcine liver (native) and decellularized porcine liver using methods A (A) and B (B).
Protein Content in the Decellularized Porcine Tissue
Mass Spectrometry analysis of the pLECM produced a total of 701 proteins that were identified and categorized by NCBI annotation as S. scrofa (pig) with a probability of over 95%. Statistically, a total of 560 proteins were shared, without statistical difference (N = 3), between the liver decellularized by methods A and B. In contrast, a total of 48 proteins were significantly higher in method A than method B, and a total of 93 proteins were significantly higher in method B compared to method A (Fig. 2a). Proteins were classified using comprehensive GO annotations. The protein class of those statistically different proteins found between liver ECM decellularized by methods A and B was analyzed; it became evident that the nature of proteins was different (Table 2). Decellularized liver ECM by method A contained higher amount of proteins with a classification pertinent to the extracellular component such as fibronectin, proteoglycans, and collagen I/IV, necessary for the regeneration of liver and critical in the differentiation and stability of hepatocytes.48–50 On the contrary, the proteins in liver ECM decellularized by method B were categorized as proteins pertinent to intracellular constituents indicating incomplete decellularization. In addition, SDS-PAGE of pLECM obtained by methods A and B results showed that the decellularized liver by method B contained lower molecular weight proteins than those seen by method A (Fig. 2b). Based on the above findings, we selected method A as the most efficient method for decellularization of porcine liver; hence, from this moment forward, method A was used for the characterization of pLECM.
Fig. 2.
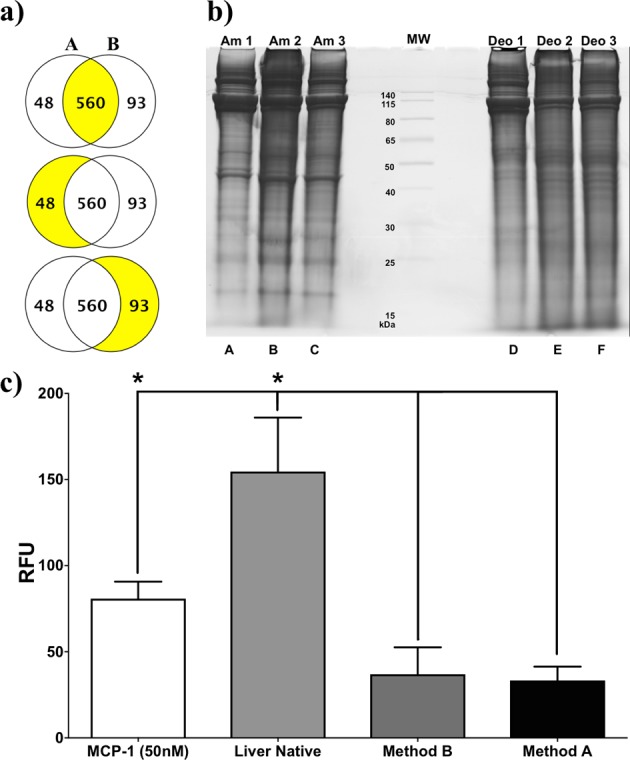
(a) Venn diagrams of proteins identified by NCBI/scaffold as unique for Sus scrofa (P < 0.05) from decellularized porcine liver using method A (left) compared to decellularized porcine liver using method B (right) and the shared proteins (P > 0.05; middle). (b) SDS-PAGE of total proteins extracted from porcine liver decellularized with method A (ammonium hydroxide; Am1, Am2, and Am3) and method B (sodium deoxycholate; Deo1, Deo2, and Deo3; N = 3). (c) Human monocyte migration (chemotaxis) in the presence of various extracellular matrix products. Porcine liver native, porcine liver decellularized (methods A and B; N = 3). *P < 0.05.
Table 2.
Highest Abundance of Proteins (Top 10) by Mass Spec for Decellularized Porcine Liver Processed by Methods A and B.
| Method A | Method B |
|---|---|
| Keratin, type II cytoskeletal 8 [Sus scrofa] (gi|227430407) | Carbamoyl phosphate synthase (ammonia), mitochondrial [Sus scrofa] (gi|545875504) |
| Keratin, type I cytoskeletal 18 [Sus scrofa] (gi|927128593) | Cytochrome P450 2C49 precursor [Sus scrofa] (gi|47523894) |
| Myosin-10-like, partial [Sus scrofa] (gi|927198158) | Uridine 5′-diphospho-glucuronosyltransferase (UDP) glucuronosyltransferase 1 family, polypeptide A6 isoform X2 [Sus scrofa] (gi|545876003) |
| Collagen α-1(I) chain [Sus scrofa] (gi|927196616) | Aldehyde dehydrogenase, mitochondrial precursor [Sus scrofa] (gi|113205888) |
| Type VI collagen α-1 chain [Sus scrofa] (gi|92020086) | Catalase [Sus scrofa] (gi|356460899) |
| Collagen α-2(I) chain precursor [Sus scrofa] (gi|343887367) | 60 kDa heat shock protein, mitochondrial [Sus scrofa] (gi|359811347) |
| Trifunctional enzyme subunit α, mitochondrial [Sus scrofa] (gi|47522754) | Prolyl 4-hydroxylase, β polypeptide [Sus scrofa] (gi|358009193) |
| Collagen α-2(VI) chain [Sus scrofa] (gi|927203606) | Alternative pig liver esterase [Sus scrofa] (gi|164414678) |
| Collagen α-1(XIV) chain isoform X1 [Sus scrofa] (gi|927116442) | Glutamate dehydrogenase 1, mitochondrial [Sus scrofa] (gi|347300243) |
| Collagen α-5(VI) chain isoform X1 [Sus scrofa] (gi|545860855) | Peroxisomal multifunctional enzyme type 2 [Sus scrofa] (gi|47523670) |
Immunological Response of Human Monocytes to pLECM
Immunological response assays provided significantly lower (P < 0.05, N = 3) chemoattraction values of human monocytes to decellularized porcine liver (pLECM) than those measured in native porcine liver (Fig. 2c).28,51,52 However, there was no statistical difference between methods A and B. Similarly, comparison of pLECM to commercially available grafts clearly showed the amount of monocyte chemoattraction of the commercially available grafts was much higher (∼2-fold and ∼10-fold) than those of decellularized liver (data not shown).
Effect of Decellularization on Collagen
PSR staining of pLECM in combination with polarized light showed that decellularization using method A preserved the morphology and the color pattern of the collagen fibers, a sign of effective decellularization, while retaining the fibrillar native ECM configuration (Fig. 3a and b). Specifically, yellow fibers were visualized on the perimeter of the liver lobule and the central vein and green fibers were seen between the sinusoidal spaces of Disse, typical of that seen in the normal liver tissue.
Fig. 3.
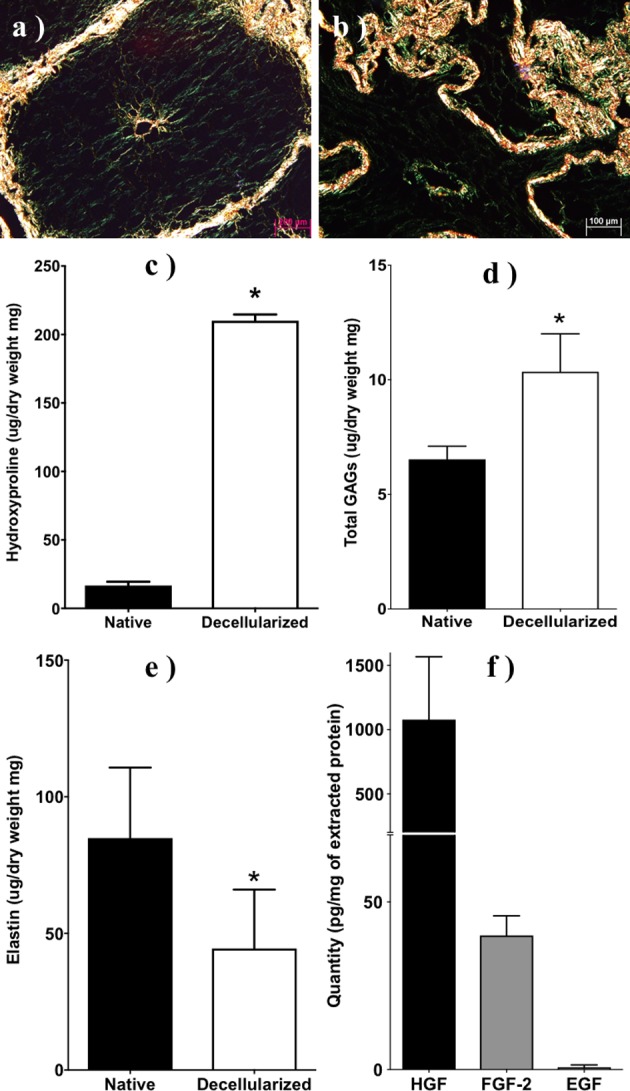
Histological sections of porcine liver stained with picrosirius red under polarized bright field (a) before and (b) after decellularization. Scale bar: 200 and 100 µm, respectively. Total concentration of (c) hydroxyproline, (d) glycosaminoglycans (GAGs), (e) elastin, (f) hepatocyte growth factor (HGF), fibroblast growth factor 2 (FGF-2), and epidermal growth factor (EGF) in native and decellularized porcine liver using method A (N = 3). *P < 0.05.
ECM Biochemistry
Biochemical analysis of selected extracellular proteins, including collagen (hydroxyproline), elastin, and glycoproteins, before and after decellularization (method A) demonstrated increased concentrations of all proteins except elastin. Specifically, the amount of collagen (normalized by dry weight) statistically (P < 0.05) increased from native to decellularized porcine liver; <1% (33 μg/mg) to >18% (183.7 μg/mg) of total dry weight, respectively (Fig. 3c). Similarly, there was a significant (P < 0.05) increase of GAGs in the decellularized porcine liver (10.35 µg/mg) compared to native (6.5 µg/mg) porcine liver (Fig. 3d). In contrast, elastin showed a significant reduction (P < 0.05) in the decellularized porcine liver 44.5 µg/mg from 84.9 µg/mg in native porcine liver (Fig. 3e). To further understand the bioactive potential of our pLECM, we examined the presence of key growth factors in liver function including HGF, EGF, and bFGF. ELISA results demonstrated the presence of HGF (1.07 µg/mg) in our pLECM (Fig. 3f) and it was the most abundant of the growth factors tested53,54 and higher than those found in rat liver.55 EGF was detected at 0.7 pg/mg in the pLECM (Fig. 3f) and was the least abundant detected from the growth factors tested. Likewise, bFGF was detected at 40 pg/mg (Fig. 3f).
Solubilization of Decellularized Porcine Liver
Lyophilized pLECM (Fig. 4a) was solubilized (Fig. 4b). To optimize and understand the effect of the solubilization variables in the pLECM, we first evaluated the effect of pepsin concentration in the digestion of pLECM. Partial digestion of pLECM with pepsin was attained and reached a saturation point at 20 mg (data not shown) for 50 mg of pLECM. Indicating that pepsin quantities below 20 mg are suboptimal for digestion and higher amounts are unnecessary. The highest yield of solubilized pLECM (27.3%) was reached using 0.01 N HCl (method A) and it plateaued at higher concentrations (Fig. 4c). Similarly, the highest yield of solubilized pLECM (24.5%) using AA (method B) was seen at a concentration 0.5 M (Fig. 4c). Both methods resulted in similar pLECM solubilization yield (P > 0.05). However, the concentration of AA (0.5 M) was 50 times higher than that required by HCl (0.01 N) to achieve the same solubilization yield. Finally, the pLECM solubilization was optimal using 50 mg ECM: 10 mL, 0.5 M AA: 20 mg pepsin.
Fig. 4.
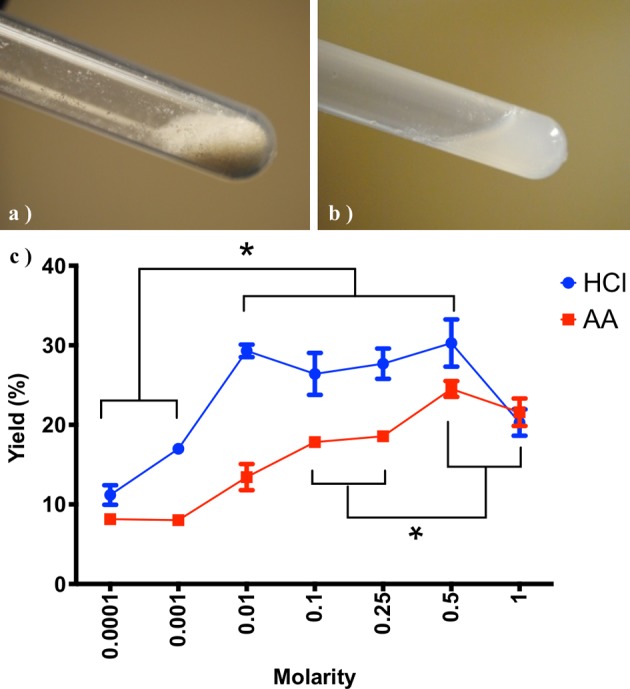
Decellularized porcine liver in (a) powder form and (b) solubilized. (c) Solubilization of porcine liver extracellular matrix using different concentrations and types of acid (N = 3). *P < 0.05.
Immunogenic Response of Solubilized pLECM
The chemoattraction values of human monocytes to solubilized decellularized porcine liver by both AA and HCl methods were significantly lower (P < 0.05, N = 3) than those measured in native porcine liver (Fig. 5). Interestingly, the chemoattractant values of solubilized pLECM by either method were not different than the pLECM alone, suggesting that the solubilization did not influence the chemotaxis of human monocytes (Fig. 5).
Fig. 5.
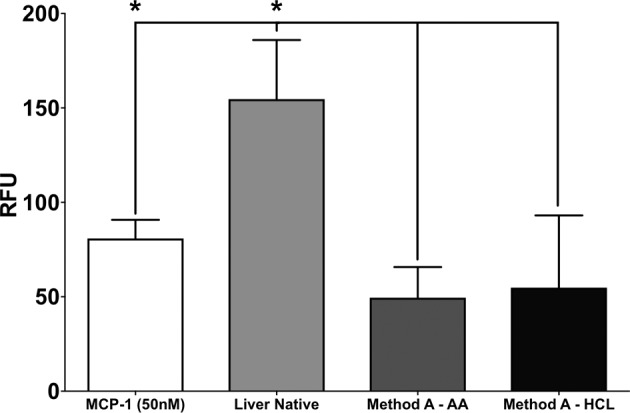
Human monocyte transmigration in the presence of various extracellular matrix (ECM) products. Porcine liver ECM (pLECM; method A), solubilized pLECM with acetic acid (AA) and hydrochloric acid (HCl), and Monocyte Chemotactic Protein 1 (MCP-1) were used as control (N = 3). *P < 0.05.
Cytotoxicity of Solubilized pLECM on HepG2/C3A Cells
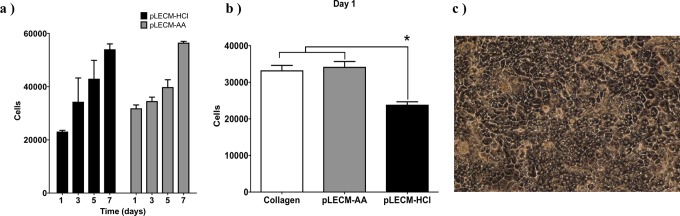
MTT results demonstrated similar increase in cell proliferation and noncytotoxic effect from the substrate to HepG2/C3A cells cultured on solubilized pLECM-AA- and pLECM-HCl-coated plates (Fig. 6a). However, the number of HepG2/C3A cells that attached after seeding (day 1) was higher in the pLECM-AA than the cells cultured in pLECM-HCl (Fig. 6b). In addition, the attachment rate of cells on pLECM-AA was similar to that of cells cultured on rat type I collagen–coated wells which is considered the gold standard in hepatocyte culture in vitro (Fig. 6b).56–59
Fig. 6.
(a) Proliferation of HepG2/C3A cells cultured on substrates made of porcine liver extracellular matrix (pLECM)–hydrochloric acid (HCl) and pLECM–acetic acid (AA). (b) Attachment (day 1) of HepG2/C3A cells cultured on different biomatrices (N = 3). *P < 0.05. (c) Primary porcine hepatocytes cultured on pLECM-AA. 100× Magnification.
Albumin and Urea Production by HepG2/C3A
Conditioned media from HepG2/C3A cultured on pLECM-AA showed statistically (P < 0.05) higher cumulative albumin when cultured in pLECM-AA (416.6 μg/1 × 106 cells/mL) substrates compared to cells cultured in pLECM-HCl (355.5 μg/1 × 106 cells/mL) substrates (Fig. 7a). Similarly, HepG2/C3A cultured on pLECM-AA showed statistically (P < 0.05) higher cumulative amounts of urea (13,469.2 μg/dL/1 × 106) compared to cells cultured on pLECM-HCl (10,062.4 μg/dL/1 × 106) substrates (Fig. 7b). Therefore, pLECM-AA was selected as the preferred solubilized substrate for the in vitro culture of cells from this point forward.
Fig. 7.
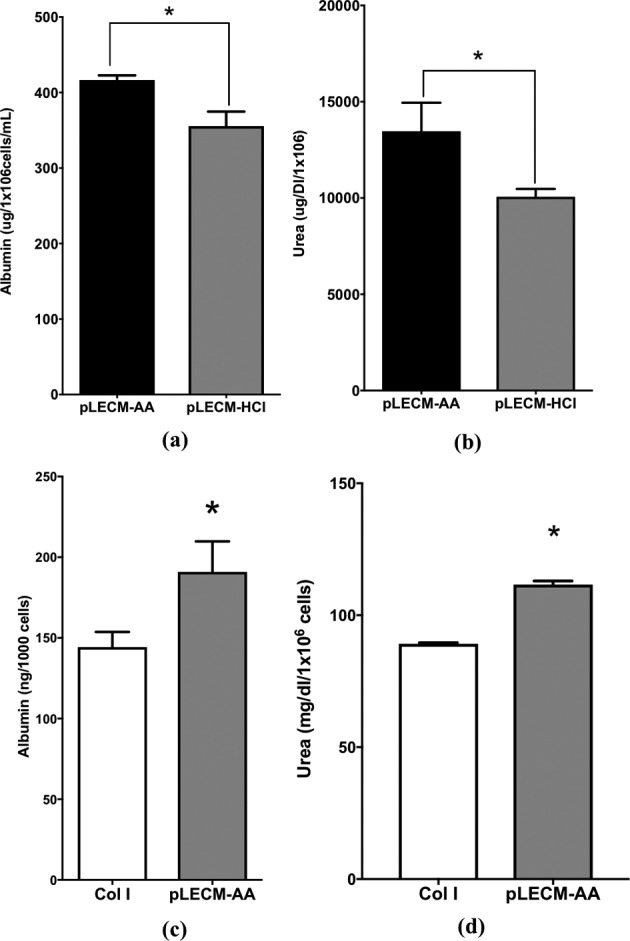
(a) Cumulative (7 d) albumin production and (b) urea production from HepG2/C3A cells cultured on porcine liver extracellular matrix (pLECM)–acetic acid (AA) and pLECM–hydrochloric acid (HCl; n = 3; *P < 0.05). (c) Albumin production from primary porcine hepatocytes cultured on pLECM-AA and type I collagen substrates. (d) Urea production from primary porcine hepatocytes cultured in pLECM-AA and type I collagen substrates (N = 3; *P < 0.05).
Isolation of Primary Porcine Hepatocytes
Hepatocytes were isolated from a porcine liver section taken from the middle lobe (∼10 g) of an intact porcine liver, using a 2-step collagenase perfusion. The average porcine hepatocyte isolation yielded ∼5 to 6 × 106 cells/g of wet liver, with an average viability (trypan blue exclusion) of ∼95%. Isolated porcine primary hepatocyte cells were then cultured on the pLECM-AA substrate at various seeding densities in order to optimize the appropriate cell number, which has been reported to be crucial in hepatocyte attachment, morphology, and cell–cell interaction in vitro.9,10,20,60–62 Based on the above parameters, we concluded that the optimal seeding density was ∼130,000 cells/cm2 for culturing hepatocytes that displayed a confluent monolayer with cuboidal morphology (Fig. 6c).
Albumin and Urea Synthesis by Primary Porcine Hepatocytes
Evaluation of albumin and urea synthesis by primary porcine hepatocytes in monolayer culture over 7 d produced more albumin (P < 0.05) when cultured on pLECM-AA substrates compared to cells cultured on standard type I collagen substrates. Porcine hepatocytes synthesized an average of 31.81 ng/1,000 cells/d of albumin when cultured on monolayers of pLECM-AA compared to 24.05 ng/1,000 cells/d of albumin when cultured on type I collagen (P < 0.05). The cumulative albumin production of primary porcine hepatocytes cultured on pLECM-AA substrates was 31.82 ng/1,000 cells, which is significantly higher (P < 0.05) than cumulative albumin produced by porcine hepatocytes cultured on type I collagen (24.04 ng/1,000 cells) substrates (Fig. 7c). Similarly, porcine hepatocytes synthesized significantly (P < 0.05) more urea when cultured on pLECM-AA substrates compared to those cells cultured on standard type I collagen substrate. Porcine hepatocytes synthesized an average of 18.4 mg/dL/1 × 106 cells/d of urea when cultured on monolayers of pLECM-AA compared to 14.8 mg/dL/1 × 106 cells/d of urea when cultured on type I collagen (P < 0.05). The cumulative levels of urea were highest when porcine hepatocyte cells were cultured on pLECM-AA (111.6 mg/dL/1 × 106 cells) compared to those cells cultured on type I collagen (89.14 mg/dL/1 × 106 cells) substrates (P < 0.05, Fig. 7d).
Bile Canaliculi Formation
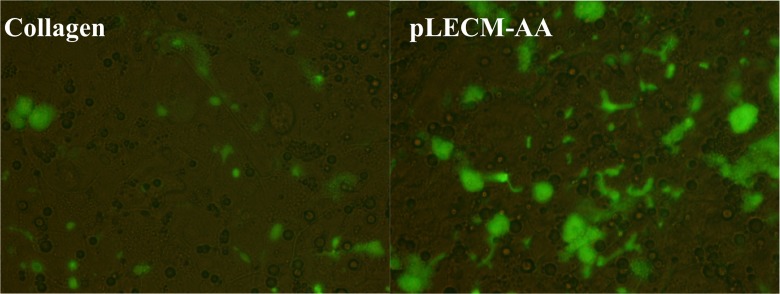
The CLF staining of porcine hepatocytes cultured in pLECM-AA showed fluorescent staining associated to functional canalicular space formation and hepatocyte polarization. Furthermore, cells cultured on pLECM-AA showed more robust fluorescent structures than those cells cultured on type I collagen substrate, and the difference between the canalicular spaces/fluorescence was more noticeable on the last day of culture (Fig. 8).
Fig. 8.
Cholyl-lysyl-fluorescein retention on hepatocytes canalicular structures when cultured on type I collagen (left) and porcine liver extracellular matrix–acetic acid (right) at day 6. 200× Magnification.
Discussion
In this study, we have successfully developed and optimized a method to create a substrate from decellularized porcine liver (pLECM) that enhances pertinent biological functions of primary porcine hepatocytes in vitro compared to the standard collagen type I substrates. Although both decellularization methods, ammonium hydroxide and deoxycolate, produced comparable results in the reduction of dsDNA and intact nuclei, pLECM-AA exhibited higher preservation of ECM components and the biochemical profile typically seen in the native liver. These ECM components are not only needed as adhesion molecules for the survival of hepatocytes but are also essential as bioactive molecules, since they can change the fate of cultured cells due to their inherent mechanical properties, migrating platform, and/or signal transduction activation.
Some liver ECM proteins increased in quantity after decellularization. Specifically, there was a significant increase in hydroxyproline and GAGs in the decellularized liver compared to native liver which can be explained as follows: the normalization was based on dry weight of tissue, which in the native liver is largely due to cells. Cells do not contain insoluble collagen fibers or GAGs; hence, the percentage of collagen per total dry weight is low in the native liver. After decellularization most of the dry weight in the tissue is ECM, something that could be described as “compaction” of the ECM, which occurs with the removal of the cells and causes the tissue to “collapse” or shrink. Hence, the higher content of collagen and GAGs is in the decellularized liver. In contrast, elastin showed a significant reduction, primarily because it is synthesized intracellularly from soluble monomers (tropoelastin).63–65 Our method of detection uses 5,10,15,20-tetraphenyl-21H,23H-porphine tetrasulfonate which is not capable of distinguishing insoluble from soluble elastin. Hence, the measured elastin in the native liver was not entirely from the ECM but from a combination of intra- and extracellular compartments. It is possible that porcine liver tissue specifically contains a considerable amount of intracellular soluble elastin (tropoelastin) that is being statistically (P < 0.05) reduced by our method of decellularization.66–69 It is important to mention that elastin is a minor connective tissue component in normal liver, but it is actively synthesized by hepatic stellate cells and portal fibroblasts in diseased liver. Elastin precursor tropoelastin, elastin polymer, and elastin degradation products all possess biological activity that may influence inflammatory cells and connective tissue cells in the organ.70
Analysis of growth factor present in the pLECM decellularized with ammonium hydroxide showed high quantities of HGF followed by fibroblast growth factor 2 (FGF-2) and small amounts of EGF after decellularization. Expression of the above growth factors plays a very important role in regeneration and hepatocyte proliferation. Specifically, a typical effect after partial hepatectomy is the interplay between hepatocytes, nonparenchymal cells, and the ECM that leads to the rapid restoration of liver mass, predominantly through the proliferation of the adult hepatocyte population.71 This process is driven by a diverse range of mitogens of which HGF, epidermal growth factor receptor (EGFR) ligands, fibroblast growth factor 1, and FGF-2 are among the most potent.72 HGF causes a strong mitogenic response and clonal expansion of hepatocytes in culture.73 When HGF and EGFR ligands are infused into the regenerating liver (e.g., after partial hepatectomy), they increase the rate of hepatocyte proliferation and can lead to a faster restoration of liver mass.74–76
Solubilization of pLECM reached comparable total protein yield in solution with 0.5 M AA and 0.01 N HCl. pLECM-AA showed higher initial attachment and higher albumin and urea production in HepG2/C3A cell cultures compared to pLECM-HCl. The need of a higher AA concentration to achieve comparable HCl yields was anticipated due to the nature of the acids: strong (HCl) versus weak (AA). Similarly, it is possible that the weak nature of the AA better preserved bioactive moieties compared to solubilization using HCl, thus the different HepG2/C3A biological response.
Another important element to study when using acellular matrices in tissue engineering is the ability to characterize their possible immunogenicity. In our in vitro chemotactic assay, the developed pLECM-AA demonstrated lower immunogenicity when exposed to human monocytes compared to native tissue. Although an in vitro immunogenicity test cannot predict with certainty the in vivo response, our results are encouraging. This assay seems to not only provide a functional way to study immunogenicity but also a functional way to quantify the quality of decellularization in vitro.
To better understand the biological influence of our pLECM-AA on hepatocytes, we cultured primary porcine hepatocytes on pLECM-AA-coated plates. Then, quantified their albumin synthesis,77–79 which is commonly used as a marker to describe the overall hepatocyte differentiated state due to its adult-liver specificity. We also measured ureogenesis (urea synthesis from ammonia),80 which is an exclusive function of the liver, and the formation of intact/functional canalicular networks. The study of these functions was done over the course of 7 d, since hepatocytes cultured in a conventional monolayer usually retain their hepatocyte characteristics for approximately 1 wk accompanied by rapid metabolic decline, phenotypic alterations, and apoptosis.81,82 Albumin and urea synthesis by primary porcine hepatocytes were higher on pLECM-AA than that on type I collagen indicating that pLECM-AA is more suitable for culturing primary porcine hepatocytes compared to type I collagen substrates.
In addition to albumin and urea synthesis, we studied bile formation, which is a vital function of the liver and involves sodium-dependent and sodium-independent uptake of bile salts from the portal blood and Adenosine triphosphate (ATP)-dependent canalicular excretion.83 In this study, we used fluorescein-labeled CLF, which closely resembles the biological behavior of bile acid that naturally occurs with cholylglycine. CLF is commonly used as an agent for determining bile transport function, since it is mainly secreted into bile canaliculi by bile salt efflux pump.84 This assay is a simple and fast method to evaluate changes of tight junction polarity and permeability in hepatocyte couplets. Retention of the fluorescent dye is an indication of canalicular space formation and an indication of hepatocyte functional polarization.85 Conversantly, the fluorescent dye is secreted to the media when hepatocyte polarity is lost.86 Our results showed higher retention of the fluorescent dye by the hepatocytes cultured on pLECM-AA than those on type I collagen, suggesting that hepatocytes cultured on pLECM-AA are possibly going through a slower epithelial–mesenchymal transition or the ECM components are conducive of canalicular structure formation. Due to the complexity of the matrix, it is not clear which molecule(s) is the main contributor in retaining the epithelial functionality of the hepatocytes. However, few candidates like keratin, heparan sulfate, plectin, and plakophilin would be worth investigating in the future.87–91
It is important to note that the cellular functions studied here are not the only relevant properties when studying hepatocytes. Hence, attention is recommended when selecting the best decellularization/solubilization method used, depending on the parameter most relevant to the investigator.
Establishment and maintenance of hepatocyte polarity is essential for many functions of hepatocytes and requires carefully orchestrated cooperation between cell adhesion molecules, cell junctions, cytoskeleton, ECM, and intracellular trafficking machinery. Having a substrate-like pLECM-AA that can prolong hepatocyte polarity is of great advantage and needed technology. This biomatrix is of special importance in the field of medical research and translational technologies that rely on accurate and stable physiological hepatocyte functions. In our future work, we are planning to study on a deeper level the metabolic effect of pLECM-AA on cells.
Although the method of decellularization and solubilization in this study was carefully designed to preserve most of the tissue properties, there is still much work to be done to identify the signaling mechanisms that promoted improved hepatocyte function on our pLECM-AA but not on pLECM-HCl and type I collagen matrix. For the time being, having a liquid pLECM-AA substrate that can be easily produced, stored, transported, printed, and applied to tissue culture plasticware represents a step forward in the ongoing advancements for liver studies that heavily rely on in vitro culture of hepatocytes.
Acknowledgments
We thank Dr. Vacanti’s laboratory group (Laboratory for Tissue Engineering and Organ Fabrication at the Massachusetts General Hospital), Ms. Kulig, and Dr. Neville for kindly providing us with the training necessary to isolate hepatocytes; also Dr. Badylak’s group and Dr. Gilbert’s laboratory for kindly providing training on decellularization; and Leon Bunegin, David Burmeister, Amit Aurora, and Nicole Rice for their expertise and feedback.
Footnotes
Ethical Approval: The procedures were approved by the Institutional Animal Care and Use Committee, US Army Institute of Surgical Research.
Statement of Human and Animal Rights: Statement of Human and Animal Rights is not applicable as livers were obtained from deceased porcine under a material transfer agreement.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Baccarani U, Adani GL, Sainz M, Donini A, Risaliti A, Bresadola F. Human hepatocyte transplantation for acute liver failure: state of the art and analysis of cell sources. Transplant Proc. 2005;37(6):2702–2704. [DOI] [PubMed] [Google Scholar]
- 2. Fisher RA, Strom SC. Human hepatocyte transplantation: worldwide results. Transplantation. 2006;82(4):441–449. [DOI] [PubMed] [Google Scholar]
- 3. Wang F, Zhou L, Ma X, Ma W, Wang C, Lu Y, Chen Y, An L, An W, Yang Y. Monitoring of intrasplenic hepatocyte transplantation for acute-on-chronic liver failure: a prospective five-year follow-up study. Transplant Proc. 2014;46(1):192–198. [DOI] [PubMed] [Google Scholar]
- 4. Kim WH, Lee JH, Han SU, Jin YM, Kwak YS, Wang HJ, Kim MW. Systematic analysis of the effects of hepatocyte transplantation on rats with acute liver failure. Hepatogastroenterology. 2000;47(32):371–374. [PubMed] [Google Scholar]
- 5. Cereghini S, Yaniv M, Cortese R. Hepatocyte dedifferentiation and extinction is accompanied by a block in the synthesis of mRNA coding for the transcription factor HNF1/LFB1. EMBO J. 1990;9(7):2257–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rowe C, Goldring CE, Kitteringham NR, Jenkins RE, Lane BS, Sanderson C, Elliott V, Platt V, Metcalfe P, Park BK. Network analysis of primary hepatocyte dedifferentiation using a shotgun proteomics approach. J Proteome Res. 2010;9(5):2658–26568. [DOI] [PubMed] [Google Scholar]
- 7. Fraczek J, Bolleyn J, Vanhaecke T, Rogiers V, Vinken M. Primary hepatocyte cultures for pharmaco-toxicological studies: at the busy crossroad of various anti-dedifferentiation strategies. Arch Toxicol. 2013;87(4):577–610. [DOI] [PubMed] [Google Scholar]
- 8. Arterburn LM, Zurlo J, Yager JD, Overton RM, Heifetz AH. A morphological study of differentiated hepatocytes in vitro. Hepatology. 1995;22(1):175–187. [PubMed] [Google Scholar]
- 9. Elaut G, Henkens T, Papeleu P, Snykers S, Vinken M, Vanhaecke T, Rogiers V. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr Drug Metab. 2006;7(6):629–660. [DOI] [PubMed] [Google Scholar]
- 10. Swift B, Brouwer KL. Influence of seeding density and extracellular matrix on bile acid transport and mrp4 expression in sandwich-cultured mouse hepatocytes. Mol Pharm. 2010;7(2):491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enns GM, Millan MT. Cell-based therapies for metabolic liver disease. Mol Genet Metab. 2008;95(1–2):3–10. [DOI] [PubMed] [Google Scholar]
- 12. Nussler A, Konig S, Ott M, Sokal E, Christ B, Thasler W, Brulport M, Gabelein G, Schormann W, Schulze M, et al. Present status and perspectives of cell-based therapies for liver diseases. J Hepatol. 2006;45(1):144–159. [DOI] [PubMed] [Google Scholar]
- 13. Bissell DM, Arenson DM, Maher JJ, Roll FJ. Support of cultured hepatocytes by a laminin-rich gel. Evidence for a functionally significant subendothelial matrix in normal rat liver. J Clin Invest. 1987;79(3):801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang YJ, Liu HL, Guo HT, Wen HW, Liu J. Primary hepatocyte culture in collagen gel mixture and collagen sandwich. World J Gastroenterol. 2004;10(5):699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fassett J, Tobolt D, Hansen LK. Type I collagen structure regulates cell morphology and EGF signaling in primary rat hepatocytes through cAMP-dependent protein kinase A. Mol Biol Cell. 2006;17(1):345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeisberg M, Kramer K, Sindhi N, Sarkar P, Upton M, Kalluri R. De-differentiation of primary human hepatocytes depends on the composition of specialized liver basement membrane. Mol Cell Biochem. 2006;283(1–2):181–189. [DOI] [PubMed] [Google Scholar]
- 17. Plant AL, Bhadriraju K, Spurlin TA, Elliott JT. Cell response to matrix mechanics: focus on collagen. Biochim Biophys Acta. 2009;1793(5):893–902. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y, He Y, Bharadwaj S, Hammam N, Carnagey K, Myers R, Atala A, Van Dyke M. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials. 2009;30(23–24):4021–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. te Velde AA, Ladiges NC, Flendrig LM, Chamuleau RA. Functional activity of isolated pig hepatocytes attached to different extracellular matrix substrates. Implication for application of pig hepatocytes in a bioartificial liver. J Hepatol. 1995;23(2):184–192. [DOI] [PubMed] [Google Scholar]
- 20. Hamilton GA, Jolley SL, Gilbert D, Coon DJ, Barros S, LeCluyse EL. Regulation of cell morphology and cytochrome P450 expression in human hepatocytes by extracellular matrix and cell-cell interactions. Cell Tissue Res. 2001;306(1):85–99. [DOI] [PubMed] [Google Scholar]
- 21. Mooney D, Hansen L, Vacanti J, Langer R, Farmer S, Ingber D. Switching from differentiation to growth in hepatocytes: control by extracellular matrix. J Cell Physiol. 1992;151(3):497–505. [DOI] [PubMed] [Google Scholar]
- 22. Lorenti A, Barbich M, Hidalgo A, Hyon SH, Sorroche P, Guinle A, Schenone A, Chamoles N, Argibay P. Culture of porcine hepatocytes: the dogma of exogenous matrix revisited. Artif Organs. 2001;25(7):546–550. [DOI] [PubMed] [Google Scholar]
- 23. Du Y, Chia SM, Han R, Chang S, Tang H, Yu H. 3D hepatocyte monolayer on hybrid RGD/galactose substratum. Biomaterials. 2006;27(33):5669–5680. [DOI] [PubMed] [Google Scholar]
- 24. Mitaka T, Sato F, Mizuguchi T, Yokono T, Mochizuki Y. Reconstruction of hepatic organoid by rat small hepatocytes and hepatic nonparenchymal cells. Hepatology. 1999;29(1):111–125. [DOI] [PubMed] [Google Scholar]
- 25. LeCluyse E, Bullock P, Madan A, Carroll K, Parkinson A. Influence of extracellular matrix overlay and medium formulation on the induction of cytochrome P-450 2B enzymes in primary cultures of rat hepatocytes. Drug Metab Dispos. 1999;27(8):909–915. [PubMed] [Google Scholar]
- 26. Lang R, Stern MM, Smith L, Liu Y, Bharadwaj S, Liu G, Baptista PM, Bergman CR, Soker S, Yoo JJ, et al. Three-dimensional culture of hepatocytes on porcine liver tissue-derived extracellular matrix. Biomaterials. 2011;32(29):7042–7052. [DOI] [PubMed] [Google Scholar]
- 27. Wang X, Cui J, Zhang BQ, Zhang H, Bi Y, Kang Q, Wang N, Bie P, Yang Z, Wang H, et al. Decellularized liver scaffolds effectively support the proliferation and differentiation of mouse fetal hepatic progenitors. J Biomed Mater Res A. 2014;102(4):1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mirmalek-Sani SH, Sullivan DC, Zimmerman C, Shupe TD, Petersen BE. Immunogenicity of decellularized porcine liver for bioengineered hepatic tissue. Am J Pathol. 2013;183(2):558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barakat O, Abbasi S, Rodriguez G, Rios J, Wood RP, Ozaki C, Holley LS, Gauthier PK. Use of decellularized porcine liver for engineering humanized liver organ. J Surg Res. 2012;173(1): e11–e25. [DOI] [PubMed] [Google Scholar]
- 30. Nakamura S, Ijima H. Solubilized matrix derived from decellularized liver as a growth factor-immobilizable scaffold for hepatocyte culture. J Biosci Bioeng. 2013;116(6):746–753. [DOI] [PubMed] [Google Scholar]
- 31. Yang YG, Sykes M. Xenotransplantation: current status and a perspective on the future. Nat Rev Immunol. 2007;7(7):519–531. [DOI] [PubMed] [Google Scholar]
- 32. Cooper DK, Ayares D. The immense potential of xenotransplantation in surgery. Int J Surg. 2011;9(2):122–129. [DOI] [PubMed] [Google Scholar]
- 33. Carpentier A, Lemaigre G, Robert L, Carpentier S, Dubost C. Biological factors affecting long-term results of valvular heterografts. J Thorac Cardiovasc Surg. 1969;58(4):467–483. [PubMed] [Google Scholar]
- 34. Dohmen PM, Lembcke A, Holinski S, Kivelitz D, Braun JP, Pruss A, Konertz W. Mid-term clinical results using a tissue-engineered pulmonary valve to reconstruct the right ventricular outflow tract during the Ross procedure. Ann Thorac Surg. 2007;84(3):729–736. [DOI] [PubMed] [Google Scholar]
- 35. O’Brien MF, Goldstein S, Walsh S, Black KS, Elkins R, Clarke D. The SynerGraft valve: a new acellular (nonglutaraldehyde-fixed) tissue heart valve for autologous recellularization first experimental studies before clinical implantation. Semin Thorac Cardiovasc Surg. 1999;11(4 Suppl 1):194–200. [PubMed] [Google Scholar]
- 36. Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17(20):2310–2316. [DOI] [PubMed] [Google Scholar]
- 37. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. [DOI] [PubMed] [Google Scholar]
- 38. Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10(6):1265–1269. [DOI] [PubMed] [Google Scholar]
- 39. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. [DOI] [PubMed] [Google Scholar]
- 40. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33(Database issue):D284–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanyal S, Qian S, Partridge J, Kosovksy M. Optimized chemotaxis conditions for primary blood monocytes or THP-1 cells using BD Falcon™ FluoroBlok™ 96-multiwell insert plates Technical Bulletin #457, BD Biosciences, Discovery Labware, Bedford, MA: 2007. [Google Scholar]
- 45. Starzl TE, Bell RH, Beart RW, Putnam CW. Hepatic trisegmentectomy and other liver resections. Surg Gynecol Obstet. 1975;141(3):429–437. [PMC free article] [PubMed] [Google Scholar]
- 46. Patel N, Solanki E, Picciani R, Cavett V, Caldwell-Busby JA, Bhattacharya SK. Strategies to recover proteins from ocular tissues for proteomics. Proteomics. 2008;8(5):1055–1070. [DOI] [PubMed] [Google Scholar]
- 47. Song Z, Peng Z, Liu Z, Yang J, Tang R, Gu Y. Reconstruction of abdominal wall musculofascial defects with small intestinal submucosa scaffolds seeded with tenocytes in rats. Tissue Eng Part A. 2013;19(13–14):1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Natesan S, Wrice NL, Baer DG, Christy RJ. Debrided skin as a source of autologous stem cells for wound repair. Stem Cells. 2011;29(8):1219–1230. [DOI] [PubMed] [Google Scholar]
- 49. Snykers S, Vanhaecke T, Papeleu P, Luttun A, Jiang Y, Vander Heyden Y, Verfaillie C, Rogiers V. Sequential exposure to cytokines reflecting embryogenesis: the key for in vitro differentiation of adult bone marrow stem cells into functional hepatocyte-like cells. Toxicol Sci. 2006;94(2):330–341; discussion 235-9. [DOI] [PubMed] [Google Scholar]
- 50. Yamamoto Y, Banas A, Murata S, Ishikawa M, Lim CR, Teratani T, Hatada I, Matsubara K, Kato T, Ochiya T. A comparative analysis of the transcriptome and signal pathways in hepatic differentiation of human adipose mesenchymal stem cells. FEBS J. 2008;275(6):1260–1273. [DOI] [PubMed] [Google Scholar]
- 51. Rieder E, Steinacher-Nigisch A, Weigel G. Human immune-cell response towards diverse xenogeneic and allogeneic decellularized biomaterials. Int J Surg. 2016;36(Pt A):347–351. [DOI] [PubMed] [Google Scholar]
- 52. Boer U, Schridde A, Anssar M, Klingenberg M, Sarikouch S, Dellmann A, Harringer W, Haverich A, Wilhelmi M. The immune response to crosslinked tissue is reduced in decellularized xenogeneic and absent in decellularized allogeneic heart valves. Int J Artif Organs. 2015;38(4):199–209. [DOI] [PubMed] [Google Scholar]
- 53. Masumoto A, Yamamoto N. Sequestration of a hepatocyte growth factor in extracellular matrix in normal adult rat liver. Biochem Biophys Res Commun. 1991;174(1):90–95. [DOI] [PubMed] [Google Scholar]
- 54. Masumoto A, Yamamoto N. Stimulation of DNA synthesis in hepatocytes by hepatocyte growth factor bound to extracellular matrix. Biochem Biophys Res Commun. 1993;191(3):1218–1223. [DOI] [PubMed] [Google Scholar]
- 55. Ren H, Shi X, Tao L, Xiao J, Han B, Zhang Y, Yuan X, Ding Y. Evaluation of two decellularization methods in the development of a whole-organ decellularized rat liver scaffold. Liver Int. 2013;33(3):448–458. [DOI] [PubMed] [Google Scholar]
- 56. Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger J, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 2013;87(8):1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gomez-Lechon MJ, Lahoz A, Gombau L, Castell JV, Donato MT. In vitro evaluation of potential hepatotoxicity induced by drugs. Curr Pharm Des. 2010;16(17):1963–1977. [DOI] [PubMed] [Google Scholar]
- 58. Soldatow VY, Lecluyse EL, Griffith LG, Rusyn I. In vitro models for liver toxicity testing. Toxicol Res (Camb). 2013;2(1):23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Knobeloch D, Ehnert S, Schyschka L, Buchler P, Schoenberg M, Kleeff J, Thasler WE, Nussler NC, Godoy P, Hengstler J, et al. Human hepatocytes: isolation, culture, and quality procedures. Methods Mol Biol. 2012;806:99–120. [DOI] [PubMed] [Google Scholar]
- 60. Qiao L, Farrell GC. The effects of cell density, attachment substratum and dexamethasone on spontaneous apoptosis of rat hepatocytes in primary culture. In Vitro Cell Dev Biol Anim. 1999;35(7):417–424. [DOI] [PubMed] [Google Scholar]
- 61. Greuet J, Pichard L, Ourlin JC, Bonfils C, Domergue J, Le Treut P, Maurel P. Effect of cell density and epidermal growth factor on the inducible expression of CYP3A and CYP1A genes in human hepatocytes in primary culture. Hepatology. 1997;25(5):1166–1175. [DOI] [PubMed] [Google Scholar]
- 62. McIntyre M, Desdouets C, Senamaud-Beaufort C, Laurent-Winter C, Lamas E, Brechot C. Differential expression of the cyclin-dependent kinase inhibitor P27 in primary hepatocytes in early-mid G1 and G1/S transitions. Oncogene. 1999;18(32):4577–4585. [DOI] [PubMed] [Google Scholar]
- 63. Hinek A, Braun KR, Liu K, Wang Y, Wight TN. Retrovirally mediated overexpression of versican v3 reverses impaired elastogenesis and heightened proliferation exhibited by fibroblasts from Costello syndrome and Hurler disease patients. Am J Pathol. 2004;164(1):119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu J, Tirlapur U, Fairbank J, Handford P, Roberts S, Winlove CP, Cui Z, Urban J. Microfibrils, elastin fibres and collagen fibres in the human intervertebral disc and bovine tail disc. J Anat. 2007;210(4):460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bailey AJ. Collagen and elastin fibres. J Clin Pathol Suppl (R Coll Pathol). 1978;12:49–58. [PMC free article] [PubMed] [Google Scholar]
- 66. Godin LM, Sandri BJ, Wagner DE, Meyer CM, Price AP, Akinnola I, Weiss DJ, Panoskaltsis-Mortari A. Decreased laminin expression by human lung epithelial cells and fibroblasts cultured in acellular lung scaffolds from aged mice. PLoS One. 2016;11(3):e0150966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maghsoudlou P, Georgiades F, Smith H, Milan A, Shangaris P, Urbani L, Loukogeorgakis SP, Lombardi B, Mazza G, Hagen C, et al. Optimization of liver decellularization maintains extracellular matrix micro-architecture and composition predisposing to effective cell seeding. PLoS One. 2016;11(5):e0155324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wong ML, Wong JL, Horn RM, Sannajust KC, Rice DA, Griffiths LG. Effect of urea and thiourea on generation of xenogeneic extracellular matrix scaffolds for tissue engineering. Tissue Eng Part C Methods. 2016;22(7):700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mazza G, Rombouts K, Rennie Hall A, Urbani L, Vinh Luong T, Al-Akkad W, Longato L, Brown D, Maghsoudlou P, Dhillon AP, et al. Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci Rep. 2015;5:13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Almine JF, Wise SG, Weiss AS. Elastin signaling in wound repair. Birth Defects Res C Embryo Today. 2012;96(3):248–257. [DOI] [PubMed] [Google Scholar]
- 71. Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39(6):1477–1487. [DOI] [PubMed] [Google Scholar]
- 72. Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132(6):1133–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Webber EM, Godowski PJ, Fausto N. In vivo response of hepatocytes to growth factors requires an initial priming stimulus. Hepatology. 1994;19(2):489–497. [PubMed] [Google Scholar]
- 75. Ishii T, Sato M, Sudo K, Suzuki M, Nakai H, Hishida T, Niwa T, Umezu K, Yuasa S. Hepatocyte growth factor stimulates liver regeneration and elevates blood protein level in normal and partially hepatectomized rats. J Biochem. 1995;117(5):1105–1112. [DOI] [PubMed] [Google Scholar]
- 76. Fujiwara K, Nagoshi S, Ohno A, Hirata K, Ohta Y, Mochida S, Tomiya T, Higashio K, Kurokawa K. Stimulation of liver growth by exogenous human hepatocyte growth factor in normal and partially hepatectomized rats. Hepatology. 1993;18(6):1443–1449. [PubMed] [Google Scholar]
- 77. Nyberg SL, Hardin J, Amiot B, Argikar UA, Remmel RP, Rinaldo P. Rapid, large-scale formation of porcine hepatocyte spheroids in a novel spheroid reservoir bioartificial liver. Liver Transpl. 2005;11(8):901–910. [DOI] [PubMed] [Google Scholar]
- 78. Nelson LJ, Treskes P, Howie AF, Walker SW, Hayes PC, Plevris JN. Profiling the impact of medium formulation on morphology and functionality of primary hepatocytes in vitro. Sci Rep. 2013;3:2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Joly A, Desjardins JF, Fremond B, Desille M, Campion JP, Malledant Y, Lebreton Y, Semana G, Edwards-Levy F, Levy MC, et al. Survival, proliferation, and functions of porcine hepatocytes encapsulated in coated alginate beads: a step toward a reliable bioartificial liver. Transplantation. 1997;63(6):795–803. [DOI] [PubMed] [Google Scholar]
- 80. Watford M. The urea cycle: a two-compartment system. Essays Biochem. 1991;26:49–58. [PubMed] [Google Scholar]
- 81. Rogiers V, Vercruysse A. Rat hepatocyte cultures and co-cultures in biotransformation studies of xenobiotics. Toxicology. 1993;82(1–3):193–208. [DOI] [PubMed] [Google Scholar]
- 82. Luttringer O, Theil FP, Lave T, Wernli-Kuratli K, Guentert TW, de Saizieu A. Influence of isolation procedure, extracellular matrix and dexamethasone on the regulation of membrane transporters gene expression in rat hepatocytes. Biochem Pharmacol. 2002;64(11):1637–1650. [DOI] [PubMed] [Google Scholar]
- 83. Meier PJ, Stieger B. Bile salt transporters. Annu Rev Physiol. 2002;64:635–661. [DOI] [PubMed] [Google Scholar]
- 84. de Waart DR, Hausler S, Vlaming ML, Kunne C, Hanggi E, Gruss HJ, Oude Elferink RP, Stieger B. Hepatic transport mechanisms of cholyl-L-lysyl-fluorescein. J Pharmacol Exp Ther. 2010;334(1):78–86. [DOI] [PubMed] [Google Scholar]
- 85. Roma MG, Orsler DJ, Coleman R. Canalicular retention as an in vitro assay of tight junctional permeability in isolated hepatocyte couplets: effects of protein kinase modulation and cholestatic agents. Fundam Appl Toxicol. 1997;37(1):71–81. [DOI] [PubMed] [Google Scholar]
- 86. LeCluyse EL, Audus KL, Hochman JH. Formation of extensive canalicular networks by rat hepatocytes cultured in collagen-sandwich configuration. Am J Physiol. 1994;266(6 Pt 1):C1764–C1774. [DOI] [PubMed] [Google Scholar]
- 87. Richter JR, de Guzman RC, Van Dyke ME. Mechanisms of hepatocyte attachment to keratin biomaterials. Biomaterials. 2011;32(30):7555–7561. [DOI] [PubMed] [Google Scholar]
- 88. Galarneau L, Loranger A, Gilbert S, Marceau N. Keratins modulate hepatic cell adhesion, size and G1/S transition. Exp Cell Res. 2007;313(1):179–194. [DOI] [PubMed] [Google Scholar]
- 89. Huang RY, Guilford P, Thiery JP. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J Cell Sci. 2012;125(Pt 19):4417–4422. [DOI] [PubMed] [Google Scholar]
- 90. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Musat AI, Sattler CA, Sattler GL, Pitot HC. Reestablishment of cell polarity of rat hepatocytes in primary culture. Hepatology. 1993;18(1):198–205. [PubMed] [Google Scholar]