Acute myeloid leukemia (AML) is a heterogeneous disease. Based on risk stratification at diagnosis, patients with AML either receive consolidation chemotherapy or undergo allogeneic hematopoietic stem cell transplantation (allo-HSCT) after attaining initial remission. It has been shown that cytogenetic abnormalities at diagnosis are associated with outcome after post remission therapy, including allo-HSCT1. Based on cytogenetic risk stratification, Koreth et al.2 undertook a systematic review and meta-analysis of prospective trials, and concluded that allo-HSCT had significant relapse-free survival and overall survival (OS) benefit for intermediate- and for poor-risk AML, but not for good-risk AML in first complete remission. A growing number of recurrent genetic abnormalities have been recognized in the revised 2016 World Health Organization (WHO) classification of AML3. Acquired recurrent genetic abnormalities at diagnosis are among the most important independent factors used for diagnosis and prognostic stratification, and deepens our understanding of the disease pathogenesis4. Whether recurrent genetic abnormalities are also important markers that imply response to allo-HSCT is unknown. To address this question, we examined samples from patients with AML who had undergone allo-HSCT to determine whether recurrent genetic abnormalities were associated with long-term outcome after transplantation.
This is a retrospective study. A total of 78 patients who received allo-HSCT for AML (derived from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) were included in our study. Median age of the patients was 51 years (range, 18–72 years) and 45 (58%) patients were males. FAB subtype data were available for 77 patients, which included M0 (n = 10; 13%), M1 (n = 23; 30%), M2 (n = 20; 26%), M3 (n = 3; 4%), M4 (n = 14; 18%), M5 (n = 5; 6%), M6 (n = 1; 1%), and M7 (n = 1; 1%). The median WBC count at diagnosis for the entire cohort was 28.55 × 109/L (range, 0.6 × 109/L–223.8 × 109/L), and five patients (6%) had WBC count of ≥100 × 109/L at diagnosis. The median bone marrow blast at diagnosis for the entire cohort was 71.5% (range, 30–100%), and 65 patients (83%) had a bone marrow blast percentage of ≥50%. Cytogenetic data were available for 77 patients. Eight patients (10%) belonged to the good-risk, 47 (60%) to the intermediate-risk, and 22 (28%) to the poor-risk groups. Donors included human leukocyte antigen (HLA)-identical matched related donors (MRD, n = 33), HLA-identical matched unrelated donors (MUD, n = 43), and haploidentical related donors (HRD, n = 2). Twenty-seven patients did not achieve complete remission (CR) before transplantation. The median numbers of recurrent genetic mutations at diagnosis was 5 (range, 0–12). Seventy-seven patients had mutations in one or more genes. NPM1 was the most frequently mutated gene (n = 21, 27%), followed by DNMT3A (n = 19, 24%), FLT3-ITD (n = 17, 22%), IDH1 (n = 11, 14%), IDH2 (n = 9, 12%), RUNX1 (n = 9, 12%), WT1 (n = 9, 12%), CEBPA (n = 8, 10%), MYH11-CBFB (n = 5, 6%), MLL-translocation (n = 5, 6%), MLL-PTD (n = 4, 5%), TET2 (n = 4, 5%), TP53 (n = 4, 5%), KIT (n = 4, 5%), U2AF1 (n = 3, 4%), STAG2 (n = 3, 4%), ASXL1 (n = 2, 3%), EZH2 (n = 2, 3%), BCR-ABL1 (n = 2, 3%), and NUP98-NSD1 (n = 2, 3%). Clinical and molecular characteristics are summarized in Supplementary Table 1.
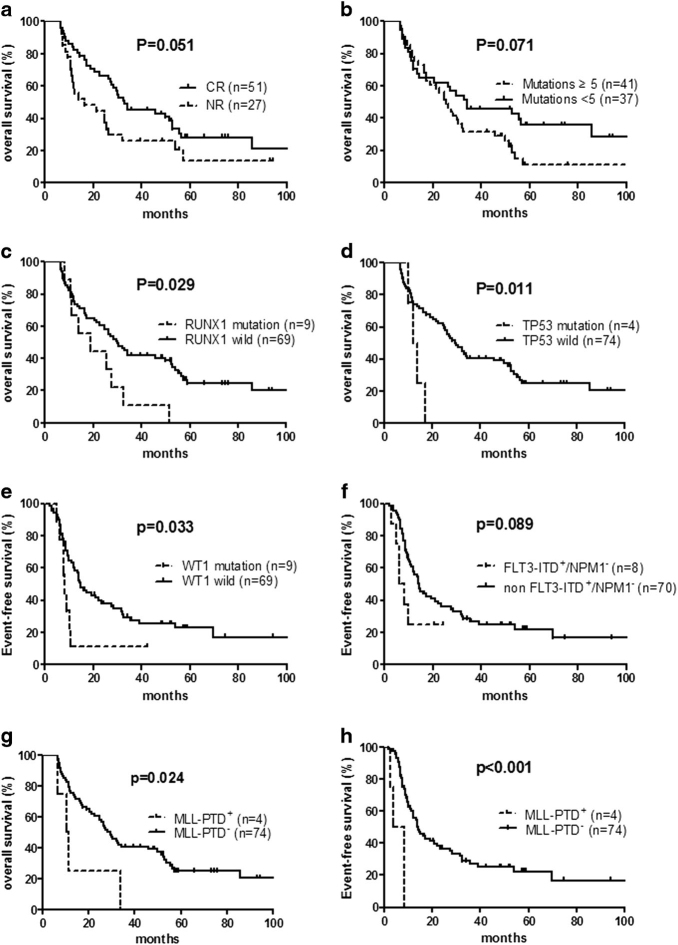
To assess the prognostic significance of these mutations, we focused on 14 genetic mutations which were detected in 5% or more of the patient population (NPM1, DNMT3A, FLT3-ITD, IDH1, IDH2, RUNX1, WT1, CEBPA, FLT3-ITD + /NPM1 -, MYH11-CBFB, MLL-PTD, TET2, TP53, and KIT). Factors including age (<60 vs. ≥60), WBC count (<100 × 109/L vs. ≥100 × 109/L), bone marrow blast percentage (<50% vs. ≥50%), cytogenetic risk (poor vs. others), donor type (MRD vs. MUD), disease state (CR vs. not in CR) as well as numbers of recurrent genetic mutations (<5 vs. ≥5). The results are summarized in Table 1. Based on univariate analyses, MLL-PTD was unfavorable for both OS (P = 0.024; Fig. 1g) and EFS (P < 0.001; Fig. 1h). Mutations in RUNX1 (P = 0.029; Fig. 1c) and TP53 (P = 0.011; Fig. 1d) negatively affected OS, while mutations in WT1 (P = 0.033; Fig. 1e) were identified as unfavorable for EFS. Patients with genotype “FLT3-ITD + /NPM1 -” showed a trend of poor EFS compared with those without the cytogenetic characteristic, but it did not reach statistical significance (P = 0.089; Fig. 1f). OS also appeared shorter in patients who did not achieve CR before transplantation (P = 0.051; Fig. 1a) and those having ≥5 recurrent genetic mutations (P = 0.071; Fig. 1b) whereas statistical significance was not achieved. Other clinical parameters, including age, WBC count, bone marrow blast percentage, cytogenetic risk, and donor type, were not associated with survival.
Table 1.
Univariate and multivariate analysis for EFS and OS
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| P | Log rank χ 2 test | P | HR (95% CI) | |
| OS | ||||
| Age (<60 vs. ≥60 years) | 0.200 | 1.640 | 0.451 | 0.783 (0.414–1.481) |
| WBC (<100×109/L vs. ≥100×109/L) | 0.429 | 0.626 | 0.211 | 0.495 (0.164–1.490) |
| BM blast (<50% vs. ≥50%) | 0.710 | 0.138 | 0.790 | 0.908 (0.445–1.851) |
| Cytogenetic risk (poor vs. others) | 0.737 | 0.113 | 0.949 | 0.978 (0.489–1.921) |
| Disease state (CR vs. not in CR) | 0.051 | 3.792 | 0.096 | 0.619 (0.352–1.089) |
| Mutated recurrent genes (<5 vs. ≥5) | 0.071 | 3.271 | 0.348 | 0.741 (0.396–1.387) |
| RUNX1 | 0.029 | 4.768 | 0.199 | 0.569 (0.241–1.345) |
| WT1 | 0.196 | 1.675 | 0.276 | 0.624 (0.267–1.457) |
| TP53 | 0.011 | 6.482 | 0.012 | 0.202 (0.059–0.700) |
| MLL-PTD | 0.024 | 5.096 | 0.060 | 0.316 (0.095–1.050) |
| EFS | ||||
| Age (<60 vs. ≥60 years) | 0.907 | 0.014 | 0.886 | 0.953 (0.497–1.828) |
| WBC (<100×109/L vs. ≥100×109/L) | 0.571 | 0.321 | 0.331 | 0.584 (0.198–1.726) |
| BM blast (<50% vs. ≥50%) | 0.711 | 0.137 | 0.631 | 1.191 (0.548–2.428) |
| Cytogenetic risk (poor vs. others) | 0.901 | 0.015 | 0.756 | 1.115 (0.561–2.215) |
| Disease state (CR vs. not in CR) | 0.311 | 1.026 | 0.545 | 0.842 (0.482–1.470) |
| Mutated recurrent genes (<5 vs. ≥5) | 0.528 | 0.399 | 0.873 | 0.951 (0.517–1.753) |
| RUNX1 | 0.281 | 1.161 | 0.777 | 0.879 (0.359–2.152) |
| WT1 | 0.033 | 4.521 | 0.138 | 0.509 (0.208–1.242) |
| TP53 | 0.245 | 1.353 | 0.167 | 0.425 (0.126–1.430) |
| MLL-PTD | 0.000 | 14.865 | 0.016 | 0.204 (0.056–0.746) |
The variables selected in the Cox proportional hazard model: age, WBC count, bone marrow blast, cytogenetic risk, disease state, mutated recurrent genes, and mutations (including mutations with frequency ≥5% and have poor OS or EFS based on univariate analyses: RUNX1, WT1, TP53, and MLL-PTD)
BM bone marrow, CR complete remission, EFS event-free survival, HR hazard ratio, OS overall survival, WBC white blood cell
Fig. 1. Kaplan–Meier curves of OS and EFS.
a Patients did not achieve CR before transplantation tended to have shorter OS than those transplanted in CR. b Patients with ≥5 mutations appeared to have shorter OS than those with <5 mutations. c, d Patients with RUNX1 and TP53 mutations had worse OS than wild-type groups. e Patients with WT1 mutations had worse EFS than wild-type groups. f Patients with genotype “mutated FLT3-ITD without NPM1” showed poor EFS compared with those without these mutations. g, h Patients with MLL-PTD mutations had worse OS and EFS than wild-type groups. Overall survival (OS) and event-free survival (EFS) were stratified by univariate prognostic factors. P-value was estimated by the log-rank test
We conducted multivariate COX regression analyses to identify independent risk factor for OS and/or EFS in the cohort. The four recurrent genetic mutations with demonstrated adverse effects on OS and/or EFS (RUNX1, WT1, TP53, and MLL-PTD, each of which showed significant associations with OS/EFS) and age (<60 vs. ≥60), WBC count (<100 × 109/L vs. ≥100 × 109/L), bone marrow blast percentage (<50% vs. ≥50%), cytogenetic risk (poor vs. others), disease state (CR vs. not in CR), as well as number of recurrent genetic mutations (<5 vs. ≥5) were incorporated in the analysis. Age (<60 vs. ≥60), WBC count (<100 × 109/L vs. ≥100 × 109/L), bone marrow blast (<50% vs. ≥50%), cytogenetic risk (poor vs. others), and number of recurrent genetic mutations (<5 vs. ≥5) had similar OS and EFS. TP53 mutation was an independent risk factor for OS (HR, 0.202; 95% confidence interval, CI, 0.059–0.700, P = 0.012). MLL-PTD was an independent risk factor for EFS (HR, 0.204; 95% CI, 0.056–0.746, P = 0.016). The results of the multivariate analysis are summarized in Table 1.
In this study, we examined the prognostic significance of recurrent genetic mutations and other clinical parameters in post allo-HSCT AML patients. Previous studies had controversial findings about how FLT3-ITD mutations affected the prognosis of AML patients after allo-HSCT5,6. RUNX1 mutations were found in 8 and 16% of younger and older patients with cytogenetically normal-AML (CN-AML), respectively, and they had lower CR rates and shorter disease-free survival (DFS), OS, and EFS than wild-type RUNX1 patients7. It was found in a study that patients with WT1 mutations had shorter DFS and OS than patients with wild-type WT1(ref. 8). Compared with these studies, our results show that mutations in these genes were not associated with survival. We assumed that allogeneic hematopoietic stem cell transplantation in our cohort might have reversed the unfavorable influences of mutations in these genes.
A recent study observed no significant difference in either OS or DFS between MLL-PTD + and MLL-PTD − patients9. The authors postulated that intensive consolidation therapy, which included autologous stem cell transplantation in first complete remission, might have contributed to the better outcome of this historically poor-prognosis group of CN-AML patients. TP53 gene mutations have been associated with monosomal karyotype and complex karyotype in myeloid malignancies10. The karyotype of the leukemic cells is by far the strongest prognostic factor for both response to induction therapy and survival11,12. A recent analysis of 858 AML patients demonstrated poor OS in patients with TP53 mutation13. In our study, we demonstrated that the TP53 and MLL-PTD mutations were independent predictors for inferior survival in post allo-HSCT patients. Our analyses indicated that evaluating the mutational status of the TP53 and MLL-PTD genes would be necessary before planning allo-HSCT for AML patients.
Older age was traditionally associated with poorer outcomes in AML patients, but age has not been shown to be the most important predictor for either transplant-related mortality or resistance to therapy. Our study concurred with the previous study by showing that the OS and EFS of patients ≥60-year-old were similar to those <60-year-old and age was not an independent prognostic factor in multivariate analysis. Quality of life is an important outcome for hematopoietic cell transplantation recipients. Particularly physical functioning and functional well-being may provide independent prognostic information beyond standard clinical measures in allo-HSCT recipients14.
Previous studies suggest that heavier mutation burdens might be associated with poorer prognosis in myelodysplastic syndromes15. It is possible that mutation burden also influences the survival of AML patients. We found that OS appeared to be shorter in patients having mutated recurrent genes ≥5 based on univariate analyses, but did not show statistical significance. Further studies are warranted to validate this hypothesis.
Our study has two important limitations. First, the relatively small number of patients is the major limitation of our study. Retrospective study designs are generally considered inferior to prospective study designs; this is the second limitation of our retrospective analysis. Nevertheless, our data suggest that assessing TP53 and MLL-PTD mutational status may be valuable for predicting survival in post allo-HSCT AML patients.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81500118, 61501519), the China Postdoctoral Science Foundation funded project (project No.2016M600443), and PLAGH project of Medical Big Data (Project No. 2016MBD-025).
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Supplementary information
The online version of this article (10.1038/s41408-017-0027-6) contains supplementary material
Contributor Information
Jinlong Shi, Phone: +86-10-66936921, Email: jinlong_301@163.com.
Lin Fu, Phone: +86-10-82267651, Email: fulin022@126.com.
References
- 1.Kim DH, et al. Parameters for predicting allogeneic PBSCT outcome of acute myeloid leukemia: cytogenetics at presentation versus disease status at transplantation. Ann. Hematol. 2005;84:25–32. doi: 10.1007/s00277-004-0942-z. [DOI] [PubMed] [Google Scholar]
- 2.Koreth J, et al. Allogeneic stem cell transplantation for acute myeloid leukemia in first complete remission: systematic review and meta-analysis of prospective clinical trials. JAMA. 2009;301:2349–2361. doi: 10.1001/jama.2009.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savani BN. Transplantation in AML CR1. Blood. 2010;116:1822–1823. doi: 10.1182/blood-2010-06-291500. [DOI] [PubMed] [Google Scholar]
- 6.Sengsayadeth S, et al. Allo-SCT for high-risk AML-CR1 in the molecular era: impact of FLT3|[sol]|ITD outweighs the conventional markers. Bone Marrow Transplant. 2012;47:1535–1537. doi: 10.1038/bmt.2012.88. [DOI] [PubMed] [Google Scholar]
- 7.Mendler JH, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J. Clin. Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paschka P, et al. Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B Study. J. Clin. Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitman SP, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: a Cancer and Leukemia Group B Study. Blood. 2007;109:5164–5167. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breems DA, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J. Clin. Oncol. 2008;26:4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- 11.Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract. Res. Clin. Haematol. 2001;14:497–529. doi: 10.1053/beha.2001.0152. [DOI] [PubMed] [Google Scholar]
- 12.Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 13.Stengel A, et al. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2016;31:705–711. doi: 10.1038/leu.2016.263. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton BK, et al. Prognostic significance of pre-transplant quality of life in allogeneic hematopoietic cell transplantation recipients. Bone Marrow Transplant. 2015;50:1235. doi: 10.1038/bmt.2015.122. [DOI] [PubMed] [Google Scholar]
- 15.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.