Abstract
Background
Patients with primary sclerosing cholangitis associated with inflammatory bowel disease (PSC-IBD) have a very high risk of developing colorectal neoplasia. Alterations in the gut microbiota and/or gut bile acids could account for the increase in this risk. However, no studies have yet investigated the net result of cholestasis and a potentially altered bile acid pool interacting with a dysbiotic gut flora in the inflamed colon of PSC-IBD.
Aim
The aim of this study was to compare the gut microbiota and stool bile acid profiles, as well as and their correlation in patients with PSC-IBD and inflammatory bowel disease alone.
Methods
Thirty patients with extensive colitis (15 with concomitant primary sclerosing cholangitis) were prospectively recruited and fresh stool samples were collected. The microbiota composition in stool was profiled using bacterial 16S rRNA sequencing. Stool bile acids were assessed by high-performance liquid chromatography tandem mass spectrometry.
Results
The total stool bile acid pool was significantly reduced in PSC-IBD. Although no major differences were observed in the individual bile acid species in stool, their overall combination allowed a good separation between PSC-IBD and inflammatory bowel disease. Compared with inflammatory bowel disease alone, PSC-IBD patients demonstrated a different gut microbiota composition with enrichment in Ruminococcus and Fusobacterium genus compared with inflammatory bowel disease. At the operational taxonomic unit level major shifts were observed within the Firmicutes (73%) and Bacteroidetes phyla (17%). Specific microbiota-bile acid correlations were observed in PSC-IBD, where 12% of the operational taxonomic units strongly correlated with stool bile acids, compared with only 0.4% in non-PSC-IBD.
Conclusions
Patients with PSC-IBD had distinct microbiota and microbiota-stool bile acid correlations as compared with inflammatory bowel disease. Whether these changes are associated with, or may predispose to, an increased risk of colorectal neoplasia needs to be further clarified.
Keywords: Gut microbiota, bile acids, primary sclerosing cholangitis, inflammatory bowel disease
Key summary
Summary of the established knowledge on this subject
Primary sclerosing cholangitis is a chronic cholestatic disease of unknown etiology and frequently associated with inflammatory bowel disease.
Emerging evidence suggests that alterations in the microbiome may be associated with this special phenotype.
No studies have yet investigated the net result of cholestasis and a potentially altered BA pool interacting with a dysbiotic gut flora in the inflamed colon of PSC-IBD.
What are the significant and/or new findings of this study?
Patients with PSC-IBD presented demonstrated a different gut microbiota composition, and specific microbiota-fecal BA correlations.
Despite no significant differences in the specific BA in stool, the overall combination of stool BA was discriminant between PSC-IBD and IBD.
Introduction
Primary sclerosing cholangitis (PSC) is a rare chronic cholestatic liver disorder of unclear aetiology.1 It is characterized by chronic inflammation of the biliary epithelium, that eventually leads to fibrosis, resulting in multifocal strictures of the intrahepatic and extrahepatic bile ducts. It can lead to cirrhosis, and end-stage liver disease requiring orthotopic liver transplantation (OLT).1 Furthermore, PSC is also associated with an increased risk of cholangiocarcinoma, gallbladder cancer and colorectal cancer.2,3 The strongest risk factor for having PSC is a history of inflammatory bowel disease (IBD). Around 60–80% of PSC patients will also have IBD, most commonly ulcerative colitis (UC). Ironically, despite having mild or quiescent extensive colitis, patients with PSC-IBD have the highest risk of developing colitis-associated neoplasia, which, in comparison with IBD, tends to be located preferentially in the right side of the colon.2,4 The factors that contribute to the increased risk of colorectal neoplasia in PSC remain unknown.5,6 A potential role for altered luminal concentration and/or composition of secondary bile acids (BAs) has been suggested, but never confirmed.7 Data from basic and clinical studies have long supported the hypothesis that the intestinal microbiota may have a role in PSC pathogenesis.8–10 Recently, studies using next-generation sequencing have reported a distinct fecal or mucosal microbiota composition in PSC-IBD patients.11–18 There is a close interplay between gut flora and BA metabolism. Besides their role in nutrient absorption and lipid digestion, BAs are important signaling molecules, acting in inflammation and metabolism, through activation of BA receptors such as the G-protein-coupled BA transmembrane receptor TGR5, and the nuclear BA receptor Farnesoid X receptor (FXR).19 BAs have antimicrobial properties, and through FXR-activation they regulate the expression of host genes whose products promote innate defence against luminal bacteria.20,21 On the other hand, BA metabolism is a property of the gastrointestinal microflora; BAs are transformed from primary BA (cholic acid (CA) and chenodeoxycholic acid (CDCA)) to secondary BA (litocholic acid (LCA) and deoxycholic acid (DCA)) by deconjugation, 7-alpha de-hydroxylation and epimerization (CDCA → ursodeoxycholic acid (UDCA)) by the gut microbiota; therefore the degree of activation of the BA receptors is also largely influenced by the gut microbiota.22–24 Nothing is known about the net result of cholestasis associated with PSC, and a potentially altered BA pool interacting with a dysbiotic gut flora in the inflamed colon of PSC-IBD patients. In this article, we have explored the BA profiles, the gut microbiota, and their correlation in PSC-IBD as compared with IBD patients.
Methods
Ethical considerations
This study was approved by the Portuguese National Committee for Data Protection and the local ethics committee. All patients signed an informed consent form.
Subjects and samples
Between October 2014–July 2015, 15 patients with PSC-IBD and 15 patients with IBD were prospectively recruited. The inclusion criteria were age greater than 18 years old, confirmed diagnosis of PSC based on histology and/or abnormal cholangiogram (Endoscopic Retrograde Cholangio-Pancreatography or Magnetic resonance cholangiopancreatography), a confirmed diagnosis of IBD by conventional endoscopic and histological criteria, and the presence of extensive colitis. Patients with a personal history of colectomy, a diagnosis of secondary sclerosing cholangitis or a history of OLT were excluded. All patients provided clinical and demographic information, and completed a semi-quantitative food frequency questionnaire (FFQ) validated for the Portuguese population.25 Clinical activity was scored according to the Mayo score for ulcerative colitis,26 and the Harvey-Bradshaw index for Crohn’s disease.27 Endoscopic activity was scored according to the Mayo endoscopic score for UC26 and the Simple Endoscopic Score for Crohn's Disease (SES-CD) for CD.28 All study participants collected serum sample, and a stool sample for BA analysis and microbiota analysis. All PSC patients on UDCA therapy were required to stop it two weeks before specimen collection. A minimum interval of three months was required between antibiotic intake or bowel preparation (for colonoscopy) and sample collection. During colonoscopy disease severity was recorded, and biopsies for colorectal neoplasia screening were obtained, according to current guidelines.
Serum BA profiles
A fasting serum sample was obtained from each patient. Individual amidated BAs in serum (1 ml) were determined by high-performance liquid chromatography (HPLC),29 after solid-phase extraction using Sep-Pak C18 cartridges (Waters Corp., Milford, Massachusetts, USA).30 Only the conjugate fraction of BAs was measured in serum.
Stool BA profiles
A morning stool sample was obtained and dried to obtain a lyophilized extract. To lyophilized faecal samples weighing 1 g, 80% methanol was added. All samples were sonicated for 30 min, refluxed for two hours, and then cooled and filtered.31 The residue was re-suspended in chloroform/methanol (1:1, v/v), refluxed for one hour, and filtered. The combined extracts were taken to dryness, and re-suspended in 10 ml methanol (MeOH). An aliquot of 1 ml was added with 2 µl of 1 mg/ml nordeoxycholic acid, and was diluted in 10 ml deionized water and deposed on a 300 mg HLB Oasis column, washed with 10 volumes deionized, 1 volume cyclohexane, and the BAs were then eluted with 5 ml MeOH and were taken to dryness and resuspended in 250 µl MeOH. Four microliters were injected on liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) as previously described.32 Results are reported in nmol/g of dried stool for total BAs and in proportion of the median after calibration of the method, with weighted mixtures and normalization relative to the internal standard.32 The conjugate and non-conjugated species were quantified.
BA analysis
BAs were not normally distributed according to the Shapiro-Wilk test; therefore, their distributions were compared using non-parametric tests. The relative proportion of a given BA corresponds to its concentrations divided by the total of BAs. BA results are presented as the median proportion. For example, the total primary stool BA is the sum of CA and CDCA and their respective glyco-, tauro-, and sulphoderivatives. Linear discriminant analysis (LDA) was conducted to illustrate the classification of disease groups (IBD only and PSC-IBD) using stool BAs. LDA is a dimension reduction statistical technique that looks for a combination of features (continuous variables) that maximize the separation between classes. LDA was performed using the MASS package in R software.
Stool DNA extraction
Approximately 200 mg of stool were transferred into bead tubes (MO-BIO, Carlsbad, California, USA) and homogenized using the bead-beating method. Homogenized stool samples were further processed using the Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s protocol (Qiagen, Valencia, California, USA). Total DNA concentration was determined with Qubit 2.0 Fluorometer (Life Technologies, Norwalk, Connecticut, USA).
16S ribosomal RNA (rRNA) sequencing
The phylogenetically informative V3–V4 region of 16S ribosomal RNA (rRNA) gene was amplified using universal primer set 347F/803R.33 The primers were synthesized by IDT (Integrated DNA Technology, Coralville, Iowa, USA). We used a dual-barcoding approach to label the 16S rRNA amplicons from each sample as described previously.34 The 16S rRNA amplicons were further pooled with equal molarity and submitted for MiSeq 2 × 300 pair-end sequencing at high depth. The paired sequence readings were merged and filtered by size (>400 bp) and quality score (>Q30) using paired-end assembler for DNA sequences (PANDAseq).35 The processed readings were further split by dual barcodes for each sample and assigned taxonomic classification using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline 1.9.0.36 Repeated measurements of the same sample were made to assess sequencing reproducibility. After processing, QIIME provided detailed OTU tables containing the microbiota composition and abundance for each individual sample.
Data analysis
First, we measured the diversity of the overall microbiota communities within or across each sample. The overall species richness within each patient group, so-called alpha-diversity, was measured using the Chao1 and Shannon Index on rarefied tables at 8000 sequences per sample.37 Beta-diversity was measured using unweighted and weighted UniFrac distance matrices on the rarefied tables. The permutational analysis of variance (PERMANOVA) test (number of permutations = 999), was performed using QIIME command compare_categories.py to test the overall microbiota differences between groups by PSC and IBD status.38 Secondly, at the taxa level, the LDA effect size (LefSe) analysis was used with default parameters to select taxa features from phylum to genus level that were associated to PSC status.39 Only features with LDA score >2.0 were kept. A Kruskal-Wallis test on the LefSe selected differential taxa at the genus level was performed, and corresponding p-values were adjusted for multiple comparisons. Finally, the Kruskal-Wallis test was also performed at individual OTUs to select OTUs with significant differential abundance with respect to the PSC-IBD status. All singleton OTUs were removed prior to all analysis.
Correlation networks
We calculated both Pearson’s and Spearman’s correlations between the most abundant (mean relative abundance >0.1%) 65 genera in the gut microbiota and the stool BA levels in PSC and non-PSC IBD. To reduce the bias in the correlation analysis due to non-normality, we removed the variables with more than eight null value results, and removed the measurements beyond the 5% quantile of the distribution. We computed the raw probabilities. The p-values of the Pearson’s correlation were calculated using the corr.test function in R software with false discovery rate (FDR) adjustment for multiple comparisons. Spearman’s correlations of the selected pairs with significant p-values in the Pearson correlations were also computed to check the consistency of the correlations. We listed the genus-BA pairs with both significant Pearson’s correlation (adjusted p < 0.05) and strong Spearman’s correlations (p < 0.05) in the Supplementary Material, Table 2.
Results
Study population
Thirty patients with IBD, of whom 15 had concomitant PSC, were prospectively enrolled. All patients enrolled had pancolitis; two out of the four patients with CD also had ileal involvement. Two of the 15 PSC patients had concomitant liver cirrhosis (Child-Pugh A, six points). No patient had a prior history of abdominal or liver surgery. There were no significant differences in the overall daily intake of macro or micronutrients as assessed by the food-frequency questionnaire (data not shown). Patients with PSC-IBD presented, as expected, significantly higher levels of cholestasis markers, and were more frequently medicated with ursodeoxycholic acid. No further significantly different clinical variables were found, except for body mass index (BMI) that was significantly lower in PSC-IBD patients (Table 1). The median interval between stool collection and colonoscopy was 17.5 days (9–62). The additional demographic and clinical characteristics of PSC and IBD patients are described in Table 1.
Table 1.
Demographic and clinical characteristic of patients.
| PSC-IBD (n = 15) | IBD (n = 15) | p valuea | |
|---|---|---|---|
| Male (n, %) | 5 (33%) | 10 (67%) | 0.07 |
| Age (years) | |||
| Median, IQR | 42(24) | 45 (13) | 0.6 |
| Smoking status (n, %) | |||
| Never | 12 (80%) | 12 (80%) | 1.0 |
| Ever | 3 (20%) | 3 (20%) | |
| Type of IBD (n, %) | |||
| UC | 11 (73%) | 12 (80%) | 0.6 |
| CD | 4 (27%) | 3 (20%) | |
| PSC duration | |||
| Median years (IQR) | 7.8 (11.7) | – | – |
| IBD duration | |||
| Median years, (IQR) | 11.4 (5.26) | 11.1 (15.7) | 0.8 |
| PSC Mayo score | |||
| Median, (min, max) | –0.57 (–1.6, 1.7) | – | – |
| ALP (UI/l) (median, IQR) | 200 (166) | 54 (28) | <0.001 |
| GGT (UI/l) (median, IQR) | 332 (414) | 28 (20) | <0.001 |
| CRP (mg/dl) (median, IQR) | 0.2 (0.7) | 1.1 (1.3) | 0.061 |
| Disease clinical activity (n, %) | |||
| Remission-mild | 13 (87%) | 15 (100%) | 0.5 |
| Moderate-severe | 2 (13%) | 0 | |
| Disease endoscopic activity (n, %)b | |||
| Remission-mild | 9 (64%) | 13 (87%) | 0.2 |
| Moderate-severe | 5 (36%) | 2 (13%) | |
| Mean BMI (Kg/m2) | 24 ± 4.5 | 30.1 ± 6.4 | 0.005 |
| Presence of colorectal dysplasia (n, %) | 3/14 (21%) | 1/15 (6%) | 0.2 |
| Medications (n, %) | |||
| 5-ASA | 12 (80%) | 11 (73%) | 1.0 |
| Thiopurines | 5 (33%) | 8 (53%) | 0.3 |
| Anti-TNF | 3 (20%) | 3 (20%) | 1.0 |
| UDCA | 10 (67%) | – | <0.001 |
5-ASA: 5-aminosalicilate; ALP: alkaline phosphatase; anti-TNF: anti-tumor necrosis factor; BMI: body mass index; CRP: C-reactive protein; GGT: gammaglutamyl transpeptidase; IQR: interquartile range; PSC: primary sclerosing cholangitis; PSC-IBD: primary sclerosing cholangitis associated with inflammatory bowel disease; UC: ulcerative colitis; UDCA: ursodeoxycholic acid.
Variable distribution was compared using the Student’s t test, the Mann–Whitney test or the χ2 test, as appropriate; bin the PSC-IBD group one patient refused colonoscopy.
Serum BA
The total BA (µmol/l) pool was significantly expanded in PSC-IBD (p-value = 0.007, Mann–Whitney) (Supplementary Material, Table 1). No significant differences were seen in the proportion of individual BAs between groups. There was a positive correlation between PSC duration and total serum BAs (ρ = 0.66, p = 0.009).
Stool BA profiles
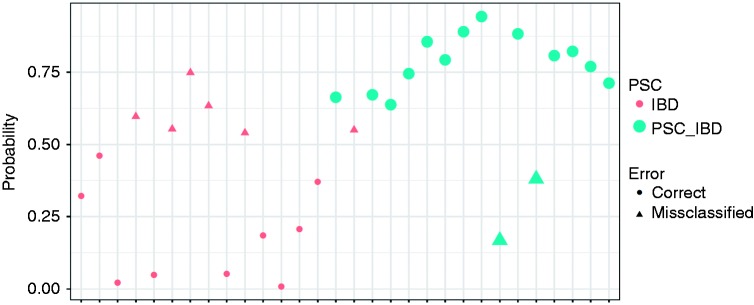
The median total stool BAs were significantly reduced in PSC-IBD (167.2 µmol/l in PSC-IBD versus 282.4 µmol/l in IBD, p = 0.021). Overall there were no significant differences in the proportions of each BA (Table 2), although the overall combination of stool BA allowed a good separation between PSC-IBD and IBD, as visualized in the linear discriminant analysis (Figure 1). Using the main BAs (CA, CDCA, LCA, DCA and UDCA), the classification accuracy of the LDA was 73%, with a sensitivity and specificity of 86.7% and 60% respectively (Figure 1). When we used all individual BAs (taurine and glycine conjugates and sulphated BA), the accuracy of the LDA for classifying PSC-IBD versus IBD was 100% (Supplementary Material, Figure 1). Additional LDA analysis was conducted using the top four most discriminatory stool BA (Supplementary Material, Figure 1). PSC-IBD patients presented a higher proportion of conjugated BA, although this did not reach statistical significance. DCA, a secondary BA, was also elevated, albeit non-significantly, in PSC-IBD. The proportion of UDCA in stool was not different in the PSC patients who were medicated with UDCA versus those who were not (1.075 nmol/g versus 1.35 nmol/g, respectively, p-value=0.7, Mann–Whitney U Test). Likewise, the results for all stool BA comparisons did not change after excluding the two CD patients with ileal involvement (data not shown). There was a negative correlation between the concentration of secondary BAs and endoscopic disease activity (ρ = –539, p = 0.003); this was also observed when the analysis was stratified by patient group (data not shown).
Table 2.
Stool bile acids (BAs) in primary sclerosing cholangitis associated with inflammatory bowel disease (PSC-IBD) and inflammatory bowel disease (IBD) patients.
| BAs | PSC-IBD | IBD | p value |
|---|---|---|---|
| Primary BAs | 9.5 (18) | 4.2 (15.2) | 0.29 |
| Secondary BAs | 89.4 (24.6) | 91.2 (15.1) | 0.57 |
| CA | 4.6 (6.45) | 1.49 (4.77) | 0.06 |
| CDCA | 4.73 (10.4) | 2.72 (11.0) | 1.0 |
| DCA | 52.5 (23.5) | 43.6 (14.3) | 0.55 |
| LCA | 34.1 (33.8) | 46.2 (1.9) | 0.14 |
| UDCA | 1.1 (1.9) | 1.8 (3.9) | 0.37 |
| Tauro/glyco conjugates | 0.47 (0.67) | 0.34 (0.69) | 0.98 |
| Sulfated BAs | 2.1 (3.1) | 2.4 (16.3) | 0.41 |
| Conjugated BAs | 4.5 (13.7) | 2.7 (6.9) | 0.23 |
CA: cholic acid; CDCA: chenodeoxycholic acid; DCA: deoxycholic acid; LCA: litocholic acid; UDCA: ursodeoxycholic acid.
BAs are expressed as percentage median (interquartile range) of total BAs. Distributions were compared with non-parametric tests (Mann–Whitney). Due to a small amount of minor BA species in stool (muricholic acid, hycholic acid, hyodeoxycholic acid, or ursodeoxhycolic acid) which are considered by some authors as ‘tertiary BAs’, the sum of primary and secondary BAs is not 100%.
Figure 1.
Results of the linear discriminant analysis allowing to see the discrimination of primary sclerosing cholangitis associated with inflammatory bowel disease (PSC-IBD) versus inflammatory bowel disease (IBD) alone, based on the combination of the main bile acids (BAs) present in stool (cholic acid (CA), chenodeoxycholic acid (CDCA), litocholic acid (LCA), deoxycholic acid (DCA) and ursodeoxycholic acid (UDCA)). On the x axis each marker represents a patient. On the y axis is represented the probability of being correctly classified as PSC-IBD using the BA analytes. The green markers represent patients with PSC-IBD and the pink markers represent patients with IBD. The circles represents patients that were correctly assigned to their disease group. The classification accuracy of the linear discriminant analysis (LDA) was 73%, with a sensitivity and specificity of 86.7% and 60% respectively.
Survey of gut microbiota
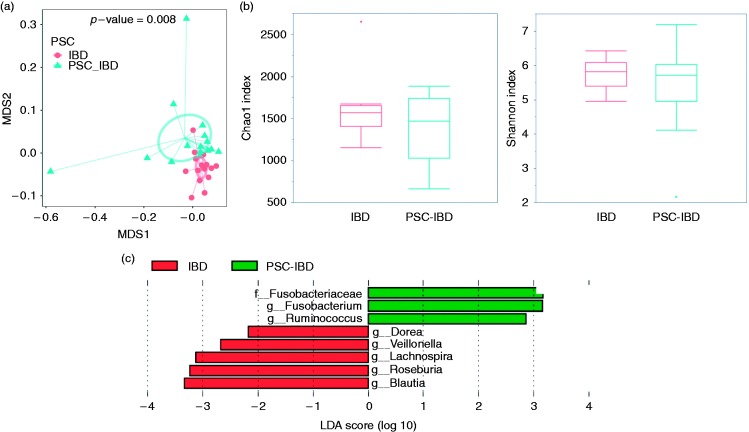
Using 16S rRNA sequencing, we surveyed the microbiome composition of 30 stool samples. The duplicate measurements showed Pearson correlation over 99% at genus level, confirming the reproducibility of the experimental approach. PSC-IBD presented lower alpha-diversity, albeit not significantly different (Chao1 899.3 for IBD vs 832.0 for PSC-IBD, p-value = 0.36; Shannon index 5.7 for IBD vs 5.3 for PSC-IBD, p = 0.23) (Figure 2(b)). Patients with PSC and concomitant cirrhosis (n = 2) presented significantly lower bacterial alpha-diversity (p = 0.005) as compared with those with PSC without cirrhosis (data not shown). The overall microbiota dissimilarities among all samples grouped by PSC and IBD status were accessed using the UniFrac distance matrices (Figure 2(a)). The overall qualitative microbial composition of patients with PSC-IBD was different as observed in the multidimensional scaling (MDS) plot (Figure 2(a): unweighted UniFrac, PERMANOVA: pseudo-F statistic: 2.99, p value=0.008). At the individual taxa level (Figure 2(c)), we found seven genera differentially expressed in PSC-IBD vs IBD (logarithmic LDA score>2 by LEfSe analysis): Ruminococcus and Fusobacterium were more abundant in PSC-IBD, while Dorea, Veillonella, Lachnospira, Blautia, and Roseburia were less abundant. All of those genera were found to be significant (p < 0.05) when their relative abundance was compared using a Kruskal-Wallis test (p values adjusted for multiple comparisons). No significant differences in the microbial overall composition (ß-diversity) were observed by PSC disease severity (as measured by the PSC Mayo score), BMI, UDCA use, IBD type, or IBD disease activity (data not shown).
Figure 2.
Overall microbiota dissimilarities between samples grouped by primary sclerosing cholangitis (PSC) and inflammatory bowel disease (IBD) status. (a) Dissimilarities were measured using UniFrac unweighted distances and visualized using a multidimensional plot (MDS) plot. The smaller circle represents patients with IBD, while the larger circle the samples from PSC-IBD patients. (b) The boxplots show the mean and variance of the richness of the microbial community between different disease status (Chao1 in the left and Shannon index on the right); no significant differences are seen (p value: 0.36 and 0.23 respectively). (c) Top discriminative bacteria in primary sclerosing cholangitis associated with inflammatory bowel disease (PSC-IBD) and IBD patients as determined by LEfSe analysis (linear discriminant analysis (LDA)). On the right are represented the increased taxa in PSC-IBD, while on the left the decreased taxa in PSC-IBD, as compared with IBD.
Differential OTUs by PSC status
Based on 97% similarity of the 16S sequencing reads, the open-reference OTU picking using QIIME pipeline assigned all sequencing reads into individual OTUs. After removing singletons, we compared 3839 OTUs and selected 143 OTUs which were significantly (p < 0.05 by Kruskal–Wallis test, not adjusted) differential and presented a >2 fold changes in the mean abundance between IBD and PSC-IBD (Supplementary Material, Figure 2). Compared with IBD only, the relative abundance of 32 OTUs were increased and 111 OTUs were decreased in PSC-IBD. At the phylum level, we found that most of the shifts associated with PSC occurred within the Firmicutes (73%) and Bacteroidetes phyla (17%). Consistent with the LEfSe analysis at the genus level, that found Blautia and Ruminococcus as the two most significant differential genera by PSC status, we found that all 16 OTUs of Blautia genus were reduced while four of five OTUs of Ruminococcus genus and Ruminococcaceae family were enriched in PSC samples.
Correlation between microbiota genera and stool BAs
Correlations between microbiota genus and the stool BA were calculated as described in our method section to test the interactions between gut microbiota and stool BA. Without stratifying by PSC status, we found four genera, including Blautia and Veillonella to be correlated to specific types of BAs. In PSC-IBD, bacteria with significant correlations with BA metabolites mostly belonged to the Firmicutes phylum, specifically within the Clostridia and Bacilli classes. Different correlations were observed in IBD (Supplementary Material, Table 2). Compared with IBD, seven genera appeared and two genera disappeared in PSC-IBD. The total relative abundance of genera correlated to BA was 12% in PSC-IBD, compared with 0.4% in IBD. Two Firmicutes, Lachnospira and Veillonella, which were significantly reduced in PSC-IBD, showed strong correlations with multiple BA, only in PSC-IBD.
Discussion
Herein, for the first time we have analyzed the stool BA profiles and their correlation with the faecal microbiota composition in patients with PSC-IBD as compared with IBD alone. The serum BA pool was increased and the stool BA pool was significantly reduced in PSC-IBD as compared with IBD alone. No significant differences in the individual stool BA components were found, but their overall composition differed from IBD (Figure 1). A significantly different microbiota composition based on the unweighted UniFrac distances was found between IBD and PSC-IBD, indicating differences in taxon composition for rare taxa (Figure 2(a)). Specifically, PSC-IBD patients presented an enrichment in bacteria belonging to the genera Ruminococcus and Fusobacterium as compared with IBD alone (Figure 2(c)). Finally, specific microbiota-stool BA correlations were observed in PSC-IBD (Supplementary Material, Table 2).
In the past, some authors have hypothesized that the increased risk of right-sided colorectal neoplasia in PSC-IBD could be linked to an increase in secondary BA, although this had never been demonstrated.2,7 Normally, most of the BAs secreted by the liver are efficiently reabsorbed in the terminal ileum, through the sodium-dependent BA transporter (ASBT), leaving only approximately 5% of the total BAs to reach the colonic lumen. In the right colon, primary BAs are transformed in to secondary BAs mostly by bacterial mediated deconjugation, oxidation/reduction, epimerization, and dehydroxylation.40 Therefore, faecal BA are mainly deconjugated, secondary BAs. A small fraction of secondary BA is passively absorbed through the colonic mucosa, whilst the rest will be extruded with faeces.41 During obstructive cholestasis, the expression of the apical BA transporter, which permits intracellular absorption of BAs, is down-regulated, as a compensatory mechanism.42 This could hypothetically lead to a relative increase in the proportion of BAs entering the proximal colon in PSC-IBD patients, where they would be converted from primary into secondary BAs. Interestingly, secondary BAs have been shown to have anti-inflammatory properties but at the same have been shown to bear carcinogenic properties.19,32,43–45 Herein, we observed a significant reduction in the total stool BAs in PSC-IBD as compared with IBD, which was expected taking into consideration the obstructive cholestatic nature of PSC. However, we did not find an increase in the relative proportion of the stool secondary BAs in PSC-IBD patients, as previously hypothesized.7,12 No significant differences in individual proportion of serum or stool BAs were found, which could perhaps be due to the small sample size. The proportion of DCA, a secondary BA was increased in PSC-IBD, although this did not reach statistical significance. Furthermore, the proportion of conjugate BAs was also non-significantly increased in PSC-IBD as compared with IBD, which could indirectly indicate a decrease in the deconjugation activity of the microbiota. The decrease in Bacteroides, Clostridium, Bifidobacterium, and Lactobacillus genus, observed at the OTU level, and known to be involved in BA deconjugation, could hypothetically be involved in this finding.46 In this cohort, patients with PSC-IBD demonstrated an enrichment in bacteria from the Ruminococcus and Fusobacterium taxa, and a decrease in bacteria from the genus Dorea, Veillonella, Lachnospira, Blautia, and Roseburia. At the OTU level most shifts were observed within the Firmicutes (73%) and Bacteroidetes phylae (17%). Some of our findings are in consonance with recently published results on PSC microbiota also showing an increase of Fusobacterium13 (a bacterial taxon that has been linked with adenomas and colorectal cancer) and in Ruminococcus in stool from patients with PSC-IBD or a decrease in Roseburia genus. However, others are not; Kummen et al. reported PSC patients to have a significant increase in Veillonella genus in comparison with healthy controls and patients with IBD.14 In this cohort, Veillonella genus was positively correlated with disease severity and was more abundant in patients that had undergone OLT. Indeed, this genus has been reported to be increased in fibrotic conditions such as liver or lung fibrosis or cystic fibrosis.47–50 In cirrhosis, Veillonella has also been associated with complications such as hepatic encephalopathy.51 In another large cohort of patients with PSC-IBD, Veillonella was only significantly elevated in patients with PSC that presented concomitant liver cirrhosis.13 Of note, in our PSC population, only two patients presented early liver cirrhosis, no patient presented severe PSC as measured by the PSC Mayo score or had undergone liver transplant. Also worthy of note is the well described disconnect between mucosa and stool microbiota, as in a prior work Blautia was increased in the mucosa from PSC-IBD patients as compared with healthy controls.12
No study had yet looked at the correlations between stool BA and the stool microbiota in PSC-IBD. However, BA pool size and composition have been shown to be important factors in regulating the gut microbiota.24,52,53 Herein, despite our relatively small sample size, and after correcting for multiple comparisons, we were able to observe unique correlations between stool microbiota and stool BAs in PSC-IBD. Within IBD alone, this broad BA-microbiota correlation disappeared. In particular, in PSC-IBD, the taxa that significantly correlated to the stool BA corresponded to ∼12% of the total microbiota, while in IBD, this was less than 1%. Without any functional data, we may only speculate that these results suggest that under PSC conditions, the BA changes may have dominant effects on defining the gut microbiota shifts, potentially towards a more pro-carcinogenic profile. Interestingly, bacteria from the genus of both Fusobacterium and Ruminococcus, are known to be involved in oxidation, epimerization and desulfation of BAs.46
The major limitation of this study is our small sample size, which prevented us from adjusting for potential confounders in the microbiota and BA analysis. To overcome this, we tried to make our cohort as uniform as possible. All patients had pancolitis, and no patient had prior abdominal surgery or history of liver transplantation; all patients had mild to moderate PSC, as measured by the Mayo score, and dietary intake was also similar within groups as assessed by the food frequency questionnaire. Furthermore, all patients stopped UDCA intake for two weeks and had no antibiotics or bowel preparation within at least three months of sample collection, all external factors that could potentially impact microbiota composition. While it may be argued that a two-week interval to stop UDCA may not be enough to remove its effects, we did not observe any differences in the faecal BA composition or in the microbiota composition between those who were medicated with UDCA as compared with those who were not, consistent with what has been previously reported.13
In summary, in this exploratory study, patients with PSC-IBD had a distinct stool BA and stool microbiota composition, as well as specific microbiota-stool BA correlations when compared with IBD. Whether these changes are associated with or may predispose to the specific PSC-IBD phenotype including the increased risk of colorectal neoplasia needs to be further clarified and warrants further research.
Acknowledgements
The authors wish to thank the OCS genome technology center of New York University Langone Medical Center for the Illumina Miseq molecule library preparation and sequencing service. JT, SHI, MC, JFC, and JH have contributed to the study design and concept. JT, CP, MC, XB, PMS, JPS, CV, and AO have contributed to the acquisition of data. JT, RH, KP, HB, CR, DR, LH and JH analyzed, and interpreted the data. JT and JH drafted the manuscript. All authors have reviewed the final version of the manuscript and provided critical comments. JT and JH obtained funding support.
Declaration of conflicting interests
J Torres has received consulting fees from Takeda. J-F Colombel has served as consultant, advisory board member or speaker for Abbvie, AB Science, Amgen, Bristol Meyers Squibb, Celltrion, Danone, Ferring, Genentech, Giuliani SPA, Given Imaging, Janssen, Immune Pharmaceuticals, Medimmune, Merck & Co., Millenium Pharmaceuticals Inc., Neovacs, Nutrition Science Partners Ltd, Pfizer Inc., Prometheus Laboratories, Protagonist, Receptos, Sanofi, Schering Plough Corporation, Second Genome, Shire, Takeda, Teva Pharmaceuticals, Tigenix, UCB Pharma, Vertex, and Dr August Wolff GmbH & Co. SH Itzkowitz received grant support and fees for serving on a scientific advisory board from Exact Sciences. The remaining authors have no conflicts of interest to declare.
Ethics approval
This study was approved by the Portuguese National Committee for Data Protection and by the local ethics committee at each participating center.
Funding
This study was supported by funds from GEDII (Portuguese Group for the Study of Inflammatory Bowel Disease) (JT) and from the National Institute of Health (NIDDK 1K01DK094986-01(JH) and NIDDK R03 DK106481-01(JH)).
Informed consent
All patients participating in this study signed an informed consent form.
References
- 1.Eaton JE, Talwalkar JA, Lazaridis KN, et al. Pathogenesis of primary sclerosing cholangitis and advances in diagnosis and management. Gastroenterology 2013; 145: 521–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres J, Pineton de Chambrun G, Itzkowitz S, et al. Review article: Colorectal neoplasia in patients with primary sclerosing cholangitis and inflammatory bowel disease. Aliment Pharmacol Ther 2011; 34: 497–508. [DOI] [PubMed] [Google Scholar]
- 3.Folseraas T, Boberg KM. Cancer risk and surveillance in primary sclerosing cholangitis. Clin Liver Dis 2016; 20: 79–98. [DOI] [PubMed] [Google Scholar]
- 4.Goldstone R, Itzkowitz S, Harpaz N, et al. Dysplasia is more common in the distal than proximal colon in ulcerative colitis surveillance. Inflamm Bowel Dis 2012; 18: 832–837. [DOI] [PubMed] [Google Scholar]
- 5.Loftus EV, Harewood GC, Loftus CG, et al. PSC-IBD: A unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut 2005; 54: 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton JE, Juran BD, Atkinson EJ, et al. A comprehensive assessment of environmental exposures among 1000 North American patients with primary sclerosing cholangitis, with and without inflammatory bowel disease. Aliment Pharmacol Ther 2015; 41: 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shetty K, Rybicki L, Brzezinski A, et al. The risk for cancer or dysplasia in ulcerative colitis patients with primary sclerosing cholangitis. Am J Gastroenterol 1999; 94: 1643–1649. [DOI] [PubMed] [Google Scholar]
- 8.Tabibian JH, O'Hara SP, Lindor KD. Primary sclerosing cholangitis and the microbiota: Current knowledge and perspectives on etiopathogenesis and emerging therapies. Scand J Gastroenterol 2014; 49: 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabibian JH, O'Hara SP, Trussoni CE, et al. Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology 2016; 63: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabibian JH, Talwalkar JA, Lindor KD. Role of the microbiota and antibiotics in primary sclerosing cholangitis. Biomed Res Int 2013; 2013 Article ID 389537, 7pp. DOI: 10.1155/2013/389537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossen NG, Fuentes S, Boonstra K, et al. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J Crohns Colitis 2015; 9: 342–348. [DOI] [PubMed] [Google Scholar]
- 12.Torres J, Bao X, Goel A, et al. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment Pharmacol Ther 2016; 43: 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabino J, Vieira-Silva S, Machiels K, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016; 65: 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kummen M, Holm K, Anmarkrud JA, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 2016; 66: 611–619. [DOI] [PubMed] [Google Scholar]
- 15.Rühlemann MC, Heinsen F-A, Zenouzi R, et al. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut 2016; 66: 753–754. [DOI] [PubMed] [Google Scholar]
- 16.Quraishi MN, Sergeant M, Kay G, et al. The gut-adherent microbiota of PSC-IBD is distinct to that of IBD. Gut 2016; 66: 386–388. [DOI] [PubMed]
- 17.Kevans D, Tyler AD, Holm K, et al. Characterization of intestinal microbiota in ulcerative colitis patients with and without primary sclerosing cholangitis. J Crohns Colitis 2016; 10: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasawa K, Suda W, Tsunoda T, et al. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut. Epub ahead of print 26 September 2016. DOI: 10.1136/gutjnl-2016-312533. [DOI] [PMC free article] [PubMed]
- 19.Pavlidis P, Powell N, Vincent RP, et al. Systematic review: Bile acids and intestinal inflammation-luminal aggressors or regulators of mucosal defence? Aliment Pharmacol Ther 2015; 42: 802–817. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann AF, Eckmann L. How bile acids confer gut mucosal protection against bacteria. Proc Natl Acad Sci U S A 2006; 103: 4333–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inagaki T, Moschetta A, Lee Y-K, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A 2006; 103: 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones ML, Martoni CJ, Ganopolsky JG, et al. The human microbiome and bile acid metabolism: Dysbiosis, dysmetabolism, disease and intervention. Expert Opin Biol Ther 2014; 14: 467–482. [DOI] [PubMed] [Google Scholar]
- 23.Sagar NM, Cree IA, Covington JA, et al. The interplay of the gut microbiome, bile acids, and volatile organic compounds. Gastroenterol Res Pract 2015; 2015 Article ID 398585, 6 pp. DOI: 10.1155/2015/398585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam KBMS, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011; 141: 1773–1781. [DOI] [PubMed] [Google Scholar]
- 25.Lopes C, Aro A, Azevedo A, et al. Intake and adipose tissue composition of fatty acids and risk of myocardial infarction in a male Portuguese community sample. J Am Diet Assoc 2007; 107: 276–286. [DOI] [PubMed] [Google Scholar]
- 26.Lewis JD, Chuai S, Nessel L, et al. Use of the noninvasive components of the Mayo score to assess clinical response in ulcerative colitis. Inflamm Bowel Dis 2008; 14: 1660–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermeire S, Schreiber S, Sandborn WJ, et al. Correlation between the Crohn's disease activity and Harvey-Bradshaw indices in assessing Crohn's disease severity. Clin Gastroenterol Hepatol 2010; 8: 357–363. [DOI] [PubMed] [Google Scholar]
- 28.Vuitton L, Marteau P, Sandborn WJ, et al. IOIBD technical review on endoscopic indices for Crohn's disease clinical trials. Gut 2016; 65: 1447–1455. [DOI] [PubMed] [Google Scholar]
- 29.Labbé D, Gerhardt MF, Myara A, et al. High-performance liquid chromatographic determination of tauro- and glyco-conjugated bile acids in human serum. J Chromatogr 1989; 490: 275–284. [DOI] [PubMed] [Google Scholar]
- 30.Feldmann D, Fenech C, Cuer JF. Evaluation of a sample-preparation procedure for bile acids in serum and bile. Clin Chem 1983; 29: 1694–1694. [PubMed] [Google Scholar]
- 31.Setchell KD, Lawson AM, Tanida N, et al. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J Lipid Res 1983; 24: 1085–1100. [PubMed] [Google Scholar]
- 32.Duboc H, Rajca S, Rainteau D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013; 62: 531–539. [DOI] [PubMed] [Google Scholar]
- 33.Ahn J, Yang L, Paster BJ, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One 2011; 6: e22788–e22788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Franzen O, Pei Z, et al. P-237 Multiple double-barcoding 16S sequencing on the MiSeq Platform to study the gut microbiome in Ashkenazi Jews with Crohn’s disease. Inflamm Bowel Dis; 19: S119–S121.
- 35.Masella AP, Bartram AK, Truszkowski JM, et al. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinformatics 2012; 13: 31–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev 2008; 32: 557–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong M, McHardy I, Ruegger P, et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J 2014; 8: 2193–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12: R60–R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Augustin O, Sanchez de Medina F. Intestinal bile acid physiology and pathophysiology. World J Gastroenterol 2008; 14: 5630–5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006; 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 42.Hruz P, Zimmermann C, Gutmann H, et al. Adaptive regulation of the ileal apical sodium dependent bile acid transporter (ASBT) in patients with obstructive cholestasis. Gut 2006; 55: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duboc H, Taché Y, Hofmann AF. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig Liver Dis 2014; 46: 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein H, Bernstein C, Payne CM, et al. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 2005; 589: 47–65. [DOI] [PubMed] [Google Scholar]
- 45.Bernstein C, Holubec H, Bhattacharyya AK, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol 2011; 85: 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013; 3: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lv LX, Fang DQ, Shi D, et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol 2016; 18: 2272–2286. [DOI] [PubMed] [Google Scholar]
- 48.Madan JC, Koestler DC, Stanton BA, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: Interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio 2012; 3: e00251.12–e00251.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Ji F, Guo J, Shi D, et al. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci Rep 2016; 6: 34055–34055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei X, Yan X, Zou D, et al. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol 2013; 13: 175–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014; 60: 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014; 30: 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kakiyama G, Pandak WM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 2013; 58: 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]